Key Points
GATA1 mutations are common in neonates with Down syndrome but are often unsuspected and detectable only with sensitive methods.
Multilineage blood abnormalities in all Down syndrome neonates in the absence of GATA1 mutations suggests that trisomy 21 itself perturbs hemopoiesis.
Abstract
Transient abnormal myelopoiesis (TAM), a preleukemic disorder unique to neonates with Down syndrome (DS), may transform to childhood acute myeloid leukemia (ML-DS). Acquired GATA1 mutations are present in both TAM and ML-DS. Current definitions of TAM specify neither the percentage of blasts nor the role of GATA1 mutation analysis. To define TAM, we prospectively analyzed clinical findings, blood counts and smears, and GATA1 mutation status in 200 DS neonates. All DS neonates had multiple blood count and smear abnormalities. Surprisingly, 195 of 200 (97.5%) had circulating blasts. GATA1 mutations were detected by Sanger sequencing/denaturing high performance liquid chromatography (Ss/DHPLC) in 17 of 200 (8.5%), all with blasts >10%. Furthermore low-abundance GATA1 mutant clones were detected by targeted next-generation resequencing (NGS) in 18 of 88 (20.4%; sensitivity ∼0.3%) DS neonates without Ss/DHPLC-detectable GATA1 mutations. No clinical or hematologic features distinguished these 18 neonates. We suggest the term “silent TAM” for neonates with DS with GATA1 mutations detectable only by NGS. To identify all babies at risk of ML-DS, we suggest GATA1 mutation and blood count and smear analyses should be performed in DS neonates. Ss/DPHLC can be used for initial screening, but where GATA1 mutations are undetectable by Ss/DHPLC, NGS-based methods can identify neonates with small GATA1 mutant clones.
Introduction
Children with Down syndrome (DS) have a 150-fold increased risk of acute myeloid leukemia (ML-DS) during the first 5 years of life, compared with children without DS, despite a lower incidence of other cancers.1,2 ML-DS cells harbor acquired, N-terminal–truncating mutations in the key hematopoietic transcription factor gene GATA1.3-9 GATA1 mutations are also present in the neonatal preleukemic disorder transient abnormal myelopoiesis (TAM), which is unique to neonates with trisomy 21.3-5,7-9 TAM often precedes ML-DS and the same GATA1 mutations are usually present in both disorders,3 clonally linking the two conditions.
Retrospective clinical studies suggest that TAM affects ∼10% of neonates with DS10 and has a mortality of ∼20%.11-13 Furthermore, an estimated 20% to 30% of babies with TAM subsequently develop ML-DS.2,11,12,14 Thus, TAM is an important clinical problem. Nevertheless, a key difficulty is the lack of clear clinical, hematologic, and molecular diagnostic criteria for TAM.
The World Health Organization (WHO) defines TAM as increased peripheral blood blast cells in neonates with DS.15 Retrospective studies have used differing clinical and/or hematologic criteria to define TAM.11-14 In these studies, presentation varied from disseminated leukemic infiltration with hepatic fibrosis to largely asymptomatic disease where diagnosis was based on various nonspecific blood count abnormalities, such as circulating blasts, leukocytosis, and/or thrombocytopenia, which have all been reported in DS neonates without TAM.16 Importantly, no retrospective studies have systematically screened neonates for GATA1 mutations.11-14 Thus, current definitions of TAM specify neither the percentage of blasts considered abnormal in DS neonates nor the role of GATA1 mutation analysis in diagnosis. As a result, asymptomatic TAM may be missed in some neonates, while in others TAM may be overdiagnosed by relying on nonspecific clinical and hematologic features. Indeed, the only large systematic GATA1 mutation screen, performed by Sanger sequencing (Ss) of polymerase chain reaction (PCR) products from dried blood spots, found a prevalence of GATA1 mutations in DS neonates of only 3.8%,17 in contrast to the 5% to 10% prevalence of TAM diagnosed by clinical and hematologic criteria.10
The aims of this study were to more precisely define the population at risk of developing ML-DS and understand how to best define TAM. We prospectively, systematically determined GATA1 mutation status, blood counts, blood cell morphology, blast cell frequency, and clinical findings in 200 DS neonates recruited to the Oxford-Imperial Down Syndrome Cohort Study (OIDSCS). This provides a base to accurately study the natural evolution and impact of interventional treatment of this preleukemic disorder for the first time.
Patients, materials, and methods
Study population
Neonates with a clinical diagnosis of DS confirmed by karyotyping were prospectively enrolled in OIDSCS between October 2006 and March 2012 in 18 UK hospitals. Of 213 DS babies born in OIDSCS hospitals during this period, 94% (200) were recruited. Reasons for nonrecruitment were lack of available blood samples (n = 7) or parental consent (n = 6). OIDSCS recruits represent ∼5% of total DS births in England over this period (http://www.wolfson.qmul.ac.uk/ndscr/reports/NDSCRreport11.pdf). Parents gave written informed consent in accordance with the Declaration of Helsinki. The study was approved by the Thames Valley Research Ethics Committee (06MRE12-10; NIHR portfolio no. 6362).
Laboratory and clinical data
Complete blood counts (CBCs) and blood smears were processed in treating hospitals. Peripheral blood smears were assessed by 2 independent observers (I.R. and G.H.) blinded to GATA1 mutation status and recorded on OIDSCS proformas (supplemental Figure 1, available on the Blood Web site). Differential counts were expressed as the percentage (%) of leukocytes on a 200-cell manual differential count. Results on DS neonates were compared with neonatal laboratory normal ranges used at Imperial College Healthcare NHS Trust derived from 80 healthy term (37-42 weeks) and 43 preterm (31-36 weeks) neonates screened for suspected sepsis but found to have no clinical, microbiologic, or hematologic evidence of sepsis. Morphologic evaluation of red cells and leukocytes was performed using European Working Group in Childhood (EWOG-MDS) criteria.18 Giant platelets and megakaryocyte fragments were defined by their diameter (giant platelets, diameter >4 μM but <8 μM; megakaryocyte fragments, diameter >8 μM).19 Polycythemia was defined as hematocrit >0.6520 and thrombocytopenia as platelets <150 × 109/L21,22 as per current guidelines.
TAM.
Diagnosis of TAM (n = 17) was made clinically by local teams in 13 of 17 patients or by OIDSCS review of CBCs and blood smears (4 of 17 patients). Because the WHO classification for TAM15,23 mentions only “increased” peripheral blood blasts without defining normal ranges in neonates with or without DS, we used 2 approaches to define increased blast cells in TAM. First, we used the normal neonatal range for circulating blasts at Imperial College Healthcare NHS Trust (see above) for healthy term and preterm neonates without DS (range 0%-4%; median 0%). Second, to allow for possible effects of sepsis on blast counts, we performed manual blast counts on blood smears from 80 sick preterm neonates without DS referred for hematologic review because of suspected sepsis at Imperial College Healthcare NHS Trust (range 0%-8%; median 1%). We then prospectively defined TAM in OIDSCS as neonates with DS and peripheral blood blasts >10% and a GATA1 mutation detected by Ss/DHPLC analysis followed by NGS.
Mutational analysis of the GATA1 gene
Conventional Ss and DHPLC analysis.
GATA1 analysis was performed on all consented peripheral blood samples (188 of 200; 94%) by Ss direct high-pressure liquid chromatography (DHPLC) (WAVE; Transgenomic, Omaha, NE) as previously described.24
Targeted NGS.
Next-generation resequencing (NGS) of GATA1 exon 2 was performed on an independent aliquot of DNA from all cases with available DNA (n = 104). Samples with targeted NGS-identified mutations were confirmed by pyrosequencing in a third independent DNA aliquot (supplemental Methods). GATA1 exon 2 was amplified using Phusion High Fidelity DNA Polymerase (NEB, UK) with forward 5′-TTTGAGAAGCTTAAAGGAGGGAAGAGGAGCAG-3′ and reverse 5′-TTTGAGAAGCTTCCAGCCATTTCTGA-3′ primers. PCR conditions were: 98°C for 30 seconds, 35 cycles of 98°C for 10 seconds, 66.3°C for 30 seconds, and 72°C for 15 seconds. After the last cycle, additional steps were 72°C for 5 minutes and 4°C for 10 minutes. PCR products were purified (PCR purification kit; Qiagen, Hilden, Germany) and quality-checked (Agilent 2100 Bioanalyser; Agilent Technologies, Waldbronn, Germany). PCR products were digested overnight with HindIII followed by another round of purification. Purified product was ligated with T4 ligase overnight at 16°C, and 1 µg of ligated DNA was fragmented (Covaris S2; Covaris, Woburn, MA). After shearing, fragment distribution was measured (Agilent 2100 Bioanalyser). Libraries were constructed using the NEB Next DNA Sample Prep Master Mix Set 1 Kit (NEB) and ligated with 3 µL of Illumina Adapters. Ligated libraries were size-selected using Agencourt AMPure magnetic beads (Beckman Coulter, Brea, CA) and purified fragment distribution measured (Agilent 2100 Bioanalyser). Each library was PCR-enriched with 25 µM each of the following primers:
Multiplex PCR primer 1.0: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
Index primer: CAAGCAGAAGACGGCATACGAGAT[INDEX]CAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT.
Eight base-pair index tags were developed and validated at the Wellcome Trust Centre for Human Genetics (Oxford). Enrichment and adapter extension of each preparation was obtained using 5 µL of size-selected library in a 50-µL PCR reaction. Cycling conditions were as recommended by Illumina (Hayward, CA). After 10 cycles of amplification, product was purified using AMPureXp beads (Beckman Coulter). Final size distribution was determined using a Tapestation 1DK system (Agilent Technologies). Library concentration was determined by Agilent qPCR Library Quantification Kit on a MX3005P instrument (Agilent Technologies). Sequencing was performed on an Illumina HiSeq2000 as 50 or 100 base-paired end reads.
A custom Perl script was used to filter unprocessed reads (fastq files) by selecting reads with an Illumina Phred score for each base of >20. Filtered reads were mapped to GATA1 exon 2 reference sequence (NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) using Novoalign (http://www.novocraft.com). SAMtools,25 R (http://www.r-project.org), and custom Perl scripts were used to generate and plot base pair frequencies within exon 2 using the mapped sequence reads. The resulting plots were used to identify possible mutations. Reads covering the region of mutation were then analyzed to establish their sequence. To quantitate mutant clone size, a custom Perl script was used to analyze original unprocessed reads by filtering using less stringent parameters (based on an average Phred score of >20 across the whole read) and counting numbers of reads containing mutated versus wild type sequence. Sensitivity and specificity of NGS were tested using serial DNA dilutions of a male ML-DS cell line with a hemizygous GATA1 mutation and normal cord blood (supplemental Table 1). Mutation quantitation from NGS (3-5 × 105 mapping reads analyzed/sample) was compared with pyrosequencing. The limit of detection of mutant GATA1 sequence was ∼1% by pyrosequencing and ∼0.3% by NGS.
Blast cell immunophenotyping
Mononuclear cells (MNC) were isolated by Ficoll density gradient separation from peripheral blood samples from 7 DS neonates (2 with “silent TAM” and 5 with no GATA1 mutations detected by NGS), 8 anonymized samples from neonates without DS, and 7 samples with a confirmed diagnosis of TAM as defined in the OIDSC study (see supplemental Methods).
Statistical analysis
Statistical analysis was performed using Prism software. Populations were compared using 1-way ANOVA, 2-sided t tests, or Fisher exact test; P < .05 indicate statistically significant differences.
Results
Peripheral blood blasts and GATA1 mutation analysis by Ss/DHPLC in DS neonates
Previous studies defined TAM using either increased peripheral blood blasts, where blast % either was not defined or varied (as in the WHO classification15 ), or detection of GATA1 mutations by standard Ss/DHPLC.7,8 Neither approach has been tested in prospective studies and the relation between blast % and GATA1 mutation status is unknown. Therefore, to identify consistent criteria for diagnosis of TAM, we evaluated blasts on peripheral blood smears and correlated this with GATA1 analysis by Ss/DHPLC.
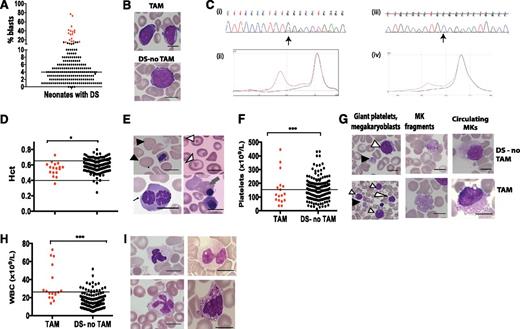
Surprisingly, 97.5% of DS neonates (195 of 200) had blasts on blood smears (range, 1%-77%) (Figure 1A-B). By contrast, only 8.5% (17 of 200) had GATA1 mutations by Ss/DHPLC (Figure 1C); all 17 had blasts >10% (Table 1; Figure 1A). Blast cell frequency in neonates without GATA1 mutations was lower (median 4%; P < .0001), but 6 neonates without GATA1 mutations had blasts >10% (11%-15%) (Figure 1A; supplemental Table 2). None of these 6 neonates had clinical features associated with TAM, and no exon 2 GATA1 mutations were detected even using targeted NGS (supplemental Table 3). While these cases may carry clones with large GATA1 deletions or low-abundance GATA1 exon 3 mutations, the low frequency of such mutations reported in other studies24 makes this unlikely. None of these 6 individuals has developed ML-DS (median follow-up, 35 months). Blast cell morphology was indistinguishable between those with or without GATA1 mutations (Figure 1B). Thus, although a blast threshold of >10% had 100% sensitivity for detection of GATA1 mutations by Ss/DHPLC, the specificity of this diagnostic criterion was only 74%. Because automated hematology analyzers also failed to identify blasts in 11 of 17 neonates with GATA1 mutations, we suggest that a practical and sensitive definition of TAM is the presence of blasts >10% on blood smears and a GATA1 mutation detected by Ss/DHPLC. Using this definition, we determined the clinical and hematologic features of DS neonates with TAM compared with those without TAM.
Hematologic abnormalities and GATA1 mutation analysis by Ss/DHPLC in neonates with DS. (A) Percentage of blasts on blood films from the first week of life in 200 neonates with DS, 17 with TAM (red circles) and 183 without TAM (black circles). (B) Photomicrographs of typical blast cells in a neonate with TAM (top) and in a DS neonate without TAM (bottom). (C) GATA1 mutation analysis in TAM by Ss and DHPLC. (Ci,ii) Mutation analysis of sample DST11. The mutation is detected by both Ss and DHPLC. (Ci) Sanger sequence trace. The arrow points the start of a double sequence trace indicative of an acquired GATA1 mutation. (Cii) DHPLC trace from the same sample (red line, mutant; black line, normal). (Ciii,iv) Mutation analysis of sample DST9. The mutation is detected by DHPLC but not by Ss. (Ciii) Sequence trace. (Civ) DHPLC trace from the same sample (red line, mutant; black line, normal). (D,F,H) Scatter graphs of hematocrit (D), platelet counts (F), and leukocytes (H) in 200 DS neonates in the first week of life, 17 with TAM (red circles) and 183 without TAM (black circles). The horizontal lines show the upper and/or lower limits of the normal neonatal laboratory range (see supplemental Methods). (E,G,I) Photomicrographs of erythrocyte (E), platelet (G), and leukocyte (I) morphologic abnormalities in neonates with DS. (E) Top left: macrocytes (black arrowheads); top right: target cells (white arrowheads); bottom left: dyserythropoietic erythroblasts (fine black arrow); bottom right: basophilic stippling (gray arrow). (G) Examples of giant platelets (GP) (black arrowhead) and megakaryoblasts (white arrowhead), megakaryocyte fragments (MK fragment), and circulating megakaryocytes (MKs) in blood films from DS neonates without TAM (top row) and with TAM (bottom row). (I) Top left: hypogranular neutrophil; top right: pseudo-Pelger neutrophil; bottom left: monocyte with stellate nucleus; bottom right, dysplastic basophil. Scale bars indicate 10 μm. WBC, white blood cell.
Hematologic abnormalities and GATA1 mutation analysis by Ss/DHPLC in neonates with DS. (A) Percentage of blasts on blood films from the first week of life in 200 neonates with DS, 17 with TAM (red circles) and 183 without TAM (black circles). (B) Photomicrographs of typical blast cells in a neonate with TAM (top) and in a DS neonate without TAM (bottom). (C) GATA1 mutation analysis in TAM by Ss and DHPLC. (Ci,ii) Mutation analysis of sample DST11. The mutation is detected by both Ss and DHPLC. (Ci) Sanger sequence trace. The arrow points the start of a double sequence trace indicative of an acquired GATA1 mutation. (Cii) DHPLC trace from the same sample (red line, mutant; black line, normal). (Ciii,iv) Mutation analysis of sample DST9. The mutation is detected by DHPLC but not by Ss. (Ciii) Sequence trace. (Civ) DHPLC trace from the same sample (red line, mutant; black line, normal). (D,F,H) Scatter graphs of hematocrit (D), platelet counts (F), and leukocytes (H) in 200 DS neonates in the first week of life, 17 with TAM (red circles) and 183 without TAM (black circles). The horizontal lines show the upper and/or lower limits of the normal neonatal laboratory range (see supplemental Methods). (E,G,I) Photomicrographs of erythrocyte (E), platelet (G), and leukocyte (I) morphologic abnormalities in neonates with DS. (E) Top left: macrocytes (black arrowheads); top right: target cells (white arrowheads); bottom left: dyserythropoietic erythroblasts (fine black arrow); bottom right: basophilic stippling (gray arrow). (G) Examples of giant platelets (GP) (black arrowhead) and megakaryoblasts (white arrowhead), megakaryocyte fragments (MK fragment), and circulating megakaryocytes (MKs) in blood films from DS neonates without TAM (top row) and with TAM (bottom row). (I) Top left: hypogranular neutrophil; top right: pseudo-Pelger neutrophil; bottom left: monocyte with stellate nucleus; bottom right, dysplastic basophil. Scale bars indicate 10 μm. WBC, white blood cell.
GATA1 mutation analysis and peripheral blood blasts in DS neonates with TAM
| Patient . | Mutation detection by Ss/DHPLC . | Nature and position of mutation detected by Ss . | Nature and position of mutation detected by NGS* . | Effect of mutation . | Bloodblast % . | Clone size by NGS (%) . | Clinical features . |
|---|---|---|---|---|---|---|---|
| DST1 | Ss and DHPLC | G>C point mutation, position 48649737 | Confirmed | Loss of splice donor site | 17 | 56.7 | Jaundice, hepatosplenomegaly, IUGR, A&W 48 mo |
| DST2 | Ss and DHPLC | G>A point mutation, position 48649737 | Confirmed | Loss of splice donor site | 77 | 45.28 | CHD, jaundice, hepatomegaly, A&W 30 mo |
| DST3 | Ss and DHPLC | Deletion 17bp GCGGCACTGGCCTACTA, position 48649688 | Confirmed | Frameshift | 37 | 49.63 | CHD, jaundice, rash, bacterial sepsis, A&W 33 mo |
| DST4 | Ss and DHPLC | dup 20bp 48649670 | 20 bp duplication ACAGCCACCGCTGCAGCTGC, position 48649670 | Frameshift | 35 | 5.96 | Jaundice, preterm (gestation at birth 34 wk), A&W 32 mo |
| DST5 | Ss and DHPLC | A>T mutation at position 48649739 | Confirmed | Loss of splice donor site | 40 | 27.9 | CHD, jaundice, hepatosplenomegaly, coagulopathy, transient spontaneous remission, sudden clinical deterioration with increasing blasts age 2 mo, diagnosed as TAM/ML-DS, Rx: AraC, CCR 24 mo |
| DST6 | DHPLC only | N/A | Clone 1: T>A mutation at position 48649738; clone 2: 1 bp deletion (G) at position 48649666 | Clone 1: loss of splice donor site; clone 2: frameshift | 32 | 10.49 | CHD, jaundice, A&W 57 mo |
| DST7 | Ss and DHPLC | G>A point mutation, position, 48649520 | Confirmed | Premature stop codon | 41 | 3.79 | Jaundice, hepatosplenomegaly, effusions, bacterial sepsis, IUGR, Rx: Ara C, A&W 30 mo |
| DST8 | DHPLC only | N/A | G>T point mutation, position 48649715 | Premature stop codon | 15 | 15.7 | IUGR, A&W 45 mo |
| DST9 | DHPLC only | N/A | 7 bp insertion GGTGAGC position 48649670 | Frameshift | 20 | 3.81 | Nothing of note, A&W 22 mo |
| DST10 | Ss and DHPLC | Insertion CAGTGCCTACT, position 48649704 | Confirmed | Frameshift | 42 | 15.48 | Jaundice, spontaneous remission by 6 wk, ML-DS at 22 mo, Rx: 4 cycles AML chemotherapy, CCR 28 mo |
| DST11 | Ss and DHPLC | Duplication 7bp CCCCTCT, position 48649625 | Confirmed | Frameshift | 38 | 17.06 | Hepatosplenomegaly, rash, CHD, A&W 41 mo |
| DST12† | Ss and DHPLC | 2 bp deletion AG, position 48649606 | Clone 1: 2 bp deletion (AG), position 48649606; clone 2: 8 bp duplication CACCGCTG position 48649675 | Clone 1: frameshift; clone 2, frameshift | 23 | Clone 1: 20.21; clone 2: 0.54 | CHD, jaundice, hepatomegaly, rash, liver failure, spontaneous remission by 2 wk, developed ML-DS age 4 mo, Rx: 4 cycles AML chemotherapy, CCR 34 mo |
| DST13 | Ss and DHPLC | Insertion GCAGCTGGAGCACAGCC, position 48649676 | Confirmed | Frameshift | 50 | 17.07 | Jaundice, hepatomegaly, liver failure, Rx AraC, A&W 36 mo |
| DST14 | Ss and DHPLC | C>T point mutation, position 48649565 | Confirmed | Premature stop codon | 73 | 82.31 | CHD, jaundice, hepatosplenomegaly, coagulopathy, Rx AraC, A&W 49 mo |
| DST15 | Ss and DHPLC | Deletion 14bp GTAACTCCATTGAG, position 48649737 | Confirmed | Loss of splice donor site | 17 | 33 | CHD, jaundice, hepatomegaly, coagulopathy, Rx AraC, A&W 43 mo |
| DST16 | Ss and DHPLC | 2 bp deletion (AG), position 48649600 | ND | Frameshift | 33 | ND | CHD, A&W 51 mo |
| DST17 | DHPLC only | N/A | 2 bp deletion (AG) at position 48649552 | Frameshift | 16 | 3 | Jaundice, A&W 37 mo |
| Patient . | Mutation detection by Ss/DHPLC . | Nature and position of mutation detected by Ss . | Nature and position of mutation detected by NGS* . | Effect of mutation . | Bloodblast % . | Clone size by NGS (%) . | Clinical features . |
|---|---|---|---|---|---|---|---|
| DST1 | Ss and DHPLC | G>C point mutation, position 48649737 | Confirmed | Loss of splice donor site | 17 | 56.7 | Jaundice, hepatosplenomegaly, IUGR, A&W 48 mo |
| DST2 | Ss and DHPLC | G>A point mutation, position 48649737 | Confirmed | Loss of splice donor site | 77 | 45.28 | CHD, jaundice, hepatomegaly, A&W 30 mo |
| DST3 | Ss and DHPLC | Deletion 17bp GCGGCACTGGCCTACTA, position 48649688 | Confirmed | Frameshift | 37 | 49.63 | CHD, jaundice, rash, bacterial sepsis, A&W 33 mo |
| DST4 | Ss and DHPLC | dup 20bp 48649670 | 20 bp duplication ACAGCCACCGCTGCAGCTGC, position 48649670 | Frameshift | 35 | 5.96 | Jaundice, preterm (gestation at birth 34 wk), A&W 32 mo |
| DST5 | Ss and DHPLC | A>T mutation at position 48649739 | Confirmed | Loss of splice donor site | 40 | 27.9 | CHD, jaundice, hepatosplenomegaly, coagulopathy, transient spontaneous remission, sudden clinical deterioration with increasing blasts age 2 mo, diagnosed as TAM/ML-DS, Rx: AraC, CCR 24 mo |
| DST6 | DHPLC only | N/A | Clone 1: T>A mutation at position 48649738; clone 2: 1 bp deletion (G) at position 48649666 | Clone 1: loss of splice donor site; clone 2: frameshift | 32 | 10.49 | CHD, jaundice, A&W 57 mo |
| DST7 | Ss and DHPLC | G>A point mutation, position, 48649520 | Confirmed | Premature stop codon | 41 | 3.79 | Jaundice, hepatosplenomegaly, effusions, bacterial sepsis, IUGR, Rx: Ara C, A&W 30 mo |
| DST8 | DHPLC only | N/A | G>T point mutation, position 48649715 | Premature stop codon | 15 | 15.7 | IUGR, A&W 45 mo |
| DST9 | DHPLC only | N/A | 7 bp insertion GGTGAGC position 48649670 | Frameshift | 20 | 3.81 | Nothing of note, A&W 22 mo |
| DST10 | Ss and DHPLC | Insertion CAGTGCCTACT, position 48649704 | Confirmed | Frameshift | 42 | 15.48 | Jaundice, spontaneous remission by 6 wk, ML-DS at 22 mo, Rx: 4 cycles AML chemotherapy, CCR 28 mo |
| DST11 | Ss and DHPLC | Duplication 7bp CCCCTCT, position 48649625 | Confirmed | Frameshift | 38 | 17.06 | Hepatosplenomegaly, rash, CHD, A&W 41 mo |
| DST12† | Ss and DHPLC | 2 bp deletion AG, position 48649606 | Clone 1: 2 bp deletion (AG), position 48649606; clone 2: 8 bp duplication CACCGCTG position 48649675 | Clone 1: frameshift; clone 2, frameshift | 23 | Clone 1: 20.21; clone 2: 0.54 | CHD, jaundice, hepatomegaly, rash, liver failure, spontaneous remission by 2 wk, developed ML-DS age 4 mo, Rx: 4 cycles AML chemotherapy, CCR 34 mo |
| DST13 | Ss and DHPLC | Insertion GCAGCTGGAGCACAGCC, position 48649676 | Confirmed | Frameshift | 50 | 17.07 | Jaundice, hepatomegaly, liver failure, Rx AraC, A&W 36 mo |
| DST14 | Ss and DHPLC | C>T point mutation, position 48649565 | Confirmed | Premature stop codon | 73 | 82.31 | CHD, jaundice, hepatosplenomegaly, coagulopathy, Rx AraC, A&W 49 mo |
| DST15 | Ss and DHPLC | Deletion 14bp GTAACTCCATTGAG, position 48649737 | Confirmed | Loss of splice donor site | 17 | 33 | CHD, jaundice, hepatomegaly, coagulopathy, Rx AraC, A&W 43 mo |
| DST16 | Ss and DHPLC | 2 bp deletion (AG), position 48649600 | ND | Frameshift | 33 | ND | CHD, A&W 51 mo |
| DST17 | DHPLC only | N/A | 2 bp deletion (AG) at position 48649552 | Frameshift | 16 | 3 | Jaundice, A&W 37 mo |
A&W, alive and well; AraC, cytosine arabinoside; CCR, complete clinical remission; CHD, congenital heart disease; N/A, not applicable; ND, not done; Rx, treatment.
Coordinates refer to human genome, build GRCh37 (hg19).
DST12: 4 copies of RUNX1 in 6% of BM cells at diagnosis of ML-DS by FISH (no other additional cytogenetic abnormalities were detected at diagnosis of TAM or ML-DS in the other cases).
Clinical characteristics
A total of 13 of 17 (76.5%) neonates with TAM (as defined above) presented with clinical or hematologic signs that led to a clinician diagnosis of TAM, which was subsequently confirmed by OIDSCS criteria (Table 2). The remaining 4 of 17 cases were unsuspected by clinical teams and diagnosed through OIDSCS blood smear review with increased blasts (16%-41%) and GATA1 mutation analysis. Hepatosplenomegaly, jaundice, and rash were more common in TAM but were not specific to TAM (Table 2). Thus, TAM cannot be recognized by clinical signs alone. The spectrum of congenital abnormalities was similar to previous reports.26
Clinical and hematologic data of neonates recruited to the OIDSC Study
| . | Number of neonates with DS . | |||
|---|---|---|---|---|
| TAM (n = 17) . | DS without TAM (n = 183) . | P . | Normal range* . | |
| Clinical characteristics | ||||
| Gender | 9:8 | 96:87 | 1.0000 | — |
| M:F | (1·1:1) | (1.1:1) | ||
| Gestation at birth, wk (range) | 37·0 (34·4-39·6) | 38·0 (30.9-42.6) | .1717 | — |
| Preterm (<37 wk) | 5 (29.4%) | 39 (21.3%) | .5408 | — |
| Small for gestational age† | 2 (11.8%) | 10 (5.5%) | .2749 | — |
| Hepato(spleno)megaly | 7 (41.2%) | 6 (3.3%) | <.0001 | — |
| Jaundice | 13 (76.4%) | 78 (42.9%) | .0098 | — |
| Rash | 3 (17.6%) | 1 (0.6%) | .0020 | — |
| Pleural/pericardial effusion/ ascites | 1 (5.9%) | 3 (1.6%) | .3010 | — |
| Congenital heart disease | 8 (47.1%) | 90 (49.2%) | 1.0000 | — |
| Other congenital anomalies‡ | 2 (11.8%) | 20 (10.9%) | 1.0000 | — |
| Hematologic characteristics | ||||
| Median Hb g/dL (range) | 18.4 (11.7-22.2) | 20·5 (7.7-28.0) | .0011 | 16.6 (12.7-20.3) |
| Median hematocrit (range) | 0·562 (0.357-0.647) | 0.599 (0.243-0.822) | .0122 | 0.503 (0.408-0·610) |
| Anemia | 1 (5.9%) | 3 (1.6%) | .3010 | Hct >0.400 |
| Median MCV (fL) | 108 (93.3-133) | 108 (88.7-123.8) | .2104 | 103·5 (89.6-117.7) |
| Nucleated red cells/100 WBC | 8 (1-122) | 5 (0-186) | .0187 | 1 (0-29) |
| Median platelets ×109/L§ | 117 (36-446) | 148.5 (9-432) | .9014 | 253 (150-388) |
| MPV (fL)§ | 9·75 (7.4-13.5) | 10.6 (7.3-12.8) | .6448 | 10.4 (8.1-12.5) |
| Thrombocytopenia | 10 (54.6%) | 91 (50.6%) | .6152 | Platelets <150 × 109/L |
| WBC ×109/L | 25.8 (19.7-73.2) | 14.1 (4.6-51.72) | <.0001 | 11.6 (4.9-26.7) |
| Blasts (%) | 35 (15-77) | 4 (0-15) | <.0001 | 0 (0-4) |
| Neutrophils ×109/L | 13·80 (7.0-31.0) | 9.57 (0.12-38.1) | .0372 | 6.2 (1.36-20.1) |
| Metamyelocytes ×109/L | 0.79 (0-5.66) | 0.35 (0-5.18) | .2986 | N/A |
| Myelocytes ×109/L | 0.25 (0-3.96) | 0 (0-1.94) | <.0001 | N/A |
| Monocytes ×109/L | 1.41 (0.3-6.7) | 1.11 (0.08-6.96) | .3082 | 0.78 (0-2.67) |
| Basophils ×109/L | 0·37 (0-2.0) | 0.17 (0-2·3) | <.0001 | 0 (0-0.15) |
| Eosinophils ×109/L | 0·24 (0-1.12) | 0.22 (0-1·18) | .9543 | 0·20 (0-1.87) |
| Lymphocytes ×109/L | 6.0 (0.23-19.2) | 2.94 (0.4-8.5) | <.0001 | 3.8 (0.86-9.7) |
| . | Number of neonates with DS . | |||
|---|---|---|---|---|
| TAM (n = 17) . | DS without TAM (n = 183) . | P . | Normal range* . | |
| Clinical characteristics | ||||
| Gender | 9:8 | 96:87 | 1.0000 | — |
| M:F | (1·1:1) | (1.1:1) | ||
| Gestation at birth, wk (range) | 37·0 (34·4-39·6) | 38·0 (30.9-42.6) | .1717 | — |
| Preterm (<37 wk) | 5 (29.4%) | 39 (21.3%) | .5408 | — |
| Small for gestational age† | 2 (11.8%) | 10 (5.5%) | .2749 | — |
| Hepato(spleno)megaly | 7 (41.2%) | 6 (3.3%) | <.0001 | — |
| Jaundice | 13 (76.4%) | 78 (42.9%) | .0098 | — |
| Rash | 3 (17.6%) | 1 (0.6%) | .0020 | — |
| Pleural/pericardial effusion/ ascites | 1 (5.9%) | 3 (1.6%) | .3010 | — |
| Congenital heart disease | 8 (47.1%) | 90 (49.2%) | 1.0000 | — |
| Other congenital anomalies‡ | 2 (11.8%) | 20 (10.9%) | 1.0000 | — |
| Hematologic characteristics | ||||
| Median Hb g/dL (range) | 18.4 (11.7-22.2) | 20·5 (7.7-28.0) | .0011 | 16.6 (12.7-20.3) |
| Median hematocrit (range) | 0·562 (0.357-0.647) | 0.599 (0.243-0.822) | .0122 | 0.503 (0.408-0·610) |
| Anemia | 1 (5.9%) | 3 (1.6%) | .3010 | Hct >0.400 |
| Median MCV (fL) | 108 (93.3-133) | 108 (88.7-123.8) | .2104 | 103·5 (89.6-117.7) |
| Nucleated red cells/100 WBC | 8 (1-122) | 5 (0-186) | .0187 | 1 (0-29) |
| Median platelets ×109/L§ | 117 (36-446) | 148.5 (9-432) | .9014 | 253 (150-388) |
| MPV (fL)§ | 9·75 (7.4-13.5) | 10.6 (7.3-12.8) | .6448 | 10.4 (8.1-12.5) |
| Thrombocytopenia | 10 (54.6%) | 91 (50.6%) | .6152 | Platelets <150 × 109/L |
| WBC ×109/L | 25.8 (19.7-73.2) | 14.1 (4.6-51.72) | <.0001 | 11.6 (4.9-26.7) |
| Blasts (%) | 35 (15-77) | 4 (0-15) | <.0001 | 0 (0-4) |
| Neutrophils ×109/L | 13·80 (7.0-31.0) | 9.57 (0.12-38.1) | .0372 | 6.2 (1.36-20.1) |
| Metamyelocytes ×109/L | 0.79 (0-5.66) | 0.35 (0-5.18) | .2986 | N/A |
| Myelocytes ×109/L | 0.25 (0-3.96) | 0 (0-1.94) | <.0001 | N/A |
| Monocytes ×109/L | 1.41 (0.3-6.7) | 1.11 (0.08-6.96) | .3082 | 0.78 (0-2.67) |
| Basophils ×109/L | 0·37 (0-2.0) | 0.17 (0-2·3) | <.0001 | 0 (0-0.15) |
| Eosinophils ×109/L | 0·24 (0-1.12) | 0.22 (0-1·18) | .9543 | 0·20 (0-1.87) |
| Lymphocytes ×109/L | 6.0 (0.23-19.2) | 2.94 (0.4-8.5) | <.0001 | 3.8 (0.86-9.7) |
N/A, not applicable; WBC, white blood cells.
Derived from anonymized data from 80 normal healthy term (37-42 wk) and 43 preterm (31-36 wk) neonates (see supplemental Methods).
Defined as birthweight <10th percentile for gestational age.
Duodenal atresia, esophageal atresia, tracheo-esophageal fistula, imperforate anus, Hirschsprung's, renal anomalies
Platelet count not available in 3 neonates due to platelet clumping and automated MPV measurable for 6 of 17 neonates with TAM, 59 of 183 DS neonates without TAM, and 122 neonates without DS.
Hematologic data
All DS neonates had abnormalities of >1 hematopoietic lineage and 95.0% had abnormalities in >3 lineages (Table 2).
Erythrocyte abnormalities.
Overall, DS neonates had higher hematocrit (Hct), hemoglobin (Hb), and circulating erythroblasts than normal. Although median Hct (P = .0122) and Hb (P = .0011) were slightly lower in TAM than in DS neonates without TAM, there was considerable overlap, and only 1 neonate with TAM was anemic (Figure 1D; Table 2; supplemental Table 2). Dyserythropoiesis was common in DS neonates (increased mean cell volume [MCV], macrocytes, dyserythropoietic erythroblasts, target cells, and basophilic stippling) (Figure 1E; Table 2) and was not significantly greater in TAM (supplemental Table 4). There was no correlation between Hct, MCV, or erythroblasts and heart disease, intrauterine growth restriction (IUGR), or maternal complications (supplemental Table 5). This suggests that abnormal erythropoiesis in DS neonates is a consequence of trisomy 21 rather than GATA1 mutation or secondary causes.
Platelet abnormalities.
Median platelet counts were lower than normal in DS and not reduced further in TAM (P = .9014) (Table 2; Figure 1F). The frequency of thrombocytopenia (platelets <150 × 109/L) was similar in neonates with and without TAM (P = .6162) and in DS neonates with or without potential secondary causes (eg, sepsis)27 (supplemental Table 2), although median platelets were slightly lower in DS neonates with IUGR (supplemental Table 5). Median mean platelet volume (MPV) was similar in neonates with and without TAM (Table 2). Platelet morphology was abnormal in 193 of 200 (96.7%) DS neonates (giant platelets, circulating megakaryocytes, and/or megakaryocyte fragments) (Figure 1G), consistent with trisomy 21–mediated effects on platelet production. Megakaryocyte fragments were strongly associated with TAM (16 of 17 [94.1%] vs 84 of 183 [45.9%] DS neonates without TAM; P = .0002; supplemental Table 4). Although not specific, absence of megakaryocyte fragments had a negative predictive value for TAM of 99.0%.
Leukocyte abnormalities.
Although leukocyte counts were higher in TAM than DS neonates without TAM (25.8 vs 14.1 × 109/L; P < .0001), leukocyte counts were in the normal range in 9 of 17 (52.9%) neonates with TAM (Figure 1H). Neutrophils, myelocytes, basophils, monocytes, and blasts were increased in neonates with and without TAM (Table 3). Leukocytosis was not due to sepsis as only 10 of 200 had culture-positive sepsis (supplemental Table 5). Dysplastic neutrophils and monocytes (EWOG-MDS criteria18 ) were common in DS neonates with and without TAM, including hypogranular neutrophils, pseudo-Pelger forms, stellate monocytes, and dysplastic basophils (Figure 1I; supplemental Table 4), implicating trisomy 21, rather than mutant GATA1, in these changes.
Clinical and hematologic features of DS neonates with silent GATA1 mutations (silent TAM) compared with DS neonates without GATA1 mutations by targeted NGS
| Clinical and hematologic characteristics . | Number (%) of neonates with DS . | ||||
|---|---|---|---|---|---|
| No GATA1 mutations (by targeted NGS) (n = 70) . | Silent TAM (n = 18) . | Silent TAM vs no GATA1 mutations (P value) . | TAM (n = 17) . | Silent TAM vs TAM (P value) . | |
| Gender (male:female) | 30:40 | 10:8 | .4282 | 9:8 | 1.00 |
| Median gestation at birth (wk) | 38.1 | 38.3 | .7122 | 37.0 | .0431 |
| Hepatosplenomegaly | 4 (5.7%) | 0 | .5775 | 7 (341.2%) | .0072 |
| Jaundice | 33 (47.1%) | 11 (61.1%) | .6024 | 13 (76.4%) | .7283 |
| Rash | 1 (1.4%) | 0 | 1.00 | 3 (17.6%) | .2273 |
| Pleural/pericardial effusion and/or ascites | 1 (1.4%) | 0 | 1.00 | 1 (5.9%) | .4848 |
| Congenital heart disease | 37 (52.9%) | 9 (50%) | 1.000 | 8 (47.1%) | .4935 |
| Death | 1 (1.4%) | 1 (5.6%) | .3691 | 0 (0%) | 1.000 |
| ML-DS* | 0 | 1 (5.6%) | .2069 | 3 (17.6%) | .3377 |
| Median follow up (mo) | 34 | 33 | .3587 | 40 | .1043 |
| Hct | 0.592 | 0.617 | .2414 | 0.562 | .0024 |
| Median (range) | (0.243-0.80) | (0.509-0.736) | (0.357-.0.65) | ||
| MCV (fL) | 108.0 | 108.4 | .1715 | 108 | .3777 |
| Median (range) | 88.7-122.1) | (94.4-122.2) | (93.3-133) | ||
| Platelets ×109/L | 166 | 127 | .1840 | 117 | .5274 |
| Median (range) | (26-432) | (50-253) | (36-1208) | ||
| WBC ×109/L | 14.8 | 13.5 | .1074 | 25.8 | <.0001 |
| Median (range) | (4.7-44.2) | (5.5-29.1) | (19.7-73.2) | ||
| Blasts (%) | 4 | 4.5 | .9842 | 35 | <.0001 |
| Median (range) | (0-15) | (1-10) | 15-77) | ||
| Neutrophils ×109/L | 10.58 | 8.5 | .0427 | 13.80 | .0285 |
| Median (range) | (1.5-38.1) | (2.1-23.3) | (7.00-31.00) | ||
| Monocytes ×109/L | 1.19 | 1.10 | .1650 | 1.41 | .1398 |
| Median (range) | (0.38-6.0) | (0.23-2.28) | (0.67-5.28) | ||
| Basophils ×109/L | 0.2 | 0.19 | .6835 | 0.37 | .0197 |
| Median (range) | (0-1.07) | (0-0.6) | (0-1.27) | ||
| Clinical and hematologic characteristics . | Number (%) of neonates with DS . | ||||
|---|---|---|---|---|---|
| No GATA1 mutations (by targeted NGS) (n = 70) . | Silent TAM (n = 18) . | Silent TAM vs no GATA1 mutations (P value) . | TAM (n = 17) . | Silent TAM vs TAM (P value) . | |
| Gender (male:female) | 30:40 | 10:8 | .4282 | 9:8 | 1.00 |
| Median gestation at birth (wk) | 38.1 | 38.3 | .7122 | 37.0 | .0431 |
| Hepatosplenomegaly | 4 (5.7%) | 0 | .5775 | 7 (341.2%) | .0072 |
| Jaundice | 33 (47.1%) | 11 (61.1%) | .6024 | 13 (76.4%) | .7283 |
| Rash | 1 (1.4%) | 0 | 1.00 | 3 (17.6%) | .2273 |
| Pleural/pericardial effusion and/or ascites | 1 (1.4%) | 0 | 1.00 | 1 (5.9%) | .4848 |
| Congenital heart disease | 37 (52.9%) | 9 (50%) | 1.000 | 8 (47.1%) | .4935 |
| Death | 1 (1.4%) | 1 (5.6%) | .3691 | 0 (0%) | 1.000 |
| ML-DS* | 0 | 1 (5.6%) | .2069 | 3 (17.6%) | .3377 |
| Median follow up (mo) | 34 | 33 | .3587 | 40 | .1043 |
| Hct | 0.592 | 0.617 | .2414 | 0.562 | .0024 |
| Median (range) | (0.243-0.80) | (0.509-0.736) | (0.357-.0.65) | ||
| MCV (fL) | 108.0 | 108.4 | .1715 | 108 | .3777 |
| Median (range) | 88.7-122.1) | (94.4-122.2) | (93.3-133) | ||
| Platelets ×109/L | 166 | 127 | .1840 | 117 | .5274 |
| Median (range) | (26-432) | (50-253) | (36-1208) | ||
| WBC ×109/L | 14.8 | 13.5 | .1074 | 25.8 | <.0001 |
| Median (range) | (4.7-44.2) | (5.5-29.1) | (19.7-73.2) | ||
| Blasts (%) | 4 | 4.5 | .9842 | 35 | <.0001 |
| Median (range) | (0-15) | (1-10) | 15-77) | ||
| Neutrophils ×109/L | 10.58 | 8.5 | .0427 | 13.80 | .0285 |
| Median (range) | (1.5-38.1) | (2.1-23.3) | (7.00-31.00) | ||
| Monocytes ×109/L | 1.19 | 1.10 | .1650 | 1.41 | .1398 |
| Median (range) | (0.38-6.0) | (0.23-2.28) | (0.67-5.28) | ||
| Basophils ×109/L | 0.2 | 0.19 | .6835 | 0.37 | .0197 |
| Median (range) | (0-1.07) | (0-0.6) | (0-1.27) | ||
ML-DS was diagnosed in 3 neonates with TAM (DST5, age 2 mo; DST10, age 22 mo; DST12, age 4 mo) and in 1 neonate with silent TAM (DS 108). This neonate had 5% blasts and mild thrombocytopenia at birth (79 × 109/L) but had a normal CBC and smear at age 9 mo. Isolated thrombocytopenia (23 × 109/L) was noted at age 11 mo shortly after a viral illness and was attributed to immune thrombocytopenia (no blasts were seen on the blood smear). Thrombocytopenia persisted, and by age 15 mo occasional blasts were seen on the smear. Progressive pancytopenia led to the diagnosis of ML-DS at age 18 mo (BM blasts 35%; no additional cytogenetic abnormalities). All patients in whom ML-DS developed remain in complete clinical remission after treatment with modified AML chemotherapy.
GATA1 mutation analysis by NGS
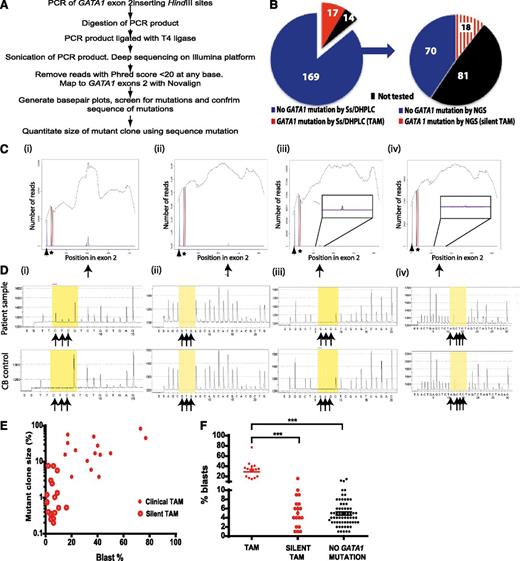
The mutant/wild-type fraction required to identify GATA1 mutant clones is ∼30% by Ss and ∼5% by DHPLC (Table 1; K.A. and P.V., unpublished data). To determine whether smaller-sized mutant GATA1 clones explained the high prevalence of blasts in DS neonates without TAM, we performed targeted NGS of GATA1 exon 2 (97% of GATA1 mutations are in exon 2).24 The flow diagram in Figure 2A outlines the methodology. The bioinformatic pipeline for mapping reads (3-5 × 105 mapped reads analyzed/sample), identifying mutations and quantitating mutant/wild-type fraction is described in “Patients, materials, and methods.” To confirm mutation identification/quantitation by NGS, independent DNA aliquots were analyzed by pyrosequencing. Detection limits of GATA1 mutation were ∼1% to 2% by pyrosequencing and ∼0.3% by NGS (supplemental Table 1). GATA1 mutations were not detected in normal cord blood controls (n = 2) included in each run (data not shown).
GATA1 mutation analysis in DS neonates with TAM and silent TAM. (A) Flow diagram of preparation and analysis of samples for deep sequencing. (B) Pie charts of GATA1 mutation analysis of the 200 babies in the cohort by standard Ss/DHPLC (left) and NGS (right). (C) Examples of base-pair plots from NGR analysis of patient samples (mutation indicated by arrows) with (D) corresponding pyrosequencing traces below (mutant peaks indicated by arrows). On the x-axis is the position along the GATA1 exon 2 amplicon (432 base pairs). On the y-axis is the read depth at different positions along the amplicon. Therefore, the black line trace shows the number of reads mapping to GATA1 sequence at different positions along the amplicon. At the position of the black arrowhead, a mutation was introduced into the PCR primer (mapping outside GATA1 exon 2) so that all PCR products would have a unique tag. This introduced mutation is detected by the blue line. All PCR products have this introduced mutation as the height of the blue line is to the level of black trace (total number of mapping reads). Sequence analysis also shows there are 2 common single-nucleotide polymorphism at positions rs62600348 T>G and rs66717003 T>G (indicated by the star) in the amplicon that map to position 48649449 and 48649456 within GATA1 exon 2. Nucleotide 0 is the first nucleotide of GATA1 exon 1 including exons and introns. NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) was used. The location of GATA1 mutation is indicated by the black arrow. (Ci,Di) Patient sample DST11 with a 7 bp duplication at position 48649625 previously detected by Ss/DHPLC. (Cii,Dii) Patient sample DST9 with an insertion of 7 bp at position 48649670 previously detected by DHPLC only. (Ciii,Diii) Patient sample DS158 with a 2 bp deletion at position 48649552 detected by NGS only and confirmed by pyrosequencing. (Civ,Div) Patient sample DS79 with a point mutation at position 48649565 detected by NGS only but not detectable by pyrosequencing. (E) Relationship between GATA1 mutant clone size (y-axis) as determined by NGR and % blasts detected by morphology. (F) Distribution of % blasts in TAM (n = 17) (filled red circles, left), silent TAM (n = 88) (open red cell circles, middle) and in samples without a GATA1 mutation detected by NGR (n = 70) (filled black circles, right).
GATA1 mutation analysis in DS neonates with TAM and silent TAM. (A) Flow diagram of preparation and analysis of samples for deep sequencing. (B) Pie charts of GATA1 mutation analysis of the 200 babies in the cohort by standard Ss/DHPLC (left) and NGS (right). (C) Examples of base-pair plots from NGR analysis of patient samples (mutation indicated by arrows) with (D) corresponding pyrosequencing traces below (mutant peaks indicated by arrows). On the x-axis is the position along the GATA1 exon 2 amplicon (432 base pairs). On the y-axis is the read depth at different positions along the amplicon. Therefore, the black line trace shows the number of reads mapping to GATA1 sequence at different positions along the amplicon. At the position of the black arrowhead, a mutation was introduced into the PCR primer (mapping outside GATA1 exon 2) so that all PCR products would have a unique tag. This introduced mutation is detected by the blue line. All PCR products have this introduced mutation as the height of the blue line is to the level of black trace (total number of mapping reads). Sequence analysis also shows there are 2 common single-nucleotide polymorphism at positions rs62600348 T>G and rs66717003 T>G (indicated by the star) in the amplicon that map to position 48649449 and 48649456 within GATA1 exon 2. Nucleotide 0 is the first nucleotide of GATA1 exon 1 including exons and introns. NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) was used. The location of GATA1 mutation is indicated by the black arrow. (Ci,Di) Patient sample DST11 with a 7 bp duplication at position 48649625 previously detected by Ss/DHPLC. (Cii,Dii) Patient sample DST9 with an insertion of 7 bp at position 48649670 previously detected by DHPLC only. (Ciii,Diii) Patient sample DS158 with a 2 bp deletion at position 48649552 detected by NGS only and confirmed by pyrosequencing. (Civ,Div) Patient sample DS79 with a point mutation at position 48649565 detected by NGS only but not detectable by pyrosequencing. (E) Relationship between GATA1 mutant clone size (y-axis) as determined by NGR and % blasts detected by morphology. (F) Distribution of % blasts in TAM (n = 17) (filled red circles, left), silent TAM (n = 88) (open red cell circles, middle) and in samples without a GATA1 mutation detected by NGR (n = 70) (filled black circles, right).
All samples with sufficient DNA were studied by targeted NGS. In DNA from 104 DS neonates (16/17 TAM; 88/169 non-TAM), mutations were detected in 34 cases: 16/16 with TAM and 18/88 (20.4%) neonates without TAM where direct Ss/DHPLC had not detected GATA1 mutations (Figure 2B-D; supplemental Table 3). Pyrosequencing confirmed GATA1 mutations in 17/34 (data not shown). In the remaining 17 of 34, mean estimated mutant clone size was 1.5% (range, 0.245%-5.96%) consistent with GATA1 mutant fractions below the sensitivity of pyrosequencing detection. The size of the GATA1 mutant clone correlated with blast % (P < .0001; Figure 1E) from the same sample.
Clinical and hematologic features of silent TAM.
Clinical and hematologic features of the 18 of 88 neonates with GATA1 mutant clones identified only by NGS were indistinguishable from the 70 DS neonates without detectable GATA1 mutations by NGS (Table 3). We therefore suggest the term “silent TAM” where GATA1 mutations are detectable by NGS but not by Ss/DHPLC. As expected, mutant clones were smaller and blast frequency lower in silent TAM compared with TAM (Figure 1E). Since blasts % in silent TAM (median 5%) was similar to DS neonates negative for NGS-detected GATA1 mutations (median 4%) (Figure 2F), morphology-based enumeration of blasts cannot distinguish silent TAM from babies without a GATA1 mutation. Preliminary data suggest that immunophenotyping of circulating blasts from DS neonates using standard diagnostic panels11,28 also fails to identify silent TAM (supplemental Table 6).
ML-DS in DS neonates with TAM or silent TAM.
After a median follow-up of >33 months, ML-DS was diagnosed in 4 cases: 3 out of 17 with TAM (DST5, DST10, DST12) and 1 out of 18 with silent TAM (DS108). None of the 70 neonates without NGS-detected GATA1 mutations has developed ML-DS (Table 3). In 3 of 4 ML-DS cases, a single mutant GATA1 mutation was detected by NGS at birth and the same DNA substitution was present at diagnosis of ML-DS; in 1 of 4 (DST12) cases, both mutations detected at birth were also found at diagnosis of ML-DS (Table 1), confirming the clonal relationship between TAM (silent or overt) and ML-DS. Only 1 further case of ML-DS was diagnosed from the 18 study centers during the study period. It is not known whether or not this child had clinically or hematologically silent TAM because they were not recruited to OIDSCS (the family moved abroad) and neither GATA1 mutation analysis nor blood smear review was performed at birth.
Discussion
OIDSCS is the first prospective study to systematically determine blood counts, blood cell morphology, and GATA1 mutation status in neonates with DS with the aims of (1) identifying the population at risk of developing ML-DS; (2) defining the clinical and hematologic features associated with a mutant GATA1 clone in DS neonates; and (3) determining how best to define TAM. We demonstrate for the first time that almost all DS neonates have multiple quantitative and morphologic hematologic abnormalities, independent of their GATA1 mutation status, providing strong correlative evidence that trisomy 21 itself causes trilineage perturbation of neonatal, as well as fetal, hematopoiesis.29-34
GATA1 mutation analysis showed that 8.5% of babies had a GATA1 mutation detected by Ss/DHPLC, similar to estimates from retrospective studies (5%-10%).10 Since an additional 20.4% (18 of 88 neonates with sufficient available DNA) had GATA1 mutations detectable only by targeted NGS, the overall frequency of GATA1 mutations was 3-fold higher (29%) than previous estimates; importantly, two-thirds of cases were clinically and hematologically silent. Thus, GATA1 mutation analysis using NGS is the most reliable way of detecting all babies with mutant GATA1 clones. In keeping with our previous work,3 a significant proportion of neonates (5 of 35) had >1 GATA1 mutation, suggesting that the N-terminal–truncated GATA1 protein confers a selective growth advantage to mutant GATA1-containing clones or, alternatively, that trisomic cells have a “mutator phenotype.”35,36 Although the low frequency of nonhematologic malignancies in DS argues against this, trisomy 21 might induce high mutation rates at specific genomic loci (eg, GATA1), as in the kataegis phenotype,37 rather than causing widely dispersed mutations.
What implications do these data have for the definition and diagnosis of TAM? Most clinicians screen DS newborns with a CBC and use clinical and hematologic findings to flag possible diagnoses of TAM, which then undergo GATA1 mutation analysis by conventional Ss/DHPLC. Our data show that TAM cannot be reliably diagnosed by clinical or hematologic features alone. Furthermore, no hematologic features are specific for TAM. Indeed, 4 out of 17 cases of TAM in our study were unsuspected by clinical teams, either because a normal CBC led to routine smears not being evaluated, or because blasts were wrongly attributed to “prematurity.” This highlights the practical difficulty in defining TAM solely by a specific % of blasts, especially given interobserver variation in blast cell enumeration on smears.38 The threshold of 10% used in OIDSCS was a useful, rapid way of identifying all possible cases but was not specific, and our data (Figure 1) indicate that even if a larger cohort was studied, it would be difficult to define TAM with high sensitivity and specificity on a blast % threshold.
An alternative way of considering the issue of the diagnosis of TAM is to ask, why it is important to diagnose TAM? First, an accurate definition of TAM allows the population at risk of transforming to ML-DS to be identified. This facilitates regular clinical and laboratory follow up by pediatric hemato-oncologists and ensures appropriate management of cytopenias that may precede ML-DS, including the timing of antileukemic therapy. Given the clear etiologic link between GATA1 mutations and ML-DS,3-5,7-9 it is not surprising that babies with GATA1 mutations detectable by Ss/DHPLC or NGS were at risk of ML-DS. Our data clearly show that the best way to identify all of those at risk is to comprehensively screen for GATA1 mutations by Ss/DHPLC and NGS. The second reason for making a definitive diagnosis of TAM is that it may facilitate management of early complications of TAM, such as effusions and liver dysfunction. In fulminant cases, the diagnosis is usually clinically straightforward, the blast % is high, and GATA1 mutations should be easy to detect by Ss/DHPLC as the circulating disease burden is high. Our findings may have a modest impact on the immediate management of such cases of “classical” TAM but will prevent misdiagnosis in neonates with clinical and hematologic features mimicking TAM (Table 3).
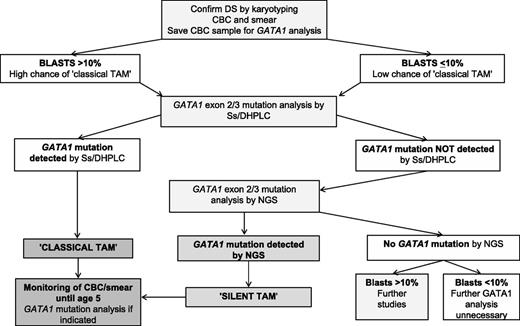
We favor a diagnostic algorithm (Figure 3) where evaluation of blood smears, as well as CBCs, as recommended by the American Academy of Pediatrics,39 is a useful, immediate screening step to identify DS neonates with “classical” TAM who may require early treatment (especially where GATA1 analysis is unavailable or delayed). We suggest all DS neonates should also have GATA1 mutation analysis by Ss/DHPLC to quickly identify those with large mutant GATA1 clones. For DS neonates without mutations detected by Ss/DHPLC, we suggest that NGS be used to identify low-abundance GATA1 mutations. By comprehensively detecting GATA1 mutations, pediatric hematology follow-up can be limited to those at risk of transformation rather than all babies with blasts (ie, approximately all babies with DS).
Algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Suggested algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Evaluation of a blood smear and CBC can be used as an immediate screening step to identify DS neonates with “classical” TAM who may require early treatment (especially where GATA1 analysis is unavailable or delayed). As a next step, GATA1 mutation analysis by Ss/DHPLC will quickly identify DS neonates with large mutant GATA1 clones. For DS neonates without mutations detected by Ss/DHPLC, NGS is the most reliable way of identifying low-abundance GATA1 mutations, allowing pediatric hematology follow-up to be limited to those at risk of transformation rather than all DS babies with peripheral blood blasts. Monitoring of all DS children with GATA1 mutations until the age of 5 years is recommended. This can be done using serial CBC/smears with GATA1 mutation analysis as indicated (eg, for persistent cytopenias). For the small number of DS babies with blasts >10% who have no detectable GATA1 mutations by NGS, more detailed studies to exclude the presence of rare GATA1 deletions are suggested.
Algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Suggested algorithm for diagnosis and monitoring of mutant GATA1 clones in DS neonates. Evaluation of a blood smear and CBC can be used as an immediate screening step to identify DS neonates with “classical” TAM who may require early treatment (especially where GATA1 analysis is unavailable or delayed). As a next step, GATA1 mutation analysis by Ss/DHPLC will quickly identify DS neonates with large mutant GATA1 clones. For DS neonates without mutations detected by Ss/DHPLC, NGS is the most reliable way of identifying low-abundance GATA1 mutations, allowing pediatric hematology follow-up to be limited to those at risk of transformation rather than all DS babies with peripheral blood blasts. Monitoring of all DS children with GATA1 mutations until the age of 5 years is recommended. This can be done using serial CBC/smears with GATA1 mutation analysis as indicated (eg, for persistent cytopenias). For the small number of DS babies with blasts >10% who have no detectable GATA1 mutations by NGS, more detailed studies to exclude the presence of rare GATA1 deletions are suggested.
Using this diagnostic approach, GATA1 mutations will frequently be clinically and hematologically silent. Therefore, we suggest the term “silent TAM” for DS neonates with small GATA1 mutant clones detectable only by NGS. This approach illustrates the dilemma facing clinicians in the current era of high-throughput genetic sequencing. In describing a new disease entity, silent TAM, we acknowledge that we cannot yet know the best management for these infants. However, we know that silent TAM is clinically important because it can give rise to ML-DS. Furthermore, TAM and silent TAM offer a unique opportunity to address the significance of small preleukemic clones in DS. Because ML-DS virtually always presents within a predictable time window spanning the first 5 years of life,40 we suggest that all DS neonates with GATA1 mutations should be monitored until the age of 5 years with regular CBCs, smears, and GATA1 mutation analysis by NGS. We acknowledge that it is currently unknown whether this would allow early diagnosis and treatment of ML-DS or improve outcome. Although CBC data, particularly platelet counts, may provide a simpler, albeit less specific, early indicator of impending ML-DS, whether this would be equally effective in improving outcome is also unknown. Nevertheless, monitoring of mutant GATA1 clones using such sensitive methods at last provides the opportunity to design effective protocols to eradicate GATA1 mutant clones during the neonatal period and potentially prevent ML-DS. NGS, rather than Ss/DHPLC, is required to pick up small clones at birth and at follow-up. NGS technology is now widely available. Even though it is becoming cheaper, the suggested approach would limit NGS to those who do not have mutations detected by a simpler method. In the future, it may be that NGS becomes the most cost-effective method to detect GATA1 mutations.
Finally, what is the risk of transformation conferred by harboring GATA1 mutations? In previous retrospective studies of clinically diagnosed TAM, the risk of ML-DS was ∼20% to 30%.11-14 Given that population studies show that 1% to 2% of children with DS develop ML-DS,1 and that we found GATA1 mutations in 29% of DS neonates, this suggests that GATA1 mutation(s) may confer a risk of ML-DS of ∼5% to 10%. This is consistent with our data in which 4 of 35 (11.4%) neonates with GATA1 mutation(s) have developed ML-DS within a median follow-up period of >33 months. By contrast, none of the neonates without a GATA1 mutation has developed ML-DS despite a median follow-up of 40 months. Ultimately, longitudinal follow-up of all DS neonates in OIDSCS will allow accurate assessment of the relationship between mutant GATA1 clone size and development of ML-DS and thus fully define the natural history of TAM and silent TAM.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all the parents who generously allowed their children to be included in the study and the haematology laboratory, nursing, medical, and clerical staff in all the participating centres for their valuable contributions.
I.R., P.V., K.A., G.J., K.P., G.C., and H.R. are funded by Leukaemia & Lymphoma Research (LLR) Specialist Programme Award 08030 and A.N. and A.R. by LLR Clinical Fellowships. I.R. is also funded by the Imperial College London Comprehensive Biomedical Research Centre and P.V. by the NIHR Biomedical Research Centres funding scheme, the Medical Research Council (MRC) Disease Team Award, and the MRC Molecular Haematology Unit. N.B. is funded by the Kay Kendall Leukaemia Fund and Leuka. A.G. is funded by the Wellcome Trust (programme grant 091182). The funding sources had no role in study design or conduct and management of the study and no role in data collection, data analysis, data interpretation, or writing of the report.
Authorship
Contribution: I.R. and P.V. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis; I.R., P.V., and G.H. contributed to all parts of the study; K.A., G.J., H.R., A.N., G.V., K.P., E.M., S.M.G., A.R., G.C., C.S., N.B., A.G., and P.C. performed the research, analyzed data, and revised the paper. All other authors contributed patient samples, acquired and interpreted data, and revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Irene Roberts, Centre for Haematology Commonwealth Building, Hammersmith Campus Imperial College London, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: irene.roberts@imperial.ac.uk; and Paresh Vyas, MRC Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, University of Oxford and Department of Haematology, Oxford University Hospitals NHS Trust, OX3 9DS Oxford, United Kingdom; e-mail: paresh.vyas@imm.ox.ac.uk.