Key Points
The coding genome of fludarabine-refractory CLL patients is characterized by 16 mutations/case and 4 copy number aberrations per case on average.
Fludarabine-refractory CLL cases are enriched in FAT1 mutations occurring in 10% of patients, suggesting a role in the refractoriness event.
Abstract
Fludarabine refractoriness (FR) represents an unsolved clinical problem of chronic lymphocytic leukemia (CLL) management. Although next-generation sequencing studies have led to the identification of a number of genes frequently mutated in FR-CLL, a comprehensive evaluation of the FR-CLL genome has not been reported. Toward this end, we studied 10 FR-CLLs by combining whole-exome sequencing and copy number aberration (CNA) analysis, which showed an average of 16.3 somatic mutations and 4 CNAs per sample. Screening of recurrently mutated genes in 48 additional FR-CLLs revealed that ∼70% of FR-CLLs carry ≥1 mutation in genes previously associated with CLL clinical course, including TP53 (27.5%), NOTCH1 (24.1%), SF3B1 (18.9%), and BIRC3 (15.5%). In addition, this analysis showed that 10.3% of FR-CLL cases display mutations of the FAT1 gene, which encodes for a cadherin-like protein that negatively regulates Wnt signaling, consistent with a tumor suppressor role. The frequency of FAT1-mutated cases was significantly higher in FR-CLL than in unselected CLLs at diagnosis (10.3% vs 1.1%, P = .004), suggesting a role in the development of a high-risk phenotype. These findings have general implications for the mechanisms leading to FR and point to Wnt signaling as a potential therapeutic target in FR-CLL.
Introduction
The clinical course of patients with chronic lymphocytic leukemia (CLL), the most common leukemia in adults, spans from an asymptomatic disease never requiring therapy to a rapidly progressive disorder requiring intensive treatment.1,2
Over the last decade, major advances have been made in our understanding of the pathogenesis of CLL and in the range of available therapeutic options for CLL patients. The current first-line standard of care for fit patients is a fludarabine-based approach usually including cyclophosphamide and rituximab (FCR). This combination therapy offers overall response rates of 95%, complete responses of 44%, and a median progression-free survival of 52 months.3 Nevertheless, fludarabine refractoriness (FR), accounting for ∼10% of treated patients, remains a challenging clinical problem associated with poor survival.3-5 Until recently, the molecular basis of CLL chemorefractoriness was incompletely characterized, and the only known genetic lesion with an established pathogenic role was inactivation of the TP53 gene occurring in 30% to 40% of refractory patients.6,7 In the last few years, next-generation sequencing (NGS) and targeted resequencing studies have led to the identification of novel genes that are recurrently mutated in CLL, including some that appear to be preferentially altered in FR patients, namely NOTCH1, SF3B1, and BIRC3.8-12 However, a comprehensive and unbiased description of the mutational profile and copy number aberration (CNA) load of FR-CLL has thus far not been provided.
Furthermore, all NGS studies performed to date analyzed heterogeneous cohorts of CLL patients in terms of both biological and clinical features. This study was aimed at characterizing the genetic background of a clinically homogeneous group of FR-CLL patients.
Materials and methods
Study population
Peripheral blood from 9 FR-CLL patients sampled prior to the fludarabine-based treatment that resulted in chemo-refractoriness and from 1 patient with del17p13—known to induce FR—was used in whole-exome sequencing (WES) and single-nucleotide polymorphism array (SNP) experiments (discovery panel; Table 1). Paired germline DNA was also analyzed.
Characteristics of the discovery panel cases at refractoriness
| Characteristics . | FR-CLL_1 . | FR-CLL_2 . | FR-CLL_3 . | FR-CLL_4 . | FR-CLL_5 . | FR-CLL_6 . | FR-CLL_7 . | FR-CLL_8 . | FR-CLL_9 . | FR-CLL_10 . |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Female | Male | Male | Male | Male | Male | Male | Female | Male | Male |
| Age, years | 75 | 75 | 62 | 69 | 62 | 73 | 58 | 48 | 58 | 67 |
| Binet stage | B | B | C | B | B | B | A progressive | B | B | A progressive |
| Lymphocytes (×109/L) | 26 | 44 | 129 | 102 | 66 | 21 | 10 | 142 | 32 | 97 |
| CD5+/CD19+ (%) | 80 | 92 | 90 | 96 | 90 | 80 | 73 | 95 | 90 | 91 |
| Hb, g/dL | 10.9 | 11.2 | 12.2 | 14.7 | 13.5 | 15.6 | 15.7 | 12 | 15.6 | 12.4 |
| Platelets (×109/L) | 142 | 237 | 46 | 119 | 231 | 310 | 118 | 201 | 231 | 121 |
| B2M, mg/L | 2.9 | 1.9 | 3.5 | 3.2 | 2.6 | 3 | NA | 1.54 | 3.27 | NA |
| LDH, U/L | 326 | 421 | 382 | 404 | 256 | 500 | 271 | 332 | 105 | NA |
| IGHV mutation status | UM | M | UM | M | UM | UM | UM | M | UM | UM |
| IGHV | 3-49*03 | 3-52*01 | 3-30-3*01 | 2-5*10 | 5-51*01 | 1-2*02 | 4-30*2*01 | 1-46*01 | 1-69*01 | 4-59*01 |
| IGHD | 3-10*01 | 2-2*03 | 3-3*02 | 1-7*01 | 3-3*01 | 6-19*01 | 3-10*01 | 3-22*01 | 3-3*01 | 2-21*02 |
| IGHJ | 1*01 | 5*02 | 6*02 | 4*02 | 6*02 | 4*02 | 6*02 | 1*01 | 6*02 | 3*02 |
| IGHV_homology | 98.09 | 97.09 | 100.00 | 95.87 | 100.00 | 99.21 | 100.00 | 88.50 | 100.00 | 99.30 |
| FISH | del11q22-q23; del13q14 | del13q14 | Normal | del17p13; del13q14 | del17p13; del11q22-23; del13q14 | del17p13 | Normal | +12 | del17p12; +12 | del17p13 |
| TP53 mutational status | WT | WT | M | M | WT | WT | WT | WT | WT | M |
| NOTCH1 mutational status | WT | WT | M | WT | WT | WT | WT | M | WT | WT |
| BIRC3 disruption | WT | WT | WT | WT | Δ | WT | WT | WT | WT | WT |
| ZAP-70 expression* | N | N | P | P | P | N | P | N | P | P |
| CD38 expression† | N | N | P | N | N | P | P | P | N | P |
| Timing of refractoriness | Acquired | Primary | Primary | Primary | Primary | Primary | Acquired | Primary | Primary | Primary |
| Treatment resulting in refractoriness | FCR | FR | FCR | FCR | FA | FCR | FCL | FC | —‡ | FR |
| Previous treatment | FC | — | — | — | — | — | Fluda-based | — | Campath | Leukeran |
| Time to first treatment (months) | 36 | 30 | 0 | 0 | 2 | 1 | 35 | 1 | 10 | 31 |
| Survival (months)§ | 32 | 32 | 30 | 30 | 14 | 14 | 19 | 51 | 21 | 8 |
| Status | Alive | Alive | Alive | Alive | Alive | Death | Alive | Alive | Alive | Death |
| Control DNA source | Granulocytes | Granulocytes | Saliva | Saliva | Granulocytes | Granulocytes | Saliva | Saliva | Saliva | Saliva |
| Characteristics . | FR-CLL_1 . | FR-CLL_2 . | FR-CLL_3 . | FR-CLL_4 . | FR-CLL_5 . | FR-CLL_6 . | FR-CLL_7 . | FR-CLL_8 . | FR-CLL_9 . | FR-CLL_10 . |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Female | Male | Male | Male | Male | Male | Male | Female | Male | Male |
| Age, years | 75 | 75 | 62 | 69 | 62 | 73 | 58 | 48 | 58 | 67 |
| Binet stage | B | B | C | B | B | B | A progressive | B | B | A progressive |
| Lymphocytes (×109/L) | 26 | 44 | 129 | 102 | 66 | 21 | 10 | 142 | 32 | 97 |
| CD5+/CD19+ (%) | 80 | 92 | 90 | 96 | 90 | 80 | 73 | 95 | 90 | 91 |
| Hb, g/dL | 10.9 | 11.2 | 12.2 | 14.7 | 13.5 | 15.6 | 15.7 | 12 | 15.6 | 12.4 |
| Platelets (×109/L) | 142 | 237 | 46 | 119 | 231 | 310 | 118 | 201 | 231 | 121 |
| B2M, mg/L | 2.9 | 1.9 | 3.5 | 3.2 | 2.6 | 3 | NA | 1.54 | 3.27 | NA |
| LDH, U/L | 326 | 421 | 382 | 404 | 256 | 500 | 271 | 332 | 105 | NA |
| IGHV mutation status | UM | M | UM | M | UM | UM | UM | M | UM | UM |
| IGHV | 3-49*03 | 3-52*01 | 3-30-3*01 | 2-5*10 | 5-51*01 | 1-2*02 | 4-30*2*01 | 1-46*01 | 1-69*01 | 4-59*01 |
| IGHD | 3-10*01 | 2-2*03 | 3-3*02 | 1-7*01 | 3-3*01 | 6-19*01 | 3-10*01 | 3-22*01 | 3-3*01 | 2-21*02 |
| IGHJ | 1*01 | 5*02 | 6*02 | 4*02 | 6*02 | 4*02 | 6*02 | 1*01 | 6*02 | 3*02 |
| IGHV_homology | 98.09 | 97.09 | 100.00 | 95.87 | 100.00 | 99.21 | 100.00 | 88.50 | 100.00 | 99.30 |
| FISH | del11q22-q23; del13q14 | del13q14 | Normal | del17p13; del13q14 | del17p13; del11q22-23; del13q14 | del17p13 | Normal | +12 | del17p12; +12 | del17p13 |
| TP53 mutational status | WT | WT | M | M | WT | WT | WT | WT | WT | M |
| NOTCH1 mutational status | WT | WT | M | WT | WT | WT | WT | M | WT | WT |
| BIRC3 disruption | WT | WT | WT | WT | Δ | WT | WT | WT | WT | WT |
| ZAP-70 expression* | N | N | P | P | P | N | P | N | P | P |
| CD38 expression† | N | N | P | N | N | P | P | P | N | P |
| Timing of refractoriness | Acquired | Primary | Primary | Primary | Primary | Primary | Acquired | Primary | Primary | Primary |
| Treatment resulting in refractoriness | FCR | FR | FCR | FCR | FA | FCR | FCL | FC | —‡ | FR |
| Previous treatment | FC | — | — | — | — | — | Fluda-based | — | Campath | Leukeran |
| Time to first treatment (months) | 36 | 30 | 0 | 0 | 2 | 1 | 35 | 1 | 10 | 31 |
| Survival (months)§ | 32 | 32 | 30 | 30 | 14 | 14 | 19 | 51 | 21 | 8 |
| Status | Alive | Alive | Alive | Alive | Alive | Death | Alive | Alive | Alive | Death |
| Control DNA source | Granulocytes | Granulocytes | Saliva | Saliva | Granulocytes | Granulocytes | Saliva | Saliva | Saliva | Saliva |
FA, fludarabine, alemtuzumab; FCR, fludarabine, cyclophosphamide, rituximab; FR, fludarabine, rituximab; M, mutated; N, negative; NA, not available; P, positive; Δ, deleted.
Considered positive if the percentage of positive leukemic cells was >20% by FACS analysis.
Considered positive if the percentage of positive leukemic cells was >30%.
FR-CLL_9 underwent a non–fludarabine-based treatment being a bona fide chemorefractory patient because he carried a del17p13.
Survival from refractoriness.
Refractoriness to fludarabine or to a fludarabine-containing regimen was defined as nonresponse or progression within 6 months from the last antileukemic therapy, in agreement with the International Workshop on CLL-National Cancer Institute (IWCLL-NCL) Working Group criteria.13 We included both primary (ie, refractory to the first fludarabine-based treatment, N = 8) and acquired refractory patients (ie, patients who responded to the first fludarabine-based treatment but became refractory to the second fludarabine-based therapy, N = 2).
Patients provided informed consent in accordance with local ethical committee requirements and the Declaration of Helsinki. All samples satisfied the diagnostic criteria for CLL and had >80% of clonal CD19+/CD5+ cells.13 IGHV gene analysis, CD38 and ZAP-70 expression, and fluorescence in situ hybridization (FISH) analyses were performed as previously described.14-17 This study was approved by the Ethical Committees of Policlinico Umberto I, “Sapienza” University of Rome (rif.2182/16.06.2011).
Mutation analysis of TP53 exons 4 to 9 was carried out by DNA direct sequencing (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems, Foster City, CA), as reported.18
Matched germline DNA, extracted from saliva or granulocytes, proved to be negative for patient-specific IGHV-D-J rearrangements. Bacterial contamination of saliva DNA was excluded by performing the Quantifiler assay (Applied Biosystems).
Forty-eight well-characterized FR-CLL cases (screening panel; supplemental Table 1 on the Blood Web site) with >70% clonal CD19+/CD5+ cells were used to screen the genes identified by WES and to extend the SNP array analysis.
Whole-exome capture and massively parallel sequencing
Exome capture and massively parallel sequencing were performed at the Fasteris SA HiSeq Service (Plan-les-Ouates, Switzerland). Purified tumor and germline genomic DNA (3 µg) from the 10 FR-CLL cases was enriched in protein coding sequences using the in-solution exome capture SureSelect Human All Exon 50Mb kit (Agilent Technologies, Santa Clara, CA), according to the manufacturer’s protocol. The captured targets were subjected to massively parallel sequencing using the Illumina HiSeq 2000 analyzer (Illumina, San Diego, CA) with the paired-end 2 × 100-bp read option, following the manufacturer’s instructions.
The performance of WES is reported in supplemental Table 3. Additional information is provided in supplemental Methods.
Sequence mapping and identification of tumor-specific variants
Paired-end reads obtained by high-throughput sequencing were aligned to the human genome reference hg19/NCBI GRCh37 using the Burrows-Wheeler alignment tool version 0.5.9. Sequence variants, ie, differences from the reference sequence, were identified separately for each tumor and germline sample. The frequency of each variant was estimated from the total number of reads covering the position of that variant. Using the SAVI algorithm,19 an empirical prior was constructed from which we obtained a corresponding high-credibility interval (posterior probability ≥1-10−5) for the frequency of each variant and a high-credibility interval for the corresponding change in frequency between the tumor and the normal samples. The nonsilent variants were kept for validation if fulfilling the following criteria: (1) not reported in dbSNP137, (2) with variant frequency >15% in tumor and <3% in normal, and (3) at least 1% change in frequency in tumor from the normal (with high posterior probability ≥1-10−5).
Validation of candidate somatic mutations by Sanger sequencing
Candidate nonsilent somatic mutations were subjected to validation by conventional Sanger-based resequencing of polymerase chain reaction (PCR) products obtained from both tumor and paired germline high-molecular-weight genomic DNA using primers specific for the exon encompassing the variant (supplemental Methods).
In silico analysis of the mutated genes
Genes affected by validated nonsilent mutations were assigned to functional categories and pathways by means of DAVID software (http://david.abcc.ncifcrf.gov/) and the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp).20,21 Also, it was verified whether they were listed in the Catalogue of Somatic Mutations in the Cancer (COSMIC) database22 and in the Cancer Gene Census database (http://www.sanger.ac.uk/genetics/CGP/Census/).
Last, confirmed, somatic nonsilent mutations were tested for their consequences by the PolyPhen-2 algorithm (http://genetics.bwh.harvard.edu/pph2/).23
Screening of recurrently mutated genes
The novel recurrently mutated genes (FAT1, UTRN, FPGT, and TSC1) identified by WES of the discovery panel were sequenced in the screening panel. The FAT1 sequence was also analyzed in 174 CLL cases at diagnosis. The coding sequences and splice sites were analyzed by PCR amplification and Sanger sequencing (ABI PRISM 3100) of the whole genome–amplified DNA generated by the Repli-g Mini kit (Qiagen, Germantown, MD). PCR primers were designed as described in supplemental Methods. Sequence analysis was performed by using the Mutation Surveyor Version 3.97 software (SoftGenetics, State College, PA), and after subtracting synonymous mutations and SNPs, the remaining candidate somatic mutations were confirmed on high-molecular-weight genomic DNA from both tumor and normal (when available) samples by performing PCR amplification and bidirectional direct sequencing.
CNA analysis by high-density SNP arrays
Genome-wide CNA analysis was carried out on the 10 FR-CLL patients included in the discovery panel and on 29 cases belonging to the screening panel. Experiments were performed by using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA; GEO accession number GSE51711) and standard protocols. The identification of CNAs, as well as the recognition of minimal common regions of aberration, was performed as previously reported.24,25
Statistical analysis
Student t tests and Fisher exact tests were performed using the tools at http://www.physics.csbsju.edu/stats/. For the definition of an outlier, the Tukey method was applied: data points are suspected outliers if they are 1.5 × interquartile range above the third quartile or below the first quartile.
Results
Mutational spectrum of FR-CLL in the discovery panel
Ten CLL patients (Table 1) sampled prior to the fludarabine-based treatment that resulted in chemo-refractoriness were studied by WES to assess the mutational load of the FR-CLL coding genome and to identify predictors of FR.
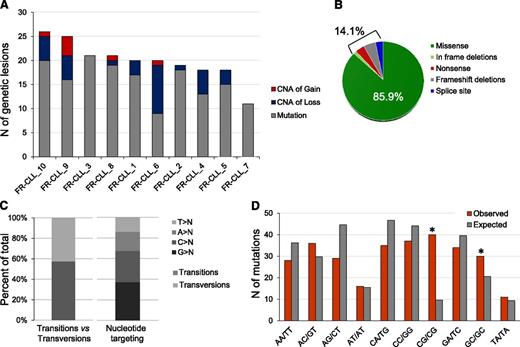
WES bioinformatic analysis predicted 170 nonsilent variants, of which 161 were confirmed to be of somatic origin by Sanger sequencing (validation rate, 94.7%), whereas the remaining 9 were absent in both tumor and normal genomic DNA. By also including 2 NOTCH1 mutations, evaluated by Sanger sequencing due to the poor capture of this region by exome arrays, FR-CLL displayed 16.3 nonsilent mutations per case on average (range, 9-23; Figure 1A).
Overall load and features of genomic lesions in the discovery panel. (A) Combined load of somatically acquired nonsilent mutations and CNA identified in the FR-CLL discovery panel. (B) Type of mutations. (C) Nucleotide targeting of point mutations. (D) Mutation frequency at specific dinucleotides (red bars). Asterisks denote statistically significant differences in over-represented changes compared with the expected frequencies (gray bars), as assessed by a Poisson distribution after correction for multiple hypotheses.
Overall load and features of genomic lesions in the discovery panel. (A) Combined load of somatically acquired nonsilent mutations and CNA identified in the FR-CLL discovery panel. (B) Type of mutations. (C) Nucleotide targeting of point mutations. (D) Mutation frequency at specific dinucleotides (red bars). Asterisks denote statistically significant differences in over-represented changes compared with the expected frequencies (gray bars), as assessed by a Poisson distribution after correction for multiple hypotheses.
Mutations were predominantly missense substitutions (85.9%) and infrequently frameshift/in frame deletions (7.3%), nonsense (3.7%), or affecting splice sites (3.1%) (Figure 1B and supplemental Table 4). As previously outlined in other tumors,24,28,29 transitions prevailed over transversions (N = 87:64; ratio, 1.35), G and C nucleotides were preferentially targeted (67.5% events at G/C base pairs vs 32.5% at A/T), and a significant bias toward alterations at 5′-CpG-3′ (P < .001) dinucleotides was found (Figure 1C-D).
These events affected 151 genes with 8 recurrently mutated genes (>1 sample): TP53 (4/10 patients), SF3B1 (3/10 patients), and ATM, NOTCH1, FAT1, FPGT, UTRN, TSC1 (each mutated in 2/10 patients).
In silico analysis showed that 126 mutated genes are annotated in the COSMIC database as affected by mutations in cancer and 13 are reported in the Cancer Gene Census database (supplemental Table 4). The DAVID software revealed a significant enrichment in specific functional groups: regulation of cell adhesion, extracellular matrix component, cell-cell junction, regulation of morphogenesis, regulation of membrane potential, protein tyrosine kinase activity, and metal ion transport (supplemental Table 5). The latter included many members of the solute carrier family; notably, among them, SLCO1A2 has a role in drug transport.30
Last, 58% of the mutations identified were predicted as deleterious by the PolyPhen-2 algorithm (supplemental Table 4).
CNA analysis and overall load of genomic lesions in the discovery panel
FR-CLL patients and their paired normal DNA were also analyzed by SNP 6.0 arrays. Forty somatic CNAs were identified (average 4 lesions per sample; range, 0-13; supplemental Table 6) with a prevalence of deletions (N = 33, 82.5%) over gains, consistent with previous reports.8
This analysis highlighted 27 novel CNAs beyond those detected by FISH, although none were recurrent. Specifically, the known 13q14 was the most common minimally deleted region (MDR), which encompassed the RB1 gene in 3 of 4 cases, in keeping with its association with a relatively poor prognosis.31
The combination of sequencing and CNA data revealed that the coding genome of FR-CLL patients is affected by a total of 20.3 lesions per sample (range, 11-26), with a prevalence of point mutations (Figure 1).
Recurrently mutated genes in FR-CLL
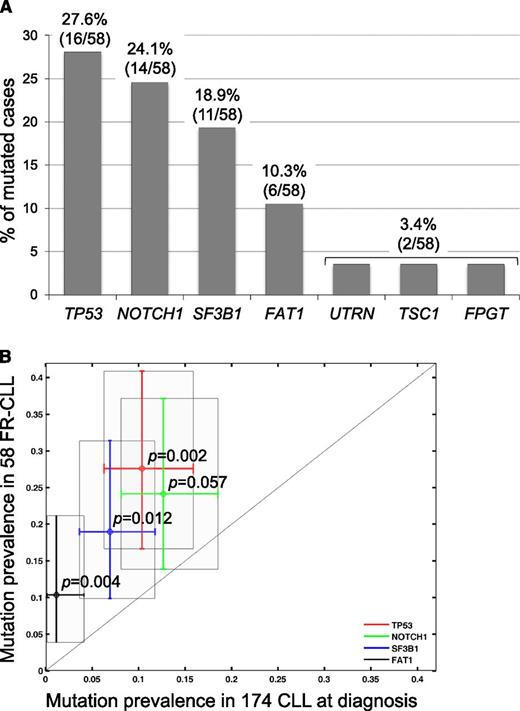
To verify the frequency of the mutations observed in the 4 newly identified genes, namely, FAT1, FPGT, UTRN, and TSC1, their coding exons and consensus splice sites were sequenced by Sanger in the screening panel for a total number of 58 FR-CLL cases analyzed. Although no additional alterations were found in FPGT, UTRN, and TSC1 (3.4%), FAT1 was recurrently mutated (6/58, 10.3%; Figure 2A).
Screening of the recurrently mutated genes. (A) Percentage of FR-CLL primary cases harboring mutations in candidate genes after targeted resequencing of a screening dataset (total N = 58 FR-CLL, including discovery and screening cases). The number of mutated cases over total analyzed is as follows: TP53, 16/58 (27.6%); NOTCH1 14/58 (24.1%); SF3B1 11/58 (18.9%); FAT1 6/58 (10.3%); UTRN 2/58 (3.4%); TSC1 2/58 (3.4%); FPGT 2/58 (3.4%). (B) Prevalence of TP53 (P = .002), NOTCH1 (P = .057), SF3B1 (P = .012), and FAT1 (P = .004) mutations in 58 FR-CLL vs 174 unselected CLL cases at diagnosis.
Screening of the recurrently mutated genes. (A) Percentage of FR-CLL primary cases harboring mutations in candidate genes after targeted resequencing of a screening dataset (total N = 58 FR-CLL, including discovery and screening cases). The number of mutated cases over total analyzed is as follows: TP53, 16/58 (27.6%); NOTCH1 14/58 (24.1%); SF3B1 11/58 (18.9%); FAT1 6/58 (10.3%); UTRN 2/58 (3.4%); TSC1 2/58 (3.4%); FPGT 2/58 (3.4%). (B) Prevalence of TP53 (P = .002), NOTCH1 (P = .057), SF3B1 (P = .012), and FAT1 (P = .004) mutations in 58 FR-CLL vs 174 unselected CLL cases at diagnosis.
In this extended cohort, the screening of TP53, NOTCH1, and SF3B1 was also performed. The combined analysis of the discovery and screening cohorts confirmed the common involvement of selected genes as follows: (1) TP53 was the most frequently mutated gene (27.5%); (2) NOTCH1 mutations, mainly represented by the ΔCT, were detected in 14 patients (24.1%); (3) SF3B1 was mutated in 18.9% of cases; and (4) BIRC3 mutations occurred in 15.5% of cases (Figure 2A), as detailed in recent papers by our group.8-10 Furthermore, compared with 174 CLL cases at diagnosis, TP53, NOTCH1, SF3B1, and FAT1 mutations significantly prevailed in the FR-CLL cohort (Figure 2B).
Overall, 68.9% of FR-CLLs displayed ≥1 mutation in these genes, and 17.2% of cases carried >1 mutated gene. The percentage of altered cases further increased (84%) when considering also TP53 and BIRC3 deletions only (N = 7 and N = 9, respectively; the latter data were available in 48/58 cases). Notably, we could not detect significant differences in mutational load and pattern between IGHV mutated and unmutated cases (supplemental Figure 1), as well as between primary and acquired refractory cases (data not shown).
FAT1 is mutated in a significant number of FR-CLL
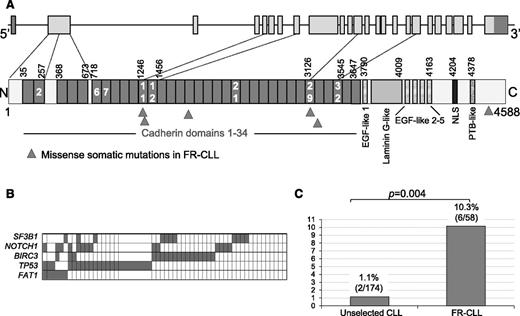
FAT1 is an ortholog of the Drosophila fat gene, which encodes a tumor suppressor essential for controlling cell proliferation during development. This member of the cadherin superfamily contains 34 tandem cadherin-type repeats, 5 epidermal growth factor–like repeats, and 1 laminin A-G domain and presumably functions as an adhesion molecule and/or signaling receptor.
In our cohort, we detected 6 heterozygous missense mutations in FAT1, which predominantly affect the cadherin domains (5/6, 83%), with 2 mutations in the cadherin domain 11. The remaining mutation affected the intracytoplasmic domain (Figure 3A). In all cases, the somatic origin of the mutation was confirmed, and the aminoacidic change was predicted to be deleterious by PolyPhen-2 with a high score (Table 2).
Features of FAT1 mutations. (A) Schematic representation of the FAT1 (upper) gene and (lower) protein, with its conserved functional domains. Triangles indicate the position of the mutations found in FR-CLL. (B) Mutual relationship of FAT1 mutations with other gene alterations in FR-CLL. In the heat map, rows correspond to identical genes and columns represent individual patients, color-coded based on the gene status (white indicates WT; gray indicates mutations and/or deletion of TP53, mutations and/or deletion of BIRC3, mutations of SF3B1, and mutations of NOTCH1). (C) Prevalence of FAT1 mutations in FR-CLL vs unselected CLL at diagnosis.
Features of FAT1 mutations. (A) Schematic representation of the FAT1 (upper) gene and (lower) protein, with its conserved functional domains. Triangles indicate the position of the mutations found in FR-CLL. (B) Mutual relationship of FAT1 mutations with other gene alterations in FR-CLL. In the heat map, rows correspond to identical genes and columns represent individual patients, color-coded based on the gene status (white indicates WT; gray indicates mutations and/or deletion of TP53, mutations and/or deletion of BIRC3, mutations of SF3B1, and mutations of NOTCH1). (C) Prevalence of FAT1 mutations in FR-CLL vs unselected CLL at diagnosis.
Characteristics of FAT1 mutations and FAT1 mutated cases
| Sample ID . | FR_CLL-1 . | FR_CLL-8 . | FR-CLL_13931 . | FR-CLL_4380 . | FR-CLL_6496 . | FR-CLL_6012 . |
|---|---|---|---|---|---|---|
| Panel | Discovery | Discovery | Screening | Screening | Screening | Screening |
| Timing of refractoriness | Acquired | Primary | Acquired | Acquired | Acquired | Primary |
| Gender | Female | Female | Male | Female | Male | Male |
| Age, years | 75 | 48 | 78 | 76 | 80 | 82 |
| Binet stage | B | B | C | C | B | B |
| Lymphocytes | 26.4 | 142.0 | 21.6 | 21.0 | 30.8 | 13.7 |
| CD5+/CD19+ | 80% | 95% | 90% | 98% | 90% | 95% |
| IGHV | 3-49*03 | 1-46*01 | 3-30*01 | 4-59*01 | 4-39*01 | 4-34*02 |
| IGHD | 3-10*01 | 3-22*01 | 3-22*01 | NA | 6-13*01 | 6-13*01 |
| IGHJ | 1*01 | 1*01 | 4*02 | 6*03 | 5*02 | 4*02 |
| IGHV homology | 98.09 | 88.50 | 100.00 | 98.09 | 100.00 | 95.43 |
| IGHV status | UM | M | UM | UM | UM | M |
| FISH | del11q22-23; del13q14 | +12 | del17p13; del11q22-q23; del13q14 | +12; del17p13 | +12 | del17p13; del13q14 |
| TP53 mutational status | WT | WT | M | WT | WT | M |
| NOTCH1 mutational status | WT | M | M | WT | M | WT |
| SF3B1 mutational status | M | WT | WT | WT | WT | WT |
| BIRC3 disruption | WT | WT | WT | WT | M | WT |
| MYD88 mutational status | WT | WT | WT | WT | WT | WT |
| CD38 | N | P | P | N | P | N |
| ZAP70 | N | N | P | P | NA | N |
| Matutes score | 5 | 4 | 5 | 4 | 5 | 5 |
| AA change | G1331S | Y3162H | L1246R | G4567R | R1856C | E3260A |
| Domain | Cadherin 11 | Cadherin 29 | Cadherin 11 | Intracytoplasmic | Cadherin 16 | Cadherin 30 |
| PolyPhen-2 class | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| PolyPhen-2 probability | 1 | 0.997 | 0.998 | 1 | 1 | 0.96 |
| Sample ID . | FR_CLL-1 . | FR_CLL-8 . | FR-CLL_13931 . | FR-CLL_4380 . | FR-CLL_6496 . | FR-CLL_6012 . |
|---|---|---|---|---|---|---|
| Panel | Discovery | Discovery | Screening | Screening | Screening | Screening |
| Timing of refractoriness | Acquired | Primary | Acquired | Acquired | Acquired | Primary |
| Gender | Female | Female | Male | Female | Male | Male |
| Age, years | 75 | 48 | 78 | 76 | 80 | 82 |
| Binet stage | B | B | C | C | B | B |
| Lymphocytes | 26.4 | 142.0 | 21.6 | 21.0 | 30.8 | 13.7 |
| CD5+/CD19+ | 80% | 95% | 90% | 98% | 90% | 95% |
| IGHV | 3-49*03 | 1-46*01 | 3-30*01 | 4-59*01 | 4-39*01 | 4-34*02 |
| IGHD | 3-10*01 | 3-22*01 | 3-22*01 | NA | 6-13*01 | 6-13*01 |
| IGHJ | 1*01 | 1*01 | 4*02 | 6*03 | 5*02 | 4*02 |
| IGHV homology | 98.09 | 88.50 | 100.00 | 98.09 | 100.00 | 95.43 |
| IGHV status | UM | M | UM | UM | UM | M |
| FISH | del11q22-23; del13q14 | +12 | del17p13; del11q22-q23; del13q14 | +12; del17p13 | +12 | del17p13; del13q14 |
| TP53 mutational status | WT | WT | M | WT | WT | M |
| NOTCH1 mutational status | WT | M | M | WT | M | WT |
| SF3B1 mutational status | M | WT | WT | WT | WT | WT |
| BIRC3 disruption | WT | WT | WT | WT | M | WT |
| MYD88 mutational status | WT | WT | WT | WT | WT | WT |
| CD38 | N | P | P | N | P | N |
| ZAP70 | N | N | P | P | NA | N |
| Matutes score | 5 | 4 | 5 | 4 | 5 | 5 |
| AA change | G1331S | Y3162H | L1246R | G4567R | R1856C | E3260A |
| Domain | Cadherin 11 | Cadherin 29 | Cadherin 11 | Intracytoplasmic | Cadherin 16 | Cadherin 30 |
| PolyPhen-2 class | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| PolyPhen-2 probability | 1 | 0.997 | 0.998 | 1 | 1 | 0.96 |
M, mutated; N, negative; NA, not available; P, positive; UM, unmutated.
FAT1 mutations occurred both in primary (N = 2) and acquired (N = 4) refractory cases, and they were not associated to any prognosis-related feature (IGHV status, ZAP-70 and CD38 expression, FISH lesions; Table 2). More importantly, FAT1 mutations never occurred as sole aberration (Figure 3B).
To establish the prevalence of FAT1 mutations in the early clinical phases of the disease, we analyzed a consecutive cohort of 174 CLL cases sampled at diagnosis (supplemental Table 2); only 2/174 patients (1.1%, P = .004) carried FAT1 mutations, suggesting that these lesions accumulate or are selected in more advanced disease phases (Figure 3C).
Overall CNA burden in FR-CLL and identification of novel CNAs
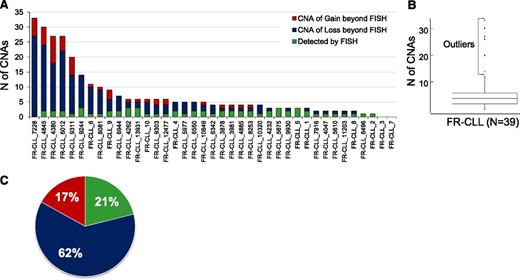
To gain a comprehensive outlook on the CNA profile of FR-CLL, we examined 29 additional FR-CLL cases (total number = 39) by SNP arrays. This analysis showed that the burden of CNAs is heterogeneous across FR-CLL patients, ranging from 0 to 33 aberrations (average, 7.4/case, or 4.1/case if we exclude outlier samples). Overall, 95% of patients were affected by CNAs, with roughly 71% displaying >3 lesions and 8 patients (20%) showing >10 lesions (Figure 4A). Particularly, 6 patients (Figure 4B and supplemental Table 7) were characterized by an outlier number of CNAs clustering within 1 single chromosome: 3 outliers (6012, 7228, and 4845) displayed >10 switches of CN state on 1 or 2 chromosomes; and the remaining 3 (4380, 9311, and 9244) carried 6 to 8 CN oscillations in 1 chromosome (supplemental Figure 2). This peculiar profile resembles the chromothripsis phenomenon.27
Distribution and characteristics of the CNAs identified in the discovery and screening panels. (A) Distribution of the CNAs identified in FR-CLL belonging to the discovery and screening panels. (B) Identification of cases with an outlier number of CNAs. (C) Type of CNAs.
Distribution and characteristics of the CNAs identified in the discovery and screening panels. (A) Distribution of the CNAs identified in FR-CLL belonging to the discovery and screening panels. (B) Identification of cases with an outlier number of CNAs. (C) Type of CNAs.
We also observed that the aberrations detected by FISH represented a small portion (21%) of all the CNAs and that 85% of cases displayed additional CNAs (Figure 4C), which were further analyzed as follows. First, we investigated the CNAs already documented in CLL.27,32-35 Del22q11.22 was observed in 4 samples (10.2%); 3 cases (7.6%) carried a del14q, with 2 displaying the same breakpoint at 14q24.1 and 1 at 14q24.3. Deletion of 6q23.3-q24.1 was found in 2/39 cases and the MDR encompassed TNFAIP3. Two samples displayed the loss of 15q15.1, with the smallest region enclosing exon 24 of MGA and the first 4 exons of MAPKB1 (Table 3). Finally, gains of 8q24.21, involving the oncogene MYC in all cases, were common and were found in 4/39 cases (10.2%).
Frequency and description of novel CNAs
| Cytoband . | Type of CNA . | Number of patients (%) . | MCR size . | Genes in MCR . | Significant by GISTIC algorithm . |
|---|---|---|---|---|---|
| 22q11.22 | Loss | 4 (10.2) | 304 kb | 4 (ZNF280B, ZNF280A, PRAME, GGTLC2) | Yes |
| 3p14.2-p14.1 | Loss | 3 (7.6) | 4.1 Mb | 11 | |
| 3p14-p12.3 | Loss | 3 (7.6) | 8.5 Mb | 8 (EIF4E3,GPR27,PROK2,SHQ1,PPP4R2,PDZRN3,CNTN3,ROBO2) | |
| 4p15.2-p14 | Loss | 3 (7.6) | 10 Mb | 7 (SLC34A2,RBPJ,CCKAR,TBC1D19,STIM2,PCDH7,ARAP2) | |
| 7q34 | Loss | 3 (7.6) | 274 kb | 4 (UBN2,C7orf55,LUC7L2,KLRG2) | |
| 9p21.3 | Loss | 3 (7.6) | 2.6 Mb | 11 (including KLHL9, members of IFNA family, CDKN2A,CDKN2B) | Yes |
| 9p21.3-p21.1 | Loss | 3 (7.6) | 8.6 Mb | 15 | |
| 14q24.3-q31.3 | Loss | 3 (7.6) | 7.8 Mb | 7 (NRXN3,C14orf145,TSHR,GTF2A1,STON2,SEL1L,FLRT2) | |
| 3p12.3-q11.2 | Loss | 2 (5.1) | 13 Mb | 11 (VGLL3 and EPHA3) | Yes |
| 3q13.32-q13.33 | Loss | 2 (5.1) | 1.8 Mb | 18 (including CD80, GSK3B) | |
| 4q21.1-q21.23 | Loss | 2 (5.1) | 8.4 Mb | 34 | |
| 4q34 | Loss | 2 (5.1) | 3.1 Mb | 7 (GALNTL6,GALNT7,HMGB2,SAP30,SCRG1,HAND2,FBXO8) | |
| 6q23.3-q24 | Loss | 2 (5.1) | 2.9 Mb | 16 (including TNFAIP3) | |
| 9q34.3 | Loss | 2 (5.1) | 160 kb | 9 (TRAF2,FBXW5,C8G,LCN12,PTGDS,FLJ45224,C9orf142,CLIC3,ABCA2) | Yes |
| 9p22.1-p21.3 | Loss | 2 (5.1) | 1.4 Mb | 13 (including MLLT3 and members of IFNA family) | |
| 10q12.3-q13.3 | Loss | 2 (5.1) | 4.7 Mb | 15 | |
| 10q23.13 | Loss | 2 (5.1) | 215 kb | 7 (CH25H,LIPA,IFIT2,IFIT3,IFIT1L,IFIT1,IFIT5) | Yes |
| 10q23.32 | Loss | 2 (5.1) | 421 kb | 3 (BTAF1,CPEB3,MARCH5) | Yes |
| 15q13.3-q15.1 | Loss | 2 (5.1) | 9.1 Mb | 41 | |
| 15q15.1 | Loss | 2 (5.1) | 41 kb | MAPKB1, MGA | Yes |
| 8q24.21 | Gain | 4 (10.2) | 5.1 Mb | 9 (MTSS1,ZNF572,KIAA0196,NSMCE2,TRIB1,FAM84B,MYC,GSDMC,FAM49) | Yes |
| 6p25.3-p25.2 | Gain | 2 (5.1) | 3.7 Mb | 22 |
| Cytoband . | Type of CNA . | Number of patients (%) . | MCR size . | Genes in MCR . | Significant by GISTIC algorithm . |
|---|---|---|---|---|---|
| 22q11.22 | Loss | 4 (10.2) | 304 kb | 4 (ZNF280B, ZNF280A, PRAME, GGTLC2) | Yes |
| 3p14.2-p14.1 | Loss | 3 (7.6) | 4.1 Mb | 11 | |
| 3p14-p12.3 | Loss | 3 (7.6) | 8.5 Mb | 8 (EIF4E3,GPR27,PROK2,SHQ1,PPP4R2,PDZRN3,CNTN3,ROBO2) | |
| 4p15.2-p14 | Loss | 3 (7.6) | 10 Mb | 7 (SLC34A2,RBPJ,CCKAR,TBC1D19,STIM2,PCDH7,ARAP2) | |
| 7q34 | Loss | 3 (7.6) | 274 kb | 4 (UBN2,C7orf55,LUC7L2,KLRG2) | |
| 9p21.3 | Loss | 3 (7.6) | 2.6 Mb | 11 (including KLHL9, members of IFNA family, CDKN2A,CDKN2B) | Yes |
| 9p21.3-p21.1 | Loss | 3 (7.6) | 8.6 Mb | 15 | |
| 14q24.3-q31.3 | Loss | 3 (7.6) | 7.8 Mb | 7 (NRXN3,C14orf145,TSHR,GTF2A1,STON2,SEL1L,FLRT2) | |
| 3p12.3-q11.2 | Loss | 2 (5.1) | 13 Mb | 11 (VGLL3 and EPHA3) | Yes |
| 3q13.32-q13.33 | Loss | 2 (5.1) | 1.8 Mb | 18 (including CD80, GSK3B) | |
| 4q21.1-q21.23 | Loss | 2 (5.1) | 8.4 Mb | 34 | |
| 4q34 | Loss | 2 (5.1) | 3.1 Mb | 7 (GALNTL6,GALNT7,HMGB2,SAP30,SCRG1,HAND2,FBXO8) | |
| 6q23.3-q24 | Loss | 2 (5.1) | 2.9 Mb | 16 (including TNFAIP3) | |
| 9q34.3 | Loss | 2 (5.1) | 160 kb | 9 (TRAF2,FBXW5,C8G,LCN12,PTGDS,FLJ45224,C9orf142,CLIC3,ABCA2) | Yes |
| 9p22.1-p21.3 | Loss | 2 (5.1) | 1.4 Mb | 13 (including MLLT3 and members of IFNA family) | |
| 10q12.3-q13.3 | Loss | 2 (5.1) | 4.7 Mb | 15 | |
| 10q23.13 | Loss | 2 (5.1) | 215 kb | 7 (CH25H,LIPA,IFIT2,IFIT3,IFIT1L,IFIT1,IFIT5) | Yes |
| 10q23.32 | Loss | 2 (5.1) | 421 kb | 3 (BTAF1,CPEB3,MARCH5) | Yes |
| 15q13.3-q15.1 | Loss | 2 (5.1) | 9.1 Mb | 41 | |
| 15q15.1 | Loss | 2 (5.1) | 41 kb | MAPKB1, MGA | Yes |
| 8q24.21 | Gain | 4 (10.2) | 5.1 Mb | 9 (MTSS1,ZNF572,KIAA0196,NSMCE2,TRIB1,FAM84B,MYC,GSDMC,FAM49) | Yes |
| 6p25.3-p25.2 | Gain | 2 (5.1) | 3.7 Mb | 22 |
CNAs occurring in >2 samples. Genes in MCRs encompassing <10 genes are also listed.
Next, we focused on novel recurrent aberrations. The following regions were deleted in 3 samples (7.6%): 4p15.2-p14, with the MDR including RBPJ; 7q34, encompassing UBN2, LUV7L2, and KLRG2; 9p21, with the involvement of interferon-α family members and CDKN2A/B; and 3p14.1-p12.3, enclosing ROBO2, which was concomitantly mutated in FR-CLL_5. Among the lesions detected in 2 samples (5.1%), it is worth mentioning del3p12.3-q11.2, with VGLL3 and EPHA3 included in the MDR; del10q23.13, with the involvement of the Interferon-induced protein family members; del9q34.3, where TRAF2 and FBXW5 map, which was homozygous in 1 sample.
Among the recurrent gains, the 6p25.3-p25.2 region, including IRF4, was amplified in 2 cases (Table 3).
Furthermore, as mentioned above, the outliers were characterized by multiple aberrations of small size. These lesions generated several MDRs (supplemental Table 8): del14q32.13-q32.2, with the MDR including TCL6 and TCL1A/1B, was detected in other 2 samples besides the outlier; and del8p23.1, with the smallest deletion encompassing BLK, was found in 3 outliers.
Comparison between TP53 WT and TP53 disrupted cases
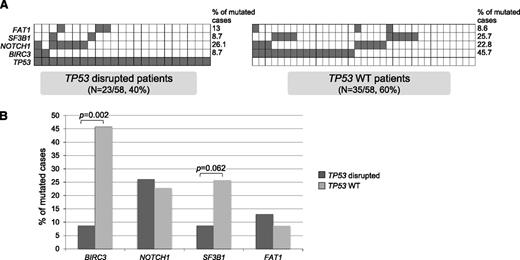
Last, we focused on the characteristics of the TP53 wild-type (WT) cases (N = 35, 60.3%), where the refractoriness determinants have remained unknown until recently. In this subgroup, BIRC3 was mutated/deleted in 45.7% of cases, SF3B1 was mutated in 25.7% of cases, NOTCH1 in 22.8% of cases, and FAT1 in 8.6% of patients. The comparison with TP53-disrupted cases showed that the TP53 WT group is significantly enriched in BIRC3 alterations (P = .002) and to a lesser extent in SF3B1 mutations (P = .062; Figure 5).
Comparison between TP53 WT and TP53 disrupted cases. (A) Mutual relationship of TP53 disruption with other gene alterations in FR-CLL. In the heat map, rows correspond to identical genes and columns represent individual patients, color-coded based on the gene status (white indicates WT; gray indicates mutations and/or deletion of TP53, mutations and/or deletion of BIRC3, mutations of SF3B1, and mutations of NOTCH1). (B) Percentage of TP53 disrupted and TP53 WT CLL primary cases harboring mutations in the indicated genes.
Comparison between TP53 WT and TP53 disrupted cases. (A) Mutual relationship of TP53 disruption with other gene alterations in FR-CLL. In the heat map, rows correspond to identical genes and columns represent individual patients, color-coded based on the gene status (white indicates WT; gray indicates mutations and/or deletion of TP53, mutations and/or deletion of BIRC3, mutations of SF3B1, and mutations of NOTCH1). (B) Percentage of TP53 disrupted and TP53 WT CLL primary cases harboring mutations in the indicated genes.
As per the CNA load, TP53 WT cases carried a lower number of CNAs (5.5 vs 9.5), although this difference did not reach statistical significance. Remarkably, the TP53-disrupted subset included 4 of 6 cases with an outlier number of CNAs.
Discussion
The study of FR is of clinical and biological relevance in CLL because it involves ∼10% of patients after first-line treatment and virtually all patients after multiple lines of therapy.5 Besides the well-established role of TP53 disruption, novel genes have been recently associated with refractoriness.8-12
To shed light into the genome of FR, we analyzed, by WES and SNP arrays, FR-CLL cases sampled prior to the fludarabine-based treatment to identify the lesions that may be responsible for and/or predictors of refractoriness, as opposed to those selected by chemotherapy.
This approach showed that FR-CLL cases are affected by 16 mutations per case on average, resembling the load of 8 to 20 mutations per case documented in other CLL series, although they were not homogeneous for clinical features.8,36-38 Furthermore, in keeping with the pattern of mutations recently described,36,37 we observed that, despite the usual prevalence of transitions over transversions, this difference was not particularly pronounced and was further reduced in mutated IGHV cases.
By WES and subsequent Sanger screening, we identified a new player: FAT1, mutated in roughly 10% of cases. We showed that FAT1 mutations significantly prevail in FR-CLL compared with CLL at diagnosis, thus suggesting that FAT1 might contribute to the development of an aggressive clinical phenotype, conceivably in combination with other alterations. From a clinical standpoint, 4 FAT1-mutated patients had an acquired chemo-refractoriness, and we could test 2 of them for the presence of the mutation at the chemosensitive stage (data not shown). Notably, the mutations were also clonally represented at this time point, indicating that this alteration is not acquired/selected after treatment.
Consistent with our results, FAT1 mutations were found in roughly 1% of cases also in other NGS studies of unselected CLL,37,38 reinforcing the notion that FAT1 lesions accumulate in FR-CLL and may be involved in the development of this high-risk phenotype. However, by measuring the overall survival from the date of treatment that resulted in chemorefractoriness, we could not demonstrate an additional adverse prognostic impact of FAT1 mutations (data not shown). Presumably, this is due to the a priori selection of patients with a poor outcome harboring a fludarabine refractory phenotype; as a proof of principle, no prognostic impact was highlighted for the other gene mutations (TP53, NOTCH1, BIRC3, and SF3B1) associated to a poor outcome (data not shown).
FAT1 is frequently mutated in glioblastoma, colorectal, head, and neck cancer (7-20%).39 Similar to these malignancies, FR-CLL–associated mutations mainly affected the cadherin and cytoplasmic domains. However, contrary to solid tumors, we did not find any CNAs encompassing this region. FAT1 is prevalently involved in the canonical Wnt/β-catenin pathway where it binds β-catenin, preventing its nuclear localization; when inactivated, FAT1 is unable to sequester β-catenin at the cell membrane and promotes Wnt signaling and tumor growth, thus supporting its role as tumor suppressor.39
The role of the Wnt pathway in CLL has been explored in several papers.40-43 Gene expression analysis showed that several members of the Wnt family, as well as the transcription factor LEF1, are overexpressed in CLL cells compared with peripheral blood B cells from healthy donors.40,41 On the other hand, it was reported that Wnt/β-catenin inhibitor genes were hypermethilated.42 Moreover, β-catenin, as well as LEF1/β-catenin inhibitors, prolonged CLL cell survival, indicating that the Wnt pathway could play a role in CLL pathogenesis.40-43 Indeed, other genes belonging to this pathway were mutated in our discovery panel (PTPRU, MED12, and INVS) in a mutual exclusive fashion.44-46 Notably, MED12 has been recently recognized as a driver in CLL by Landau et al,47 and in this context, a pivotal role in CLL pathogenesis has been ascribed to the Wnt pathway. Therefore, mutations might represent one additional mechanism that alters Wnt signaling in CLL, apart from the deregulation of its members.46 Moreover, FAT1 is involved in the planar cell polarity pathway, suggested to have a role in the pathogenesis of CLL,48 and cell-cell adhesion, which may be a secondary mechanism by which FAT1 disruption promotes tumor growth.49
Overall, our screening showed that FAT1 mutations occurred along with TP53, NOTCH1, BIRC3, and SF3B1 disruptions, confirming that in FR-CLL there is a high degree of overlap between mutations, at variance from the mutual exclusivity detected at diagnosis.50 Indeed, 84% of chemorefractory cases displayed ≥1 alteration of the abovementioned genes, with roughly 33% of cases harboring >1 alteration. In particular, we were able to detect 1 of these alterations in 74% of TP53 WT cases. This finding indicates that the disruption of these genes may have a role in FR and strongly suggests that they and their co-occurrence should be investigated prospectively in a larger TP53 WT cohort of CLL prior to fludarabine-based treatment to confirm their impact on response to therapy and prognosis.51 Indeed, the clinical significance of these novel mutations in the context of therapeutic trials needs to be further investigated, mostly to establish their relevance in patients’ management.11,12,52
From another perspective, only 16% of FR-CLL (9/58) samples were devoid of these recurrent abnormalities. In-depth analysis of this “genetically orphan” group revealed an enrichment of cases with trisomy 12 compared with the remaining samples (5/9 vs 13/49).
Last, FR-CLL cases were devoid of MYD88 mutations, presumably because our cohort was enriched in unmutated IGHV cases and consistent with its supposed association to a favorable prognosis.36,38
Mutational analysis was then integrated with CNA data. We showed that FR-CLL cases are affected by 4 CNAs per sample, with the majority of cases carrying ≥3 CNAs. By meta-analysis, we compared our series with the most comprehensive papers about CLL studied by SNP 6.0 arrays.27,34 Ouillette et al34 described 198 untreated and 57 relapsed patients: the latter cases were characterized by a higher burden of lesions, with 33% of samples carrying ≥3 lesions. In this setting, the identification of ≥2 CNAs defined a group of aggressive CLLs with a reduced overall survival. Edelmann et al27 studied a broad cohort of patients at first progression and described the frequency of known and novel lesions. Overall, in these cohorts, only a minority of cases (16-20%) carried ≥3 CNAs, and the average load was 1.8 to 2.3 per sample.27,34 Therefore, this scenario appears substantially different from the genetic complexity observed in our series.
The composite genomic picture of FR-CLL was underlined by the identification of 6 cases whose CNA profile appears compatible with the chromothripsis event: first detected in 1 CLL patient in 2011,53 to date only Edelmann et al27 described it in a broader subgroup of cases (19/353, 5%). Notably, in our cohort, a higher percentage of samples (15%) was presumed to have chromotripsis. Although these patients clustered in the TP53-disrupted subgroup, we could not document a strong association with TP53 alteration. Furthermore, the examination of the features of these patients revealed that they were older than the remaining cases (78 vs 69 years old, P = .031).
Our study identified a number of CNAs beyond those detected by FISH: some of them have been already reported in CLL but in a lower percentage of cases, namely gain 8q24, including MYC in all cases (10% vs 3.7-5%), del14q (7.6% vs 2%), and gain 6p25.3, encompassing IRF4 (5% vs 1%); in contrast, del22q11.22 and del6q were detected at slightly lower frequencies in FR-CLL cases compared with previously described CLL cohorts.27,32,33,54
In addition, novel CNAs were identified, which may have a functional impact on the regulation of the NOTCH pathway (RBPJ deletions),55 the TP53 and/or RB pathway (CDKN2A/B deletions), and the nuclear factor-κB pathway (TRAF2 deletion),56 recently reported to be involved in FR-CLL.10,57
In conclusion, this is the first study on a clinically homogeneous group of chemo-refractory CLL cases described from both a mutational and CNA perspective. This effort revealed FAT1 as a new participant to the FR phenomenon, thus paving the way for a thorough examination of the role of the Wnt pathway in the FR event. Moreover, we documented a high degree of complexity of FR-CLL, demonstrated by the high load of CNAs and the substantial proportion of cases whose genomic profile is compatible with chromothripsis. Overall, these data refine the portrait of the coding genome of FR-CLL and prompt an in-depth investigation of larger cohorts of TP53 WT and genetically orphan FR-CLL subgroups.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank V. Miljkovic and the Genomics Technologies Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University for hybridization of the Affymetrix SNP 6.0 arrays and Simona Tavolaro for fruitful discussions. Automated DNA sequencing of the validation phase was performed at Genewiz, Inc. (South Plainfield, NJ). The authors thank Fasteris SA (Plan-les-Ouates, Swizerland) for assistance with the whole-exome capture and sequencing procedure.
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Special Program Molecular Clinical Oncology, 5x1000; My First AIRC grant 13470; Progetto Giovani Ricercatori 2010 grant GR-2010-2317594, Ministero della Salute; Progetto Futuro in Ricerca 2012, Ministero dell’Istruzione, dell’Università e della Ricerca; Compagnia di San Paolo grant PMN_call_2012_0071; Fondazione Cariplo grant 2012-0689; Ministero dell’Università e Ricerca, Fondo per gli Investimenti della Ricerca di Base; Associazione Italiana per la Ricerca sulla leucemia, Compagnia di San Paolo, and NIH grant RO1-CA177319 (to R.D.-F.). M. Messina was partly supported by Associazione Cristina Bassi Onlus. G.F. is a fellow of the American Italian Cancer Foundation. R.R. acknowledges funding from the following sources: NIH grant U54 CA121852-05 and the Stewart Foundation.
Authorship
Contribution: M. Messina performed experiments, interpreted data, and wrote the manuscript; I.D.G., D.R., and S.C. provided well-characterized pathological samples, interpreted data, and revised the manuscript; H.K. and R.R. developed bioinformatics tools and performed bioinformatic analysis; S.R. and V.S. performed the screening of recurrently mutated genes and analysis of sequencing data; A.B.H. and G.F. contributed to the copy number experiment and analysis; M. Marinelli performed molecular analyses; F.R.M. provided clinical samples and data; A.P. performed the statistical analysis of mutations clinical impact; and A.G., G.G., R.D.-F., L.P., and R.F. designed the study and critically revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robin Foà, Hematology, Department of Cellular Biotechnologies and Hematology, Sapienza University, Via Benevento 6, 00161 Rome, Italy; e-mail: rfoa@bce.uniroma1.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal