Key Points
E2F1 regulates Trib2 expression and C/EBPα modulates E2F1-induced Trib2 activity at the granulocyte macrophage progenitor stage.
Pharmacological inhibition of the cell cycle resulting in a block in E2F1 or Trib2 knockdown abrogates AML cell proliferation.
Abstract
The loss of regulation of cell proliferation is a key event in leukemic transformation, and the oncogene tribbles (Trib)2 is emerging as a pivotal target of transcription factors in acute leukemias. Deregulation of the transcription factor E2F1, normally repressed by CCAAT enhancer-binding protein α (C/EBPα)-p42, occurs in acute myeloid leukemia (AML), resulting in the perturbation of cell cycle and apoptosis, emphasizing its importance in the molecular pathogenesis of AML. Here we show that E2F family members directly regulate Trib2 in leukemic cells and identify a feedback regulatory loop for E2F1, C/EBPα, and Trib2 in AML cell proliferation and survival. Further analyses revealed that E2F1-mediated Trib2 expression was repressed by C/EBPα-p42, and in normal granulocyte/macrophage progenitor cells, we detect C/EBPα bound to the Trib2 promoter. Pharmacological inhibition of the cell cycle or Trib2 knockdown resulted in a block in AML cell proliferation. Our work proposes a novel paradigm whereby E2F1 plays a key role in the regulation of Trib2 expression important for AML cell proliferation control. Importantly, we identify the contribution of dysregulated C/EBPα and E2F1 to elevated Trib2 expression and leukemic cell survival, which likely contributes to the initiation and maintenance of AML and may have significant implications for normal and malignant hematopoiesis.
Introduction
The tribbles (Trib) family of pseudokinase genes (Trib1, Trib2, and Trib3) have recently emerged as important regulators of acute leukemia and hematopoietic development.1 Trib proteins’ diverse roles include cell proliferation, survival, motility, and metabolism.2 There is a strong correlation between dysregulated Trib2 expression and acute myeloid and lymphoid leukemia (AML and ALL). Elevated Trib2 expression associates with a specific subset of AML characterized by dysregulated CCAAT enhancer-binding protein α (C/EBPα) and with T-cell ALL (T-ALL), both linked with NOTCH1 mutations.3-5 Mice reconstituted with hematopoietic stem cells (HSCs) retrovirally expressing Trib2 developed AML. The proteasomal-mediated degradation and inhibition of C/EBPα was required for the leukemogenic activity of Trib2.4-6 Although Trib2 appears to integrate a wide variety of signaling pathways, the molecular understanding of how its expression is controlled in normal and malignant hematopoiesis is limited.
We identified Trib2 as a NOTCH1-regulated transcript in T-ALL cells undergoing growth arrest. As well as NOTCH1,3,5 PITX17 and TAL18 were also found to up-regulate Trib2 in T-ALL. Both PITX1 and TAL1 are recurrently activated transcription factors in T-ALL and importantly require the expression of Trib2 for leukemic cell growth and survival.8 In AML cells, MEIS1 was found to bind to the Trib2 promoter, and Trib2 could confer growth-enhancing properties to NUP98-HOXD13/MEIS1-AML cells.9 Additionally, Trib2 expression is regulated by microRNAs in leukemia cells.10,11 It is clear therefore that Trib2 expression is controlled in a cell type–, cell context–, and cell cycle–dependent manner.
The role of Trib2 in cell cycle and cell proliferation is supported by an established role for Drosophila Tribbles (dTRIB) in the cell cycle. dTRIB coordinates cell division during gastrulation by promoting turnover of the cell cycle protein String/CDC25, thereby inhibiting premature mitosis.12-14 A further study suggested that dTRIB controls mitosis through inhibition of String and positive regulation of WEE1, as coexpression of WEE1 and Tribbles showed strong synergistic interactions that resulted in severe eye and wing developmental defects.15 Studies of mammalian Trib2 thus far indicate the genetic interactions of dTRIB and other Drosophila homolog proteins are conserved in the mammalian system. dTRIB promotes the degradation of Slbo, the Drosophila homolog of the C/EBP family of transcription factors, thereby inhibiting Slbo-dependent border cell migration during oogenesis,16 and inactivation of C/EBPα by Trib2 has been shown in the hematopoietic system to block myeloid cell differentiation.4 Likewise, dTRIB has a positive genetic interaction with N (the Drosophila homolog of NOTCH),17 and this is conserved in mammals as Trib2 was found to be a direct transcriptional target gene of NOTCH1.5
The transcription factor C/EBPα is a critical regulator of myeloid cell proliferation and differentiation. C/EBPα is frequently dysregulated in AML as a result of mutation, posttranscriptional modifications, posttranslational inhibition, epigenetic regulation, and proteasomal degradation. It is also an important tumor suppressor as it regulates a variety of cell cycle proteins.18 C/EBPα mRNA encodes for 2 major protein isoforms, C/EBPα-p42 and C/EBPα-p30, produced by alternative translational start codons. We have previously shown that Trib2 degrades C/EBPα-p42 and blocks differentiation, with an increase in the C/EBPα-p30 oncogenic protein. In addition to Trib1- and Trib2-mediated dysregulation,19 the normal protein expression ratio of p42:p30 is skewed in AML as a result of gene mutation.20 Importantly, the p30 isoform lacks a transactivation domain in the N terminus important for antimitotic activity. This N-terminal domain is crucial for growth arrest by E2F1-mediated regulation of c-Myc expression, and C/EBPα-p42 interferes with E2F1 transactivation of the c-Myc promoter.21-23 E2F1 is a master regulator of cell cycle progression. C/EBPα inhibition of E2F1 additionally occurs via protein-protein interaction in non–DNA-binding residues in the C-terminal region of C/EBPα present in both p30 and p42.22 Mutation of the C-terminal residues of C/EBPα that interact with E2F1 (BRM2) leads to a transplantable disorder of the myeloid lineage with expansion of myeloid progenitors. This revealed a role of C/EBPα-mediated E2F1 repression in controlling the proliferation of myeloid progenitors.24 Importantly, however, null mutations in CEBPA do not occur in AML, and C/EBPα-deficient HSCs do not lead to AML,25,26 demonstrating that C/EBPα is required for AML to develop.
The E2F family of cell cycle regulators are generally classed as transcriptional activators (E2F1, E2F2, E2F3a, and E2F3b) or repressors (E2F4, E2F5, E2F6, E2F7, and E2F8), but this simple classification lacks in vivo validation.27 E2F1-3–deficient hematopoietic cells have a defect in myeloid cell differentiation, with an accumulation of granulocyte/macrophage progenitor (GMP) cells and a decrease in CD11b+ myeloid cells in the bone marrow. E2F1-3s are thus required for cell survival and proliferation at distinct stages during myeloid differentiation.28 AML with CEBPA mutations has increased E2F3 and decreased miR-34a levels, which C/EBPα normally regulates in myeloid cells.29 Additional studies support a molecular network involving miR-223, C/EBPα, and E2F1 as major components of the granulocyte differentiation program, which is deregulated in AML.30
We show here that E2F1, as well as E2F2 and E2F3, is a potent inducer of Trib2 expression and that E2F1 cooperates with C/EBPα-p30 to further activate the Trib2 promoter in preleukemic cells, resulting in elevated Trib2 expression. Conversely, in normal myeloid progenitor cells, C/EBPα-p42 is found bound to the Trib2 promoter and inhibits E2F1-mediated activation of Trib2. We demonstrate that inhibition of the E2F1-Trib2 regulatory loop results in cell cycle arrest and inhibition of leukemic cell proliferation. In addition, we show that targeted inhibition of this pathway does not affect the growth and survival of normal hematopoietic cells. Our work demonstrates that Trib2 expression is regulated by E2F1 in vivo, and C/EBPα-p30 cooperates with E2F1 to activate Trib2 expression, thus preventing C/EBPα-p42–mediated E2F1 repression, ultimately contributing to the uncontrolled proliferation and cell cycle progression seen in AML.
Methods
Trib2-dependent reporter gene assay
3T3 cells were transfected with luciferase constructs alone or in combination with other expression plasmids and luminescence readings taken (see supplemental Methods on the Blood Web site for details).
Electromobility shift assay
Nonradiolabeled electromobility shift assays (EMSAs) using the LightShift Chemiluminescent EMSA Kit (Thermo Scientic) were performed on nuclear extracts prepared from cells using wild-type (WT) and mutant probes. Specificity was assessed by cold competition (see supplemental Methods for details).
Chromatin immunoprecipitation
Cells were fixed and resuspended in lysis buffer. Isolated nuclei were resuspended in shearing buffer (Active Motif) and sonicated on ice. Protein/DNA complexes were immunoprecipitated with the indicated antibody, and DNA was eluted as per the manufacturer’s instructions (Active Motif) and column purified (QIAquick Purification Kit; Qiagen). Eluted DNA was subjected to polymerase chain reaction (PCR) using promoter-specific primer sets (see supplemental Methods for details).
Mice
Trib2−/− (B6;129S5-Trib2tm1Lex) mice, backcrossed onto C57B6, were bred and housed in the University of Glasgow. Cebpa transgenic mice31 were bred and housed in the University of Copenhagen (see supplemental Methods for details). All mouse work was performed according to national and international guidelines and approved by the local United Kingdom and Danish Animal Ethical Committees.
Dataset analysis
Gene expression values for Trib2 vs E2F1 (GSE14468 and GSE1159) were plotted, and the line of best fit was calculated using linear regression (see supplemental Methods for details).
Cell cycle and proliferation analysis
Cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes) for 10 minutes and treated. Drug-treated cells were also fixed and stained with Propidium Iodide (PI)/RNase staining buffer (BD Pharmingen) (see supplemental Methods for details).
Annexin V/4′6 diamidino-2-phenylindole staining
Total bone marrow cells from WT or Trib2-deficient mice and green fluorescent protein–sorted U937 cells were suspended in binding buffer (containing annexin V-phycoerythrin [BD Biosciences] and 4′6 diamidino-2-phenylindole [DAPI; Sigma-Aldrich]) and were analyzed by flow cytometry.
Patient AML samples
Blood samples from newly diagnosed AML patients and normal peripheral blood samples were collected following protocol approved by the Ethics Review Board and with informed consent. Mononuclear cells were isolated using Histopaque (Sigma-Aldrich) according to the manufacturer’s instructions. This study was conducted in accordance with the Declaration of Helsinki.
See supplemental Methods for all other details.
Results
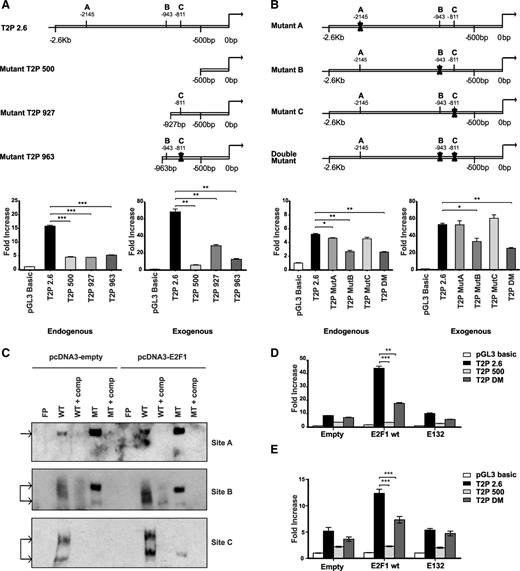
To investigate how Trib2 expression is regulated, we cloned 2 promoter regions of Trib2 into the pGL3 luciferase reporter: (1) 500 bp upstream from the transcriptional start site (−500 to +1 bp) and (2) including the entire 5′ untranslated region (UTR) and upstream region (−2.6 kb to +1 bp). The promoter activity was assessed in 293T cells, 3T3 cells, and RAW264.7 cells. Increased Trib2 promoter activity was seen in 293T and 3T3 cell lines but not the RAW cell line with the 2.6-kb promoter construct compared with the 500-bp promoter (Figure 1A). We analyzed the promoter region of Trib2 using TESS bioinformatic software and found 3 putative consensus E2F binding sites within the Trib2 promoter. Two sites were located within the 1.3-kb 5′UTR (site B at −943 bp and site C at −811 bp), and 1 site was further upstream (site A at −2.1 kb; Figure 1B). We assessed the activation of the Trib2 promoter by cotransfection with E2F1 in 3T3 and K562 myeloid cells. E2F1 expression significantly increased the promoter activity of the 2.6-kb construct in both cell types (Figure 1C,E). We performed the promoter activity assay using an E2F1 DNA binding mutant (E132 mutant) and showed that, in the absence of direct DNA binding, E2F1 is unable to activate the Trib2 promoter in both 3T3 and K562 cells (Figure 1D-E). Together these data suggest that Trib2 is a target of E2F1.
E2F1 binds and activates the Trib2 promoter. (A) 293T, 3T3, and RAW264.7 cells were cotransfected with empty pGL3 Basic or 500-bp Trib2 promoter or 2.6-kb Trib2 promoter constructs and luciferase activity was measured. (B) Schematic presentation of the Trib2 promoter region 2.6 kb upstream from the transcriptional start site and the location of 3 putative E2F binding sites. (C) 3T3 cells were cotransfected with E2F1 or pcDNA empty expression vector and Trib2 luciferase reporter plasmids (pGL3 control, 500-bp, or 2.6-kb promoter region) and luciferase activity was measured. (D) 3T3 cells were cotransfected with E2F1 or E2F1 E132 DNA binding deficient mutant expression vectors and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter) and luciferase activity was measured. Data presented are mean ± standard deviation (SD) of duplicate cultures and representative of 3 independent experiments. **P < .005 and ***P < .001 using an unpaired Student t test. (E) K562 cells were cotransfected with E2F1 or E2F1 E132 DNA binding deficient mutant expression vectors or empty control vector and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter) and luciferase activity was measured. Data presented are mean ± SD and representative of 3 independent experiments. **P < .005 using an unpaired Student t test.
E2F1 binds and activates the Trib2 promoter. (A) 293T, 3T3, and RAW264.7 cells were cotransfected with empty pGL3 Basic or 500-bp Trib2 promoter or 2.6-kb Trib2 promoter constructs and luciferase activity was measured. (B) Schematic presentation of the Trib2 promoter region 2.6 kb upstream from the transcriptional start site and the location of 3 putative E2F binding sites. (C) 3T3 cells were cotransfected with E2F1 or pcDNA empty expression vector and Trib2 luciferase reporter plasmids (pGL3 control, 500-bp, or 2.6-kb promoter region) and luciferase activity was measured. (D) 3T3 cells were cotransfected with E2F1 or E2F1 E132 DNA binding deficient mutant expression vectors and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter) and luciferase activity was measured. Data presented are mean ± standard deviation (SD) of duplicate cultures and representative of 3 independent experiments. **P < .005 and ***P < .001 using an unpaired Student t test. (E) K562 cells were cotransfected with E2F1 or E2F1 E132 DNA binding deficient mutant expression vectors or empty control vector and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter) and luciferase activity was measured. Data presented are mean ± SD and representative of 3 independent experiments. **P < .005 using an unpaired Student t test.
To identify the role of the predicted E2F1 sites in the Trib2 promoter regions, we constructed a number of deletion and mutagenized promoter constructs in the luciferase reporter (schematic representation in Figure 2A-B, upper panels, respectively). We tested the promoter activity in the presence and absence of exogenous E2F1 expression. Activation of the 500-bp promoter region can be modestly increased by expression of E2F1 (as seen in Figures 1C and 2A), despite the lack of a consensus E2F1 binding site in that region. This indicates a potential nonconsensus E2F1 binding site or E2F1-mediated activation via protein-protein interactions. Using deletion constructs containing only site B (−963-bp construct with site C mutated) or only site C (−927-bp construct; Figure 2A, upper left) revealed that in the absence of site A and site B or C, E2F1 is able to further activate the Trib2 promoter but is unable to restore complete activation as seen in the full-length promoter construct (Figure 2A, lower). We then performed site-directed mutagenesis of sites A, B, and C in the 2.6-kb promoter construct. Luciferase activity assay revealed that mutation of site A or C alone did not abrogate or inhibit E2F1-induced promoter activity, whereas mutation of B alone or both sites B and C together reduced Trib2 promoter activity (Figure 2B, lower). EMSA analysis showed that, on transfection of K562 cells with E2F1, there was an increase in binding to the WT probes for the E2F1-specific sites A, B, and C and a lack of E2F1-inducible binding to 3 distinct mutant probes for sites A, B and C (Figure 2C). Mutation of B and C together significantly reduced Trib2 promoter activity in both 3T3 (Figure 2D) and K562 cells (Figure 2E) but not to as great an extent as the 500-bp promoter (lacks sites A, B, and C). Together, these data reveal that E2F1 binds at sites A, B, and C in the Trib2 promoter in fibroblasts and myeloid cells.
E2F1 binding is localized to 2 consensus binding sites on the Trib2 promoter. (A) (Upper) Schematic presentation of full-length 2.6-kb and deletion mutant Trib2 promoter constructs with the indicated E2F1 binding sites. 3T3 cells were (lower left) transfected with Trib2 luciferase reporter plasmids (pGL3 control, 2.6-kb, 500-bp, 927-bp, or 963-bp promoter region) and (lower right) cotransfected with E2F1 or pcDNA empty expression vector, and luciferase activity was measured. (B) (Upper) Schematic presentation of full-length 2.6 kb with site-directed mutations of the 3 putative E2F1 binding sites. 3T3 cells were (lower left) transfected with Trib2 luciferase reporter plasmids (pGL3 control, 2.6-kb, MutA, MutB, MutC, or double mutant DM) and (lower right) cotransfected with E2F1 or pcDNA empty expression vector, and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 3 independent experiments. *P < .05, **P < .005, and ***P < .001. (C) Nuclear extracts prepared from K562 cells transfected with pcDNA3 empty or E2F1 were assayed for E2F1 binding for sites A, B, and C by EMSA. Arrow indicates E2F1 binding. FP, free probe; comp, cold competition; MT, mutant type. (D) 3T3 cells were cotransfected with E2F1 WT or E2F1 E132 DNA binding deficient mutant or empty expression vector and Trib2 luciferase reporter constructs (pGL3 control, 2.6-kb, 500-bp, or DM promoter region), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. **P < .005 and ***P < .001 using an unpaired Student t test. (E) K562 cells were cotransfected with E2F1 WT or E2F1 E132 DNA binding deficient mutant or empty expression vector and Trib2 luciferase reporter constructs (pGL3 control, 2.6-kb, 500-bp, or DM promoter region), and luciferase activity was measured. Data presented are mean ± SD and representative of 2 independent experiments. ***P < .001 using an unpaired Student t test.
E2F1 binding is localized to 2 consensus binding sites on the Trib2 promoter. (A) (Upper) Schematic presentation of full-length 2.6-kb and deletion mutant Trib2 promoter constructs with the indicated E2F1 binding sites. 3T3 cells were (lower left) transfected with Trib2 luciferase reporter plasmids (pGL3 control, 2.6-kb, 500-bp, 927-bp, or 963-bp promoter region) and (lower right) cotransfected with E2F1 or pcDNA empty expression vector, and luciferase activity was measured. (B) (Upper) Schematic presentation of full-length 2.6 kb with site-directed mutations of the 3 putative E2F1 binding sites. 3T3 cells were (lower left) transfected with Trib2 luciferase reporter plasmids (pGL3 control, 2.6-kb, MutA, MutB, MutC, or double mutant DM) and (lower right) cotransfected with E2F1 or pcDNA empty expression vector, and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 3 independent experiments. *P < .05, **P < .005, and ***P < .001. (C) Nuclear extracts prepared from K562 cells transfected with pcDNA3 empty or E2F1 were assayed for E2F1 binding for sites A, B, and C by EMSA. Arrow indicates E2F1 binding. FP, free probe; comp, cold competition; MT, mutant type. (D) 3T3 cells were cotransfected with E2F1 WT or E2F1 E132 DNA binding deficient mutant or empty expression vector and Trib2 luciferase reporter constructs (pGL3 control, 2.6-kb, 500-bp, or DM promoter region), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. **P < .005 and ***P < .001 using an unpaired Student t test. (E) K562 cells were cotransfected with E2F1 WT or E2F1 E132 DNA binding deficient mutant or empty expression vector and Trib2 luciferase reporter constructs (pGL3 control, 2.6-kb, 500-bp, or DM promoter region), and luciferase activity was measured. Data presented are mean ± SD and representative of 2 independent experiments. ***P < .001 using an unpaired Student t test.
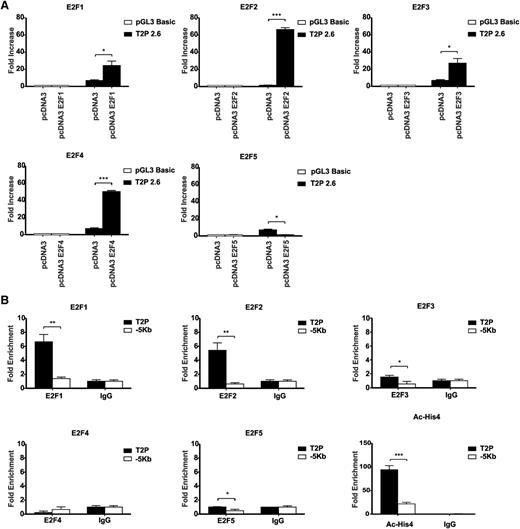
We next tested whether this was specific to E2F family members. Luciferase assays in 3T3 cells using the full-length 2.6-kb Trib2 promoter construct revealed that E2F1, E2F2, E2F3, and E2F4 activated the Trib2 promoter, whereas E2F5 was unable to induce Trib2 promoter activity (Figure 3A). To test the ability of these E2F family members to bind to Trib2 promoter in a myeloid leukemia cell, we performed chromatin immunoprecipitation (ChIP) analysis using antibodies for endogenous E2F1, E2F2, E2F3, E2F4, and E2F5 in K562 cells (express high levels of Trib2 protein; Figure 7E). In comparison with the IgG-negative control, there was enrichment of E2F1, E2F2, and E2F3 on the Trib2 promoter region containing sites B and C, but not at a −5-kb Trib2 promoter region used as a negative control (Figure 3B). The Trib2 promoter region was shown to be transcriptionally active as demonstrated by the presence of acetylated histone 4. There was no significant enrichment of E2F4 or E2F5 on the Trib2 promoter. These data show that E2F1, E2F2, E2F3, and E2F4 are capable of activating the Trib2 promoter; in leukemia cells, only E2F1, 2, and 3 actually bind the Trib2 promoter. These data demonstrate that E2F proteins directly regulate Trib2 expression.
Other E2F family members can bind and activate Trib2 promoter. (A) 3T3 cells were cotransfected with E2F1, E2F2, E2F3, E2F4, E2F5, or pcDNA empty expression vector and Trib2 luciferase reporter plasmids (pGL3 basic control or 2.6-kb promoter region), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 3 independent experiments. *P < .05 and ***P < .001. (B) K562 cells were chromatin immunoprecipitated using anti-E2F1, anti-E2F2, anti-E2F3, anti-E2F4, anti-E2F5, antiacetylated histone 4 (Ac-His4), or normal IgG control antibodies. PCR was performed using primers directed against the E2F binding region containing sites B and C and using primers against a −5-kb region as a negative control. Graphs represent fold enrichment of DNA compared with the IgG control and are representative of 3 independent experiments, and error bars denote ± SD of each sample. *P < .05, **P < .005, and ***P < .001.
Other E2F family members can bind and activate Trib2 promoter. (A) 3T3 cells were cotransfected with E2F1, E2F2, E2F3, E2F4, E2F5, or pcDNA empty expression vector and Trib2 luciferase reporter plasmids (pGL3 basic control or 2.6-kb promoter region), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 3 independent experiments. *P < .05 and ***P < .001. (B) K562 cells were chromatin immunoprecipitated using anti-E2F1, anti-E2F2, anti-E2F3, anti-E2F4, anti-E2F5, antiacetylated histone 4 (Ac-His4), or normal IgG control antibodies. PCR was performed using primers directed against the E2F binding region containing sites B and C and using primers against a −5-kb region as a negative control. Graphs represent fold enrichment of DNA compared with the IgG control and are representative of 3 independent experiments, and error bars denote ± SD of each sample. *P < .05, **P < .005, and ***P < .001.
To further analyze the role of E2F1 in Trib2 regulation, we assessed E2F1-induced Trib2 mRNA expression in cells transfected with E2F1. 293T, E2F1−/− mouse embryonic fibroblasts (MEFs), and K562 cells were transfected with control pcDNA vector, E2F1, or intracellular activated Notch1 ([ICNX] as a positive control5 ), and Trib2 mRNA expression was measured. There was a significant increase in Trib2 mRNA following overexpression of E2F1 (Figure 4A). Indeed there was significant reduction in Trib2 mRNA expression in E2F1−/− MEFs compared with WT MEFs (Figure 4B). E2F1−/− MEFs express little to no Trib2 protein expression (Figure 4C), and on transfection with E2F1 (and ICNX as positive control), we saw an increase in Trib2 protein expression (Figure 4D, left, lane 2). Similarly, transfection of K562 cells with E2F1 leads to an increase in Trib2 protein expression (Figure 4D, right). Conversely, small interfering RNA (siRNA) directed against E2F1 in 293T, HeLa, and K562 cells resulted in decreased Trib2 mRNA expression levels compared with siRNA scramble controls (Figure 4E-G), and analysis of a previously published transcriptional profile of E2F1-3−/− CD11b+ myeloid cells28 revealed a 4.6-fold reduction in Trib2.
E2F1 regulates Trib2 mRNA and protein expression levels. (A) (Left) E2F1−/− (KO) MEFs, (center) 293T, or (right) K562 cells transfected with E2F1, pcDNA control, or ICNX as indicated were measured for Trib2 mRNA expression by real-time PCR analyses. Data are presented relative to control pcDNA transfected cells ± SD of duplicate cultures. *P < .05, **P < .005, and ***P < .001. (B) WT and E2F1−/− (KO) SV40 MEFs were measured for (left) E2F1 and (right) Trib2 mRNA expression by real-time PCR analyses. (C) Western blot analyses of E2F1 and Trib2 protein levels in WT and E2F1−/− (KO) MEFs. Actin shown as protein loading control. (D) (Left) E2F1−/− (KO) MEFs or (right) K562 cells were transfected with pcDNA3 empty vector, E2F1, ICNX, or Trib2 as indicated, and western blot analyses were performed for Trib2, E2F1, and actin as a loading control. (E) 293T, (F) HeLa, and (G) K562 cells were transfected with scrambled control siRNA and siE2F1 (1 or 2 different E2F1 siRNAs as indicated), and real-time PCR analysis was performed for E2F1 and Trib2. Data are presented relative to control transfected cells and representative of 2 independent experiments. Error bars denote ± SD of each sample measured in triplicate. *P < .05, **P < .005, and ***P < .001.
E2F1 regulates Trib2 mRNA and protein expression levels. (A) (Left) E2F1−/− (KO) MEFs, (center) 293T, or (right) K562 cells transfected with E2F1, pcDNA control, or ICNX as indicated were measured for Trib2 mRNA expression by real-time PCR analyses. Data are presented relative to control pcDNA transfected cells ± SD of duplicate cultures. *P < .05, **P < .005, and ***P < .001. (B) WT and E2F1−/− (KO) SV40 MEFs were measured for (left) E2F1 and (right) Trib2 mRNA expression by real-time PCR analyses. (C) Western blot analyses of E2F1 and Trib2 protein levels in WT and E2F1−/− (KO) MEFs. Actin shown as protein loading control. (D) (Left) E2F1−/− (KO) MEFs or (right) K562 cells were transfected with pcDNA3 empty vector, E2F1, ICNX, or Trib2 as indicated, and western blot analyses were performed for Trib2, E2F1, and actin as a loading control. (E) 293T, (F) HeLa, and (G) K562 cells were transfected with scrambled control siRNA and siE2F1 (1 or 2 different E2F1 siRNAs as indicated), and real-time PCR analysis was performed for E2F1 and Trib2. Data are presented relative to control transfected cells and representative of 2 independent experiments. Error bars denote ± SD of each sample measured in triplicate. *P < .05, **P < .005, and ***P < .001.
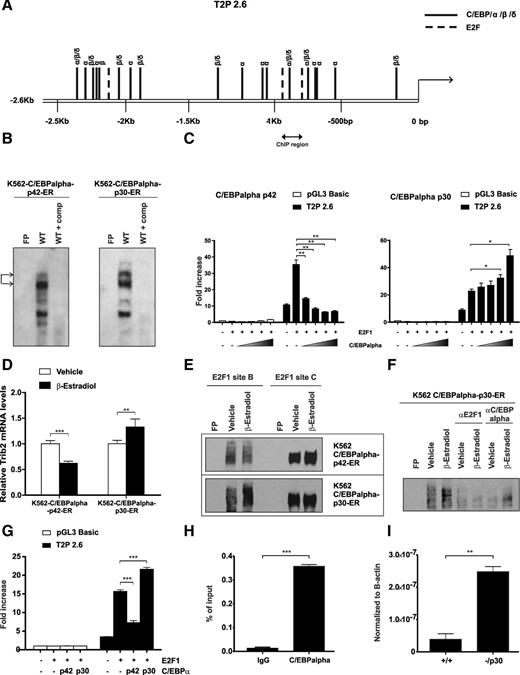
We also identified putative C/EBPα binding sites in the Trib2 promoter using the TESS bioinformatics analysis tool. Indeed E2F1 binding sites B and C surround a region of the Trib2 promoter that contains a high confidence C/EBP binding site (−900 bp) (Figure 5A). C/EBPα-mediated inhibition of E2F1 is pivotal for granulopoiesis. As our previous work has shown that Trib2 degraded C/EBPα-p42 leaving increased expression of the oncogenic C/EBPα-p30 form in AML, we tested whether there was an E2F1-C/EBPα feedback loop regulating Trib2 expression. EMSA revealed that both C/EBPα-p42 and -p30 bind to this region at −900 bp using a C/EBPα binding site–specific probe (Figure 5B). Trib2 promoter assays showed a dose-dependent decrease in activity following cotransfection of increasing doses of C/EBPα-p42 compared with E2F1 alone (Figure 5C, left). Significantly, cotransfection of E2F1 and C/EBPα-p30 resulted in a dose-dependent increase in E2F1-induced Trib2 promoter activity (Figure 5C, right). These data show that the E2F1 regulation of Trib2 can be modulated by the presence of C/EBPα-p42 and C/EBPα-p30. Importantly, Trib2 mRNA expression decreased in myeloid K562 cells on C/EBPα-p42 expression, and reciprocally, Trib2 expression increased on C/EBPα-p30 expression (Figure 5D). This was not due to an increase in E2F protein expression by C/EBPα-p30 (supplemental Figure 1). This regulation was seen in K562 nuclear extracts at the DNA binding sites, as EMSA analysis showed that C/EBPα-p42 and -p30 modulated the binding of E2F1 to the Trib2 promoter at site B only (Figure 5E). The slower-migrating protein complex induced by C/EBPα-p30 contained both E2F1 and C/EBPα-p30 as seen by the disappearance of this complex in a supershift assay, indicating that these proteins are components of this complex (Figure 5F). Additionally, the E2F1-induced Trib2 promoter activity in K562 cells was modulated by C/EBPα-p42 and -p30 expression (Figure 5G). To investigate the physiological binding of C/EBPα to the Trib2 promoter in the myeloid lineage, we performed ChIP analysis for C/EBPα in normal murine GMPs at −900 bp. We detected C/EBPα bound to the Trib2 promoter in the region spanning the identified E2F1 binding sites B and C (Figure 5H) revealing that in normal GMPs, C/EBPα is recruited onto the Trib2 promoter. This is consistent with the ability of C/EBPα-p42 to inhibit E2F1 in normal myelopoiesis. Interestingly, real-time quantitative PCR analysis of normal WT GMPs compared with GMPs isolated from preleukemic mutant C/EBPα knockin mice that retain p30 expression but have lost p42 expression (Lp30, −/p30) reveals a significant increase in Trib2 expression in the mutant GMPs (Figure 5I). Therefore, in the presence of C/EBPα-p30 in the preleukemic GMP population, Trib2 mRNA expression is increased. Together, these data reveal that C/EBPα localizes to the Trib2 promoter in GMP progenitor cells, and the balance of C/EBPα-p42 and C/EBPα-p30 modulates the ability of E2F1 to activate Trib2 expression in normal and preleukemic myeloid cells.
C/EBPα p42 and p30 oppositely modulate Trib2 expression via binding of the promoter. (A) Putative binding sites of C/EBP family of transcription factors in the Trib2 promoter. Several binding sequences are shared among members α, β, and δ of the C/EBPα family. E2F putative binding sites are also indicated (long dash line), and the region is targeted for ChIP-PCR analysis. (B) Nuclear extracts prepared from K562-C/EBPα-p42-ER or K562-C/EBPα-p30-ER cells induced with β-estradiol for 24 hours were assayed for C/EBPα binding by EMSA. Arrows indicate C/EBPα binding. FP, free probe; comp, cold competition. (C) 3T3 cells were cotransfected with empty control, E2F1 alone, or in combination with increasing amount of (left) C/EBPα-p42 or (right) C/EBPα-p30 and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. *P < .05 and **P < .005. (D) K562-C/EBPα-p42-ER and K562-C/EBPα-p30-ER cells induced with β-estradiol or vehicle control for 24 hours were measured for Trib2 mRNA expression by real-time PCR analyses. Data are presented relative to vehicle control and representative of 2 independent experiments. Error bars denote ±SD of each sample measured in triplicate. **P < .005 and ***P < .001. (E) Nuclear extracts prepared from (upper) K562-C/EBPα-p42-ER and (lower) K562-C/EBPα-p30-ER cells transfected with E2F1 and induced with β-estradiol or vehicle control for 24 hours were assayed for E2F1 binding for WT sites B and C by EMSA. FP, free probe. (F) Supershift EMSA assay was performed in nuclear extracts prepared from K562-C/EBPα-p30-ER cells transfected with E2F1 and induced with β-estradiol or vehicle control for 24 hours using antibody specific for E2F1 or C/EBPα. Oligonucleotide for WT site B was used as a probe. FP, free probe. (G) K562 cells were cotransfected with empty control, E2F1 alone, or E2F1 in combination with C/EBPα-p42 or C/EBPα-p30 and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter region), and luciferase activity was measured. Data presented are mean ± SD and representative of 2 independent experiments. ***P < .001 using an unpaired Student t test. (H) Normal murine GMP cells were chromatin immunoprecipitated using anti-C/EBPα or normal IgG control antibodies. PCR was performed using primers against the region indicated in A. Error bars denote ±SD of each sample measured in triplicate. ***P < .001. (I) Real-time PCR analysis of Trib2 mRNA levels in WT (+/+) GMP and −/p30 GMP cells. **P < .005.
C/EBPα p42 and p30 oppositely modulate Trib2 expression via binding of the promoter. (A) Putative binding sites of C/EBP family of transcription factors in the Trib2 promoter. Several binding sequences are shared among members α, β, and δ of the C/EBPα family. E2F putative binding sites are also indicated (long dash line), and the region is targeted for ChIP-PCR analysis. (B) Nuclear extracts prepared from K562-C/EBPα-p42-ER or K562-C/EBPα-p30-ER cells induced with β-estradiol for 24 hours were assayed for C/EBPα binding by EMSA. Arrows indicate C/EBPα binding. FP, free probe; comp, cold competition. (C) 3T3 cells were cotransfected with empty control, E2F1 alone, or in combination with increasing amount of (left) C/EBPα-p42 or (right) C/EBPα-p30 and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter), and luciferase activity was measured. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. *P < .05 and **P < .005. (D) K562-C/EBPα-p42-ER and K562-C/EBPα-p30-ER cells induced with β-estradiol or vehicle control for 24 hours were measured for Trib2 mRNA expression by real-time PCR analyses. Data are presented relative to vehicle control and representative of 2 independent experiments. Error bars denote ±SD of each sample measured in triplicate. **P < .005 and ***P < .001. (E) Nuclear extracts prepared from (upper) K562-C/EBPα-p42-ER and (lower) K562-C/EBPα-p30-ER cells transfected with E2F1 and induced with β-estradiol or vehicle control for 24 hours were assayed for E2F1 binding for WT sites B and C by EMSA. FP, free probe. (F) Supershift EMSA assay was performed in nuclear extracts prepared from K562-C/EBPα-p30-ER cells transfected with E2F1 and induced with β-estradiol or vehicle control for 24 hours using antibody specific for E2F1 or C/EBPα. Oligonucleotide for WT site B was used as a probe. FP, free probe. (G) K562 cells were cotransfected with empty control, E2F1 alone, or E2F1 in combination with C/EBPα-p42 or C/EBPα-p30 and Trib2 luciferase reporter constructs (pGL3 control or 2.6-kb promoter region), and luciferase activity was measured. Data presented are mean ± SD and representative of 2 independent experiments. ***P < .001 using an unpaired Student t test. (H) Normal murine GMP cells were chromatin immunoprecipitated using anti-C/EBPα or normal IgG control antibodies. PCR was performed using primers against the region indicated in A. Error bars denote ±SD of each sample measured in triplicate. ***P < .001. (I) Real-time PCR analysis of Trib2 mRNA levels in WT (+/+) GMP and −/p30 GMP cells. **P < .005.
To target the E2F1 pathway and determine the effect on Trib2 and the leukemic cell, we used known cyclin-dependent kinase inhibitors (CDKis), flavopiridol (a pan CDK inhibitor),32 pentoxifylline (PTX, a nonspecific phosphodiesterase inhibitor), and dibutyryl cAMP (a cAMP analog).33 We treated U937 cells (AML cells that express high levels of Trib2 protein; Figure 7E) with escalating doses of flavopiridol and PTX and assessed cell death (Figure 6A). Both flavopiridol (IC50 of 94 nM) and PTX (IC50 of 4 mM) were cytotoxic to U937, whereas up to 4 mM dibutyryl cAMP was not cytotoxic. At a lower-dose (2 mM PTX, 4 mM dibutyryl cAMP, and 62.5 nM flavopiridol) treatment at 24 hours, U937 cells underwent G1 cell cycle arrest without increased apoptosis (sub-G1 DNA content) as assessed by propidium iodide staining (Figure 6B) and had reduced proliferation as assessed by CFSE staining at 96 hours (Figure 6C). Using low drug doses that inhibit cell proliferation but have minimal cytotoxicity and apotosis, we assessed the effect on cell cycle proteins E2F1 and Trib2. Western blot analysis determined that PTX, dibutyryl cAMP, and flavopiridol led to a reduction in Cdk6, Cdk2, phospho-RB (phosphorylated-retinoblastoma), E2F1, and Trib2 with 24-hour treatment (Figure 6D). Importantly, global protein expression of RB is not decreased, and C/EBPα levels remain unchanged, if not slightly increased at the lower concentrations when cell cycle arrest occurred. These results show that the inhibition of CDKs results in cell cycle arrest and reduced proliferation, inhibition of E2F1, and a decrease in Trib2. To determine the effect in normal cells, we treated WT and Trib2 knockout (KO; Trib2−/−) total bone marrow (BM) cells and HSCs (Lineage−Sca-1+c-Kit+) with PTX, dibutyryl cAMP, and flavopiridol. There was very little cytotoxic effect on WT BM cells and HSCs as assessed by Annexin V/DAPI staining (Figure 6E-F), and no difference was observed in cell death in Trib2−/− BM and HSC cells compared with WT cells (Figure 6E-F). No cytotoxic effect was seen in colony-forming assays of untreated and treated HSCs from WT and Trib2−/− animals (Figure 6G). These data show that there is limited cell toxicity of targeting this pathway using these drugs in normal cells or cells that lack Trib2 expression.
CDK inhibition decreases Trib2 resulting in a block in AML cell proliferation. (A) PTX and flavopiridol dose response of U937 cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay at 24 hours. Data are expressed as percentage over untreated control. Values are expressed as mean ± SD (N = 2, performed in triplicate). (B) Cell cycle analysis of PTX, dibutyryl cAMP, and flavopiridol dose response in U937 cells at 24 hours. (C) Cell proliferation of PTX and flavopiridol dose response of U937 cells assessed by CFSE staining 96 hours after treatment. (D) Western blot analysis of protein lysates from U937 cells treated for 24 hours with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol. Shown are Cdk6, Cdk2, pRB (phosphor-RB), E2F1, Trib2, and C/EBPα-p42. Actin, total RB, and histone deacetylase 1 (HDAC1) are shown as protein loading controls. (E) WT and Trib2−/− (KO) total bone marrow cells were treated with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (F) WT and Trib2−/− (KO) HSCs were treated with different concentrations of (left) PTX and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (G) WT and Trib2−/− (KO) HSCs were treated with 2 mM PTX or 62.5 nM of flavopiridol (FLV) for 24 hours and plated in a CFC assay. Data presented are mean ± SD of duplicate cultures.
CDK inhibition decreases Trib2 resulting in a block in AML cell proliferation. (A) PTX and flavopiridol dose response of U937 cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay at 24 hours. Data are expressed as percentage over untreated control. Values are expressed as mean ± SD (N = 2, performed in triplicate). (B) Cell cycle analysis of PTX, dibutyryl cAMP, and flavopiridol dose response in U937 cells at 24 hours. (C) Cell proliferation of PTX and flavopiridol dose response of U937 cells assessed by CFSE staining 96 hours after treatment. (D) Western blot analysis of protein lysates from U937 cells treated for 24 hours with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol. Shown are Cdk6, Cdk2, pRB (phosphor-RB), E2F1, Trib2, and C/EBPα-p42. Actin, total RB, and histone deacetylase 1 (HDAC1) are shown as protein loading controls. (E) WT and Trib2−/− (KO) total bone marrow cells were treated with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (F) WT and Trib2−/− (KO) HSCs were treated with different concentrations of (left) PTX and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (G) WT and Trib2−/− (KO) HSCs were treated with 2 mM PTX or 62.5 nM of flavopiridol (FLV) for 24 hours and plated in a CFC assay. Data presented are mean ± SD of duplicate cultures.
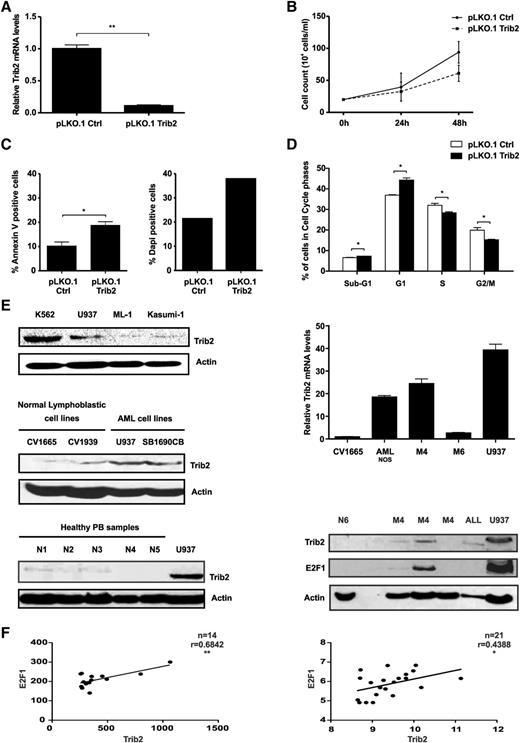
AML cells express high levels of Trib2 protein and correlate with E2F family members mRNA expression. (A) Real-time analysis of Trib2 mRNA levels in U937 cells following transduction with pLKO.1 control (Ctrl) or pLKO.1 Trib2. (B) Cell growth analysis in U937 cells transduced with pLKO.1 ctrl or pLKO.1 Trib2 assessed by trypan blue exclusion. (C) Cell death analysis in U937 transduced with pLKO.1 ctrl or pLKO.1 Trib2 assessed by (left) Annexin V and (right) DAPI staining. (D) Cell cycle analysis in sorted U937 cells transduced with pLKO.1 ctrl or pLKO.1 Trib2. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. (E) Western blot analysis of Trib2 protein expression in (top left) myeloid leukemia lines (K562, U937, ML-1, and Kasumi-1), (middle left) normal cell lines (CV1665 and CV1939), and (bottom left) normal peripheral blood (PB and N1-5). (Top right) Real-time analysis of Trib2 mRNA levels in AML patient samples. (Bottom right) Western blot analysis of Trib2 and E2F1 protein expression in normal PB (N6), AML patient samples (denoted by their subtypes), ALL patient samples, and U937 cells. Actin is shown as a protein loading control. (F) Correlation analysis of Trib2 vs E2F1 in the top 95th and 96th percentiles of patient samples from the dataset GSE115934 (N = 14) and GSE1446835 (N = 21). P < .05 and correlation coefficient (Pearson r, close to +1) show significant positive correlations.
AML cells express high levels of Trib2 protein and correlate with E2F family members mRNA expression. (A) Real-time analysis of Trib2 mRNA levels in U937 cells following transduction with pLKO.1 control (Ctrl) or pLKO.1 Trib2. (B) Cell growth analysis in U937 cells transduced with pLKO.1 ctrl or pLKO.1 Trib2 assessed by trypan blue exclusion. (C) Cell death analysis in U937 transduced with pLKO.1 ctrl or pLKO.1 Trib2 assessed by (left) Annexin V and (right) DAPI staining. (D) Cell cycle analysis in sorted U937 cells transduced with pLKO.1 ctrl or pLKO.1 Trib2. Data presented are mean ± SD of duplicate cultures and representative of 2 independent experiments. (E) Western blot analysis of Trib2 protein expression in (top left) myeloid leukemia lines (K562, U937, ML-1, and Kasumi-1), (middle left) normal cell lines (CV1665 and CV1939), and (bottom left) normal peripheral blood (PB and N1-5). (Top right) Real-time analysis of Trib2 mRNA levels in AML patient samples. (Bottom right) Western blot analysis of Trib2 and E2F1 protein expression in normal PB (N6), AML patient samples (denoted by their subtypes), ALL patient samples, and U937 cells. Actin is shown as a protein loading control. (F) Correlation analysis of Trib2 vs E2F1 in the top 95th and 96th percentiles of patient samples from the dataset GSE115934 (N = 14) and GSE1446835 (N = 21). P < .05 and correlation coefficient (Pearson r, close to +1) show significant positive correlations.
To prove that inhibition of Trib2 expression plays a role in leukemic cell proliferation, we used lentiviral technology to knockdown Trib2 expression in AML cells (Figure 7A; supplemental Figure 2). Trib2 knockdown inhibited the growth of AML cells (Figure 7B), induced apopotosis and decreased cell viability (Figure 7C), and induced G1 cell cycle arrest (Figure 7D), indicating that Trib2 expression is required for the survival and proliferation of AML cells. Therefore, to assess the prevalence of high Trib2 expression in AML, we analyzed primary AML patient samples by mRNA and western blot analysis and compared the levels to normal peripheral blood samples (N1-6), normal lymphoblast cell lines (CV1665 and CV1939), and myeloid leukemic cell lines (K562, U937, ML-1, Kasumi, and SB1690CB). We detected high Trib2 expression in AML patient samples compared with normal controls (Figure 7E). High E2F1 expression correlated with high Trib2 protein expression (Figure 7E). AML patient datasets were investigated for a correlative relationship between elevated Trib2 expression and E2F1 expression. Using 2 previously published AML patient microarray datasets,34,35 we found a significant positive correlation between E2F1 and the high Trib2-expressing patient samples (Figure 7F). As positive controls, we determined that there was a significant positive correlation between Trib2 and NOTCH1 and a significant negative correlation between Trib2 and C/EBPα (data not shown). Therefore, these data strongly support the role of E2F1-regulated Trib2 expression in AML and its function in the dysregulation of myeloid cell proliferation. Our data support the finding that the perturbation of C/EBPα by Trib2 functions in a positive feedback loop to enhance E2F1-mediated Trib2 expression in AML.
Discussion
Trib2 expression may be differentially regulated depending on the cell type (myeloid or lymphoid, immature or mature), cell context (normal or malignant), and the cell cycle (quiescent or self-renewing or proliferating). Here we described the regulation of Trib2 in normal myeloid and leukemic cells via an E2F1 and C/EBPα feedback mechanism. We demonstrated a correlation between the expression levels of E2F1 and Trib2 in AML cells and showed that E2F1 is recruited to the Trib2 promoter, activating Trib2 expression. E2F1-regulated Trib2 activation was negatively and positively modulated via C/EBPα-p42 and C/EBPα-p30 expression, respectively. We demonstrated the recruitment of C/EBPα-p42 to the Trib2 promoter in normal myeloid cells and the elevation of Trib2 expression in C/EBPα-p30 expressing preleukemic myeloid cells. Our results reveal a positive feedback loop for E2F1, Trib2, and C/EBPα-p30 in AML. Our data suggest that the regulation of Trib2 expression in AML cells is pivotal for AML cell proliferation and survival.
The identification of E2F direct gene targets via consensus and nonconsensus binding sites has been globally assessed using ChIP technologies.36 Indeed the recruitment of E2F1 to promoters does not always require DNA binding, as other transcription factors have been shown to recruit E2F1, such as nuclear factor-κB, Myc, and C/EBPα.27 C/EBPα-p30 has been shown to recruit E2F1 to the promoter of PIN1, and increased PIN1 expression was proposed to contribute to the differentiation block in AML.37 We showed and validated the presence of E2F1 binding sites in the Trib2 promoter. Using site-directed and deletional mutagenesis and EMSA analysis, we conclude that E2F1 binds directly to the Trib2 promoter and is modulated by C/EBPα specifically at site B at −943 bp. We demonstrate the recruitment of C/EBPα to this region in normal GMP cells. Using a ChIP assay, we were unable to distinguish between C/EBPα-p42 and C/EBPα-p30 binding to the Trib2 promoter in normal or preleukemic cells. It has been suggested that C/EBPα-p30 has independent functions of C/EBPα-p42 and that it can target and modulate a unique set of genes in addition to its role as a dominant negative of C/EBPα-p42.38,39 Given that we have shown that C/EBPα-p30 preleukemic GMP cells have elevated Trib2 expression above that present in normal GMPs, these data support the positive feedback loop that we propose between E2F1, C/EBPα, and Trib2.
The central mechanisms identified for the negative regulation of cell proliferation by C/EBPα are repression of E2F,22 interference with Cdk2 and Cdk4 function,40 and p21 stabilization (a Cdk inhibitor).41 Flavopiridol is a small molecule cyclin-dependent kinase inhibitor that induces cell cycle arrest, apoptosis, and clinical responses in AML patients. Following the inhibition of Cdks, phosphorylation of RB is inhibited, and E2F is released and drives cell cycle arrest and apoptosis. Studies have shown that flavopiridol induces apoptosis in leukemic blasts from patients with poor-risk AML or ALL42 and represses E2F1 expression in leukemic blasts from patients with refractory AML.43 PTX has been shown to induce cell cycle arrest and apoptosis of AML and ALL cells44,45 and has been used in combination therapy for patients with myelodysplastic syndromes and AML.46,47 Elevated cAMP, as a result of PTX and dibutyryl cAMP treatment, has been shown to target CDK and E2F1 in leukemic cells, leading to cell cycle arrest,33 apoptosis,48 and leukemic cell differentiation.49 Notwithstanding, these drugs also target other pathways, for instance, nuclear factor-κB and antiapoptotic proteins.44 Therefore, the inhibition of CDK has been shown to induce a number of effects in different subtypes of AML, which all prove to be detrimental to the leukemic cell (differentiation, cell cycle arrest, and apoptosis). The inhibition of CDK activity in high Trib2-expressing AML cells resulted in the arrest of cells in G1 phase of the cell cycle and a subsequent decrease in cell in S phase. Using concentrations of drug that induced minimal apoptosis and a block in proliferation, there was a decrease in E2F1 expression and a subsequent decrease in Trib2 expression, and these effects ultimately led to apoptotic cell death (sub-G1 increase) of the AML cell. The fact that we did not observe any proliferative defect following treatment in nonleukemic WT and Trib2-deficient BM cells corroborates the role of Trib2 in the control of leukemic cell proliferation.
Here we identify a positive regulatory loop controlled by E2F1 that contributes to the aberrant expression of Trib2 in AML cells. Our findings provide a novel link between oncogenic Trib2 function and AML cell proliferation. Whether E2F-mediated regulation of Trib2 occurs also in ALL remains to be investigated and of great interest.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Kevin Ryan (Beatson Institute for Cancer Research, Glasgow, Scotland) for E2F plasmids and Stefan Meyer (University of Manchester) for cell lines CV1665, CV1939, and SB1690CB. MEFs derived from E2F1-deficient or WT mouse embryos were a kind gift from Dr Lili Yamasaki (Columbia University). The authors thank the technical staff at the Paul O’Gorman Leukaemia Research Centre and at the biological services at the University of Glasgow.
The authors thank the Kay Kendall Foundation (KKL501) and the Howat Foundation for flow cytometry facility funding. K.K. was supported by funds from the Howat Foundation, Children with Cancer United Kingdom, and financial support from Science Foundation Ireland President of Ireland Young Researcher Award and Marie Curie EU-FP7-PEOPLE-IRG. This work was also supported by the Childrens Leukaemia Research Project, Irish Research Council for Science, Engineering and Technology fellowship (to M. Hannon), and Health Research Board (to J.T.). Work in the Porse laboratory was supported by the Danish Cancer Society, the Danish Research Council for Strategic Research, and through a center grant from the NovoNordisk Foundation (The Novo Nordisk Foundation Section for Stem Cell Biology in Human Disease).
Authorship
Contribution: K.K. designed the study; L.R., M.S., M. Hannon, M. Hasemann, A.-K.F., J.C., J.T., C.O., and K.K. performed the research; K.K., L.R., M.S., M. Hannon, M. Hasemann, A.F., and B.P. analyzed the data; B.P. contributed an essential mouse model and reagents; M.R.C. provided access to patient samples; L.R. and M.S. made the figures; K.K. wrote the paper; and K.K., L.R., M.S., B.P., and M. Hasemann revised and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen Keeshan, Paul O’Gorman Leukaemia Research Centre, Institute of Cancer Sciences, University of Glasgow, Glasgow, Scotland; e-mail: Karen.keeshan@glasgow.ac.uk.



![Figure 6. CDK inhibition decreases Trib2 resulting in a block in AML cell proliferation. (A) PTX and flavopiridol dose response of U937 cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide; 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay at 24 hours. Data are expressed as percentage over untreated control. Values are expressed as mean ± SD (N = 2, performed in triplicate). (B) Cell cycle analysis of PTX, dibutyryl cAMP, and flavopiridol dose response in U937 cells at 24 hours. (C) Cell proliferation of PTX and flavopiridol dose response of U937 cells assessed by CFSE staining 96 hours after treatment. (D) Western blot analysis of protein lysates from U937 cells treated for 24 hours with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol. Shown are Cdk6, Cdk2, pRB (phosphor-RB), E2F1, Trib2, and C/EBPα-p42. Actin, total RB, and histone deacetylase 1 (HDAC1) are shown as protein loading controls. (E) WT and Trib2−/− (KO) total bone marrow cells were treated with different concentrations of (left) PTX, (center) dibutyryl cAMP, and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (F) WT and Trib2−/− (KO) HSCs were treated with different concentrations of (left) PTX and (right) flavopiridol and assessed for apoptosis by AnnexinV/DAPI staining. (G) WT and Trib2−/− (KO) HSCs were treated with 2 mM PTX or 62.5 nM of flavopiridol (FLV) for 24 hours and plated in a CFC assay. Data presented are mean ± SD of duplicate cultures.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/15/10.1182_blood-2013-07-511683/4/m_2389f6.jpeg?Expires=1769147921&Signature=lEd0cHfyuD8vGzevS4Vo-2ONvcgM~opp1fBqQM64Up5V6wBDaG8EbP0Wu9XLP-Wwg5OMgekOvSl~n1N4E5~EG5xpkpczB6YPB9Nx9~yUULrTcXlaHg29pROMbyXqmNJI~4gwo9QINrOFeox5-NJASnPz3fxNdfP5MUeRizK9odX8rCs~uL-5liw7csI65jNVYtzTEeVB7m2wHGXWUnrqXthtaWVfetKYhPillAy1UONYQC~zHCIKp~DUA0McvROS596cWy1rPlA1XD-kxwNPpWbxfA0o8h8wwXat-~1npAShO3EUOoC4MMzKMCmo1e5S5Fs5MxXSWTgYdbgCX3hHdg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal