Key Points
HLA mismatches at the allele and antigen level (possibly with the exception of HLA-DQB1) should be treated equally in donor selection.
HLA mismatches at >1 locus (including HLA-DQB1) have additive detrimental effects.
Abstract
To validate current donor selection strategies based on previous international studies, we retrospectively analyzed 2646 transplantations performed for hematologic malignancies in 28 German transplant centers. Donors and recipients were high resolution typed for HLA-A, -B, -C, -DRB1, and -DQB1. The highest mortality in overall survival analysis was seen for HLA-A, -B, and DRB1 mismatches. HLA-DQB1 mismatched cases showed a trend toward higher mortality, mostly due to HLA-DQB1 antigen disparities. HLA incompatibilities at >1 locus showed additive detrimental effects. HLA mismatching had no significant effect on relapse incidence and primary graft failure. Graft source had no impact on survival end points, neither in univariate nor in multivariate analysis. Higher patient age, advanced disease, transplantations before 2004, patient C2C2 killer cell immunoglobulin-like receptor (KIR)-ligand phenotype, and unavailability of a national donor adversely influenced outcomes in multivariate analysis. Our study confirms the association of HLA-A, -B, -C, and -DRB1 incompatibilities with adverse outcome in hematopoietic stem cell transplantation (HSCT). The relevance of HLA-DQB1 disparities in single mismatched transplantations remains unclear. Similar hazard ratios for allele and antigen mismatches (possibly with an exception for HLA-DQB1) highlight the importance of allele level typing and matching in HSCT. The number of incompatibilities and their type significantly impact survival.
Introduction
Hematopoietic stem cell transplantation (HSCT) from unrelated volunteer donors is the most widely used treatment alternative when no suitable sibling donor is identified. HSCT is potentially curative for a variety of malignant diseases of the hematopoietic system and certain life-threatening nonmalignant conditions. Transplantation of hematopoietic stem cells is a field of constant evolution and change. Major improvements have been the introduction of reduced intensity conditioning regimes (RIC), which decrease toxicity, and as a consequence transplant-related mortality (TRM), as well as the introduction of high-resolution HLA typing for donor selection.1,2 As HSCT becomes a safer therapy for many patients, increasing numbers of transplantations are performed worldwide. In this context, 3 main trends can be observed over the last decade: (1) the proportion of older transplant patients increases; (2) peripheral blood stem cells (PBSCs) are presently the main graft source; and (3) the demand for unrelated donors is increasing.3,4
Notwithstanding these changes, the paramount criterion for donor selection has remained HLA matching and, despite some variation in detail, a complete match at all relevant loci is unanimously recommended. However, the diversity of HLA remains a challenge in donor search and selection, sometimes forcing acceptance of mismatched donors. The aim of our study was to validate current donor selection strategies based on previous international studies and to investigate whether allelic HLA mismatches differentially impact outcome of HSCT compared with antigen mismatches at individual loci. As killer cell immunoglobulin-like receptor (KIR)-ligand interactions were reported to influence HSCT outcome, these were considered as well.5,6
Patients and methods
Patients
Adult patients diagnosed with a malignant hematological disorder and for whom the unrelated donor search was performed in the search unit in Ulm were included in this analysis. All patients received a first allogeneic transplant using bone marrow or PBSCs without T-cell depletion between 1997 and 2010 (Table 1). Retransplanted patients were censored at the time of second transplantation. Disease stage definitions were adopted from a previous report of the European Group for Blood and Marrow Transplantation (EBMT) study group defining the EBMT risk score for outcome after HSCT.7 Myeloablative conditioning was defined as total body irradiation ≥10 Gy and/or cyclophosphamide ≥120 mg/m2 and/or busulfan ≥16 mg/kg.8,9 Patients treated with less intense conditioning regimens were considered RIC. All patients received T cell–replete grafts. Patient and donor consents for HLA typing and for the analysis of clinical data were obtained in accordance with the Declaration of Helsinki. The study was approved by the ethical review board of the University of Ulm (project 263/09).
Patient characteristics
| Category . | n . | Percent . |
|---|---|---|
| Number of patients | 2646 | 100 |
| Number of centers | 28 | |
| Age category | ||
| 18-29 | 333 | 12.6 |
| 30-39 | 348 | 13.2 |
| 40-49 | 554 | 20.9 |
| 50-59 | 770 | 29.1 |
| 60-69 | 577 | 21.8 |
| 70-79 | 64 | 2.4 |
| Diagnosis | ||
| AML | 810 | 30.6 |
| ALL | 350 | 13.2 |
| AL | 135 | 5.1 |
| CML | 211 | 8.0 |
| MDS | 486 | 18.4 |
| NHL | 437 | 16.5 |
| MM | 217 | 8.2 |
| Ethnicity | ||
| White | 2636 | 99.6 |
| Asian | 7 | 0.3 |
| African | 3 | 0.1 |
| Disease stage | ||
| Early | 1047 | 39.6 |
| Intermediate | 922 | 34.8 |
| Advanced | 677 | 25.6 |
| Conditioning regimen | ||
| Myeloablative | 1692 | 63.9 |
| Reduced intensity | 954 | 36.1 |
| Stem cell source | ||
| BM | 346 | 13.1 |
| PBSC | 2300 | 86.9 |
| Donor-recipient sex match | ||
| Male-male | 1227 | 46.4 |
| Male-female | 664 | 25.1 |
| Female-male | 362 | 13.7 |
| Female-female | 393 | 14.9 |
| Year of transplanation | ||
| 1997-2000 | 299 | 11.3 |
| 2001-2005 | 589 | 22.3 |
| 2006-2010 | 1758 | 66.4 |
| Category . | n . | Percent . |
|---|---|---|
| Number of patients | 2646 | 100 |
| Number of centers | 28 | |
| Age category | ||
| 18-29 | 333 | 12.6 |
| 30-39 | 348 | 13.2 |
| 40-49 | 554 | 20.9 |
| 50-59 | 770 | 29.1 |
| 60-69 | 577 | 21.8 |
| 70-79 | 64 | 2.4 |
| Diagnosis | ||
| AML | 810 | 30.6 |
| ALL | 350 | 13.2 |
| AL | 135 | 5.1 |
| CML | 211 | 8.0 |
| MDS | 486 | 18.4 |
| NHL | 437 | 16.5 |
| MM | 217 | 8.2 |
| Ethnicity | ||
| White | 2636 | 99.6 |
| Asian | 7 | 0.3 |
| African | 3 | 0.1 |
| Disease stage | ||
| Early | 1047 | 39.6 |
| Intermediate | 922 | 34.8 |
| Advanced | 677 | 25.6 |
| Conditioning regimen | ||
| Myeloablative | 1692 | 63.9 |
| Reduced intensity | 954 | 36.1 |
| Stem cell source | ||
| BM | 346 | 13.1 |
| PBSC | 2300 | 86.9 |
| Donor-recipient sex match | ||
| Male-male | 1227 | 46.4 |
| Male-female | 664 | 25.1 |
| Female-male | 362 | 13.7 |
| Female-female | 393 | 14.9 |
| Year of transplanation | ||
| 1997-2000 | 299 | 11.3 |
| 2001-2005 | 589 | 22.3 |
| 2006-2010 | 1758 | 66.4 |
AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; AL, unclassified acute leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; MM, multiple myeloma; BM, bone marrow.
HLA typing
HLA typing of donors and patients carried out after May 2005 required testing for HLA-A, -B, -C, -DRB1, and -DQB1 at high resolution (n = 1804; 68.2%). Confirmatory typings for patients and donors from searches completed before May 2005 (n = 842; 31.8%) included low-resolution typing for HLA-A and -B and high-resolution typing for HLA-DRB1 and -DQB1. In these cases, HLA class I high-resolution typing was carried out retrospectively.
Exons 2 and 3 were analyzed for HLA class I typing, and exon 2 only for HLA class II typing. High-resolution HLA typing was performed by sequencing analysis (sequence-based typing) for HLA class I, and by the polymerase chain reaction (PCR)-sequence-specific oligonucleotide probes method or sequence-based typing for HLA class II. Ambiguities involving the most frequent null (not expressed) alleles were resolved if necessary by the PCR-sequence-specific primer technique.10 Like in studies performed by the Center for International Blood and Marrow Transplant Research (CIBMTR),11-13 differences of the first 2 digits were classified as antigen mismatches irrespective of their vector14 ; other differences were classified as allele mismatches with some exceptions, which, as in the CIBMTR studies, were grouped according to their serologic reactivity.15
To better characterize patients without a suitable donor in the national registry, patients with rare HLA phenotypes were identified using high-resolution 5-locus HLA-haplotypes, derived from German registry donors.16,17 Patients were assigned to 2 groups: if the patient HLA phenotype was explainable by any pair of haplotypes available in the registry haplotype database, the phenotype was considered common; other HLA phenotypes, consisting of at least one rare putative haplotype, where the patient's HLA type could not be formed from haplotypes listed in the database, were considered as rare.
Definitions
Overall survival (OS) was defined as time to death of any cause and was censored at the time of last follow-up. Disease-free survival (DFS) was defined as time to relapse of primary disease or death from any cause and was also censored at the time of last follow-up. Relapse incidence (RI) was defined as the cumulative probability of relapse at any given time point; TRM was defined as mortality in complete remission of disease. Primary graft failure (PGF) was defined as failure to engraft 28 days after transplantation, with engraftment defined as absolute neutrophil count >0.5 × 109/L for the first of ≥3 consecutive days.8
Statistical analysis
For univariate analysis of OS, the Kaplan-Meier method and log-rank testing were used. Multivariate analysis of OS and DFS was performed using extended Cox models.18 For TRM and RI, competing risks analysis was used.19 PGF was analyzed using logistic regression. Statistical models covered covariates defining the EBMT risk score: patient age, disease stage, time to transplantation, and donor-recipient gender combination.7 In addition to these, HLA matching status, patient and donor cytomegalovirus (CMV) status, year of transplantation, conditioning regimen intensity, donor origin (national vs international), and treatment with anti-thymocyte globulin (ATG) were evaluated. Missing data for ATG and CMV status were treated as separate categories in multivariate analysis (missing: ATG treatment 19.0%, and CMV status 34.9%).20 To account for possible natural killer cell alloreactivity, the KIR-ligand incompatibility model (predicted from the HLA phenotype in HLA-B and -C mismatched cases), as well as the patient HLA-C phenotype (C1 group, Asn80; C2 group, Lys80), were included in the statistical models.5,6 Adjustments for diagnosis and center effect were made.7,21 Subanalysis of single HLA mismatches was performed, but as numbers in some subgroups were too small for a robust analysis, combined models, including all HLA mismatches, were calculated as well. Confounding effects for presence of multiple HLA mismatches were considered in these models and adjusted.22 All models were checked for interactions and proportional hazards assumption. Violations of the proportional hazards assumption by the disease stage and conditioning regimen intensity were adjusted using time-dependent covariates. Because of multiple comparisons, the significance level in multivariate analyses was set to .01.
Results
The patient cohort characteristics are summarized in Table 1. A total of 2646 patients were included in this analysis, with a median post-transplant follow-up time of 24 months. Transplantations were performed between 1997 and 2010. Median patient age was 51 years (range: 18-75 years). The conditioning regime was myeloablative in 63.9% of the cases. Most patients received PBSCs as a stem cell source (86.9%).
OS
Analysis of OS showed higher risk for death if any HLA mismatch was present (Table 2). Highest hazard ratios (HRs) were seen for HLA-A, -B, and -DRB1 mismatches. Classification of mismatches in allele or antigen mismatches (Table 3) showed that the HLA-B allele and HLA-C antigen mismatches were most strongly associated with adverse outcome. Comparison of allele vs antigen mismatches within each locus showed slightly lower estimates for the HLA-C allele and HLA-DQB1 allele mismatches. These differences were not statistically significant (HLA-C allele vs antigen mismatch: HR 0.96, confidence interval [CI] 0.70-1.30, P = .780; HLA-DQB1 allele vs antigen mismatch: HR 0.79, CI 0.54-1.17, P = .240). HLA-DQB1 mismatching did not reach statistical significance, although a trend toward higher risk could be seen (HR 1.23, CI 1.00-1.51, P = .050; Table 2). Separate analysis of allele and antigen mismatches showed that an impact of HLA-DQB1 mismatching might be limited to antigen differences (HR 1.39, CI 1.04-1.85, P = .028; Table 3).
Analysis of the impact of HLA mismatches in all transplantations after multivariate modeling
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A MM | 282 | 1.43 | 1.19-1.72 | <.001 |
| HLA-B MM | 283 | 1.52 | 1.20-1.93 | <.001 |
| HLA-C MM | 620 | 1.35 | 1.17-1.56 | <.001 |
| HLA-DRB1 MM | 126 | 1.42 | 1.10-1.82 | .006 |
| HLA-DQB1 MM | 191 | 1.23 | 1.00-1.51 | .050 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A MM | 282 | 1.25 | 1.05-1.50 | .014 |
| HLA-B MM | 283 | 1.32 | 1.05-1.66 | .016 |
| HLA-C MM | 620 | 1.32 | 1.15-1.51 | <.001 |
| HLA-DRB1 MM | 126 | 1.29 | 1.01-1.63 | .040 |
| HLA-DQB1 MM | 191 | 1.18 | 0.97-1.43 | .110 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| HLA-A MM | 282 | 1.51 | 1.16-1.98 | .003 |
| HLA-B MM | 283 | 1.50 | 1.19-1.89 | <.001 |
| HLA-C MM | 620 | 1.45 | 1.19-1.76 | <.001 |
| HLA-DRB1 MM | 126 | 1.57 | 1.12-2.21 | .009 |
| HLA-DQB1 MM | 191 | 1.28 | 0.96-1.71 | .091 |
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A MM | 282 | 1.43 | 1.19-1.72 | <.001 |
| HLA-B MM | 283 | 1.52 | 1.20-1.93 | <.001 |
| HLA-C MM | 620 | 1.35 | 1.17-1.56 | <.001 |
| HLA-DRB1 MM | 126 | 1.42 | 1.10-1.82 | .006 |
| HLA-DQB1 MM | 191 | 1.23 | 1.00-1.51 | .050 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A MM | 282 | 1.25 | 1.05-1.50 | .014 |
| HLA-B MM | 283 | 1.32 | 1.05-1.66 | .016 |
| HLA-C MM | 620 | 1.32 | 1.15-1.51 | <.001 |
| HLA-DRB1 MM | 126 | 1.29 | 1.01-1.63 | .040 |
| HLA-DQB1 MM | 191 | 1.18 | 0.97-1.43 | .110 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| HLA-A MM | 282 | 1.51 | 1.16-1.98 | .003 |
| HLA-B MM | 283 | 1.50 | 1.19-1.89 | <.001 |
| HLA-C MM | 620 | 1.45 | 1.19-1.76 | <.001 |
| HLA-DRB1 MM | 126 | 1.57 | 1.12-2.21 | .009 |
| HLA-DQB1 MM | 191 | 1.28 | 0.96-1.71 | .091 |
Analysis of OS, DFS, and TRM for any mismatch at each locus. Significance level set to P < .01. Significant results are bold.
Analysis of the impact of HLA mismatches in all transplantations after multivariate modeling
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A allele MM | 121 | 1.37 | 1.04-1.80 | .026 |
| HLA-A antigen MM | 164 | 1.41 | 1.12-1.77 | .003 |
| HLA-B allele MM | 237 | 1.48 | 1.15-1.91 | .003 |
| HLA-B-antigen MM | 50 | 1.48 | 1.00-2.20 | .052 |
| HLA-C allele MM | 106 | 1.28 | 0.98-1.67 | .072 |
| HLA-C antigen MM | 528 | 1.34 | 1.15-1.57 | <.001 |
| HLA-DRB1 allele MM | 108 | 1.46 | 1.12-1.90 | .005 |
| HLA-DRB1 antigen MM | 18 | 1.12 | 0.57-2.17 | .750 |
| HLA-DQB1 allele MM | 107 | 1.10 | 0.84-1.45 | .480 |
| HLA-DQB1 antigen MM | 85 | 1.39 | 1.04-1.85 | .028 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A allele MM | 121 | 1.22 | 0.94-1.59 | .140 |
| HLA-A antigen MM | 164 | 1.23 | 0.98-1.53 | .070 |
| HLA-B allele MM | 237 | 1.33 | 1.04-1.69 | .024 |
| HLA-B-antigen MM | 50 | 1.20 | 0.81-1.77 | .370 |
| HLA-C allele MM | 106 | 1.20 | 0.93-1.55 | .150 |
| HLA-C antigen MM | 528 | 1.32 | 1.14-1.52 | <.001 |
| HLA-DRB1 allele MM | 108 | 1.38 | 1.07-1.77 | .014 |
| HLA-DRB1 antigen MM | 18 | 0.87 | 0.45-1.69 | .670 |
| HLA-DQB1 allele MM | 107 | 1.06 | 0.81-1.37 | .690 |
| HLA-DQB1 antigen MM | 85 | 1.34 | 1.02-1.77 | .036 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| HLA-A allele MM | 121 | 1.22 | 0.80-1.86 | .360 |
| HLA-A antigen MM | 164 | 1.64 | 1.21-2.22 | .002 |
| HLA-B allele MM | 237 | 1.62 | 1.26-2.08 | <.001 |
| HLA-B-antigen MM | 50 | 1.07 | 0.61-1.89 | .810 |
| HLA-C allele MM | 106 | 1.55 | 1.09-2.21 | .015 |
| HLA-C antigen MM | 528 | 1.38 | 1.12-1.69 | .002 |
| HLA-DRB1 allele MM | 108 | 1.68 | 1.17-2.40 | .005 |
| HLA-DRB1 antigen MM | 18 | 0.71 | 0.22-2.30 | .570 |
| HLA-DQB1 allele MM | 107 | 1.27 | 0.88-1.82 | .200 |
| HLA-DQB1 antigen MM | 85 | 1.25 | 0.82-1.89 | .300 |
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A allele MM | 121 | 1.37 | 1.04-1.80 | .026 |
| HLA-A antigen MM | 164 | 1.41 | 1.12-1.77 | .003 |
| HLA-B allele MM | 237 | 1.48 | 1.15-1.91 | .003 |
| HLA-B-antigen MM | 50 | 1.48 | 1.00-2.20 | .052 |
| HLA-C allele MM | 106 | 1.28 | 0.98-1.67 | .072 |
| HLA-C antigen MM | 528 | 1.34 | 1.15-1.57 | <.001 |
| HLA-DRB1 allele MM | 108 | 1.46 | 1.12-1.90 | .005 |
| HLA-DRB1 antigen MM | 18 | 1.12 | 0.57-2.17 | .750 |
| HLA-DQB1 allele MM | 107 | 1.10 | 0.84-1.45 | .480 |
| HLA-DQB1 antigen MM | 85 | 1.39 | 1.04-1.85 | .028 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| HLA-A allele MM | 121 | 1.22 | 0.94-1.59 | .140 |
| HLA-A antigen MM | 164 | 1.23 | 0.98-1.53 | .070 |
| HLA-B allele MM | 237 | 1.33 | 1.04-1.69 | .024 |
| HLA-B-antigen MM | 50 | 1.20 | 0.81-1.77 | .370 |
| HLA-C allele MM | 106 | 1.20 | 0.93-1.55 | .150 |
| HLA-C antigen MM | 528 | 1.32 | 1.14-1.52 | <.001 |
| HLA-DRB1 allele MM | 108 | 1.38 | 1.07-1.77 | .014 |
| HLA-DRB1 antigen MM | 18 | 0.87 | 0.45-1.69 | .670 |
| HLA-DQB1 allele MM | 107 | 1.06 | 0.81-1.37 | .690 |
| HLA-DQB1 antigen MM | 85 | 1.34 | 1.02-1.77 | .036 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| HLA-A allele MM | 121 | 1.22 | 0.80-1.86 | .360 |
| HLA-A antigen MM | 164 | 1.64 | 1.21-2.22 | .002 |
| HLA-B allele MM | 237 | 1.62 | 1.26-2.08 | <.001 |
| HLA-B-antigen MM | 50 | 1.07 | 0.61-1.89 | .810 |
| HLA-C allele MM | 106 | 1.55 | 1.09-2.21 | .015 |
| HLA-C antigen MM | 528 | 1.38 | 1.12-1.69 | .002 |
| HLA-DRB1 allele MM | 108 | 1.68 | 1.17-2.40 | .005 |
| HLA-DRB1 antigen MM | 18 | 0.71 | 0.22-2.30 | .570 |
| HLA-DQB1 allele MM | 107 | 1.27 | 0.88-1.82 | .200 |
| HLA-DQB1 antigen MM | 85 | 1.25 | 0.82-1.89 | .300 |
Analysis of OS, DFS, and TRM with allele and antigen mismatches separated. Significance level set to P < .01. Significant results are bold.
n, number of individuals within the respective category; MM, mismatch.
Subanalysis of single mismatches (Table 4) revealed similar HRs for HLA class I mismatches compared with the model including multiple mismatches (Table 3).
Subanalysis of single mismatched transplantations after multivariate modeling
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Matched | 1511 | 1.00 | ||
| A-allele-MM | 61 | 1.44 | 1.02-2.04 | .039 |
| A-antigen-MM | 121 | 1.31 | 1.00-1.72 | .047 |
| B-allele-MM | 76 | 1.53 | 1.11-2.09 | .009 |
| B-antigen-MM | 30 | 1.79 | 1.13-2.83 | .014 |
| C-allele-MM | 67 | 1.10 | 0.78-1.56 | .600 |
| C-antigen-MM | 300 | 1.27 | 1.06-1.53 | .009 |
| DR-allele-MM | 46 | 1.17 | 0.75-1.82 | .500 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 0.76 | 0.48-1.21 | .250 |
| DQ-antigen-MM | 41 | 1.42 | 0.93-2.17 | .100 |
| DFS | ||||
| Matched | 1511 | 1.00 | ||
| A-allele-MM | 61 | 1.25 | 0.90-1.76 | .190 |
| A-antigen-MM | 121 | 1.16 | 0.89-1.50 | .270 |
| B-allele-MM | 76 | 1.33 | 0.98-1.81 | .067 |
| B-antigen-MM | 30 | 1.39 | 0.88-2.21 | .160 |
| C-allele-MM | 67 | 1.12 | 0.81-1.55 | .480 |
| C-antigen-MM | 300 | 1.25 | 1.05-1.48 | .012 |
| DR-allele-MM | 46 | 1.14 | 0.75-1.72 | .550 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 0.79 | 0.52-1.20 | .260 |
| DQ-antigen-MM | 41 | 1.32 | 0.88-1.97 | .180 |
| TRM | ||||
| Matched | 1509 | 1.00 | ||
| A-allele-MM | 61 | 1.33 | 0.80-2.23 | .280 |
| A-antigen-MM | 121 | 1.53 | 1.07-2.20 | .021 |
| B-allele-MM | 76 | 1.66 | 1.10-2.52 | .017 |
| B-antigen-MM | 30 | 1.60 | 0.85-2.99 | .140 |
| C-allele-MM | 67 | 1.34 | 0.85-2.12 | .210 |
| C-antigen-MM | 300 | 1.29 | 0.99-1.69 | .064 |
| DR-allele-MM | 46 | 1.53 | 0.86-2.73 | .150 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 1.03 | 0.59-1.81 | .910 |
| DQ-antigen-MM | 41 | 1.16 | 0.57-2.40 | .680 |
| Locus . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Matched | 1511 | 1.00 | ||
| A-allele-MM | 61 | 1.44 | 1.02-2.04 | .039 |
| A-antigen-MM | 121 | 1.31 | 1.00-1.72 | .047 |
| B-allele-MM | 76 | 1.53 | 1.11-2.09 | .009 |
| B-antigen-MM | 30 | 1.79 | 1.13-2.83 | .014 |
| C-allele-MM | 67 | 1.10 | 0.78-1.56 | .600 |
| C-antigen-MM | 300 | 1.27 | 1.06-1.53 | .009 |
| DR-allele-MM | 46 | 1.17 | 0.75-1.82 | .500 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 0.76 | 0.48-1.21 | .250 |
| DQ-antigen-MM | 41 | 1.42 | 0.93-2.17 | .100 |
| DFS | ||||
| Matched | 1511 | 1.00 | ||
| A-allele-MM | 61 | 1.25 | 0.90-1.76 | .190 |
| A-antigen-MM | 121 | 1.16 | 0.89-1.50 | .270 |
| B-allele-MM | 76 | 1.33 | 0.98-1.81 | .067 |
| B-antigen-MM | 30 | 1.39 | 0.88-2.21 | .160 |
| C-allele-MM | 67 | 1.12 | 0.81-1.55 | .480 |
| C-antigen-MM | 300 | 1.25 | 1.05-1.48 | .012 |
| DR-allele-MM | 46 | 1.14 | 0.75-1.72 | .550 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 0.79 | 0.52-1.20 | .260 |
| DQ-antigen-MM | 41 | 1.32 | 0.88-1.97 | .180 |
| TRM | ||||
| Matched | 1509 | 1.00 | ||
| A-allele-MM | 61 | 1.33 | 0.80-2.23 | .280 |
| A-antigen-MM | 121 | 1.53 | 1.07-2.20 | .021 |
| B-allele-MM | 76 | 1.66 | 1.10-2.52 | .017 |
| B-antigen-MM | 30 | 1.60 | 0.85-2.99 | .140 |
| C-allele-MM | 67 | 1.34 | 0.85-2.12 | .210 |
| C-antigen-MM | 300 | 1.29 | 0.99-1.69 | .064 |
| DR-allele-MM | 46 | 1.53 | 0.86-2.73 | .150 |
| DR-antigen-MM | 7 | NA | NA | NA |
| DQ-allele-MM | 54 | 1.03 | 0.59-1.81 | .910 |
| DQ-antigen-MM | 41 | 1.16 | 0.57-2.40 | .680 |
Impact of single allele or antigen HLA MMs per locus on OS, DFS, and TRM. Significance level set to P < .01. Cases with more than one HLA mismatch were excluded from this analysis. Significant results are bold.
NA, number too low for analysis.
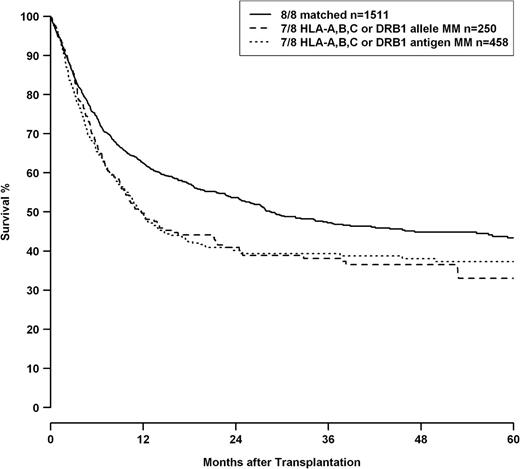
Analysis of mismatch combinations showed HRs increasing with the numbers of HLA mismatches, whereas classification in “allele or antigen mismatch” for single and double mismatches did not add any predictive value (Table 5) regarding mortality risk. Univariate analysis also did not reveal any significant difference between single allelic and antigenic HLA-A, -B, -C, or -DRB1 mismatches as shown in Figure 1 (loci combined, P = .924), whereas the difference between 8/8 and 7/8 matched transplantations was highly significant (P < .001). HLA-DQB1 mismatches were not included in this analysis because multivariate analysis indicated a trend toward worse outcome for HLA-DQB1 antigen differences compared with allelic incompatibilities for this locus.
Subanalysis of mismatch combinations after multivariate modeling
| Mismatch combination . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| Single allele MM | 304 | 1.21 | 1.01-1.44 | .044 |
| Single antigen MM | 499 | 1.30 | 1.12-1.51 | <.001 |
| Double allele MM | 46 | 1.70 | 1.15-2.53 | .008 |
| Double MM allele + antigen | 139 | 2.04 | 1.54-2.69 | <.001 |
| Double antigen MM | 62 | 1.76 | 1.25-2.48 | .001 |
| Triple MM | 61 | 2.56 | 1.71-3.85 | <.001 |
| Multiple MM | 24 | 3.11 | 1.70-5.67 | <.001 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| Single allele MM | 304 | 1.11 | 0.93-1.31 | .240 |
| Single antigen MM | 499 | 1.21 | 1.05-1.39 | .009 |
| Double allele MM | 46 | 1.60 | 1.09-2.33 | .015 |
| Double MM allele + antigen | 139 | 1.75 | 1.34-2.27 | <.001 |
| Double antigen MM | 62 | 1.75 | 1.27-2.41 | <.001 |
| Triple MM | 61 | 2.03 | 1.37-3.03 | <.001 |
| Multiple MM | 24 | 2.17 | 1.22-3.88 | .009 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| Single allele MM | 304 | 1.46 | 1.14-1.87 | .003 |
| Single antigen MM | 499 | 1.34 | 1.08-1.66 | .007 |
| Double allele MM | 46 | 2.08 | 1.21-3.57 | .008 |
| Double MM allele + antigen | 139 | 2.16 | 1.57-2.99 | <.001 |
| Double antigen MM | 62 | 1.90 | 1.18-3.07 | .009 |
| Triple MM | 61 | 2.94 | 1.91-4.51 | <.001 |
| Multiple MM | 24 | 2.93 | 1.66-5.16 | <.001 |
| Mismatch combination . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| Single allele MM | 304 | 1.21 | 1.01-1.44 | .044 |
| Single antigen MM | 499 | 1.30 | 1.12-1.51 | <.001 |
| Double allele MM | 46 | 1.70 | 1.15-2.53 | .008 |
| Double MM allele + antigen | 139 | 2.04 | 1.54-2.69 | <.001 |
| Double antigen MM | 62 | 1.76 | 1.25-2.48 | .001 |
| Triple MM | 61 | 2.56 | 1.71-3.85 | <.001 |
| Multiple MM | 24 | 3.11 | 1.70-5.67 | <.001 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| Single allele MM | 304 | 1.11 | 0.93-1.31 | .240 |
| Single antigen MM | 499 | 1.21 | 1.05-1.39 | .009 |
| Double allele MM | 46 | 1.60 | 1.09-2.33 | .015 |
| Double MM allele + antigen | 139 | 1.75 | 1.34-2.27 | <.001 |
| Double antigen MM | 62 | 1.75 | 1.27-2.41 | <.001 |
| Triple MM | 61 | 2.03 | 1.37-3.03 | <.001 |
| Multiple MM | 24 | 2.17 | 1.22-3.88 | .009 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| Single allele MM | 304 | 1.46 | 1.14-1.87 | .003 |
| Single antigen MM | 499 | 1.34 | 1.08-1.66 | .007 |
| Double allele MM | 46 | 2.08 | 1.21-3.57 | .008 |
| Double MM allele + antigen | 139 | 2.16 | 1.57-2.99 | <.001 |
| Double antigen MM | 62 | 1.90 | 1.18-3.07 | .009 |
| Triple MM | 61 | 2.94 | 1.91-4.51 | <.001 |
| Multiple MM | 24 | 2.93 | 1.66-5.16 | <.001 |
Analysis of OS, DFS, and TRM in different mismatch groups. Multiple mismatches, >3 HLA mismatches. Significance level set to P < .01. Significant results are bold.
OS of patients with 8/8 matched donors (solid line), patients with single allele mismatched donors at loci HLA-A, -B, -C, or -DRB1 (dashed line), and single antigen mismatched donors at loci HLA-A, -B, -C, or -DRB1 (dotted line). HLA-DQB1 status was omitted from this analysis because multivariate modeling indicated a possible differential impact of HLA-DQB1 allele and antigen mismatches. Comparison single allele vs single antigen mismatch, P = .924; comparison 8/8 matched vs 7/8 matched log-rank, P < .001.
OS of patients with 8/8 matched donors (solid line), patients with single allele mismatched donors at loci HLA-A, -B, -C, or -DRB1 (dashed line), and single antigen mismatched donors at loci HLA-A, -B, -C, or -DRB1 (dotted line). HLA-DQB1 status was omitted from this analysis because multivariate modeling indicated a possible differential impact of HLA-DQB1 allele and antigen mismatches. Comparison single allele vs single antigen mismatch, P = .924; comparison 8/8 matched vs 7/8 matched log-rank, P < .001.
If double mismatched cases (including HLA-DQB1 mismatches) were grouped according to the HLA class combination of their mismatches (Table 6), a higher HR was seen for combined class I and class II mismatches vs double HLA class I or double HLA class II mismatches, respectively. The direct comparison between combined HLA class I+II mismatches vs double HLA class I or double HLA class II mismatches was, however, not statistically significant (HR 1.29, CI 0.88-1.90, P = .190).
Subanalysis of double HLA mismatch combinations after multivariate modeling
| Mismatch combination . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.20 | 1.60-3.03 | <.001 |
| Double MM only HLA class I | 162 | 1.68 | 1.27-2.22 | <.001 |
| Double MM only HLA class II | 18 | 1.83 | 1.04-3.22 | .036 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.03 | 1.50-2.74 | <.001 |
| Double MM only HLA class I | 162 | 1.55 | 1.19-2.02 | .001 |
| Double MM only HLA class II | 18 | 1.57 | 0.89-2.75 | .120 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.69 | 1.75-4.14 | <.001 |
| Double MM only HLA class I | 162 | 1.90 | 1.39-2.61 | <.001 |
| Double MM only HLA class II | 18 | 1.52 | 0.63-3.67 | .360 |
| Mismatch combination . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| OS | ||||
| Complete match | 1511 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.20 | 1.60-3.03 | <.001 |
| Double MM only HLA class I | 162 | 1.68 | 1.27-2.22 | <.001 |
| Double MM only HLA class II | 18 | 1.83 | 1.04-3.22 | .036 |
| DFS | ||||
| Complete match | 1511 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.03 | 1.50-2.74 | <.001 |
| Double MM only HLA class I | 162 | 1.55 | 1.19-2.02 | .001 |
| Double MM only HLA class II | 18 | 1.57 | 0.89-2.75 | .120 |
| TRM | ||||
| Complete match | 1509 | 1.00 | ||
| Double MM HLA class I+II | 67 | 2.69 | 1.75-4.14 | <.001 |
| Double MM only HLA class I | 162 | 1.90 | 1.39-2.61 | <.001 |
| Double MM only HLA class II | 18 | 1.52 | 0.63-3.67 | .360 |
Analysis of OS, DFS, and TRM in different mismatch groups. Significance level set to P < .01. Significant results are bold.
DFS
In general, HRs for events in DFS analysis were lower than in OS. However, the observed HR-patterns were similar, with the highest impact seen for HLA-B and the lowest for HLA-DQB1 mismatches. The trend for adverse outcome of HLA-DQB1 mismatching was again mostly due to HLA-DQB1 antigen mismatches, whereas DQB1 allele mismatching had no significant impact on DFS.
Most predictive of an adverse outcome were HLA-C antigen mismatches (HR 1.32, CI 1.14-1.52, P < .001), whereas for HLA-C allele mismatches, such an association was not found (HR 1.20, CI 0.93-1.55, P = .150). The number of mismatches was a strong predictor for DFS, whereas antigen or allele differences did not show differential patterns. Classification according to combinations of class I and class II mismatches showed the highest risk for combined class I and class II mismatches and lower HRs for double class I or double class II mismatches.
TRM
The observed HRs for TRM were generally higher than those found for DFS and OS. HLA-A, -B, and -DRB1 mismatches had the highest impact, whereas the lowest impact was seen for HLA-DQB1 mismatches. The high impact of HLA mismatching on TRM was also seen in the analysis of HLA mismatch combinations. An already high risk for TRM in transplantations with single HLA allele or antigen mismatched donors increased further if ≥2 mismatches were present.
Classification of double mismatches according to the HLA class showed highest TRM for combined class I/class II mismatches and lower ratios for double class I or double class II mismatches. A direct comparison between these subgroups did, however, not reach statistical significance (HR 1.45, CI 0.88-2.38, P = .140).
RI and PGF
Univariate and multivariate analysis showed no statistically significant effect of HLA mismatching on relapse incidence, neither for all loci combined nor for individual locus mismatches (relapse incidences for all loci combined: 10/10 matched, 33.7%; 9/10 matched, 32.0%, ≤8/10 matched, 27.1%; P = .512). Interestingly, C2C2 KIR-ligand status in patients was associated with a slightly increased relapse rate (HR 1.27, CI 1.03-1.57, P = .028).
PGF rates were higher for ≥2 HLA mismatches, but this difference was not statistically significant after correction for graft source and treatment with growth factors (PGF rate 10/10 matched: 4.2%, 9/10 matched: 3.4%, ≤8/10 matched: 8.2%, P = .080). No differential impact on PGF was seen if the graft was obtained from a national or an international donor.
Clinical predictors
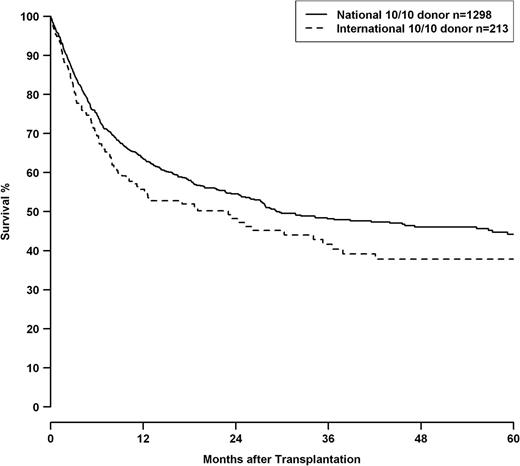
HRs for 10/10, 9/10, and 8/10 HLA mismatch groups are shown in Table 7 in context with other clinical predictors. 8/10 matched pairs showed a significantly higher risk than the 9/10 group (8/10 vs 9/10—OS: 1.47, CI 1.19-1.82, P < .001; DFS: 1.47, CI 1.20-1.80 P < .001, TRM: 1.54, CI 1.18-2.02, P = .001). Besides HLA match, adverse outcomes were associated with advancing disease stage and patient age. Transplantations before 2004 had worse outcomes. Transplantations performed with international donors had a worse outcome compared with transplantations with national donors. Male patients with female international donors showed an even higher HR in analysis of OS (HR 1.51, CI 1.18-1.92, P < .001). This effect was independent from HLA mismatching, as well as the year of transplantation, and can also be shown in a subset of transplantations performed with 10/10 matched donors (P = .047; Figure 2).
Effects of clinical predictors on survival endpoints
| Predictor . | OS . | DFS . | TRM . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | HR . | 95% CI . | P . | n . | HR . | 95% CI . | P . | n . | HR . | 95% CI . | P . | |
| HLA match | ||||||||||||
| 10/10 | 1511 | 1.00 | 1511 | 1.00 | 1509 | 1.00 | ||||||
| 9/10 | 803 | 1.26 | 1.11-1.43 | <.001 | 803 | 1.17 | 1.03-1.32 | .012 | 803 | 1.38 | 1.15-1.66 | <.001 |
| 8/10 | 247 | 1.86 | 1.51-2.29 | <.001 | 247 | 1.71 | 1.40-2.09 | <.001 | 247 | 2.13 | 1.64-2.77 | <.001 |
| Disease stage | ||||||||||||
| Early | 1047 | 1.00 | 1047 | 1.00 | 1047 | 1.00 | ||||||
| Intermediate | 922 | 1.46 | 1.26-1.70 | <.001 | 922 | 1.50 | 1.31-1.73 | <.001 | 921 | 1.60 | 1.29-1.98 | <.001 |
| Advanced | 677 | 2.18 | 1.87-2.54 | <.001 | 677 | 2.17 | 1.88-2.51 | <.001 | 676 | 1.97 | 1.59-2.43 | <.001 |
| Patient age (years) | ||||||||||||
| <40 | 734 | 1.00 | 734 | 1.00 | 733 | 1.00 | ||||||
| 40-55 | 951 | 1.39 | 1.19-1.62 | <.001 | 951 | 1.30 | 1.13-1.50 | <.001 | 950 | 1.60 | 1.27-2.00 | <.001 |
| >55 | 961 | 1.76 | 1.48-2.10 | <.001 | 961 | 1.56 | 1.33-1.82 | <.001 | 961 | 1.85 | 1.43-2.40 | <.001 |
| Donor-recipient gender | ||||||||||||
| Male-male | 1227 | 1.00 | 1227 | 1.00 | 1227 | 1.00 | ||||||
| Male-female | 362 | 1.13 | 0.95-1.34 | .170 | 362 | 1.15 | 0.98-1.35 | .087 | 362 | 0.96 | 0.75-1.22 | .720 |
| Female-female | 393 | 0.91 | 0.77-1.08 | .270 | 393 | 0.92 | 0.78-1.08 | .300 | 392 | 0.67 | 0.51-0.88 | .004 |
| Female-male | 664 | 0.99 | 0.86-1.14 | .870 | 664 | 0.98 | 0.86-1.13 | .810 | 663 | 0.91 | 0.74-1.12 | .390 |
| Year of transplantation | ||||||||||||
| 1997-2003 | 455 | 1.00 | 455 | 1.00 | 455 | 1.00 | ||||||
| 2004-2010 | 2191 | 0.76 | 0.64-0.91 | .002 | 2191 | 0.79 | 0.67-0.93 | .005 | 2189 | 0.61 | 0.49-0.76 | <.001 |
| Donor-recipient CMV status* | ||||||||||||
| Negative-negative | 520 | 1.00 | 520 | 1.00 | 520 | 1.00 | ||||||
| Negative-positive | 183 | 1.40 | 1.08-1.81 | .099 | 183 | 1.34 | 1.05-1.71 | .017 | 183 | 1.23 | 0.84-1.79 | .290 |
| Positive-negative | 479 | 1.20 | 0.98-1.47 | .081 | 479 | 1.16 | 0.96-1.40 | .140 | 478 | 1.27 | 0.95-1.70 | .100 |
| Positive-positive | 539 | 1.13 | 0.93-1.38 | .210 | 539 | 1.11 | 0.93-1.34 | .260 | 539 | 1.13 | 0.85-1.51 | .400 |
| Patient KIR-ligand status | ||||||||||||
| C1C1 or C1C2 | 2244 | 1.00 | 2244 | 1.00 | 2242 | 1.00 | ||||||
| C2C2 | 402 | 1.26 | 1.09-1.47 | .027 | 402 | 1.30 | 1.13-1.50 | .002 | 402 | 1.14 | 0.91-1.42 | .240 |
| Donor origin | ||||||||||||
| National | 2091 | 1.00 | 2091 | 1.00 | 2090 | 1.00 | ||||||
| International | 555 | 1.23 | 1.07-1.41 | .004 | 555 | 1.23 | 1.08-1.41 | .002 | 554 | 1.29 | 1.06-1.56 | .010 |
| Predictor . | OS . | DFS . | TRM . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n . | HR . | 95% CI . | P . | n . | HR . | 95% CI . | P . | n . | HR . | 95% CI . | P . | |
| HLA match | ||||||||||||
| 10/10 | 1511 | 1.00 | 1511 | 1.00 | 1509 | 1.00 | ||||||
| 9/10 | 803 | 1.26 | 1.11-1.43 | <.001 | 803 | 1.17 | 1.03-1.32 | .012 | 803 | 1.38 | 1.15-1.66 | <.001 |
| 8/10 | 247 | 1.86 | 1.51-2.29 | <.001 | 247 | 1.71 | 1.40-2.09 | <.001 | 247 | 2.13 | 1.64-2.77 | <.001 |
| Disease stage | ||||||||||||
| Early | 1047 | 1.00 | 1047 | 1.00 | 1047 | 1.00 | ||||||
| Intermediate | 922 | 1.46 | 1.26-1.70 | <.001 | 922 | 1.50 | 1.31-1.73 | <.001 | 921 | 1.60 | 1.29-1.98 | <.001 |
| Advanced | 677 | 2.18 | 1.87-2.54 | <.001 | 677 | 2.17 | 1.88-2.51 | <.001 | 676 | 1.97 | 1.59-2.43 | <.001 |
| Patient age (years) | ||||||||||||
| <40 | 734 | 1.00 | 734 | 1.00 | 733 | 1.00 | ||||||
| 40-55 | 951 | 1.39 | 1.19-1.62 | <.001 | 951 | 1.30 | 1.13-1.50 | <.001 | 950 | 1.60 | 1.27-2.00 | <.001 |
| >55 | 961 | 1.76 | 1.48-2.10 | <.001 | 961 | 1.56 | 1.33-1.82 | <.001 | 961 | 1.85 | 1.43-2.40 | <.001 |
| Donor-recipient gender | ||||||||||||
| Male-male | 1227 | 1.00 | 1227 | 1.00 | 1227 | 1.00 | ||||||
| Male-female | 362 | 1.13 | 0.95-1.34 | .170 | 362 | 1.15 | 0.98-1.35 | .087 | 362 | 0.96 | 0.75-1.22 | .720 |
| Female-female | 393 | 0.91 | 0.77-1.08 | .270 | 393 | 0.92 | 0.78-1.08 | .300 | 392 | 0.67 | 0.51-0.88 | .004 |
| Female-male | 664 | 0.99 | 0.86-1.14 | .870 | 664 | 0.98 | 0.86-1.13 | .810 | 663 | 0.91 | 0.74-1.12 | .390 |
| Year of transplantation | ||||||||||||
| 1997-2003 | 455 | 1.00 | 455 | 1.00 | 455 | 1.00 | ||||||
| 2004-2010 | 2191 | 0.76 | 0.64-0.91 | .002 | 2191 | 0.79 | 0.67-0.93 | .005 | 2189 | 0.61 | 0.49-0.76 | <.001 |
| Donor-recipient CMV status* | ||||||||||||
| Negative-negative | 520 | 1.00 | 520 | 1.00 | 520 | 1.00 | ||||||
| Negative-positive | 183 | 1.40 | 1.08-1.81 | .099 | 183 | 1.34 | 1.05-1.71 | .017 | 183 | 1.23 | 0.84-1.79 | .290 |
| Positive-negative | 479 | 1.20 | 0.98-1.47 | .081 | 479 | 1.16 | 0.96-1.40 | .140 | 478 | 1.27 | 0.95-1.70 | .100 |
| Positive-positive | 539 | 1.13 | 0.93-1.38 | .210 | 539 | 1.11 | 0.93-1.34 | .260 | 539 | 1.13 | 0.85-1.51 | .400 |
| Patient KIR-ligand status | ||||||||||||
| C1C1 or C1C2 | 2244 | 1.00 | 2244 | 1.00 | 2242 | 1.00 | ||||||
| C2C2 | 402 | 1.26 | 1.09-1.47 | .027 | 402 | 1.30 | 1.13-1.50 | .002 | 402 | 1.14 | 0.91-1.42 | .240 |
| Donor origin | ||||||||||||
| National | 2091 | 1.00 | 2091 | 1.00 | 2090 | 1.00 | ||||||
| International | 555 | 1.23 | 1.07-1.41 | .004 | 555 | 1.23 | 1.08-1.41 | .002 | 554 | 1.29 | 1.06-1.56 | .010 |
Correlation of clinical predictors with OS, DFS, and TRM. Significance level set to P < .01. Significant results are bold.
Performed as subanalysis with separate model.
OS of patients with 10/10 matched donors, grouped by transplantations performed with national donors (solid line) or international donors (dashed line), logrank P = .047. Of the 213 patients with international donors, 211 were of white and 2 were of Asian ethnicity.
OS of patients with 10/10 matched donors, grouped by transplantations performed with national donors (solid line) or international donors (dashed line), logrank P = .047. Of the 213 patients with international donors, 211 were of white and 2 were of Asian ethnicity.
Predicted KIR-ligand incompatibility in HLA-C mismatched cases did not influence outcomes, but a patient HLA-C2C2 phenotype was associated with worse outcomes, both in matched and mismatched cases (Table 7).
HLA phenotype classification indicated a significantly higher proportion of patients with rare HLA phenotypes among recipients of grafts from an international donor compared with patients with a national donor (n = 71 of 555, 12.8% vs n = 85 of 2091, 4.1%; χ2; P < .001). No significant impact was seen for ATG treatment, CMV status, and graft source on survival outcomes.
Discussion
We confirmed that a distinction between allele and antigen mismatches (as indicators for the amount of structural disparities between HLA molecules) does not correlate with differential outcome. It is rather the presence or absence of any HLA mismatch at the allele level that must be considered as clinically important for donor selection.23,24 This supports the current practice of allele-level HLA typing and matching for HSCT. Among allelic differences, permissive mismatch combinations might exist. This applies especially to HLA-C, where HRs for single-allele mismatches are constantly lower across different studies compared with single-antigen mismatched cases. Further work and significantly higher numbers of transplants are necessary to elucidate such combinations.25
An adverse effect of a patient C2C2 KIR-ligand phenotype was found, which is in accordance with previous findings.6,26,27
In our study, patients transplanted with donors from the national registry showed better survival than patients with international donors. Based on the analysis of PGF, factors determining transplant product quality do not appear to be a reason for this observation. Usually, an international donor is only selected if no well-matched national donor is available. Our findings indicate that the fraction of patients with rare HLA phenotypes is significantly higher in patients who received grafts from international donors. Adverse outcome has been reported for patients with rare HLA phenotypes by Jöris et al.28 Differences in the extended haplotype (ie, immunologically relevant loci on chromosome 6 beyond the routinely typed classical HLA loci) may influence outcome even if patients and donors are 10/10 matched as suggested by Petersdorf and Hansen.29 Such differences may be more frequent in patients with rare HLA phenotypes and when donors from a different ethnic background are selected.30 In addition, non-HLA polymorphisms may influence transplant outcome,31,32 and further diversity may be added if donors from a different immunogenetic background are selected. The national vs international donor effect may be a phenomenon that specifically applies to the German setting, because for most of the patients, a national donor can be found, possibly segregating the remaining patients as an immunogenetic risk population. The situation is quite different in countries with smaller donor registries or a more HLA-diverse population, which predominantly use international donors. Additional risk may be conferred by prolonged searches, possibly leading to progression of the disease. This finding, although significant, does not alter search algorithms as the effect of a single HLA mismatch is stronger than the hazard added by selecting an international donor (Table 7). Thus, a matched international donor would still be preferred over a single mismatched national donor.
In our analysis, we found a trend toward higher risk for HLA-DQB1 mismatches, which could be mainly attributed to HLA-DQB1 antigen differences. This did not reach statistical significance, as numbers of HLA-DQB1 mismatches were relatively small due to linkage disequilibrium with well-matched HLA-DRB1. Furthermore, DRB*3,4,5 polymorphisms in non–haplotype-matched unrelated HSCT might confer additional risk in the presence of an HLA-DQB1 mismatch.33 Future studies in an extended cohort and typing for HLA-DRB*3,4,5 may allow a better understanding of the relative impact of HLA-DQB1 allele or antigen mismatches.
Due to weak linkage disequilibrium between DPB1 and the HLA loci we analyzed, the fraction of donors matched for DPB1 or with a permissive or nonpermissive mismatch should be very similar in all subgroups of our study population.34 Therefore, we may assume that relative risk estimates obtained in our analysis are still accurate, although we could not include the HLA-DPB1 matching status in our multivariate analysis.
Analysis of mismatch combinations revealed higher HRs with increasing numbers of disparities and suggests additive effects of these incompatibilities.35 In general, the impact of HLA mismatching on TRM is stronger and the effect on DFS is smaller compared with HRs observed in OS. Similarly to other studies, there was no significant effect of HLA incompatibilities on relapse incidence.12,13 The competing event relapse (or death from relapse) therefore slightly blurs the analysis of OS and in particular of DFS, leading mostly to lower HRs in these analyses within the corresponding mismatch groups. PGF was not significantly associated with HLA mismatching in our study cohort. Findings in nonmalignant diseases differ regarding this issue.36 The graft source (bone marrow vs PBSCs) did not influence survival end points, which is consistent with a previous observation in patients with acute leukemia.37 The cohort composition with respect to graft source (limited to bone marrow and PBSCs) in large studies probably does not impair comparability of results.12,13 The vector of mismatches was not considered, because no significant differences in outcome for the end points analyzed in our study were reported if HLA mismatches were grouped accordingly.14
We saw higher HRs for patients with 2 incompatibilities when these involved a combined HLA class I+II mismatch, contrary to a double class I or II mismatch situation. Although the direct comparison of these 2 groups did not reach statistical significance, this finding is of interest, as the majority of HLA class II disparities in our study cohort affected HLA-DQB1. The pattern of HRs for double HLA mismatch combinations in our study suggests that despite its controversial impact in single mismatch cases, HLA-DQB1 appears to have a significant effect in double mismatched situations and should be considered accordingly in donor selection. The higher risk for combined HLA class I and II mismatches is descriptive at present, as direct comparison with double HLA class I or double HLA class II mismatches did not reach statistical significance. This may be due to insufficient numbers in our study and should be re-evaluated in a larger study. Nevertheless, pending such a study, it might be advisable to avoid combined HLA class I and II incompatibilities if alternatives are at hand.
In our analysis, HLA-A, and particularly HLA-B, mismatches, were strongly associated with adverse outcome. Comparisons between different other large studies, which were conducted by the CIBMTR, did not always show similar results across these cohorts.11-13 Reasons for such differences may be statistical limitations due to sample size or variations regarding patient characteristics, especially patient age, diagnosis, follow-up-period, changes in treatment over time, and, in particular, the ethnic composition of the different study cohorts.
The relatively high number of HLA-C antigen mismatches in our study can be explained mainly by the fact that HLA typing and matching requirements before May 2005 in Germany considered HLA-A and -B at low resolution only. Retrospective typing disclosed therefore a considerable number of allele and antigen mismatches for HLA-C. In addition, confirmatory typing in general is more likely to show mismatches at the HLA-C locus, as the majority of unrelated donors are usually not typed for HLA-C on registration.
HLA-B antigen mismatches are rare because if a mismatch search had to be initiated, often an HLA-A mismatch was preferred over one for HLA-B. This is due to the strong HLA-B/C linkage disequilibrium, which would often reveal an additional HLA-C mismatch if an HLA-B mismatch was selected for further testing.38
In all transplants included in this analysis, HLA class II molecules were high-resolution tested at the time of confirmatory typing, explaining the low number of HLA-DRB1 mismatches observed, in particular for HLA-DRB1 antigens, as such mismatches were totally avoided if possible. Similar HLA mismatch patterns were observed in other large studies.11-13 In contrast to these studies, the vast majority of patients in our cohort were transplanted in recent years (2006-2010; 66.4%). As a consequence, most of the patients included in our study were transplanted with PBSCs as a graft source (86.9%). Retrospective studies published thus far by the CIBMTR investigated the impact of HLA matching on HSCT outcome, mostly including patients transplanted before 2007 with bone marrow11,12 or PBSCs only13 and with patient diagnoses being restricted to AML, ALL, CML, and MDS. Our study is currently the first to analyze the impact of high-resolution HLA matching on outcome of HSCT in a large European cohort including >2500 transplantations.
In previous studies, the problem of possible confounding of multiple HLA mismatches was addressed by performing subanalyses of single HLA mismatches. We performed such an analysis in our cohort and present the results in Table 4. In our view, the advantages of elimination of confounding in such a subanalysis are offset by the limitations conferred by multiple comparisons and low numbers within certain subgroups. Comparison of the single mismatch subanalysis (Table 4) with the model including all incompatible cases (Table 3) demonstrates good consistency of estimates in the groups with substantial numbers (n > 50), showing that it is feasible to create survival models with inclusion of cases with >1 mismatch, possibly leading to more robust estimates.
Our study included patients with non-Hodgkin lymphoma and multiple myeloma. These disorders have substantially contributed to the transplant activities in Germany in recent years. Most of the transplantations in our study have been performed between 2006 and 2010 and draw a more recent clinical picture than other published large studies, which thus far include patients transplanted before 2007.11-13 On the other hand, inclusion of more recent transplants in our study resulted in shorter median follow-up compared with other studies. Nevertheless, because most adverse events occur within the first 2 years following HSCT, the observations of our study can be considered accurate.
Improvements in therapy have made conditioning regimens safer, especially for older patients. As a consequence, the proportion of older patients undergoing transplantation is rising, and this might influence outcome results. In addition, the immunogenetic background of a study population could influence results, as HLA differences in genetically diverse populations may confer different risks than in genetically more homogenous cohorts.25
Limitations of our study are that analysis of graft versus host disease was not possible and data about ATG treatment and CMV status were partly missing.
Despite the mentioned methodological differences, we could confirm patterns that were described in previous reports: HLA-C antigen mismatches are strongly predictive for adverse outcome, whereas the impact of HLA-C allele mismatch remains uncertain.11-13,39 Nevertheless, when directly compared with HLA-A and HLA-B mismatches, HRs are lower for HLA-C mismatches, especially regarding OS. This observation is consistent across different studies and can be considered robust in view of the high numbers of HLA-C antigen mismatches analyzed. It might indicate manifestation of beneficial KIR receptor-ligand incompatibilities in a subset of patients with HLA-C mismatches. We saw a strong effect of DRB1 allele mismatching (Table 3), especially regarding its impact on TRM. The low number of HLA-DRB1 mismatches in our analysis, however, limits the interpretation of these results.
Our data support HLA-A/B/C/DRB1 typing and matching as minimum requirement for donor search. In general, allele and antigen mismatches should be treated as equivalent, however, keeping in mind uncertainty of the impact of HLA-C-allele mismatches. A 10/10 matched situation is desirable; 9/10 or even 8/10 matched donors may be acceptable, in particular if a single HLA-DQB1 allele mismatch is present, when no better matched unrelated donors are available.40 In cases with more mismatches, a careful risk/benefit evaluation is warranted and alternatives, such as cord blood or haploidentical transplantation, should be considered, especially if rapid transplantation is required.37,41,42 If >1 mismatch is to be taken into account, it is advisable to avoid a combined HLA class I/II incompatibility.
Furthermore, in cases where initially no perfectly matched donor can be found, the clinical risk for disease progression must be weighed against the probability of finding a better-matched donor in a prolonged search. Depending on the clinical situation of the patient, in many cases, a mismatched transplantation performed in early disease stage may be a better choice for the patient than a transplant in advanced disease, even if this would eventually be performed with a better-matched donor.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the following colleagues for contribution of their patients: Dietger Niederwieser, Vucinic Vladan, and Anja Bühner (Department of Hematology/Oncology, University of Leipzig, Leipzig, Germany); Donald Bunjes, Stefanie von Harsdorf, and Lucia Missel (Department of Internal Medicine III, University of Ulm, Ulm, Germany); Herr Wolfgang (now Regensburg), Ralf-Georg Meyer, Karin Kober, and Birgit Waldeck (Department of Medicine III, Johannes Gutenberg-University Mainz, Mainz, Germany); Martin Gramatzki and Marianne Helweg (Division of Stem Cell Transplantation and Immunotherapy, 2nd Department of Medicine, University of Kiel, Kiel, Germany); Rainer Schwerdtfeger, Herrad Baurmann, and Susanne Eichler (Centre for Bone Marrow and Blood Stem Cell Transplantation, Deutsche Klinik für Diagnostik, Wiesbaden, Germany); Renate Arnold and Sabine Diehl (Hematology/Oncology Department, Charité Campus Virchow, Berlin, Germany); Hermann Einsele, Gernot Stuhler, Ulrich Grigoleit Götz, Stephan Mielke, and Heidrun Bönig (Department of Medicine II, University Hospital Würzburg, Würzburg, Germany); Gerald Wulf, Lorenz Trümper, and Anke Hesse (Department of Hematology/Oncology, Georg-August-University Göttingen, Göttingen, Germany); Michael Pfreundschuh, Gerhard Held, and Renate Martin (Department of Internal Medicine I, Universitätsklinikum des Saarlandes, Homburg, Germany); Norbert Schmitz, Bertram Glass, Matthias Zeis, and Susanne Dingeldein (Department of Hematology/Oncology, Asklepios Klinik St. Georg, Hamburg, Germany); Hannes Wandt and Kerstin Schäfer-Eckart (Medical Clinic 5, Hematology and Oncology, Klinikum Nuremberg, Nuremberg, Germany); Christian Peschel (Medical Clinic III, Hematology and Oncology, University Clinic Rechts der Isar, Munich, Germany); Andreas Mackensen (Medical Clinic 5, Hematology and Oncology, University of Erlangen-Nuremberg, Erlangen, Germany); Walter Erich Aulitzky, Martin Kaufmann, and Nicole Eier (Hematology/Oncology Department, Robert-Bosch Clinic, Stuttgart, Germany); Christian Junghanss and Kersten Borchert (Hematology/Oncology Department, University of Rostock, Rostock, Germany); Jochen Casper (Hematology/Oncology Department, Clinic Oldenburg, Oldenburg, Germany); Nicolaus Kröger, Axel Zander, and Gitta Amtsfeld (Clinic for Stem Cell Transplantation, University Medical Center Hamburg- Eppendorf, Hamburg, Germany); Ernst Holler (Department of Hematology/Oncology, University of Regensburg, Regensburg, Germany); Wolfgang Heit, Mohammed Wattad, and Julia Mancione (Department of Hematology/Oncology, Evangelisches Kaufmännische Krankenkasse Essen-Werden, Essen, Germany); Bernd Hertenstein and Stefan Kaun (Hematology/Oncology Department, Clinic Bremen-Mitte, Bremen, Germany); Harald Biersack (now Lübeck) (Hematology/Oncology Department, Clinic Idar-Oberstein, Idar-Oberstein Germany); Hans Martin, Dieter Hölzer, and Hubert Serve (Medical Clinic II, Hematology and Oncology, J.W.-Goethe-University, Frankfurt/Main, Germany); Hans-Joachim Schmoll (Department of Oncology/Hematology, Martin Luther University, Halle-Wittenberg, Halle, Germany); Mark Ringhoffer, Martin Bentz, and Julie Schiefer (Hematology/Oncology Department, Clinic Karlsruhe, Karlsruhe, Germany); James Beck and Bernd Gruhn (Pediatric Hematology and Oncology, Jena University Hospital, Children’s Clinic, Jena, Germany); Paul- Gerhardt Schlegel (Pediatric Hematology and Oncology, University Children’s Hospital Würzburg, Germany); Harald Biersack and Rudina Marx (Department of Hematology/Oncology, University Hospital Schleswig-Holstein, Lübeck, Germany); Wolf-Karsten Hofmann, Stefan Klein, and Christine Folz (Hematology/Oncology Department, University Hospital Mannheim, Mannheim, Germany); Marlene Fischer, Sabine Simon, Renate Hanold, and Martina Baumann (Transplantation Immunology Department, German Red Cross Blood Transfusion Service, Baden-Wuerttemberg–Hessen, and Institute of Transfusion Medicine, University of Ulm, Germany); Alexander Schmidt (Deutsche Knochenmarkspenderdatei); and Susanne Morsch (Stiftung Knochenmark- und Stammzellspende Deutschland).
This work was supported by the German José Carreras Leukaemia Foundation (grant DJCLS 11/10) and the German Red Cross Blood Transfusion Service, Baden-Wuerttemberg/Hessen.
Authorship
Contribution: D.F., C.M., H.S., and J.M. contributed in study design, data analysis/interpretation, and writing of the manuscript; V.V., D.B., W.H., M.G., R.S., R.A., H.E., G.W., M.P., and B.G. treated patients, provided clinical data, and coedited the manuscript; and K.S. contributed in HLA typing and coedited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joannis Mytilineos, IKT Ulm, BSD BaWü-He, and University of Ulm, Department of Transplantation Immunology, Helmholtzstrasse 10, 89081 Ulm, Germany; e-mail: j.mytilineos@blutspende.de.