Key Points
DNA methylation changes during the development of DS-AMKL occur in sequential waves of opposing losses and gains of methylation.
Each wave of DNA methylation abnormalities targets specific gene networks that contribute to distinct biological features of the disease.
Abstract
Acute megakaryoblastic leukemia (AMKL) is more frequently observed in Down syndrome (DS) patients, in whom it is often preceded by a transient myeloproliferative disorder (TMD). The development of DS-TMD and DS-AMKL requires not only the presence of the trisomy 21 but also that of GATA1 mutations. Despite extensive studies into the genetics of DS-AMKL, the importance of epigenetic deregulation in this disease has been unexplored. We performed DNA methylation profiling at different stages of development of DS-AMKL and analyzed the dynamics of the epigenetic program. Early genome-wide DNA methylation changes can be detected in trisomy 21 fetal liver mononuclear cells, prior to the acquisition of GATA1 mutations. These early changes are characterized by marked loss of DNA methylation at genes associated with developmental disorders, including those affecting the cardiovascular, neurological, and endocrine systems. This is followed by a second wave of changes detected in DS-TMD and DS-AMKL, characterized by gains of methylation. This new wave of hypermethylation targets a distinct set of genes involved in hematopoiesis and regulation of cell growth and proliferation. These findings indicate that the final epigenetic landscape of DS-AMKL is the result of sequential and opposing changes in DNA methylation occurring at specific times in the disease development.
Introduction
Acute megakaryoblastic leukemia (AMKL) is a rare form of acute myeloid leukemia (AML), accounting for only 1% to 2% of all de novo AMLs and as much as 10% of pediatric AML.1,2 Moreover, its incidence is several-hundredfold higher in patients with Down syndrome (DS) than in children without DS.3 Four to 5% of DS fetuses develop a preleukemic transient myeloproliferative disorder (DS-TMD), diagnosed at the time of birth, that spontaneously resolves within a few weeks in the majority of patients.4,5 Within the first 4 years of life, approximately 20% of DS cases that develop TMD in utero will develop AMKL (DS-AMKL).6-9
DS-TMD and DS-AMKL invariably present with acquired mutations in the GATA1 gene that encodes a master regulator of erythromegakaryocytic development. These mutations arise in utero and result in the expression of a short isoform of the GATA1 protein, known as GATA1s.6,10 Oncogenic cooperation between GATA1 mutations and trisomy 21 is associated with the development of DS-TMD and DS-AMKL. However, several observations in animal models11-13 and in human specimens14-18 strongly suggest that additional genetic events are required to develop leukemia, but it is still unclear how they may contribute to the phenotypic progression to AMKL. Moreover, the mechanisms behind the spontaneous remission of DS-TMD and the subsequent progression to DS-AMKL are still not fully understood.
Abnormalities in DNA methylation are a hallmark of AML and have been shown to display subtype specificity.19,20 Moreover, murine models have been used to show that impaired DNA methyltransferase 1 activity results in a decrease in the leukemogenic potential of the MLL-AF9 fusion oncogene.21,22 Notably, despite the extensive research into the genetics of DS and DS-related leukemias, little is known about the state of DNA methylation in TMD and DS-AMKL and how epigenetic changes might contribute to malignant transformation. In order to address this question we performed genome-wide DNA methylation profiling on specimens obtained at different stages of the disease and analyzed the dynamics of epigenetic reprogramming in the development of DS-AMKL.
Methods
Samples
Fetal liver mononuclear cells (MNC) with either a normal karyotype or trisomy 21 were obtained from 2nd-trimester abortions. MNC cells were isolated using a standard Ficoll separation method from a cohort of 7 DS-associated acute megakaryoblastic leukemias (DS-AMKL), 6 cases of transient myeloproliferative disorder (DS-TMD), and 8 acute megakaryoblastic leukemias from adults without DS (non–DS-AMKL) received through the Children's Oncology Group and Eastern Cooperative Oncology Group leukemia tissue banks. Supplemental Table 1 (found on the Blood Web site) describes the patients’ characteristics. The diagnosis of megakaryoblastic leukemia was established by morphology and confirmed by immunophenotyping. Institutional review board approval was obtained at Weill Cornell Medical College, University of Pennsylvania, Children’s Hospital of Michigan, and the University of Michigan. Eight specimens of human CD34+ bone marrow cells were provided by the Stem Cell and Xenograft Core Facility of the University of Pennsylvania or purchased from AllCells (Emeryville, CA). These studies were performed in accordance with the Declaration of Helsinki protocols.
DNA methylation by HELP
Genomic DNA was extracted using the Puregene kit from Qiagen (Valencia, CA) or a standard phenol-chloroform isolation followed by ethanol precipitation. HELP (HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction) representations were prepared from 100 ng of DNA as previously described23,24 and hybridized onto a custom long-oligonucleotide microarray designed to cover HpaII amplifiable fragments (HAF) at gene promoter regions. HG19 array was manufactured by Roche-NimbleGen, design ID: 100128_HG19_MKF_HELP_ChIP, covering 117 521 HAFs annotated to 29 606 unique RefSeq genes. MM9 array was manufactured by Roche-NimbleGen, design ID: 100520_MM9_KF_Meth, covering 119 260 HAFs annotated to 24 377 unique RefSeq genes. All samples were processed and hybridized at the Weill Cornell Medical College Epigenomics Core Facility. Labeling, hybridization, and scanning were performed as previously described.25 Quality control and array normalization were performed following our previously described methods,26 and any arrays that did not pass our quality control were excluded from the analysis. In order to correct for the introduction of hybridization batch effect, we subjected each individual channel to batch correction using the ComBat algorithm,27 and the adjusted channels were then used to calculate the final HpaII:MspI ratio.
Microarray analysis
Supervised analysis for the different group comparisons was performed using Bioconductor and the R 2.15 statistical software, and the genefilter, stats, and gplots packages. Pairwise comparisons were performed using Student t test, followed by adjustment of the P value with the Benjamini-Hochberg approach for multiple comparisons.28 Significant differences were defined as those with adjusted P value < .05 and an absolute difference in log2(HpaII/MspI) >2, which corresponds to a minimal methylation difference of 30%.
Gene expression arrays
Normalized gene expression values for DS-AMKL, non–DS-AMKL, and DS-TMD specimens were downloaded from the GEO repository from the study by Bourquin et al29 (accession number GSE4119). Gene set enrichment analysis was performed using the Molecular Signatures Database (MSigDB) c1 collection (positional).30
Network and pathway analysis
Network and pathway analysis using the Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA) was performed using the signatures identified for the different comparisons and the Ingenuity Knowledge database as a background reference. Network significance was scored as the negative exponent of the right-tailed Fisher exact test used to estimate the likelihood that the network-eligible molecules that are part of a network are found therein by random chance alone.
Trisomic mice cells
Bone marrow cells were isolated from the long bones of either Ts1Rhr or wild-type mice at >12 months of age from 3 independent experiments. CD41+ cells were isolated from red cell lyzed whole bone marrow cells by fluorescence-activated cell sorter using an anti-CD41 antibody (Emfret, Germany). The animal study was approved by the Northwestern University Institutional Animal Care and Use Committee.
Accession numbers
All microarray data generated for this study was deposited in the Gene Expression Omnibus database under accession number GSE46167.
Results
With the goal of studying epigenetic deregulation associated with malignant transformation in the hematopoietic system of Down syndrome patients, we performed genome-wide DNA methylation analysis using the HELP assay23,24 on a custom high-density oligonucleotide array designed to query to methylation status at ∼200 000 distinct CpG sites (annotated to 29 606 RefSeq genes). Given that the HELP assay uses as a reference an MspI-restricted representation of the patient’s own genome, which serves as an internal control for copy number and sequence variants,24,31 the presence of a trisomic chromosome in some of the specimens in this study is automatically controlled for and does not have an impact on the ability of the assay to detect methylation differences, even within chromosome 21. In order to evaluate epigenetic changes during the different stages of disease development, the following samples were collected: fetal liver (FL) MNC with normal karyotype (FL-MNC, n = 4), trisomy 21 FL MNC (Tri21 FL-MNC, n = 4), MNC from Down syndrome patients with either transient myeloproliferative disorder (DS-TMD, n = 6) or acute megakaryoblastic leukemia (DS-AMKL, n = 7), and MNC cells from patients with adult non–DS-AMKL (n = 8).
DS-AMKL is epigenetically distinct from non–DS-AMKL
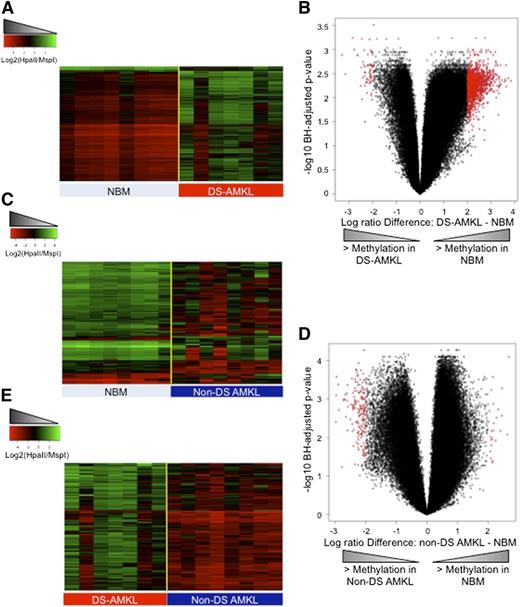
DNA methylation patterns in AML are not uniform across all subtypes, and distinct epigenetic profiles characterize the different cytogenetic and molecular subtypes of AML.19,20 Given this fact, the first goal of our study was to define the nature and extent of epigenetic abnormalities in leukemic blasts of DS-AMKL. Therefore, we compared the DNA methylation status of DS-AMKL blasts to that of normal CD34+ hematopoietic cells isolated from the bone marrows of healthy donors (n = 8). At a false discovery rate (FDR) of 5% and a minimum methylation difference of at least 30% between the 2 groups (absolute log2(HpaII/MspI) difference >2), we identified 1396 differentially methylated regions (DMRs), of which 96% (n = 1339) were aberrantly hypomethylated in the leukemic samples in relation to the normal CD34+ cells (Figure 1A-B; supplemental Table 2). In order to ensure that the detection of such profound hypomethylation was not due to an artifact generated by the comparison of the more fetal-like hematopoiesis of the DS-AMKL to a normal cohort obtained from adult CD34+ cells, we also compared DS-AMKL specimens to control FL-MNCs and found an identical pattern of profound hypomethylation in the DS-AMKLs in relation to FL-MNCs (supplemental Figure 1). By contrast, a supervised analysis between non–DS-AMKL and normal CD34+ cells revealed that epigenetic abnormalities in non–DS-AMKL are much less widespread than in DS-AMKL, with only 191 DMRs in relation to normal CD34+ cells detected at the same significance cutoff. Furthermore, this aberrant methylation in non–DS-AMKL is characterized by an opposite pattern to that seen in DS-AMKL, with a predominance of hypermethylated DMRs (87% hypermethylated DMRs vs 13% hypomethylated DMRs) (Figure 1C-D; supplemental Table 3).
DS-AMKL and non–DS-AMKL are epigenetically distinct. (A) Heatmap representation of differentially methylated regions between normal CD34+ cells (NBM) and DS-AMKL blasts. Each row represents a genomic region, and each column represents a sample. (B) Volcano plot representing the comparison of DS-AMKL blasts to normal CD34+ cells. X-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (C) Heatmap representation of differentially methylated regions between normal CD34+ cells (NBM) and non–DS-AMKL blasts. Each row represents a genomic region, and each column represents a sample. (D) Volcano plot representing the comparison of non–DS-AMKL blasts to normal CD34+ cells. X-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (E) Heatmap representation of differentially methylated regions between non–DS-AMKL and DS-AMKL blasts. Each row represents a genomic region, and each column represents a patient sample.
DS-AMKL and non–DS-AMKL are epigenetically distinct. (A) Heatmap representation of differentially methylated regions between normal CD34+ cells (NBM) and DS-AMKL blasts. Each row represents a genomic region, and each column represents a sample. (B) Volcano plot representing the comparison of DS-AMKL blasts to normal CD34+ cells. X-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (C) Heatmap representation of differentially methylated regions between normal CD34+ cells (NBM) and non–DS-AMKL blasts. Each row represents a genomic region, and each column represents a sample. (D) Volcano plot representing the comparison of non–DS-AMKL blasts to normal CD34+ cells. X-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (E) Heatmap representation of differentially methylated regions between non–DS-AMKL and DS-AMKL blasts. Each row represents a genomic region, and each column represents a patient sample.
Next we performed a direct comparison of the DNA methylation profiles of DS-AMKL and non–DS-AMKL and identified 280 differentially methylated regions (FDR <5% and methylation difference ≥30%) between the 2 types of AMKL (supplemental Table 4). These DMRs were annotated to 267 unique genes. This comparison revealed that DS-AMKL is not only hypomethylated in relation to normal CD34+ cells, but it is also significantly hypomethylated relative to non–DS-AMKL (Figure 1E). Differential methylation between these 2 forms of AMKL-targeted genes is involved in key pathways such as the Breast Cancer–dependent DNA damage response pathway (CHEK1, E2F1, FANCM, and HLTF), the extracellular signal-regulated kinase (ERK5) signaling pathway (EGF, MEF2B, PRKCZ, and SGK1), and the B-cell translocation gene (BTG)–dependent regulation of cell cycle (BTG2, E2F1, and PPP2R2C), indicating that these differences may play a functional role in the development of AMKL in these 2 distinct biological contexts. In order to ensure that any differences in DNA methylation seen between non–DS-AMKL and DS-AMKL were not simply a reflection of age-related changes in DNA methylation, we next performed a direct comparison of the DNA methylation profiles of FL-MNC and normal CD34+ cells. This analysis revealed that FL-MNC tend to have higher levels of promoter methylation than do normal CD34+ bone marrow cells, with 81% (1599 out of 1965) of the DMRs being hypermethylated in FL-MNC in relation to the normal CD34+ cells (supplemental Figure 2). Notably, only 7 of 366 DMRs with greater methylation in normal CD34+ cells overlapped with the hypermethylated DMRs from non–DS-AMKL (Fisher test P value: not significant). Thus, these higher levels of DNA methylation in FL-MNC cells lead us to conclude that the hypermethylation seen in non–DS-AMKL specimens in relation to DS-AMKL cannot be interpreted as an age-dependent effect reflecting the different age groups affected by these 2 disorders.
Aberrant hypomethylation in Down syndrome is detected early on in fetal liver mononuclear cells
In order to understand how aberrant DNA methylation patterns become established in DS-AMKL, we investigated the dynamic changes in these patterns throughout the evolution of development of this disease. We first determined whether the sole presence of an isolated extra copy of chromosome 21 is sufficient to impact the epigenome prior to the acquisition of GATA1 and other mutations affecting TP53 and JAK3. For this we compared the DNA methylation status at promoter regions of FL-MNCs with normal karyotype to those obtained from fetuses harboring trisomy 21. This analysis revealed marked differences in the genome-wide DNA methylation profiles of trisomic cells in relation to the control cells. At a significance cutoff of an FDR <5% and methylation difference ≥30%, we identified 846 DMRs annotated to 964 unique Refseq genes (supplemental Table 5). Of these DMRs, 99.4% (841 of 846) consisted of regions in which there was a loss of DNA methylation in the Tri21 FL-MNCs in comparison with the control FL-MNCs (Figure 2A-B). Analysis of the chromosomal locations of the DMRs revealed that hypomethylated regions in Tri21 FL-MNC were not restricted to chromosome 21 but instead were distributed across all chromosomes. Only chromosome 17 showed a statistically significant enrichment of DMRs over background (Benjamini-Hochberg–adjusted Fisher test, P = .024), whereas no significant enrichment was detected for chromosome 21 (Figure 2C).
Trisomy 21–associated hypomethylation is detected early on in fetal MNC. (A) Heatmap representation of differentially methylated regions between Tri21 FL-MNC and control FL-MNC. Each row represents a genomic region, and each column represents a patient sample. (B) Volcano plot representing the comparison of Tri21 to control FL-MNC. The x-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (C) Stacking barplots representing the relative proportion of chromosomes 21 (left) and 17 (right) in the whole array and the Tri21 FL-MNC DMRs. P values for Fisher test followed by correction for multiple comparisons using the Bonferroni method. (D) Heatmap representation of regions with at least 25% difference in methylation between Ts1Rhr and control CD41+ cells. Each row represents a genomic region, and each column represents a sample.
Trisomy 21–associated hypomethylation is detected early on in fetal MNC. (A) Heatmap representation of differentially methylated regions between Tri21 FL-MNC and control FL-MNC. Each row represents a genomic region, and each column represents a patient sample. (B) Volcano plot representing the comparison of Tri21 to control FL-MNC. The x-axis represents the difference in DNA methylation between the 2 groups, and the y-axis represents the statistical significance of the differences. Red dots denote DMRs with absolute log ratio difference >2 and FDR <5%. (C) Stacking barplots representing the relative proportion of chromosomes 21 (left) and 17 (right) in the whole array and the Tri21 FL-MNC DMRs. P values for Fisher test followed by correction for multiple comparisons using the Bonferroni method. (D) Heatmap representation of regions with at least 25% difference in methylation between Ts1Rhr and control CD41+ cells. Each row represents a genomic region, and each column represents a sample.
In order to confirm that the observed hypomethylation was dependent on the presence of the trisomic chromosome 21, we performed genome-wide DNA methylation analysis on CD41+ cells isolated from a partial trisomic mouse model (Ts1Rhr)32 that has been recently shown to promote DS-leukemogenesis in vivo.13 Although this system displayed the same trend toward a hypomethylation phenotype as was observed in the human Tri21 FL-MNCs, aberrant methylation in the Ts1Rhr model was not as pronounced as in the human Tri21 FL-MNCs, with only 461 regions showing at least a 25% difference in methylation between the wild-type and trisomic cells. However, despite the difference in magnitude detected, the main effect on methylation was one of predominant hypomethylation in the trisomic cells, with 89% (411 of 461) of DMRs presenting with loss of methylation in these cells in relation to their wild-type counterparts (Figure 2D).
GATA1 mutant DS-TMD blasts acquire additional epigenetic abnormalities
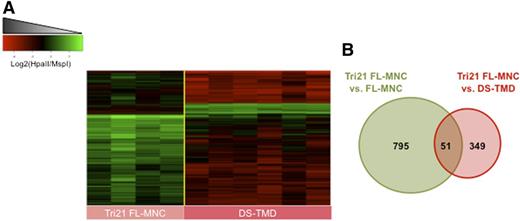
Next we determined whether acquisition of GATA1 mutations, which lead to expression of the shorter isoform GATA1s, and the development of a DS-TMD phenotype have any impact on epigenetic patterns. For this purpose we compared the DNA methylation profiles of Tri21 FL-MNC, none of which carried mutations in GATA1, to those of GATA1s mutant DS-TMD. Detailed analysis of the DMRs acquired during the DS-TMD stage revealed that these additional epigenetic changes were predominantly in the form of gains of DNA methylation. Out of 400 DMRs identified between Tri21 FL-MNCs and DS-TMD blasts (FDR <5% and methylation difference ≥30%), only 8.5% of them (n = 34) corresponded to regions with decreased DNA methylation levels in the DS-TMD specimens. The remaining 366 DMRs (91.5%) all showed an increase in DNA methylation ≥30% at the DS-TMD stage in relation to the Tri21 FL-MNCs (Figure 3A; supplemental Table 6). In order to determine whether the changes seen during the Tri21 FL-MNC to DS-TMD transition represented new epigenetic abnormalities acquired at this stage, or whether they were simply caused by a reversal of the original loss of methylation seen at the Tri21 FL-MNC stage, we compared the 2 epigenetic profiles. We found that only 51 (13%) of the DMRs observed at the DS-TMD stage corresponded to regions that had originally become hypomethylated at the Tri21 FL-MNC stage to later become hypermethylated in the GATA1s mutant DS-TMD stage. The remaining 87% of DMRs represented new epigenetic abnormalities acquired at the DS-TMD stage (Figure 3B). These findings indicate that hypermethylation acquired at the DS-TMD stage occurs in the setting of persisting global hypomethylation, with only a few regions presenting with novel gains in methylation.
Acquisition of GATA1 mutations is accompanied by a wave of hypermethylation changes at distinct loci. (A) Heatmap representation of differentially methylated regions between Tri21 FL-MNC and DS-TMD blasts. Each row represents a genomic region, and each column represents a patient sample. (B) Venn diagram representation of the overlap between the Tr1 FL-MNC vs control FL-MNC DMRs and the Tri21 FL-MNC vs DS-TMD DMRs.
Acquisition of GATA1 mutations is accompanied by a wave of hypermethylation changes at distinct loci. (A) Heatmap representation of differentially methylated regions between Tri21 FL-MNC and DS-TMD blasts. Each row represents a genomic region, and each column represents a patient sample. (B) Venn diagram representation of the overlap between the Tr1 FL-MNC vs control FL-MNC DMRs and the Tri21 FL-MNC vs DS-TMD DMRs.
Sequential methylation changes target biologically relevant gene networks
Given that epigenetic abnormalities associated with the acquisition of GATA1 mutations are not only of a different nature, ie, gains in methylation, but also target a different set of genes than the hypomethylation seen associated with trisomy 21 in FL-MNCs, we sought to better understand the role that these epigenetic changes might play at these different stages. Using the Ingenuity Pathway Analysis software (Ingenuity Systems), we identified the top 10 gene networks targeted by aberrant hypomethylation in Tri21 FL-MNCs and those targeted by aberrant hypermethylation in DS-TMD blasts. Aberrant hypomethylation in trisomic FL-MNCs was associated mainly with gene networks involved in developmental disorders, including cardiovascular, neurological, and endocrine disorders, all of which form part of the developmental abnormalities of Down syndrome (Table 1; supplemental Table 7). Notably, when this analysis was performed using the hypomethylated DMRs identified in the Ts1Rhr mice, we also found an enrichment of gene networks involved in developmental disorders of the cardiovascular, endocrine, musculoskeletal, and digestive systems (supplemental Table 8). On the other hand, aberrant hypermethylation in DS-TMD more frequently targeted gene networks associated with hematological system development and cellular growth and proliferation, as well as cell signaling, cell death, and cell cycle, implicating these epigenetic abnormalities in the development of the leukemic phenotype (Table 2; supplemental Table 9).
Top 10 networks targeted by aberrant hypomethylation in Tri21 FL-MNC
| Number . | Molecules in network . | Top functions . | Score . |
|---|---|---|---|
| 1 | ARID1B, AS3MT, BAI1, BCS1L, C12orf52, C21orf56, CDK2AP2, CUL2, FAM107B, FAM117A, FAM158A, GMDS, HNF4A, IL11RA, JINK1/2, LSR, MREG, MRPL18, NDUFV1, NUCB1, OAZ2, PABPN1, PPP1R3B, PRAF2, RFPL2, SBNO2, SLC17A5, SLMO2, SPATA6, STOML2, TBC1D23, TEF, THOC3, TLE3, VN1R1 | Cardiovascular disease, genetic disorder, neurological disease | 56 |
| 2 | AEBP1, BBC3, CARD11, CD7, CHRFAM7A, EDAR, ELF5, ETS, FABP6, HMG20B, IP6K2, KRT16, KRT17, LRDD, LSP1, NFkB, CHRN, NR1H2, NR1H3, NR1H, PIM3, PLEC, PPP1R13B, RASGRF1, RNF31, ROMO1, RRAS, RXR, SECTM1, SRPX, TRADD, TRAF1, TUBA1C, ZHX2 | Cell death, dermatological diseases & conditions, endocrine system disorders | 45 |
| 3 | GRIA1-4, AP2A1/2, ATP1A2, ATP1A4, BABAM1, CYP11B2, EPB41L1, ERK1/2, FXYD7, GABAR-A, GABRG2, GABRR1, GNAL, GPT2, HSF4, IL27RA, MRPL53, NR5A1, NSF, NTF3, PITPNM1, PRKAG1, PTGDS, RLN3, SOX7, SOX9, SOX10, STX5, TFF2, TH, TMEM109, UCN2, VAPB | Small molecule biochemistry, developmental disorder, gastrointestinal disease | 44 |
| 4 | PSMA/PSMB, PSME/PSMF, ADIPOR1, ADRM1, TUBA1A, AMPK, ASCL2, BARD1, TUBB, CLINT1, CUL7, FES, GP1BA, Hsp70, LRP, MLLT4, MST1R, ORC1, PIK3R1/2, PIK3R6, PLXNB3, PFN1/2, PSENEN, PSME1, PSME2, RAB3B, PSEN1/2, SLC39A4, SNCG, SPDEF, TCP1, TRIM9, UBB, VASP | Cell cycle, dermatological diseases & conditions, embryonic development | 32 |
| 5 | ABLIM1, ACP2, ACTG2, ADAP1, FOS/JUN, CBFA2T3, CGA/B, CGB1, CGB2, CGB7, CGBB, COL1A1/2, NR2F1/2/6, CYP17A1, CYP2D6, DIABLO, FSH, GALP, GPSM1, HAUS4, HCG, HIST1H1A-E, HMBS, MA2K4/10/12, LHB, LIF, MAP2K1-7, RPS6KB1/3, PRKACA/B/G/G1/G2, PTMS, RXRA, TGFB1/2/3, USHBP1, WNT5B, ZBED1 | Cellular movement, reproductive system development & function, nervous system development & function | 30 |
| 6 | CAPN1/10/11/, CAPN3, CASP1-12, CCNE1, CDK2, COBRA1, CRYAA, CCNA1/2, CCNE1/2, CYP27B1, DEFA1, DNAJB8, DNAJC9, E2f, ESRRB, ESR1/2, GPC1, HBG1, HIST2H3C, HIST2H4, Hsp90, Hsp22/Hsp40/Hsp90, HVCN1, KIAA1967, LIN37, OSGIN1,PRKCA/B, PNLIPRP2, SATB1, SF3B2, TFDP1, TFF1, VDR, RXR, ZDHHC11 | Cell cycle, cellular assembly & organization, embryonic development | 30 |
| 7 | ABCC1, ACR, ADCY, ADORA2A, AGAP1, AVPR2, DRD1, PTK2/2B, FOXO1, GHRHR, GIT1, Gpcr, GPR84, GPR157, GPRC5D, INS, KCNAB2, KISS1R, NPFF, P110, PAX4, PEMT, PEPCK, PRKCZ, Rab5, Ras, SRC, TBC1D3F, THRA/B, NTRK1/2/3, USP17, USP17L2, VEGF, VN1R4 | Cell signaling, nucleic acid metabolism, molecular transport | 28 |
| 8 | CBP, AFAP1, PPP3CA/B/C, CALM1, CAMK1G, CAMK2A/B, DTNA, DVL1, EPHB2, ACTA1/2, GATA2, GRIN2D, KSR1, LEFTY2, MAPK1, MYLK3, NCS1, NFATC1-4, GRIN2A-D, PIP5K1C, RHOA-H, RCVRN, REM1, RHOT2, ROCK1/2, SIT1, SLC8A2, SNX33, TNNI3, TNNT3, TPM4, TPM1/2/3/4, TUBA1A-E, UTS2 | Skeletal & muscular system, development & function, tissue morphology, cell-to-cell signaling & interaction | 26 |
| 9 | ACE, ACTA2, Akt, CD68, CD151, CD276, COL1, COL1A1/2, COL4, Cpla2, CSF2, FGA/B/G, FMOD, ITGA5, ITGB5, KLK1/3/B1, LAG3, LAMA1/5, MAP2K1/2, MFAP5, MMP26, PAK1-4, PDGFA-D, PLA2G2F, PMEL, PP2A, PROK1, PRSS1/PRSS3, RAP1A/B, SERPINF2, TPSAB1/TPSB2, VWF | Antigen presentation, inflammatory response, cardiovascular disease | 24 |
| 10 | ABR, ACTA1/2, ADHFE1, ADH, APP, AVPR1B, CKII, CNGB1, CREB1/3/5, CRYAB, CYC1, DCTN1, DHRS2, DHRS4, DMPK, DNAJB1, ENO1, FAM162A, GDP, GSK3A/B, HSPB2, NUDC, NUMBL, MAPK1/11-14, PDGFA/B, PLCB1-4, RAC1/2/3, POLR2A-L, RPL13, RYBP, PRSS1/2/3, TTLL12, WDR45, WNT1-11 | Cellular assembly & organization, organismal development, organismal injury & abnormalities | 23 |
| Number . | Molecules in network . | Top functions . | Score . |
|---|---|---|---|
| 1 | ARID1B, AS3MT, BAI1, BCS1L, C12orf52, C21orf56, CDK2AP2, CUL2, FAM107B, FAM117A, FAM158A, GMDS, HNF4A, IL11RA, JINK1/2, LSR, MREG, MRPL18, NDUFV1, NUCB1, OAZ2, PABPN1, PPP1R3B, PRAF2, RFPL2, SBNO2, SLC17A5, SLMO2, SPATA6, STOML2, TBC1D23, TEF, THOC3, TLE3, VN1R1 | Cardiovascular disease, genetic disorder, neurological disease | 56 |
| 2 | AEBP1, BBC3, CARD11, CD7, CHRFAM7A, EDAR, ELF5, ETS, FABP6, HMG20B, IP6K2, KRT16, KRT17, LRDD, LSP1, NFkB, CHRN, NR1H2, NR1H3, NR1H, PIM3, PLEC, PPP1R13B, RASGRF1, RNF31, ROMO1, RRAS, RXR, SECTM1, SRPX, TRADD, TRAF1, TUBA1C, ZHX2 | Cell death, dermatological diseases & conditions, endocrine system disorders | 45 |
| 3 | GRIA1-4, AP2A1/2, ATP1A2, ATP1A4, BABAM1, CYP11B2, EPB41L1, ERK1/2, FXYD7, GABAR-A, GABRG2, GABRR1, GNAL, GPT2, HSF4, IL27RA, MRPL53, NR5A1, NSF, NTF3, PITPNM1, PRKAG1, PTGDS, RLN3, SOX7, SOX9, SOX10, STX5, TFF2, TH, TMEM109, UCN2, VAPB | Small molecule biochemistry, developmental disorder, gastrointestinal disease | 44 |
| 4 | PSMA/PSMB, PSME/PSMF, ADIPOR1, ADRM1, TUBA1A, AMPK, ASCL2, BARD1, TUBB, CLINT1, CUL7, FES, GP1BA, Hsp70, LRP, MLLT4, MST1R, ORC1, PIK3R1/2, PIK3R6, PLXNB3, PFN1/2, PSENEN, PSME1, PSME2, RAB3B, PSEN1/2, SLC39A4, SNCG, SPDEF, TCP1, TRIM9, UBB, VASP | Cell cycle, dermatological diseases & conditions, embryonic development | 32 |
| 5 | ABLIM1, ACP2, ACTG2, ADAP1, FOS/JUN, CBFA2T3, CGA/B, CGB1, CGB2, CGB7, CGBB, COL1A1/2, NR2F1/2/6, CYP17A1, CYP2D6, DIABLO, FSH, GALP, GPSM1, HAUS4, HCG, HIST1H1A-E, HMBS, MA2K4/10/12, LHB, LIF, MAP2K1-7, RPS6KB1/3, PRKACA/B/G/G1/G2, PTMS, RXRA, TGFB1/2/3, USHBP1, WNT5B, ZBED1 | Cellular movement, reproductive system development & function, nervous system development & function | 30 |
| 6 | CAPN1/10/11/, CAPN3, CASP1-12, CCNE1, CDK2, COBRA1, CRYAA, CCNA1/2, CCNE1/2, CYP27B1, DEFA1, DNAJB8, DNAJC9, E2f, ESRRB, ESR1/2, GPC1, HBG1, HIST2H3C, HIST2H4, Hsp90, Hsp22/Hsp40/Hsp90, HVCN1, KIAA1967, LIN37, OSGIN1,PRKCA/B, PNLIPRP2, SATB1, SF3B2, TFDP1, TFF1, VDR, RXR, ZDHHC11 | Cell cycle, cellular assembly & organization, embryonic development | 30 |
| 7 | ABCC1, ACR, ADCY, ADORA2A, AGAP1, AVPR2, DRD1, PTK2/2B, FOXO1, GHRHR, GIT1, Gpcr, GPR84, GPR157, GPRC5D, INS, KCNAB2, KISS1R, NPFF, P110, PAX4, PEMT, PEPCK, PRKCZ, Rab5, Ras, SRC, TBC1D3F, THRA/B, NTRK1/2/3, USP17, USP17L2, VEGF, VN1R4 | Cell signaling, nucleic acid metabolism, molecular transport | 28 |
| 8 | CBP, AFAP1, PPP3CA/B/C, CALM1, CAMK1G, CAMK2A/B, DTNA, DVL1, EPHB2, ACTA1/2, GATA2, GRIN2D, KSR1, LEFTY2, MAPK1, MYLK3, NCS1, NFATC1-4, GRIN2A-D, PIP5K1C, RHOA-H, RCVRN, REM1, RHOT2, ROCK1/2, SIT1, SLC8A2, SNX33, TNNI3, TNNT3, TPM4, TPM1/2/3/4, TUBA1A-E, UTS2 | Skeletal & muscular system, development & function, tissue morphology, cell-to-cell signaling & interaction | 26 |
| 9 | ACE, ACTA2, Akt, CD68, CD151, CD276, COL1, COL1A1/2, COL4, Cpla2, CSF2, FGA/B/G, FMOD, ITGA5, ITGB5, KLK1/3/B1, LAG3, LAMA1/5, MAP2K1/2, MFAP5, MMP26, PAK1-4, PDGFA-D, PLA2G2F, PMEL, PP2A, PROK1, PRSS1/PRSS3, RAP1A/B, SERPINF2, TPSAB1/TPSB2, VWF | Antigen presentation, inflammatory response, cardiovascular disease | 24 |
| 10 | ABR, ACTA1/2, ADHFE1, ADH, APP, AVPR1B, CKII, CNGB1, CREB1/3/5, CRYAB, CYC1, DCTN1, DHRS2, DHRS4, DMPK, DNAJB1, ENO1, FAM162A, GDP, GSK3A/B, HSPB2, NUDC, NUMBL, MAPK1/11-14, PDGFA/B, PLCB1-4, RAC1/2/3, POLR2A-L, RPL13, RYBP, PRSS1/2/3, TTLL12, WDR45, WNT1-11 | Cellular assembly & organization, organismal development, organismal injury & abnormalities | 23 |
Genes in the network with differential methylation are in boldface type.
Top 10 networks targeted by aberrant hypermethylation in GATA1s-positive DS-TMD
| Number . | Molecules in network . | Top functions . | Score . |
|---|---|---|---|
| 1 | ADCY, ADORA3, ADRB3, FOS/JUN, ATN1, CD9, CELSR1, CXCR7, DRD2, ESR1/2, FPR3, GNAI1/2/3, GPR83, GPR101, HTR4, HTR5A, KCNA2, KCNA3, KCNJ6, LTF, NCOR1/2, NGEF, NRL, NUPR1, PI3K, RAC1/2/3, RHOA-D, RXR, S1PR3, VIPR1, VN1R4, ZFHX3 | Cell signaling, molecular transport, nucleic acid metabolism | 23 |
| 2 | ACTA2, ACTN1, ALPI/L/P, ANKRD2, APOA2, DHTR, CALM1, CAMK1G, COL1A1, COL1, CREB1/3/5, CREBL2, DCT, ERK1/2, ACTAB/C1, FSHB, GH1/2, GSN, IGFBP5, IL27RA, KCND2, KCNIP4, LEP, LHB, MYO7A, RIMS1, RLN3, SERPINA12, SGK223, SLC15A1, TFF2, TGFB, TPM1, TSEN34 | Tissue morphology, cell morphology, cellular movement | 23 |
| 3 | AEBP1, ALDH7A1, CREBP/EP300, CLEC10A, ELF2, ETS, FGA, FGA/B/G, GAS7, IFI27L2, IFNA1, IFNB1, IGHG1-4, IL24, IL25, IL12, IRF8, KLK2, LGALS7/LGALS7B, MAFG, MHC II, NFE2, NFkB, PPFIA2, PRSS1/PRSS3, PTPN2, REN, SERPINA5, SERPINF2, PRSS1/2/3, UBP1, UGCG, VWF | Cellular growth & proliferation, lymphoid tissue structure & development, cellular development | 23 |
| 4 | PSMA/PSMB, AKT1/2/3, ATAD2, CASP1-12, CYC1, DAB2IP, DLEU1, DUB, E2F1/6, ERN1, ESR1, GREB1, HIST1H3A, HIST2H3C, HIST2H4, Hsp70, HSPA1A/HSPA1B, HSPA1L, IL1, MAP2K4/8/10/12, KIAA1967, MAPK1/11/12/13/14, PEPD, POLR2H, PPARGC1A, PRR11, POLR2A-L, SF3B2, SIRT7, SKA2, SORBS2, UCHL3, USP12, VEGF, ZBTB17 | Gene expression, endocrine system development & function, small molecule biochemistry | 21 |
| 5 | ADAMTS20, ATL3, CHUK/IKBKB/IKBKG, CPVL, CSDE1, DMRT1, DMRTC1/DMRTC1B, DNTTIP2, DUPD1, E2F1, EIF4E2, ERLIN1, ETF1, FOS, GSPT2, HIST1H2AA, HIST1H2AG, HIST1H2BA, HIST1H2BJ/HIST1H2BK, HIST1H4C, HRK, HRNR, KIAA0802, LIMD1, LPPR4, MAP3K3, MARK4, PABPC1, PRDM2, PREP, SFMBT1, TBCA, TRAF6, YWHAZ, ZNF652 | Cell death, cellular development, hematological system development & function | 15 |
| 6 | ANXA8L2, ASB2, B3GALT1, CACNG5, CLC, Cyp2c70, FOXD3, GBX2, GLI1, HEBP1, HOXA3, HOXB3, HOXD13, KCTD11, L1TD1, NAT2, NAV2, NDN, NKX2-3, PAPSS2, PDZRN3, PLA2G15, POU5F1, RORA, SMPD3, SOX3, SOX7, SOX15, TCEB1, TCEB3C, TMEM100, WNT2B, WSB1, ZCCHC8 | Cellular development, cellular growth & proliferation, cell-to-cell signaling & interaction | 15 |
| 7 | CCDC82, CCL16, CRIPT, CSTF2T, DLG4, DNAH7, EPO, FAM107B, GLCE, GPX7, HGS, HNF4A, ISOC1, LRRTM2, MCTS1, MIF4GD, MRPS7, MRPS23, MSRB2, PAPOLA, PCDHA13, PDIA5, PKD1L3, PKD2L1, PRDM14, PREB, PRL, RAI2, RBMY1A1, SEC22B, TNNI3, UBQLN4, UROS, VHL, ZG16B | Cell cycle, gene expression, hematological disease | 14 |
| 8 | ACTA1/2, AGAP1, ART1, CSNK2A1/A2/B, EPS8L2, ERK, PTK2/2B, GCK, GGA3, HMGN2, INS, LGALS12, MAPK, MAZ, Mml1, MMP20, MMP24, MMP27, MMP28, MMP23A, MMP23B, MYO5B, NEUROG1, NMUR1, PCCB, PCOLCE, PDGFA/B, PRKAR1A/1B/2A/2B, PLC, KRAS/NRAS, RASA2, SLC7A7, STAT, TCOF1 | Tissue development, cellular assembly & organization, digestive system development & function | 12 |
| 9 | ATXN1, BEND2, CCDC164, CLEC4F, DDX46, DYSF, EBNA1BP2, EGLN, EGOT, FAM203A/FAM203B, GADD45G, GCAT, GLIPR2, GSTT2/GSTT2B, IGKV, IL4, IL5, ITGB1, KLHDC2, OBFC2A, PPIF, RAD54L2, RWDD2B, SASH3, SDF2L1, SERPINA3, SKA1, SLAIN1, TOMM40L, TRIM72, UBE2I, WNK2, ZNF609, ZNF777 | Cellular movement, hematological system development & function, hypersensitivity response | 12 |
| 10 | RAB7B, ART5, BST1, BTNL2, C11orf82, CD80/CD86, CECR1, CERK, CLEC10A, CLEC3B, CYBA, NCF1/2/4, DPAGT1, GAL3ST3, IFNG, IL4I1, MCHR1, MOBKL1A, MTMR11, NAD+, Pc-Plc, PHKA2, RAB36, RAB38, RAF1, RCN1, SBSN, SCN4A, SLC6A12, TCF3, TLR4, TMEM180, TREM2 | Cell morphology, cell-to-cell signaling & interaction, cellular function & maintenance | 12 |
| Number . | Molecules in network . | Top functions . | Score . |
|---|---|---|---|
| 1 | ADCY, ADORA3, ADRB3, FOS/JUN, ATN1, CD9, CELSR1, CXCR7, DRD2, ESR1/2, FPR3, GNAI1/2/3, GPR83, GPR101, HTR4, HTR5A, KCNA2, KCNA3, KCNJ6, LTF, NCOR1/2, NGEF, NRL, NUPR1, PI3K, RAC1/2/3, RHOA-D, RXR, S1PR3, VIPR1, VN1R4, ZFHX3 | Cell signaling, molecular transport, nucleic acid metabolism | 23 |
| 2 | ACTA2, ACTN1, ALPI/L/P, ANKRD2, APOA2, DHTR, CALM1, CAMK1G, COL1A1, COL1, CREB1/3/5, CREBL2, DCT, ERK1/2, ACTAB/C1, FSHB, GH1/2, GSN, IGFBP5, IL27RA, KCND2, KCNIP4, LEP, LHB, MYO7A, RIMS1, RLN3, SERPINA12, SGK223, SLC15A1, TFF2, TGFB, TPM1, TSEN34 | Tissue morphology, cell morphology, cellular movement | 23 |
| 3 | AEBP1, ALDH7A1, CREBP/EP300, CLEC10A, ELF2, ETS, FGA, FGA/B/G, GAS7, IFI27L2, IFNA1, IFNB1, IGHG1-4, IL24, IL25, IL12, IRF8, KLK2, LGALS7/LGALS7B, MAFG, MHC II, NFE2, NFkB, PPFIA2, PRSS1/PRSS3, PTPN2, REN, SERPINA5, SERPINF2, PRSS1/2/3, UBP1, UGCG, VWF | Cellular growth & proliferation, lymphoid tissue structure & development, cellular development | 23 |
| 4 | PSMA/PSMB, AKT1/2/3, ATAD2, CASP1-12, CYC1, DAB2IP, DLEU1, DUB, E2F1/6, ERN1, ESR1, GREB1, HIST1H3A, HIST2H3C, HIST2H4, Hsp70, HSPA1A/HSPA1B, HSPA1L, IL1, MAP2K4/8/10/12, KIAA1967, MAPK1/11/12/13/14, PEPD, POLR2H, PPARGC1A, PRR11, POLR2A-L, SF3B2, SIRT7, SKA2, SORBS2, UCHL3, USP12, VEGF, ZBTB17 | Gene expression, endocrine system development & function, small molecule biochemistry | 21 |
| 5 | ADAMTS20, ATL3, CHUK/IKBKB/IKBKG, CPVL, CSDE1, DMRT1, DMRTC1/DMRTC1B, DNTTIP2, DUPD1, E2F1, EIF4E2, ERLIN1, ETF1, FOS, GSPT2, HIST1H2AA, HIST1H2AG, HIST1H2BA, HIST1H2BJ/HIST1H2BK, HIST1H4C, HRK, HRNR, KIAA0802, LIMD1, LPPR4, MAP3K3, MARK4, PABPC1, PRDM2, PREP, SFMBT1, TBCA, TRAF6, YWHAZ, ZNF652 | Cell death, cellular development, hematological system development & function | 15 |
| 6 | ANXA8L2, ASB2, B3GALT1, CACNG5, CLC, Cyp2c70, FOXD3, GBX2, GLI1, HEBP1, HOXA3, HOXB3, HOXD13, KCTD11, L1TD1, NAT2, NAV2, NDN, NKX2-3, PAPSS2, PDZRN3, PLA2G15, POU5F1, RORA, SMPD3, SOX3, SOX7, SOX15, TCEB1, TCEB3C, TMEM100, WNT2B, WSB1, ZCCHC8 | Cellular development, cellular growth & proliferation, cell-to-cell signaling & interaction | 15 |
| 7 | CCDC82, CCL16, CRIPT, CSTF2T, DLG4, DNAH7, EPO, FAM107B, GLCE, GPX7, HGS, HNF4A, ISOC1, LRRTM2, MCTS1, MIF4GD, MRPS7, MRPS23, MSRB2, PAPOLA, PCDHA13, PDIA5, PKD1L3, PKD2L1, PRDM14, PREB, PRL, RAI2, RBMY1A1, SEC22B, TNNI3, UBQLN4, UROS, VHL, ZG16B | Cell cycle, gene expression, hematological disease | 14 |
| 8 | ACTA1/2, AGAP1, ART1, CSNK2A1/A2/B, EPS8L2, ERK, PTK2/2B, GCK, GGA3, HMGN2, INS, LGALS12, MAPK, MAZ, Mml1, MMP20, MMP24, MMP27, MMP28, MMP23A, MMP23B, MYO5B, NEUROG1, NMUR1, PCCB, PCOLCE, PDGFA/B, PRKAR1A/1B/2A/2B, PLC, KRAS/NRAS, RASA2, SLC7A7, STAT, TCOF1 | Tissue development, cellular assembly & organization, digestive system development & function | 12 |
| 9 | ATXN1, BEND2, CCDC164, CLEC4F, DDX46, DYSF, EBNA1BP2, EGLN, EGOT, FAM203A/FAM203B, GADD45G, GCAT, GLIPR2, GSTT2/GSTT2B, IGKV, IL4, IL5, ITGB1, KLHDC2, OBFC2A, PPIF, RAD54L2, RWDD2B, SASH3, SDF2L1, SERPINA3, SKA1, SLAIN1, TOMM40L, TRIM72, UBE2I, WNK2, ZNF609, ZNF777 | Cellular movement, hematological system development & function, hypersensitivity response | 12 |
| 10 | RAB7B, ART5, BST1, BTNL2, C11orf82, CD80/CD86, CECR1, CERK, CLEC10A, CLEC3B, CYBA, NCF1/2/4, DPAGT1, GAL3ST3, IFNG, IL4I1, MCHR1, MOBKL1A, MTMR11, NAD+, Pc-Plc, PHKA2, RAB36, RAB38, RAF1, RCN1, SBSN, SCN4A, SLC6A12, TCF3, TLR4, TMEM180, TREM2 | Cell morphology, cell-to-cell signaling & interaction, cellular function & maintenance | 12 |
Genes in the network with differential methylation are in boldface type.
DS-AMKL blasts are epigenetically indistinguishable from DS-TMD blasts
Given the fact that many TMD cases likely go undiagnosed, it is unclear whether DS-AMKL is always preceded by a DS-TMD phase. Conversely, only about 20% of patients with diagnosed TMDs eventually develop DS-AMKL. In order to determine whether these 2 entities are epigenetically related, we performed a supervised analysis in which we compared the DNA methylation patterns of DS-TMD and DS-AMKL blasts. At an FDR cutoff of <5% we failed to detect any statistically significant DMRs between these 2 groups. In order to determine whether this phenomenon was also observed at the gene expression level, we downloaded publicly available data from Bourquin et al29 and performed a direct comparison of the gene expression profiles of DS-TMD blasts and DS-AMKL blasts. Only 7 genes were found differentially expressed at an FDR <5% and a minimum fold change of 2. These findings demonstrate that DS-TMD and DS-AMKL are virtually identical, consistent with other reports that found relatively minor differences in gene expression levels between TMD and DS-AMKL.33,34 Moreover, although additional genetic lesions are likely present at the DS-AMKL stage,14,15,35 they do not appear to have a significant impact on the transcriptional and epigenetic programs of these cells.
DS-related hypomethylation on chromosome 21 is linked to trisomic gene overexpression and affects the DS critical region and nearby neighboring regions
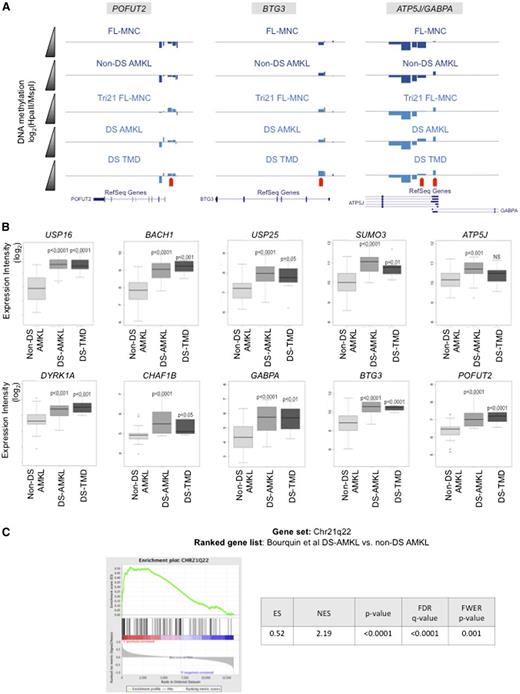
Next we focused our analysis on the DNA methylation changes seen on chromosome 21. This analysis revealed that aberrant hypomethylation on chromosome 21 observed in Tri21 FL-MNCs and persisting until the final DS-AMKL stage was more prominent on and around the DS critical region (DSCR)35-37 (supplemental Figure 3). In order to identify epigenetic changes in the DSCR that are directly related to the acquisition of an additional copy of chromosome 21 and that are independent of any additional epigenetic reprogramming secondary to malignant transformation, we examined DNA methylation differences of genes in the DSCR and neighboring regions between control FL-MNCs and Tri21 FL-MNCs. Among the genes in this region, POFUT2, BTG3, ATP5J, and GABPA were identified as hypomethylated by using our initial stringent criteria for differential methylation, ie, >30% difference in methylation (Figure 4A). In addition, CHAF1B, DYRK1A, SUMO3, USP25, BACH1, USP16, and RUNX1 also showed statistically significant changes in DNA methylation between these 2 groups, though these changes were of a smaller magnitude. Notably, DYRK1A and CHAF1B13,38 have previously been implicated in DS-leukemogenesis in human specimens and murine models of DS-AMKL.13
DNA hypomethylation is associated to overexpression of trisomic genes. (A) Visualization on the University of California at Santa Cruz (UCSC) genome browser of DNA methylation changes detected by the HELP assay on 4 genes localized in neighboring regions of the DSCR. Each custom track represents the mean HELP values measured for that group. Negative deflections below the zero line represent negative log ratios (methylated regions), and positive deflections above the zero line represent positive log ratios (hypomethylated regions). Red arrows indicate HELP probesets with detectable differences in methylation between control samples (FL-MNC and non–DS-AMKL) and DS-related samples (Tri21 FL-MNC, DS-AMKL, and DS-TMD). (B) Boxplots representing gene expression values for genes differentially methylated in and around the DS critical region. P values are for t test comparisons between each of the 2 DS-related conditions vs the non–DS-AMKL values. (C) Gene set enrichment analysis using positional gene sets MSigDB collections (c1) showing significant enrichment for genes localized in 21q22 band in DS-AMKL samples from a publicly available DS-AMKL microarray dataset.29
DNA hypomethylation is associated to overexpression of trisomic genes. (A) Visualization on the University of California at Santa Cruz (UCSC) genome browser of DNA methylation changes detected by the HELP assay on 4 genes localized in neighboring regions of the DSCR. Each custom track represents the mean HELP values measured for that group. Negative deflections below the zero line represent negative log ratios (methylated regions), and positive deflections above the zero line represent positive log ratios (hypomethylated regions). Red arrows indicate HELP probesets with detectable differences in methylation between control samples (FL-MNC and non–DS-AMKL) and DS-related samples (Tri21 FL-MNC, DS-AMKL, and DS-TMD). (B) Boxplots representing gene expression values for genes differentially methylated in and around the DS critical region. P values are for t test comparisons between each of the 2 DS-related conditions vs the non–DS-AMKL values. (C) Gene set enrichment analysis using positional gene sets MSigDB collections (c1) showing significant enrichment for genes localized in 21q22 band in DS-AMKL samples from a publicly available DS-AMKL microarray dataset.29
In order to determine whether these changes in DNA methylation correlated with changes in gene expression levels in DS-related malignancies, we analyzed the expression levels of these genes in the Bourquin et al expression data set.29 With the exception of RUNX1, all of these genes showed significantly higher expression levels in both DS-AMKL and DS-TMD in relation to non–DS-AMKL (Figure 4B). In parallel, we performed gene set enrichment analyses using positional gene set MSigDB collections (c1) and revealed that genes localized in 21q22 band are the only ones, among the whole genome, that are significantly enriched in DS-AMKL samples from this same microarray dataset29 (Figure 4C). Taken together, these results show that DNA hypomethylation of specific chromosome 21 genes, included or not in the DSCR region, is correlated with changes in gene expression levels in DS-related malignancies, and the results suggest that the hypomethylation of this region is associated with an increased expression of trisomic genes that predispose and cooperate with GATA1s and other genetic abnormalities to foster TMD and DS-AMKL.
Discussion
DS-AMKL development is associated with a series of well-characterized sequential genetic events.6 However, much less is known about the epigenetic changes that occur during this process. In the current study we report the first genome-wide DNA methylation study of DS leukemogenesis. Taking advantage of the multistep developmental process of DS-AMKL, we performed DNA methylation profiling at sequential stages of the disease. Our findings demonstrate that epigenetic deregulation in DS-AMKL leukemogenesis occurs in 2 distinct epigenetic “waves.” The first wave of epigenetic reprogramming seems to be directly linked to the additional copy of chromosome 21, which results in genome-wide hypomethylation. This hypomethylation was detected in primary human Tri21 FL-MNC specimens, in DS-AMKL in relation to non–DS-AMKL, and in the Ts1Rhr murine model. Given that the Ts1Rhr mice are partially trisomic, it is possible that the difference in magnitude in hypomethylation observed between the primary human FL-MNCs and the partial trisomic cells in these animals is due to this difference and that a larger trisomic region, or even a full trisomy 21, is required to impose the full extent of the hypomethylation phenotype observed in the human trisomy 21 specimens. Comparable hypomethylation had been previously reported in normal peripheral blood leukocytes from DS patients. However, the extent of this hypomethylation and its impact on developmentally related gene regulatory networks was not previously recognized.39 Notably, both the Ts1Rhr and Ts65Dn murine models of DS40 have been linked to decreased intestinal polyp formation in the Apcmin model.41 This reduction is analogous to the reduced polyp formation described for the Apcmin model in the Dnmt1 hypomorph background in which polyp formation was reduced in a Dnmt1 dose-dependent manner in comparison with ApcminDnmt1+/+ animals.42 Similarly, despite their increased risk for the development of acute leukemias, DS patients have an overall reduced risk for developing cancers, in particular solid tumors.43 Further studies will be required to determine whether or not the hypomethylation associated with the presence of trisomy 21 results in a protective effect against malignant transformation and if so, what the mechanism responsible for this phenomenon is. One attractive model is that hypomethylation may occur in DS due to overexpression of cystathionine β synthase, which can lead to lower S-adenosylmethionine levels.44
The second wave of aberrant DNA methylation was detected in GATA1s mutant DS-TMD MNCs and consisted mainly of gains in methylation at a distinct set of target genes. Unlike hypomethylation associated with trisomy 21, which preferentially affected gene networks associated with developmental disorders such as those seen in DS, genes affected by aberrant hypermethylation in DS-TMD blasts were specific to hematological development and regulation of key cellular processes such as growth, proliferation, cell cycle regulation, and cell death. How trisomy 21 may lead to hypomethylation while GATA1 mutations result in hypermethylation remains unclear. These findings, however, indicate that these 2 waves of epigenetic reprogramming contribute to distinct aspects of DS abnormalities (Figure 5). Additional experiments will be required to functionally determine whether the predisposing role of the additional chromosome 21 to leukemogenesis is mediated through this trisomy 21–driven hypomethylation, as well as to further define the precise nature of the epigenetic changes in sorted subpopulations (hematopoietic stem cell and MEP) obtained from the different specimen types collected in this study. Notably, an analysis of GATA1 and GATA1s occupancy by ChIP-seq45,46 failed to show substantial enrichment at regions that acquired hypermethylation in GATA1s mutant specimens (data not shown). These findings indicate that epigenetic reprogramming during DS-AMKL development is not mediated through binding of GATA1/GATA1s to its target regions. Whether transcription factors or chromatin modulators downstream of GATA1 may be responsible for the specific gains of DNA methylation remains a possibility.
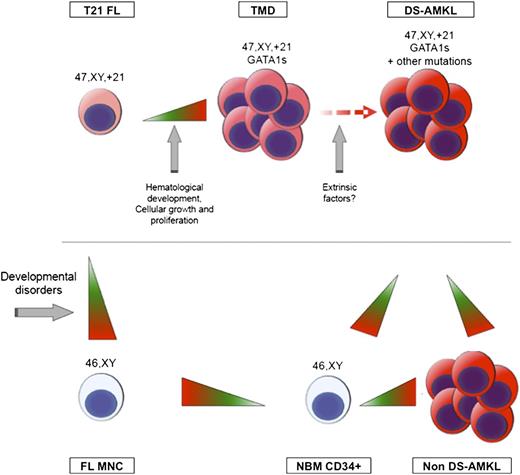
Model of sequential genetic and epigenetic events associated with DS-AMKL development. Two distinct waves of epigenetic reprogramming were detected. The first wave, which happens upon acquisition of an additional copy of chromosome 21, results in genome-wide hypomethylation and affects genes involved in developmental processes. The second wave, detected in DS-TMD blasts, results in hypermethylation of genes involved in hematological disorders and regulation of key cellular processes. No additional epigenetic changes are detected upon progression to DS-AMKL, but DS-AMKL blasts are hypomethylated in comparison with NBM CD34+ and non–DS-AMKL blasts.
Model of sequential genetic and epigenetic events associated with DS-AMKL development. Two distinct waves of epigenetic reprogramming were detected. The first wave, which happens upon acquisition of an additional copy of chromosome 21, results in genome-wide hypomethylation and affects genes involved in developmental processes. The second wave, detected in DS-TMD blasts, results in hypermethylation of genes involved in hematological disorders and regulation of key cellular processes. No additional epigenetic changes are detected upon progression to DS-AMKL, but DS-AMKL blasts are hypomethylated in comparison with NBM CD34+ and non–DS-AMKL blasts.
The general pattern of global hypomethylation with focal hypermethylation at promoter regions in cancer has been recognized for many years.47 However, the precise sequence in which this pattern is established is not clear, and whether an initial wave of hypomethylation followed by acquisition of genetic abnormalities and focal hypermethylation as described here are also observed in other tumors is yet unknown.
Previous studies of epigenetic abnormalities in myelodysplastic syndromes have shown that more aggressive forms of the disease are associated with more intense epigenetic abnormalities.48 However, this was not the case in the current study, in which the DNA methylation patterning in DS-TMD MNCs was indistinguishable from that of DS-AMKL MNCs. Likewise, a direct comparison of the transcriptional profiles of these 2 types of blasts also identified few significant differences between TMD and DS-AMKL. In 1 study, the 2 populations could be separated only by supervised clustering using a small set of differentially expressed genes.33 A second study reported only 67 genes whose expression was significantly different in the TMD vs DS-AMKL.34 Given the almost identical transcriptional and epigenetic profiles of these 2 entities, it is possible that the initial clearance of DS-TMD blasts with subsequent reappearance in the DS-AMKL phase may be due to the effect of extrinsic factors that might be correlated to different hematopoietic niches in bone marrow vs fetal hematopoiesis.
Another interesting finding is that a specific region of the chromosome 21 around the DSCR remains hypomethylated during the whole process of DS-leukemogenesis, whereas from the TMD stage onward, additional changes in methylation are dominated by hypermethylation. This region has been specifically associated to DS-AMKL development using human specimens with segmental trisomy 21,38 gene expression profiling29,49 and animal models.13 We correlated this hypomethylation to the overexpression of genes contained in this region including DYRK1A, which has been shown to cooperate with Gata1s and MPL W515L expression to promote DS-AMKL in vivo.13 Notably, only a small fraction of the chromosome 21 genes were upregulated in the DS-AMKL and DS-TMD specimens in relation to non–DS-AMKL, indicating that upregulation of these genes is not simply the result of the increased copy number due to the trisomy. Taken together, these observations strongly suggest that the observed epigenetic memory of hypomethylated DSCR genes is submitted to a selective pressure required for DS-leukemogenesis.
In summary, our study shows that the genetic changes linked to the specific stages in DS-AMKL are associated with distinct sets of changes in DNA patterning. Whether the epigenetic pattern changes contribute to disease or simply reflect the underlying genetic lesions remains to be determined. In support of a pathogenic role of the aberrant gene methylation, it is notable that the earlier changes associated with hypomethylation tended to affect developmental pathways comprising Down syndrome. The later acquired changes in methylation in cell cycle control pathways also support the idea that the changes in DNA methylation are a cause and not an effect of disease. The mechanisms through which these epigenetic changes become established are yet to be identified and require careful consideration of the genes in the critical region of chromosome 21 as well as pathways downstream of GATA1, because GATA1 target genes were not among the hypermethylated sites in TMD and DS-AMKL.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Weill Cornell Medical Center Epigenetics Core Facility for performing the HELP microarrays and Jason Berman, Soheil Meshinchi, Todd Alonzo, and Sommer Castro and the Children's Oncology Group for their assistance with DS-AMKL specimens. This article’s contents are solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
M.E.F. is supported in part by a Special Fellow Award from the Leukemia and Lymphoma Society. S.M. was supported by a Leukemia and Lymphoma Society Special Fellowship (no. 5110-10). This work was supported in part by grants from the National Cancer Institute (NCI) (CA101774) and the Samuel Waxman Cancer Research Foundation. J.W.T. is supported by an R01 grant from the NCI (CA120772). This work was partially funded by grant K08 HL093290 from the National Heart, Lung, and Blood Institute (NHLBI) (to S.T.C.) and The American Society of Hematology (to S.T.C.). This study was coordinated by the Eastern Cooperative Oncology Group (Robert L. Comis, Chair) and supported in part by Public Health Service Grants CA23318, CA66636, CA21115, CA17145, CA13650, CA15488, and CA14958 from the NCI, National Institutes of Health, and the Department of Health and Human Services. Research with DS-AMKL samples was supported by the Chair's Grant U10 CA98543 (to C.O.G.) from NCI.
Authorship
Contribution: J.C. and M.E.F. conceived and designed the study; S.M., T.C., L.C.D., and M.E.F. performed experiments; T.C., L.C.D., and M.E.F. analyzed the data; S.M. and J.C. critically revised the data; S.C., R.P.K., M.S.T., E.P., M.J.W., A.G., and J.W.T. contributed key resources and specimens; M.E.F., S.M., and J.C. wrote the manuscript; and T.C., S.C., M.J.W., R.P.K., E.P., M.S.T., A.G., and J.W.T. revised and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria E. Figueroa, Department of Pathology, University of Michigan, 109 Zina Pitcher Place, 2019 BSRB, Ann Arbor, MI 48109-2200; e-mail: marfigue@med.umich.edu; or John D. Crispino, Northwestern University Feinberg School of Medicine, Division of Hematology and Oncology, 303 E Superior St, Lurie Research Building 5-113, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu.