Key Points
Inhibitors of NF-κB activation attenuate lymphoid and myeloid leukemogenesis by BCR-ABL1 and decrease leukemic stem cells in vivo.
These results validate IKKs and NF-κB signaling as pharmacological targets for therapy of CML and Ph+ B-ALL.
Abstract
The product of the Ph chromosome, the BCR-ABL1 tyrosine kinase activates diverse signaling pathways in leukemic cells from patients with chronic myeloid leukemia (CML) and Ph+ B-cell acute lymphoblastic leukemia (B-ALL). Previous studies showed that nuclear factor κB (NF-κB) is activated in BCR-ABL1–expressing cells, but the mechanism of activation and importance of NF-κB to the pathogenesis of BCR-ABL1–positive myeloid and lymphoid leukemias are unknown. Coexpression of BCR-ABL1 and a superrepressor mutant of inhibitory NF-κB α (IκBαSR) blocked nuclear p65/RelA expression and inhibited the proliferation of Ba/F3 cells and primary BCR-ABL1–transformed B lymphoblasts without affecting cell survival. In retroviral mouse models of CML and B-ALL, coexpression of IκBαSR attenuated leukemogenesis, prolonged survival, and reduced myeloid leukemic stem cells. Coexpression of dominant-negative mutants of IκB kinase α (IKKα)/IKK1 or IKKβ/IKK2 also inhibited lymphoid and myeloid leukemogenesis by BCR-ABL1. Blockade of NF-κB decreased expression of the NF-κB targets c-MYC and BCL-X and increased the sensitivity of BCR-ABL1–transformed lymphoblasts to ABL1 kinase inhibitors. These results demonstrate that NF-κB is activated through the canonical IKK pathway and plays distinct roles in the pathogenesis of myeloid and lymphoid leukemias induced by BCR-ABL1, validating NF-κB and IKKs as targets for therapy of Ph+ leukemias.
Introduction
The BCR-ABL1 tyrosine kinase, product of the Philadelphia chromosome translocation, is the direct cause of chronic myeloid leukemia (CML) and is also implicated in ∼20% of acute B-lymphoblastic leukemias (Ph+ B-cell acute lymphoblastic leukemia [B-ALL]). ABL1 tyrosine kinase inhibitors (TKIs) such as imatinib induce cytogenetic remissions in the majority of CML patients, but relapse occurs quickly in most patients after a TKI is withdrawn,1 while those with advanced stages of CML or with B-ALL are less responsive to TKI therapy and frequently develop TKI resistance.2 Quiescent CML stem cells, which are resistant to killing by TKIs, are proposed to be the source of disease persistence and relapse.3,4 Hence, there is a need to identify and validate additional pharmacological targets in Ph+ leukemia to overcome TKI resistance and eradicate disease, leading to permanent cure.5
BCR-ABL1 activates multiple signaling networks in leukemic cells, including the RAS/mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription (STAT), c-Jun N-terminal kinase (JNK)/stress-activated protein kinase (SAPK), phosphatidylinositol 3-kinase (PI3K), nuclear factor κB (NF-κB), and c-MYC pathways,6 but a major challenge has been determining which of these pathways are essential to leukemogenesis. Mouse models of BCR-ABL1–induced leukemia can be valuable in this effort, as both CML and Ph+ B-ALL can be reproduced faithfully in mice by expressing BCR-ABL1 in bone marrow (BM) progenitors through retroviral transduction and transplantation.7 Recipients of BCR-ABL1–transduced BM develop a fatal CML-like myeloproliferative neoplasm (MPN) that originates from hematopoietic stem cells, can progress to blast crisis, and is responsive to kinase inhibitor therapy,8 providing a physiologically relevant model of CML in primary hematopoietic cells. To model B-ALL in mice, BM is transduced in the absence of myeloid cytokines, resulting in development of an aggressive B-lymphoblastic leukemia/lymphoma in transplant recipients.9 This disease originates from early lymphoid progenitors8 and is characterized by a block in B-cell differentiation at the pre-B stage.9
NF-κB is a potential downstream target of BCR-ABL1 due to its role in regulating cell survival and proliferation. The NF-κB transcription factor family consists of 5 members, p50/NF-κB1, p52/NF-κB2, RelA/p65, RelB, and c-Rel, that act as homo- or heterodimers to activate transcription of target genes.10 In quiescent or unstimulated cells, NF-κB is retained in cytoplasm by a family of inhibitory NF-κB (IκB) inhibitor proteins, including IκBα, IκBβ, IκBε, and Bcl-3. In the canonical pathway regulating innate immunity and inflammation, the stability of IκB is controlled by the IκB kinase (IKK) complex, composed of 2 catalytic subunits, IKKα (IKK1) and IKKβ (IKK2), and a regulatory subunit, IKKγ (NF-κB essential modulator [NEMO]). Upon stimulation, IKK phosphorylates IκB and causes its ubiquitination and degradation, releasing NF-κB to enter the nucleus and activate target genes. In the noncanonical pathway, IKKα plays the major role in activation of p52-RelB dimers to regulate development of secondary lymphoid organs, as well as B-cell maturation and survival.11
Previous in vitro studies have shown that NF-κB is activated in BCR-ABL1–expressing cell lines12,13 and in primary blasts from patients with advanced CML14 and Ph+ B-ALL.15 However, whether NF-κB plays a role in the pathogenesis of leukemias induced by BCR-ABL1 is unknown. It is also unclear whether the IKK complex is involved in activation of NF-κB in BCR-ABL1–expressing cells, as some studies suggest that BCR-ABL1–evoked NF-κB activation is mediated by IKK,16 while others suggest an IKK-independent mechanism.14,15 Here, we used the mouse retroviral model of CML and Ph+ B-ALL to determine the role of NF-κB signaling in myeloid and lymphoid leukemogenesis by BCR-ABL1. Our findings validate the NF-κB pathway and particularly IKKs as promising targets for therapy of Ph+ leukemia.
Materials and methods
Retroviral constructs
To coexpress BCR-ABL1 and dominant-negative inhibitors of the NF-κB pathway, complementary DNAs (cDNAs) encoding superrepressor mutant of IκBα (IκBαSR)17 and kinase-inactive mutants of IKKα (IKKαKM) and IKKβ (IKKβKM)18 were cloned into the pMIGR1 vector19 in place of green fluorescent protein (GFP). An IκBαSR cDNA without the 5′ and 3′ untranslated regions was polymerase chain reaction (PCR) amplified from the plasmid MSCV-eGFP-SR-IκBα (kind gift of Dr M. A. Shipp, Dana-Farber Cancer Institute, Boston, MA17 ), while cDNAs for FLAG-tagged IKKα-KM and IKKβ-KM constructs18 were amplified by PCR from pCR-FLAG-IKKα-KM or pCR-FLAG-IKKβ-KM plasmids (obtained from Addgene). Primer sequences are available upon request. The respective cDNAs were introduced into pMIGR1 3′ of the internal ribosome entry site (IRES) and the BCR-ABL1 cDNA (p210 isoform) subsequently inserted 5′ of the IRES.
Generation of retrovirus stocks
Replication-defective ecotropic retroviral stocks were generated by transient transfection of 293 cells using the kat packaging system and titered by transduction of NIH3T3 cells, as described. Transduction frequency was assessed by flow cytometric detection of GFP or, in the case of bicistronic viruses coexpressing BCR-ABL1 and IκBαSR or IKKα/βKM, by Southern blot analysis of proviral DNA content in genomic DNA.20 Titers determined by both methods were concordant. Viral stocks were matched by comparing the proviral copy number induced by serial dilutions and selecting concentrations that yield equivalent transduction in primary BM cells (supplemental Figure 1, available on the Blood Web site).
Transformation of cytokine-dependent hematopoietic cell lines
Ba/F3 cells were transduced with BCR-ABL1 retroviruses, selected for growth in the absence of interleukin-3 (IL-3), and growth assessed within 72 hours as described previously.21
BM transduction, transformation, and transplantation
All mouse experiments were approved by the Institutional Animal Use and Care Committee of Tufts Medical Center. Induction of B-ALL and CML-like MPN in the retroviral BM transduction/transplantation model system has been described in detail elsewhere.20 For assessment of myeloid colony formation, 1 × 104 transduced cells were plated in triplicate in cytokine-free methylcellulose (Stem Cell Technologies), and colonies were counted on day 14. Assessment of BCR-ABL1 B-lymphoid transformation by pre-B-cell colony formation in agarose22 and stromal-dependent growth23 was performed as described.
For limiting dilution secondary transplantation, primary mice (n = 2-4) with established CML-like MPN induced by BCR-ABL1/GFP, BCR-ABL1/IkBαSR, or BCR-ABL1/IKKαKM retroviruses were sacrificed, total BM isolated and pooled, and serial dilutions injected IV into cohorts (n = 5 per cell dose) of lethally irradiated secondary Balb/c recipient mice. A diagnosis of MPN was established in secondary recipients by presence of circulating CD11b+ cells > 104/μL and evidence of maturing myeloid infiltration of spleen, liver, and lungs at necropsy. Estimation of leukemia stem cell frequency and statistical analysis were performed by the extreme limiting dilution analysis method and software.24
Southern blot analysis
To determine the frequency of leukemia-initiating cells, distinct proviral integration events were quantitated by Southern blot analysis of genomic DNA from leukemia tissues, as described.21 For leukemias induced by BCR-ABL1/IκBαSR retrovirus or by BCR-ABL1/IKKα/βKM retroviruses, genomic DNA was digested with BglII or NsiI, respectively, transferred to nylon membranes and hybridized with a radioactive probe derived from the IRES. To determine proviral copy number per cell, genomic DNA was digested with BglII, and subjected to Southern blot analysis with a radioactive probe derived from human c-ABL1 gene, which detects a common 2.2-kb fragment from each BCR-ABL1 provirus, as described.21
Immunoblot analysis
Lysates were prepared from primary tumor cell suspensions from BM and spleen by direct boiling, fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotted as described previously.9 Antibodies against ABL1 were obtained from BD Pharmingen, antibodies against total and phospho-IκBα were obtained from Cell Signaling, antibodies against c-Myc were from Santa Cruz Biotechnology, and antibodies against actin were from Sigma-Aldrich.
Immunofluorescence analysis of nuclear RelA
Transformed B-lymphoid cells were washed with cold phosphate-buffered saline (PBS), fixed with 2% paraformaldehyde (Electron Microscopy Sciences) on ice for 20 minutes, and permeabilized with 100% methanol (Fisher Chemical) for 20 minutes on ice. Cells were then washed and blocked with PBS containing 0.1% Triton X-100, 5% fetal bovine serum (FBS), and mouse Fc block (BD Pharmingen) on ice for 10 minutes. After blocking, cells were incubated with 1:50 anti-p65/RelA (Santa Cruz Biotechnology) antibody at room temperature for 1 hour, followed by incubation with 1:200 secondary Alexa Fluor 555–conjugated goat anti-rabbit (Invitrogen) at room temperature for 30 minutes. Cells were then washed PBS/0.05% Triton X-100/2.5% FBS, placed on coverslips by cytospin (Shandon), and imaged using a Leica TCS SP2 confocal microscope with Leica Confocal LCS software.
Cell viability assay with TKIs
Primary B-lymphoid cells were plated in triplicate in 96-well plates in RPMI 1640 medium supplemented with 20% FBS (Gibco/Life Technologies), 200 μM l-glutamine (Cellgro), 50 μM 2-mercaptoethanol (EMD), and 1 mg/mL ciprofloxicin (Sigma-Aldrich). Imatinib or dasatinb were included in the medium at increasing concentrations. At 96 hours postincubation, cell viability was determined by the MTS assay (Promega), following the manufacturer’s instructions.
Results
NF-κB contributes to BCR-ABL1–mediated lymphoid transformation in vitro
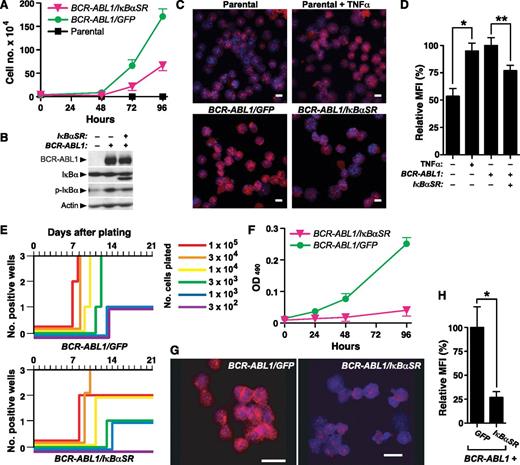
To address whether NF-κB is involved in BCR-ABL1–induced transformation and leukemogenesis, we first used IκBαSR to block NF-κB nuclear translocation and transactivation. IκBαSR contains Ser to Ala mutations at positions 32 and 36, which blocks phosphorylation of IκBαSR by its regulatory kinase(s) and prevents its subsequent degradation. Ba/F3 cells were transduced with retrovirus coexpressing BCR-ABL1 (p210 isoform) and either GFP or IκBαSR, and the growth of cells in the absence of IL-3 was assessed shortly after transduction (Figure 1A). We observed significantly decreased growth of Ba/F3 cells coexpressing BCR-ABL1 and IκBαSR that was due to a primary decrease in the proliferation rate, as there was no significant difference in apoptosis between the 2 populations (data not shown). Ba/F3 cells transduced with BCR-ABL1/IκBαSR expressed a higher mobility IκB species that was recognized by anti-IκBα antibody but not by antiphospho-IκBα antibody (confirming that the mutant IκBαSR is resistant to phosphorylation), and exhibited lower levels of endogenous IκB, consistent with reduced NF-κB activity (Figure 1B). Consistent with previous studies,14 there was increased phospho-IκBα in cells expressing BCR-ABL1 alone relative to parental cells, whereas coexpression of IκBαSR with BCR-ABL1 reduced phospho-IκBα. Expression of BCR-ABL1 increased nuclear RelA to a similar extent as in parental Ba/F3 cells stimulated with the NF-κB agonist tumor necrosis factor α (TNFα), while cells coexpressing BCR-ABL1 and IκBαSR showed significantly reduced nuclear RelA expression (Figure 1C-D).
IκBαSR inhibits in vitro B-lymphoid transformation by BCR-ABL1. (A) Ba/F3 parental cells or cells transduced with retrovirus expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR (4 × 104) were plated in triplicate in the absence of IL-3, and viable cells determined by trypan blue staining. The difference in cell number between BCR-ABL1/GFP–expressing and BCR-ABL1/IκBαSR–expressing Ba/F3 at 96 hours was significant (P < .0001, Student t test). (B) Immunoblot of extracts from the cell lines in panel A, demonstrating expression of IκBαSR. (C) Representative confocal photomicrographs of nuclear RelA expression in Ba/F3 parental cells unstimulated or treated with TNFα (20 ng/mL) for 15 minutes, and Ba/F3 cells expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Cells were stained with antibody against RelA (red) and counterstained with Hoechst dye (blue). Scale bars = 10 μm. (D) Quantification of nuclear RelA fluorescence intensity per cell in the populations shown in panel B by confocal immunofluorescence microscopy. Data are presented as MFI of nuclear RelA staining relative to cells expressing BCR-ABL1/GFP (error bars indicate SE). The differences between unstimulated and TNFα-stimulated parental Ba/F3 cells (*), and between BCR-ABL1/GFP- and BCR-ABL1/IκBαSR-expressing Ba/F3 cells (**), were significant (P = .0003 and P = .0074, respectively, Student t test). (E) Transformation of primary B-lymphoid progenitors in vitro. BM was transduced with BCR-ABL1/GFP or BCR-ABL1/IκBαSR retrovirus and plated in triplicate on stroma at decreasing numbers of cells per well, as indicated by the colored lines. Positive wells were scored vs time when the viable nonadherent cell number reached 106 per well. (F) Proliferation of primary B-lymphoid progenitors transformed by BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Cells (4 × 104) were seeded at day 0. Viable cells were determined by colorimetric assay for reduction of dimethylthiazol diphenyltetrazolium. The difference in viable cell number at 96 hours was significant (P < .0001, Student t test). (G) Representative confocal micrographs of nuclear RelA expression in B lymphoblasts expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Scale bars = 10 μm. (H) Nuclear RelA expression in the transformed cells from panel A was quantified by confocal microscopy, as described in “Materials and methods.” Data are presented as mean nuclear RelA fluorescence relative to cells expressing BCR-ABL1/GFP (error bars indicate SE). The average nuclear RelA content of BCR-ABL1/IκBαSR–transformed B-lymphoid cells was significantly decreased (*P = .0026, Student t test). MFI, mean of fluorescence intensity.
IκBαSR inhibits in vitro B-lymphoid transformation by BCR-ABL1. (A) Ba/F3 parental cells or cells transduced with retrovirus expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR (4 × 104) were plated in triplicate in the absence of IL-3, and viable cells determined by trypan blue staining. The difference in cell number between BCR-ABL1/GFP–expressing and BCR-ABL1/IκBαSR–expressing Ba/F3 at 96 hours was significant (P < .0001, Student t test). (B) Immunoblot of extracts from the cell lines in panel A, demonstrating expression of IκBαSR. (C) Representative confocal photomicrographs of nuclear RelA expression in Ba/F3 parental cells unstimulated or treated with TNFα (20 ng/mL) for 15 minutes, and Ba/F3 cells expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Cells were stained with antibody against RelA (red) and counterstained with Hoechst dye (blue). Scale bars = 10 μm. (D) Quantification of nuclear RelA fluorescence intensity per cell in the populations shown in panel B by confocal immunofluorescence microscopy. Data are presented as MFI of nuclear RelA staining relative to cells expressing BCR-ABL1/GFP (error bars indicate SE). The differences between unstimulated and TNFα-stimulated parental Ba/F3 cells (*), and between BCR-ABL1/GFP- and BCR-ABL1/IκBαSR-expressing Ba/F3 cells (**), were significant (P = .0003 and P = .0074, respectively, Student t test). (E) Transformation of primary B-lymphoid progenitors in vitro. BM was transduced with BCR-ABL1/GFP or BCR-ABL1/IκBαSR retrovirus and plated in triplicate on stroma at decreasing numbers of cells per well, as indicated by the colored lines. Positive wells were scored vs time when the viable nonadherent cell number reached 106 per well. (F) Proliferation of primary B-lymphoid progenitors transformed by BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Cells (4 × 104) were seeded at day 0. Viable cells were determined by colorimetric assay for reduction of dimethylthiazol diphenyltetrazolium. The difference in viable cell number at 96 hours was significant (P < .0001, Student t test). (G) Representative confocal micrographs of nuclear RelA expression in B lymphoblasts expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR. Scale bars = 10 μm. (H) Nuclear RelA expression in the transformed cells from panel A was quantified by confocal microscopy, as described in “Materials and methods.” Data are presented as mean nuclear RelA fluorescence relative to cells expressing BCR-ABL1/GFP (error bars indicate SE). The average nuclear RelA content of BCR-ABL1/IκBαSR–transformed B-lymphoid cells was significantly decreased (*P = .0026, Student t test). MFI, mean of fluorescence intensity.
We next asked whether NF-κB contributes to BCR-ABL1–mediated transformation and proliferation of primary B-lymphoid progenitors on stroma.23 Whereas cultures initiated with as few as 3000 progenitors transduced with BCR-ABL1/GFP retrovirus reached confluence, at least 10-fold more BCR-ABL1/IκBαSR–transduced progenitors were required for the same outcome, with a delay of ∼3 days in time to reach confluence (Figure 1E). As in Ba/F3 cells, the effect of inhibition of NF-κB on B-lymphoblast growth was primarily a decrease in cell proliferation (Figure 1F) without affecting cell survival (supplemental Figure 2). BCR-ABL1/IκBαSR–transformed primary B-lymphoid progenitors had significantly lower nuclear RelA expression than cells transformed by BCR-ABL1/GFP (Figure 1G-H). These results indicate that NF-κB contributes to transformation of B-lymphoid progenitors by BCR-ABL1.
NF-κB is required for efficient induction of lymphoid and myeloid leukemias by BCR-ABL1
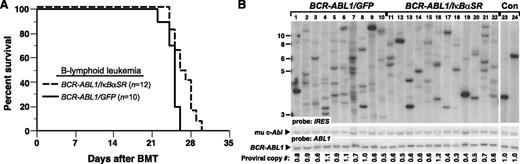
We next assessed whether inhibition of NF-κB through coexpression of IκBαSR would affect BCR-ABL1–mediated leukemogenesis in vivo. To induce B-ALL in mice, BM from non–5-fluorouracil (5-FU)–treated donors was transduced with matched-titer retroviruses expressing BCR-ABL1/GFP or BCR-ABL1/IκBαSR, followed by transplantation into irradiated syngeneic mice.9 Recipients of BM transduced with BCR-ABL1/GFP developed fatal B-ALL with a median survival of 4 weeks (Figure 2A), characterized by circulating malignant BP1+B220+ cells in peripheral blood, lymphadenopathy, moderate splenomegaly (average weight 0.3 g), and a hemorrhagic malignant pleural effusion (supplemental Figure 3). Mice transplanted with BCR-ABL1/IκBαSR–transduced marrow also developed B-ALL with similar pathological features but had a modest but significant increase in their survival (Figure 2A). Lymphoblasts from mice with B-ALL induced by BCR-ABL1/IκBαSR had decreased nuclear RelA expression relative to leukemic cells expressing BCR-ABL1/GFP (supplemental Figure 4).
IκBαSR expression prolongs survival of mice with BCR-ABL1–induced B-ALL. (A) Kaplan-Meier survival curve for recipients of BCR-ABL1/GFP– or BCR-ABL1/IκBαSR–transduced BM. The number of individual mice in each arm is indicated; all mice developed B-ALL. Mice with B-ALL induced by the BCR-ABL1/IκBαSR retrovirus survived significantly longer than control (P = .0096, Mantel-Cox test). (B) Genomic DNA from pleural effusion lymphoblasts of B-ALL mice was subjected to Southern blot analysis to quantify leukemia-initiating cells, as described in “Materials and methods.” The difference in number of proviral clones between leukemias induced by BCR-ABL1/GFP (lanes 1-10, 8.1 ± 3.5 independent clones) or BCR-ABL1/IκBαSR (lanes 11-22, 5.8 ± 2.0 independent clones) was of borderline significance (P = .0697, Student t test). The 2 control DNAs (Con, lanes 23-24) were from cell lines that each contained a single BCR-ABL1 provirus. The same blot was reprobed with a human ABL1 probe to calculate the total proviral copy number per genome, as described.21
IκBαSR expression prolongs survival of mice with BCR-ABL1–induced B-ALL. (A) Kaplan-Meier survival curve for recipients of BCR-ABL1/GFP– or BCR-ABL1/IκBαSR–transduced BM. The number of individual mice in each arm is indicated; all mice developed B-ALL. Mice with B-ALL induced by the BCR-ABL1/IκBαSR retrovirus survived significantly longer than control (P = .0096, Mantel-Cox test). (B) Genomic DNA from pleural effusion lymphoblasts of B-ALL mice was subjected to Southern blot analysis to quantify leukemia-initiating cells, as described in “Materials and methods.” The difference in number of proviral clones between leukemias induced by BCR-ABL1/GFP (lanes 1-10, 8.1 ± 3.5 independent clones) or BCR-ABL1/IκBαSR (lanes 11-22, 5.8 ± 2.0 independent clones) was of borderline significance (P = .0697, Student t test). The 2 control DNAs (Con, lanes 23-24) were from cell lines that each contained a single BCR-ABL1 provirus. The same blot was reprobed with a human ABL1 probe to calculate the total proviral copy number per genome, as described.21
To determine the effect of inhibiting NF-κB on the frequency of leukemia-initiating or leukemia stem cells in mice with BCR-ABL1–induced B-ALL, we quantified the number of unique proviral clones in leukemic cells from individual recipient mice in the 2 cohorts by Southern blotting (Figure 2B). Mice with B-ALL who were recipients of BCR-ABL1/GFP–transduced BM showed an oligoclonal to polyclonal pattern of leukemia-initiating cells, with an average of 8.1 ± 3.5 independent proviral clones per leukemic recipient. Leukemias that developed in recipients of BCR-ABL1/IκBαSR–transduced marrow exhibited a lower average number of proviral clones (5.8 ± 2.0), which was of borderline statistical significance (P = .0697, Student t test). These results demonstrate that inhibition of NF-κB attenuates B-lymphoid leukemogenesis by BCR-ABL1.
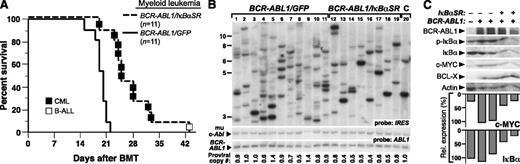
We next tested whether coexpression of IκBαSR affected the pathogenesis of the CML-like MPN induced by BCR-ABL1 in the mouse retroviral transplantation model. To induce CML-like MPN in mice, BM was harvested from donors pretreated with 5-FU and transduced with BCR-ABL1/GFP or BCR-ABL1/IκBαSR retrovirus in the presence of myeloid cytokines, followed by transplantation into irradiated syngeneic recipients. All mice transplanted with BCR-ABL1/GFP–transduced marrow developed CML-like MPN characterized by a greatly elevated leukocyte count in peripheral blood (∼1-2 × 105 cells per μL), splenomegaly (mean spleen weight 1.2 g), lung hemorrhages, and massive expansion of Gr-1+Mac-1/CD11b+ progenitors in BM, spleen, and liver (supplemental Figure 3). Mice transplanted with BCR-ABL1/IκBαSR–transduced BM developed a similar MPN, but had significantly longer survival (Figure 3A; median survival, 25 days vs 19 days for BCR-ABL1/GFP recipients; P < .0001, Mantel-Cox test). Southern blot analysis of leukemia-initiating cell frequency in the MPNs from the 2 cohorts (Figure 3B) showed that coexpression of IκBαSR with BCR-ABL1 significantly decreased the number of independent proviral clones in the malignant myeloid cells (6.8 ± 1.4 clones vs 9.9 ± 2.8 clones in BCR-ABL1/GFP recipients, P = .0096, Student t test). These results suggest that NF-κB inhibition attenuates BCR-ABL1–induced CML-like MPN in part through targeting the leukemic stem cells in this model.
Expression of IκBαSR attenuates CML-like MPN induced by BCR-ABL1. (A) Kaplan-Meier survival curve for CML-like MPN induced by BCR-ABL1/GFP or BCR-ABL1/IκBαSR. The number of individual mice in each arm is indicated. All mice receiving BCR-ABL1/GFP–transduced BM developed CML-like MPN; recipients of BCR-ABL1/IκBαSR–transduced BM developed CML-like MPN (▪) except for 1 recipient who developed B-ALL (□), and had significantly longer survival than recipients of BCR-ABL1/GFP–transduced BM (P < .0001, Mantel-Cox test). (B) Analysis of leukemia-initiating cell frequency in recipients from the 2 cohorts in panel A. Leukemias induced by BCR-ABL1/GFP (lanes 1-11) had a significantly higher number of independent clones (9.9 ± 2.8) than leukemias induced by BCR-ABL1/IκBαSR (lanes 12-19; 6.8 ± 1.4 clones, P = .0096, Student t test). The control DNA (C, lane 20) was from a cell line that each contained a single BCR-ABL1 provirus. (C) Immunoblot of protein lysates from spleens of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 2-3) or BCR-ABL1/IκBαSR (lanes 4-5) with the indicated antibodies. Lysate from untransduced mouse BM (lane 1) served as a control. The relative expression of c-MYC was quantified and normalized to actin (bar graph).
Expression of IκBαSR attenuates CML-like MPN induced by BCR-ABL1. (A) Kaplan-Meier survival curve for CML-like MPN induced by BCR-ABL1/GFP or BCR-ABL1/IκBαSR. The number of individual mice in each arm is indicated. All mice receiving BCR-ABL1/GFP–transduced BM developed CML-like MPN; recipients of BCR-ABL1/IκBαSR–transduced BM developed CML-like MPN (▪) except for 1 recipient who developed B-ALL (□), and had significantly longer survival than recipients of BCR-ABL1/GFP–transduced BM (P < .0001, Mantel-Cox test). (B) Analysis of leukemia-initiating cell frequency in recipients from the 2 cohorts in panel A. Leukemias induced by BCR-ABL1/GFP (lanes 1-11) had a significantly higher number of independent clones (9.9 ± 2.8) than leukemias induced by BCR-ABL1/IκBαSR (lanes 12-19; 6.8 ± 1.4 clones, P = .0096, Student t test). The control DNA (C, lane 20) was from a cell line that each contained a single BCR-ABL1 provirus. (C) Immunoblot of protein lysates from spleens of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 2-3) or BCR-ABL1/IκBαSR (lanes 4-5) with the indicated antibodies. Lysate from untransduced mouse BM (lane 1) served as a control. The relative expression of c-MYC was quantified and normalized to actin (bar graph).
To determine downstream signaling pathways affected by the expression of IκBαSR, protein lysates from spleens of mice with CML-like MPN induced by BCR-ABL1/GFP or BCR-ABL1/IκBαSR were subjected to immunoblot analysis. Previous reports indicated that the genes for c-Myc and the prosurvival factor Bcl-X both contain κB binding sites and are induced by BCR-ABL1 expression.25,26 Consistent with these studies, expression of both c-MYC and BCL-X proteins was induced by BCR-ABL1, whereas coexpression of IκBαSR reduced the expression of c-MYC but not BCL-X (Figure 3C). As in Ba/F3 cells, myeloid leukemia cells expressing IκBαSR had reduced levels of endogenous IκB, indicative of suppression of NF-κB signaling.
Inhibition of IKKα and IKKβ impairs BCR-ABL1–mediated B-lymphoid transformation in vitro
To investigate the mechanism of activation of NF-κB in BCR-ABL1–expressing cells, we tested whether 2 upstream kinases that negatively regulate IκBα, IKKα/IKK1 and IKKβ/IKK2, were involved. To accomplish this, we engineered retroviruses coexpressing BCR-ABL1 and kinase-inactive mutants of IKKα (IKKαKM) or IKKβ (IKKβKM), created by substitution of alanine for lysine within the ATP binding site. The ability of the resulting IKKαKM and IKKβKM mutants to block NF-κB activation has been demonstrated previously.18
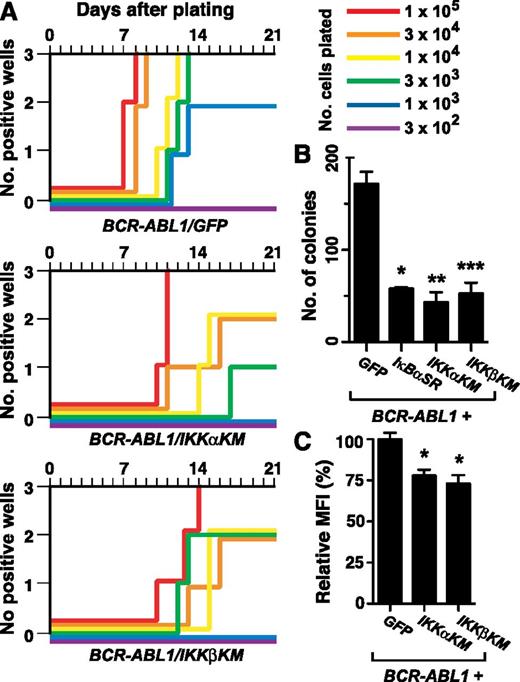
We first tested the effect of coexpression of IKKαKM or IKKβKM on BCR-ABL1–mediated B-lymphoid transformation in the stromal growth assay.23 Expression of either IKK mutant significantly impaired BCR-ABL1–mediated transformation, manifested both an increased time for culture outgrowth and a >30-fold increase in the number of transduced cells required to achieve maximal growth in all wells (Figure 4A). In a complementary transformation assay, colony formation in agarose,22 BM transduced with BCR-ABL1/IκBαSR retrovirus or either BCR-ABL1/IKK-KM retrovirus formed significantly fewer colonies (Figure 4B) that were much smaller in size (supplemental Figure 5) than those generated by transduction with the BCR-ABL1/GFP retrovirus. Lymphoblasts transformed by either BCR-ABL1/IKK-KM retrovirus had significantly reduced nuclear RelA expression relative to BCR-ABL1/GFP–expressing cells (Figure 4C; P < .0001, Student t test). Together, these results demonstrate that blocking NF-κB activation though inhibition of IKKs impairs BCR-ABL1–mediated B-lymphoid transformation.
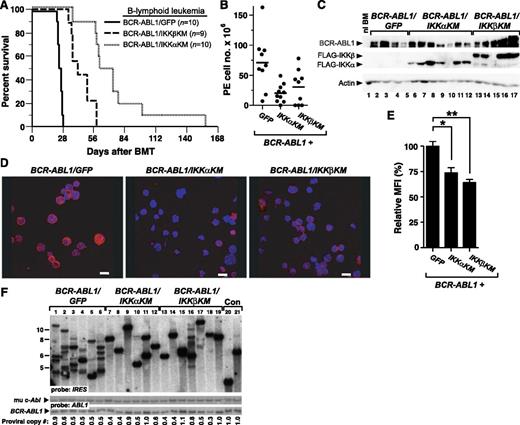
Kinase-inactive IKK mutants inhibit B-lymphoid transformation by BCR-ABL1. BM from non–5-FU–treated Balb/c donor mice was harvested and transduced with retrovirus expressing BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. (A) Assessment of stromal-dependent B-lymphoid transformation and growth. Nomenclature is as in Figure 1E. (B) B-lymphoid colony formation in agarose. Transduced cells (2 × 106 per plate, in duplicate) were seeded in agarose as described in “Materials and methods.” Colony formation was assessed at day 14. Coexpression of IκBαSR (*P = .0063), IKKαKM (**P = .0082), and IKKβKM (***P = .0101) (Student t tests) significantly reduced B-lymphoid colony formation mediated by BCR-ABL1. (C) Quantification of nuclear RelA expression in primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM (mean + SE). Cells were stained with antibody against RelA and analyzed by confocal microscopy. BCR-ABL1/IKKαKM– and BCR-ABL1/IKKβKM–transformed cells showed significantly reduced nuclear RelA compared with cells transformed by BCR-ABL1/GFP (*P < .0001, Student t test).
Kinase-inactive IKK mutants inhibit B-lymphoid transformation by BCR-ABL1. BM from non–5-FU–treated Balb/c donor mice was harvested and transduced with retrovirus expressing BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. (A) Assessment of stromal-dependent B-lymphoid transformation and growth. Nomenclature is as in Figure 1E. (B) B-lymphoid colony formation in agarose. Transduced cells (2 × 106 per plate, in duplicate) were seeded in agarose as described in “Materials and methods.” Colony formation was assessed at day 14. Coexpression of IκBαSR (*P = .0063), IKKαKM (**P = .0082), and IKKβKM (***P = .0101) (Student t tests) significantly reduced B-lymphoid colony formation mediated by BCR-ABL1. (C) Quantification of nuclear RelA expression in primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM (mean + SE). Cells were stained with antibody against RelA and analyzed by confocal microscopy. BCR-ABL1/IKKαKM– and BCR-ABL1/IKKβKM–transformed cells showed significantly reduced nuclear RelA compared with cells transformed by BCR-ABL1/GFP (*P < .0001, Student t test).
Dominant-negative IKK mutants attenuate BCR-ABL1–mediated B-ALL
We next tested the effect of IKK inhibition on B-lymphoid leukemogenesis by BCR-ABL1. As before, recipients of BCR-ABL1/GFP–transduced BM succumbed to B-ALL within 4 weeks of transplantation (Figure 5A). By contrast, recipients of BM transduced with either BCR-ABL1/IKK-KM retrovirus also developed B-ALL but had significantly longer survival than BCR-ABL1/GFP recipients (P < .0001, Mantel-Cox test) and exhibited a milder disease phenotype, with smaller malignant pleural effusions (Figure 5B) and lymph nodes (data not shown) at the time of morbidity or death. Interestingly, inhibition of IKKα appeared to have a greater effect than IKKβ, with recipients of BCR-ABL1/IKKαKM–transduced BM surviving significantly longer than those transplanted with BCR-ABL1/IKKβKM–transduced BM (Figure 5A; P = .0007, Mantel-Cox test). The transformed lymphoblasts expressed the FLAG-tagged IKK-KM mutants (Figure 5C), and had decreased nuclear RelA levels compared with BCR-ABL1/GFP–transformed cells (Figure 5D-E). Coexpression of either IKK-KM mutant with BCR-ABL1 significantly reduced the number of leukemia-initiating cells in diseased recipient mice (Figure 5F), with leukemias from BCR-ABL1/IKKαKM and BCR-ABL1/IKKβKM recipients derived from 2.0 ± 1.1 and 2.7 ± 1.7 independent proviral clones, respectively, while the average clonality of BCR-ABL1/GFP–induced leukemias was 6.2 ± 1.9 (P = .001 and P = .0057, respectively, Student t test).
Kinase-inactive IKK mutants attenuate B-lymphoid leukemogenesis by BCR-ABL1 in mice. (A) Kaplan-Meier survival curve for B-ALL induced in mice by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. The number of individual mice in each arm is indicated. All recipients developed B-ALL. Coexpression of either IKKαKM or IKKβKM significantly prolonged the survival of mice with BCR-ABL1–induced B-ALL (P < .0001, Mantel-Cox test). In addition, mice with B-ALL induced by BCR-ABL1/IKKαKM survived significantly longer than those induced by BCR-ABL1/IKKβKM (P = .0007, Mantel-Cox test). (B) Total cell number collected from malignant pleural effusions in leukemic mice from the cohorts in panel A. Compared with B-ALL induced by BCR-ABL1/GFP, recipients with B-ALL induced by BCR-ABL1/IKKαKM (P = .0039) or BCR-ABL1/IKKβKM (P = .0393, Student t test) have significantly fewer leukemic cells in malignant pleural effusions. (C) Immunoblot analysis of protein lysates from pleural effusion lymphoblasts from mice with B-ALL induced by BCR-ABL1/GFP (lanes 2-5), BCR-ABL1/IKKαKM (lanes 6-12), or BCR-ABL1/IKKβKM (lanes 13-17). BM from an untransplanted mouse was loaded in lane 1 as a control. Lysates were analyzed with antibodies against c-Abl, FLAG, and actin. (D) Representative confocal micrographs of nuclear RelA expression in B-lymphoid leukemic cells from recipients of BM transduced with BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. Cells were stained with antibody against RelA (Red) and counterstained with Hoechst dye (blue) as described in “Materials and methods.” Scale bars = 10 μm. (E) Quantification of nuclear RelA expression from the data in panel D (mean + SE). Leukemic cells expressing BCR-ABL1/GFP showed significantly higher nuclear RelA than cells expressing BCR-ABL1/IKKαKM (*P = .0003, Student t test) or BCR-ABL1/IKKβKM (**P < .0001, Student t test). (F) Analysis of genomic DNA from leukemic tissues of mice with B-ALL induced by BCR-ABL1/GFP (lanes 1-6), BCR-ABL1/IKKαKM (lanes 7-12), or BCR-ABL1/IKKβKM (lanes 13-19) by Southern blot with IRES probe to detect distinct proviral integration events. Two control DNAs (Con, lanes 20-21) were from cell lines that each contain a single BCR-ABL1 provirus. B-ALLs induced by BCR-ABL1/IKKαKM (P = .001, Student t test) and BCR-ABL1/IKKβKM (P = .0057, Student t test) showed significantly decreased frequency of leukemia-initiating cells as compared with BCR-ABL1/GFP. Because of the lower titers consistently obtained for BCR-ABL1 retroviruses coexpressing IKKα/βKM (supplemental Figure 1), we used more dilute BCR-ABL1/GFP retrovirus to match titers, resulting in a lower average number of proviral clones in the GFP control arm than in Figure 2B.
Kinase-inactive IKK mutants attenuate B-lymphoid leukemogenesis by BCR-ABL1 in mice. (A) Kaplan-Meier survival curve for B-ALL induced in mice by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. The number of individual mice in each arm is indicated. All recipients developed B-ALL. Coexpression of either IKKαKM or IKKβKM significantly prolonged the survival of mice with BCR-ABL1–induced B-ALL (P < .0001, Mantel-Cox test). In addition, mice with B-ALL induced by BCR-ABL1/IKKαKM survived significantly longer than those induced by BCR-ABL1/IKKβKM (P = .0007, Mantel-Cox test). (B) Total cell number collected from malignant pleural effusions in leukemic mice from the cohorts in panel A. Compared with B-ALL induced by BCR-ABL1/GFP, recipients with B-ALL induced by BCR-ABL1/IKKαKM (P = .0039) or BCR-ABL1/IKKβKM (P = .0393, Student t test) have significantly fewer leukemic cells in malignant pleural effusions. (C) Immunoblot analysis of protein lysates from pleural effusion lymphoblasts from mice with B-ALL induced by BCR-ABL1/GFP (lanes 2-5), BCR-ABL1/IKKαKM (lanes 6-12), or BCR-ABL1/IKKβKM (lanes 13-17). BM from an untransplanted mouse was loaded in lane 1 as a control. Lysates were analyzed with antibodies against c-Abl, FLAG, and actin. (D) Representative confocal micrographs of nuclear RelA expression in B-lymphoid leukemic cells from recipients of BM transduced with BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. Cells were stained with antibody against RelA (Red) and counterstained with Hoechst dye (blue) as described in “Materials and methods.” Scale bars = 10 μm. (E) Quantification of nuclear RelA expression from the data in panel D (mean + SE). Leukemic cells expressing BCR-ABL1/GFP showed significantly higher nuclear RelA than cells expressing BCR-ABL1/IKKαKM (*P = .0003, Student t test) or BCR-ABL1/IKKβKM (**P < .0001, Student t test). (F) Analysis of genomic DNA from leukemic tissues of mice with B-ALL induced by BCR-ABL1/GFP (lanes 1-6), BCR-ABL1/IKKαKM (lanes 7-12), or BCR-ABL1/IKKβKM (lanes 13-19) by Southern blot with IRES probe to detect distinct proviral integration events. Two control DNAs (Con, lanes 20-21) were from cell lines that each contain a single BCR-ABL1 provirus. B-ALLs induced by BCR-ABL1/IKKαKM (P = .001, Student t test) and BCR-ABL1/IKKβKM (P = .0057, Student t test) showed significantly decreased frequency of leukemia-initiating cells as compared with BCR-ABL1/GFP. Because of the lower titers consistently obtained for BCR-ABL1 retroviruses coexpressing IKKα/βKM (supplemental Figure 1), we used more dilute BCR-ABL1/GFP retrovirus to match titers, resulting in a lower average number of proviral clones in the GFP control arm than in Figure 2B.
Dominant-negative IKK mutants impair myeloid transformation and leukemogenesis by BCR-ABL1
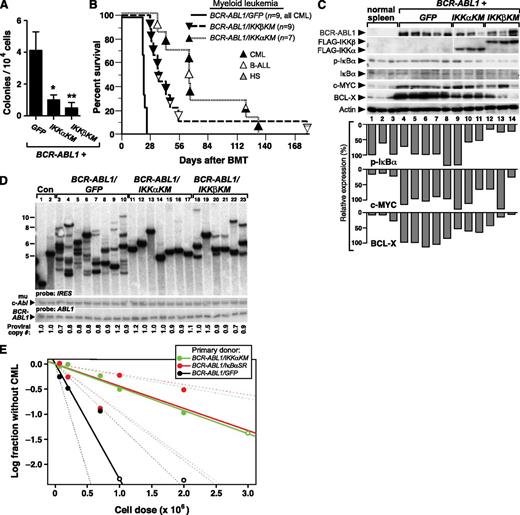
We further investigated whether expression of dominant-negative IKK mutants would affect primary myeloid transformation and leukemogenesis by BCR-ABL1. When expressed in mouse myeloid progenitors via retroviral transduction, BCR-ABL1 can induce myeloid colonies in the absence of exogenous cytokines.27 In this assay, BM progenitors transduced by either BCR-ABL1/IKK-KM retrovirus formed fewer colonies in methylcellulose culture relative to BCR-ABL1/GFP (Figure 6A), demonstrating that both IKK mutants impair cytokine-independent myelopoiesis mediated by BCR-ABL1.
Kinase-inactive IKK mutants attenuate CML-like MPN induced by BCR-ABL1 in mice. BM from 5-FU–treated donors was transduced with retrovirus expressing BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM, and subsequently either plated in methylcellulose culture (A) or transplanted into irradiated recipients to induce CML-like MPN (B). (A) Myeloid colony assay. Transduced BM cells (1 × 104) were seeded per plate in triplicate. Colony number was determined at day 14. The differences in colony number for BCR-ABL1/IKKαKM– or BCR-ABL1/IKKβKM–transduced progenitors were significant (*P = .022 and **P = .024, Student t tests). (B) Kaplan-Meier survival curve for recipients of BCR-ABL1–transduced BM in the CML model. The number of mice from the different arms is indicated, with the phenotype of disease indicated by the shading of the symbol; mice with mixed hematologic disease are indicated by mixed shading. Recipients of BM transduced by BCR-ABL1/IKKαKM (P < .0001, Mantel-Cox test) or BCR-ABL1/IKKβKM (P < .0001, Mantel-Cox test) survived significantly longer than BCR-ABL1/GFP recipients. (C) Kinase-inactive IKK mutants inhibit BCL-X expression induced by BCR-ABL1–expressing myeloid progenitors. Immunoblot analysis of protein lysates from spleens of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 4-8), BCR-ABL1/IKKαKM (lanes 9-11), and BCR-ABL1/IKKβKM (lanes12-14). Lysates from spleens of 3 untransplanted mice (lanes 1-3) were loaded as controls. The blot was analyzed with antibodies against BCR-ABL1, FLAG, IκBα, c-MYC, and BCL-X, and actin. (D) Analysis of genomic DNA from leukemic tissues of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 3-10), BCR-ABL1/IKKαKM (lanes 11-17), or BCR-ABL1/IKKβKM (lanes 18-23) by Southern blot with an IRES probe to detect distinct proviral integration events. Two control DNAs (Con, lanes 1-2) were from cell lines that each contain a single BCR-ABL1 provirus. CML-like MPN induced by BCR-ABL1/IKKαKM (P < .0001, Student t test) and BCR-ABL1/IKKβKM (P = .0003, t test) showed a significant reduction in leukemia-initiating cell frequency as compared with BCR-ABL1/GFP. Because of the lower titers consistently obtained for BCR-ABL1 retroviruses coexpressing IKKα/βKM (supplemental Figure 1), we used more dilute BCR-ABL1/GFP retrovirus to match titers, resulting in a lower average number of proviral clones in the GFP control arm than in Figure 3B. (E) Limiting dilution secondary transplantation analysis of resident BM leukemia stem cell frequency in primary mice with CML-like MPN induced by BCR-ABL1/GFP (black), BCR-ABL1/IkBαSR (red), and BCR-ABL1/IKKαKM (green) retroviruses. The frequency of secondary recipients who did not develop CML-like MPN (log scale) is indicated by the closed symbols; open symbols indicate all recipients developed MPN at that cell dose. Dashed lines indicate 95% confidence intervals.
Kinase-inactive IKK mutants attenuate CML-like MPN induced by BCR-ABL1 in mice. BM from 5-FU–treated donors was transduced with retrovirus expressing BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM, and subsequently either plated in methylcellulose culture (A) or transplanted into irradiated recipients to induce CML-like MPN (B). (A) Myeloid colony assay. Transduced BM cells (1 × 104) were seeded per plate in triplicate. Colony number was determined at day 14. The differences in colony number for BCR-ABL1/IKKαKM– or BCR-ABL1/IKKβKM–transduced progenitors were significant (*P = .022 and **P = .024, Student t tests). (B) Kaplan-Meier survival curve for recipients of BCR-ABL1–transduced BM in the CML model. The number of mice from the different arms is indicated, with the phenotype of disease indicated by the shading of the symbol; mice with mixed hematologic disease are indicated by mixed shading. Recipients of BM transduced by BCR-ABL1/IKKαKM (P < .0001, Mantel-Cox test) or BCR-ABL1/IKKβKM (P < .0001, Mantel-Cox test) survived significantly longer than BCR-ABL1/GFP recipients. (C) Kinase-inactive IKK mutants inhibit BCL-X expression induced by BCR-ABL1–expressing myeloid progenitors. Immunoblot analysis of protein lysates from spleens of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 4-8), BCR-ABL1/IKKαKM (lanes 9-11), and BCR-ABL1/IKKβKM (lanes12-14). Lysates from spleens of 3 untransplanted mice (lanes 1-3) were loaded as controls. The blot was analyzed with antibodies against BCR-ABL1, FLAG, IκBα, c-MYC, and BCL-X, and actin. (D) Analysis of genomic DNA from leukemic tissues of mice with CML-like MPN induced by BCR-ABL1/GFP (lanes 3-10), BCR-ABL1/IKKαKM (lanes 11-17), or BCR-ABL1/IKKβKM (lanes 18-23) by Southern blot with an IRES probe to detect distinct proviral integration events. Two control DNAs (Con, lanes 1-2) were from cell lines that each contain a single BCR-ABL1 provirus. CML-like MPN induced by BCR-ABL1/IKKαKM (P < .0001, Student t test) and BCR-ABL1/IKKβKM (P = .0003, t test) showed a significant reduction in leukemia-initiating cell frequency as compared with BCR-ABL1/GFP. Because of the lower titers consistently obtained for BCR-ABL1 retroviruses coexpressing IKKα/βKM (supplemental Figure 1), we used more dilute BCR-ABL1/GFP retrovirus to match titers, resulting in a lower average number of proviral clones in the GFP control arm than in Figure 3B. (E) Limiting dilution secondary transplantation analysis of resident BM leukemia stem cell frequency in primary mice with CML-like MPN induced by BCR-ABL1/GFP (black), BCR-ABL1/IkBαSR (red), and BCR-ABL1/IKKαKM (green) retroviruses. The frequency of secondary recipients who did not develop CML-like MPN (log scale) is indicated by the closed symbols; open symbols indicate all recipients developed MPN at that cell dose. Dashed lines indicate 95% confidence intervals.
To evaluate the role of IKK activation in CML, we induced CML-like MPN by transplanting BM from 5-FU–treated donors transduced with retrovirus expressing BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM. As before, all recipients of BCR-ABL1/GFP–transduced BM succumbed to CML-like MPN within 25 days of transplantation (Figure 6B). By contrast, mice receiving BM transduced with either BCR-ABL1/IKK-KM retrovirus survived significantly longer (P < .0001, Mantel-Cox test). While about half of the recipients of BCR-ABL1/IKK-KM–transduced BM developed MPN, the remaining recipients developed other hematopoietic malignancies such as B-ALL or histiocytic sarcoma, sometimes in combination with MPN. Mice that developed mixed CML-like MPN and B-ALL exhibited the cardinal clinicopathological features of both leukemias; such mice are frequently observed in recipients of BCR-ABL1–transduced BM under conditions when CML-like disease is attenuated.9 Immunoblot analysis confirmed the expression of the FLAG-tagged IKK-KM mutants in BCR-ABL1–expressing myeloid cells, and showed a consistent reduction in the level of the antiapoptotic protein BCL-XL in recipients of BM transduced with either BCR-ABL1/IKK-KM retrovirus (Figure 6C). On the other hand, reduced expression of c-MYC, which regulates proliferation in BCR-ABL1–transformed cells,28 was more variable but was observed in some MPNs induced by BCR-ABL1/IKKβKM. The level of phospho-IκBα was lower in cells expressing either dominant-negative IKK mutant, consistent with decreased NF-κB activity. Together with our previous findings with IκBαSR, this suggests the canonical IKKβ-IκBα pathway may regulate both cell proliferation and survival in CML.
There was a striking reduction in the frequency of leukemia-initiating cells in the MPN induced by BCR-ABL1/IKKαKM (1.4 ± 0.3 clones) and BCR-ABL1/IKKβKM (3.3 ± 0.5 clones) retroviruses compared with the polyclonal (7.6 ± 0.6) disease induced by BCR-ABL1/GFP (Figure 6D; P < .0001 and P = .0003, respectively, Student t test). To determine whether inhibition of NF-κB signaling reduced the frequency of resident leukemic stem cells in mice with BCR-ABL1–induced MPN, we performed limiting dilution secondary transplantation of BM from primary mice with CML-like MPN induced by BCR-ABL1/GFP, BCR-ABL1/IkBαSR, and BCR-ABL1/IKKαKM retroviruses into cohorts of lethally irradiated secondary recipients, who were followed for development of MPN. Estimation of CML stem cell frequency from these data (Figure 6E) revealed that coexpression of either IkBαSR or IKKαKM reduced the frequency of BM cells capable of initiating MPN in secondary recipients approximately fivefold (from ∼1:436 000 for GFP to ∼1:2 260 000 or 1:2 160 000 for IkBαSR or IKKαKM, respectively; P = .0052 or 0.0064, respectively, χ2 test).
NF-κB inhibition increases the sensitivity of BCR-ABL1–expressing leukemic cells to TKIs
Inhibition of NF-κB signaling through coexpression of either IκBαSR or dominant-negative IKKs attenuated lymphoid and myeloid leukemogenesis by BCR-ABL1, validating NF-κB as a potential target for therapy in Ph+ leukemia. We therefore asked whether inhibition of NF-κB might enhance the sensitivity of BCR-ABL1–expressing leukemic cells to TKI drugs such as imatinib or dasatinib. Primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IκBαSR, or BCR-ABL1/IKK-KM were cultured in vitro in the presence of increasing concentrations of TKIs. Importantly, coexpression of IκBαSR sensitized cells to imatinib, manifested by a modest but significant decrease in the IC50 from 316 nM to 118 nM (Figure 7A). Similarly, coexpression of IKKαKM or IKKβKM increased the sensitivity of BCR-ABL1–expressing lymphoblasts to imatinib (Figure 7B), with the IC50 falling from 624 nM to 171 nM and 151 nM, respectively. Coexpression of IKKαKM also sensitized the leukemic cells to the more potent second-generation TKI dasatinib, with a decrease in the IC50 from 0.78 nM to 0.26 nM (Figure 7C). We did not observe any effect of inhibition of NF-κB on the sensitivity of the malignant B lymphoblasts to glucocorticoids (dexamethasone; data not shown). These results suggest that IKK inhibition in combination with TKIs may be a rational strategy for treatment of Ph+ leukemias.
Inhibition of NF-κB sensitizes BCR-ABL1–transformed B lymphoblasts to TKIs. (A) BCR-ABL1/GFP– or BCR-ABL1/IκBαSR–transformed primary B-lymphoid progenitors (4 × 104 cells per well) were incubated with different concentration of imatinib as indicated. Cell viability was determined by MTS assay after 96 hours’ incubation. (B-C) Equal numbers of primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM were incubated with different concentration of imatinib (B) or dasatinib (C) as indicated. Cell viability was determined by (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay after 96 hours’ incubation. Sigmoidal curves were fitted and IC50 values calculated by Prism software.
Inhibition of NF-κB sensitizes BCR-ABL1–transformed B lymphoblasts to TKIs. (A) BCR-ABL1/GFP– or BCR-ABL1/IκBαSR–transformed primary B-lymphoid progenitors (4 × 104 cells per well) were incubated with different concentration of imatinib as indicated. Cell viability was determined by MTS assay after 96 hours’ incubation. (B-C) Equal numbers of primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM were incubated with different concentration of imatinib (B) or dasatinib (C) as indicated. Cell viability was determined by (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay after 96 hours’ incubation. Sigmoidal curves were fitted and IC50 values calculated by Prism software.
Discussion
Despite the considerable clinical success of TKIs in the treatment of CML, we now appreciate that TKI therapy has important limitations in this disease.29 While the majority of patients with chronic-phase CML achieve cytogenetic remission on imatinib or one of the second-generation TKIs, even those with undetectable BCR-ABL1 transcripts are at risk for rapid recurrence of leukemia when TKI therapy is discontinued.1 Ph+ B-ALL is an indication for allogeneic stem cell transplantation in first remission, and despite addition of TKIs to induction chemotherapy and transplantation treatment regimens,30 over half of allografted patients still relapse following the procedure.
These observations have triggered a major effort to identify critical signaling pathways in Ph+ leukemia whose blockade can prevent or overcome TKI resistance and perhaps lead to permanent cure of these diseases.5 In this regard, the NF-κB signaling pathway is an attractive candidate. In acute myeloid leukemia (AML), NF-κB is activated in leukemic blasts31,32 and in the more primitive leukemia-initiating or leukemic stem cells,33 but not in normal hematopoietic stem cells. Inhibition of NF-κB signaling via several pharmacological approaches, including parthenolide derivatives,34,35 proteasome inhibitors,36 and direct IKK inhibitors,37 can induce apoptosis in AML blasts. In CML, preclinical therapeutic studies of NF-κB inhibition have been more limited, but both proteasome38-40 and IKK41-43 inhibitors induce apoptosis in CML cell lines and impair myeloid colony formation from BM of CML patients. However, it is not clear whether the cytotoxic effects of proteasome inhibitors such as bortezomib on CML progenitors39,40 is due to direct inhibition of NF-κB or of another pathway, such as FOXO transcription factors.38
Here, we have used such a genetic strategy in mouse models of CML and Ph+ B-ALL to demonstrate that NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1. We used 2 validated approaches to inhibit NF-κB signaling in primary murine hematopoietic cells expressing BCR-ABL1. The superrepressor mutant of IκBα, IκBαSR, is resistant to phosphorylation by IKKs and stabilizes the IκBαSR:NF-κB complex in the cytoplasm.17 Dominant-negative, kinase-inactive mutants of IKKα and IKKβ inhibit the serine phosphorylation and kinase activity of the IKK complex and subsequently activation of NF-κB,18 probably through competition for upstream activators.
A previous report indicated that Ba/F3 cells expressing the p185 isoform of BCR-ABL1 increased NF-κB–dependent transcription without an increase in nuclear NF-κB levels,13 but we observed increased nuclear RelA/p65 in both Ba/F3 and primary B-lymphoid cells expressing p210 BCR-ABL1 (Figure 1C,G), while coexpression of IκBαSR or either IKKαKM or IKKβKM effectively inhibited NF-κB activation. In BCR-ABL1–expressing B lymphoblasts, NF-κB was inhibited 50% to 75% from maximal levels by IκBαSR, whereas coexpression of either IKKαKM or IKKβKM individually with BCR-ABL1 was slightly less effective at suppressing relative levels of nuclear RelA, possibly because of compensation from the other IKK subunit. The mechanism of activation of NF-κB by BCR-ABL1 is controversial, with studies in different cell lines suggesting either IKK-dependent16,42 or -independent12,14,15 mechanisms. Here, we observed increased serine phosphorylation of IκBα in primary murine leukemia cells expressing BCR-ABL1 and attenuation of BCR-ABL1 leukemogenesis by both IKKαKM and IKKβKM mutants, firmly linking IKK to the activation of NF-κB in BCR-ABL1+ leukemias and validating IKK as a therapeutic target in these diseases.
In physiologically accurate and quantitative mouse models of Ph+ leukemia, coexpression of IκBαSR or either IKKαKM or IKKβKM with BCR-ABL1 significantly attenuated both B-ALL and CML-like MPN, evoked by BCR-ABL1 in distinct BM target cells. It is interesting that attenuation of both lymphoid and myeloid leukemogenesis by the IKK mutants (Figures 5A and 6B) was more profound than that mediated by IκBαSR (Figures 2A and 3A) despite less effective inhibition of nuclear RelA, and that the IKKαKM mutant was more effective in this regard than IKKβKM, although IKKβ is generally considered to be the principal IKK mediating activation of NF-κB in the canonical pathway.44 In this regard, IKKα can also directly phosphorylate RelA and mediate its nuclear translocation,45 while substrates other than IκBα have been implicated in proinflammatory, proliferative, and tumor-promoting functions of IKK.46 The function of IKKα in the alternative pathway regulating mature B-cell development47 might also be relevant to its role in the pathogenesis of BCR-ABL1–induced B-ALL.
The precise mechanism of the inhibition of BCR-ABL1 leukemogenesis by NF-κB blockade will require further studies, but our data suggest differences between lymphoid and myeloid leukemias. In BCR-ABL1–expressing B lymphoblasts, the principal effect of NF-κB inhibition was on cell proliferation rather than survival, with no significant increase in apoptosis (Figure 1F and supplemental Figure 2). By contrast, in BCR-ABL1–expressing myeloid cells, NF-κB inhibition with dominant-negative IKKs caused decreased expression of BCL-X with variable effects on c-MYC (Figure 6C), consistent with a previous study in the Ph+ myeloid cell line K562.48 Our results also implicate NF-κB in the maintenance of BCR-ABL1 leukemia-initiating cells, rigorously defined in both primary and limiting dilution secondary transplant assays.49 The effect of NF-κB inhibition on these leukemic “stem” cells was more profound in the CML model than for B-ALL. Although we cannot exclude an effect of NF-κB blockade on the initial homing and engraftment of these leukemia-initiating cells, together these results suggest that the predominant role of NF-κB signaling in CML cells may be to inhibit apoptosis.
Based on our findings and previous studies, is NF-κB a promising target for therapy in Ph+ B-ALL and CML? The answer depends in part on the magnitude of the antileukemic effect mediated by genetic inhibition of the NF-κB pathway observed herein (where the suppression of NF-κB activity by these dominant-negative mutants is continuous) and the extent to which this can be reproduced in patients by NF-κB inhibitor drugs, but also on the specific pathophysiological mechanisms involved. Genetic deletion of another BCR-ABL1–induced transcription factor, STAT5, completely abolishes phenotypic CML-like MPN in this mouse model50 but does not affect the survival of BCR-ABL1–expressing leukemic stem cells. By contrast, inhibition of NF-κB attenuated but did not eliminate CML-like leukemia, but did have a significant negative effect on leukemic stem cells in this disease. In BCR-ABL1–expressing B-lymphoid blasts, inhibition of NF-κB increased their sensitivity to killing by TKIs such as imatinib. Given that relapse and long-term survival in both Ph+ B-ALL and CML are currently thought to be influenced predominantly by leukemic stem cells that persist in patients on TKI therapy and/or following allografting, our findings argue that NF-κB inhibitors might facilitate eradication of these troublesome progenitors and increase the chances of permanent cure. Testing this approach in Ph+ leukemia patients with minimal residual disease is warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health, National Cancer Institute grant CA090576 (R.A.V.E.), and by a grant from the Sackler Families Fund for Collaborative Cancer Biology Research (M.-Y.H. and R.A.V.E.).
Authorship
Contribution: M.-Y.H. and R.A.V.E. designed experiments and wrote the manuscript; and M.-Y.H. carried out the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R.A.V.E. is Chao Family Comprehensive Cancer Center, University of California, Irvine, Irvine, CA.
Correspondence: Richard A. Van Etten, Division of Hematology/Oncology, and Chao Family Comprehensive Cancer Center, University of California, Irvine, 839 Medical Sciences Court, Sprague Hall, Room 124, Irvine, CA 92697; e-mail: vanetten@uci.edu.

![Figure 7. Inhibition of NF-κB sensitizes BCR-ABL1–transformed B lymphoblasts to TKIs. (A) BCR-ABL1/GFP– or BCR-ABL1/IκBαSR–transformed primary B-lymphoid progenitors (4 × 104 cells per well) were incubated with different concentration of imatinib as indicated. Cell viability was determined by MTS assay after 96 hours’ incubation. (B-C) Equal numbers of primary B-lymphoid progenitors transformed by BCR-ABL1/GFP, BCR-ABL1/IKKαKM, or BCR-ABL1/IKKβKM were incubated with different concentration of imatinib (B) or dasatinib (C) as indicated. Cell viability was determined by (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay after 96 hours’ incubation. Sigmoidal curves were fitted and IC50 values calculated by Prism software.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/15/10.1182_blood-2014-01-547943/4/m_2401f7.jpeg?Expires=1767711323&Signature=yasiTvs26q5J3NE3CAUWDhEfys~OGx8UIrlfQH6zxxWPLtED8m4rDge2D~lYRtUasRP6lpMe0LZtP3odoKg0NsWECsjxt~VxaDDB4QlsVLZRonWcwp3sIyCTeuC1zomF6zKxLSOnXbsV7NdPWX5tv2q4OAX4S-kmxfXJe6d3wNY9JwIkuhgS9TmuoGqu3DmBLaQCNoL-20i3u7LjLljPnIgU6Rs-vyjPd2Pj09EXs-CWmqXqYq8mb5a6P5L0myxdIgultMBewyTlOUXpAeLfKBQUbqAcj2vSJG56LLSkhAs7avofjzSGHE~99jEyTsBOkA9BmIZO8FA8Vmr1XOiDrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)