Abstract
In this study, we evaluated the impact of secondary genetic lesions in acute myeloid leukemia (AML) with inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB-MYH11. We studied 176 patients, all enrolled on prospective treatment trials, for secondary chromosomal aberrations and mutations in N-/KRAS, KIT, FLT3, and JAK2 (V617F) genes. Most frequent chromosomal aberrations were trisomy 22 (18%) and trisomy 8 (16%). Overall, 84% of patients harbored at least 1 gene mutation, with RAS being affected in 53% (45% NRAS; 13% KRAS) of the cases, followed by KIT (37%) and FLT3 (17%; FLT3-TKD [14%], FLT3-ITD [5%]). None of the secondary genetic lesions influenced achievement of complete remission. In multivariable analyses, KIT mutation (hazard ratio [HR] = 1.67; P = .04], log10(WBC) (HR = 1.33; P = .02), and trisomy 22 (HR = 0.54; P = .08) were relevant factors for relapse-free survival; for overall survival, FLT3 mutation (HR = 2.56; P = .006), trisomy 22 (HR = 0.45; P = .07), trisomy 8 (HR = 2.26; P = .02), age (difference of 10 years, HR = 1.46; P = .01), and therapy-related AML (HR = 2.13; P = .14) revealed as prognostic factors. The adverse effects of KIT and FLT3 mutations were mainly attributed to exon 8 and tyrosine kinase domain mutations, respectively. Our large study emphasizes the impact of both secondary chromosomal aberrations as well as gene mutations for outcome in AML with inv(16)/t (16;16).
Key Points
More than 90% of the patients with inv(16)/t(16;16) AML harbor secondary chromosome aberrations (eg, trisomy 22) and/or mutations affecting N-RAS, K-RAS, KIT, and FLT3.
Clinical heterogeneity is reflected by genetic findings with trisomy 8 and 22, KIT, and FLT3 mutations representing prognostic markers.
Introduction
Since 2001, acute myeloid leukemia (AML) with pericentric inversion of chromosome 16, inv(16)(p13.1q22), or the less frequent translocation t(16;16)(p13.1;q22), both hereafter referred to as inv(16), is recognized by the World Health Organization as a unique entity in the category “AML with recurrent genetic abnormalities.”1 On cytogenetic analysis, inv(16) is detected in approximately 8% of adults diagnosed with AML.2 The inv(16) abnormality and the balanced translocation t(8;21)(q22;q22) define the subgroup of core-binding factor (CBF) AML with a favorable prognosis.3 Both chromosomal aberrations result in rearrangements of genes encoding subunits of CBF that is a transcription factor with a pivotal role in normal hematopoiesis.4 At the molecular level, inv(16) leads to the fusion of the core-binding factor β subunit (CBFB, PEBP2B) gene on chromosomal band 16q22 with the smooth muscle myosin heavy chain (MYH11) gene on 16p13.5 Mouse studies have shown that Cbfb/MYH11 allele causes a block in myeloid differentiation, predisposing to leukemia, but additional genetic alterations are required for the development of a leukemic phenotype.6 Such additional genetic alterations include mutations in genes encoding protein effectors controlling cell proliferation, conferring survival advantage to malignant cells, or both. Indeed, genes that encode for RAS guanosine triphosphatases, that is, NRAS (neuroblastoma rat sarcoma viral oncogene homolog) and KRAS (Kirsten rat sarcoma viral oncogene homolog) as well as 2 members of the type III tyrosine kinase family, namely, KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) and FLT3 (FMS-like tyrosine kinase), have been described as frequent secondary mutations in inv(16) AML.7-13 Notably, some studies in inv(16) AML have reported on KIT mutations as an unfavorable prognostic marker,8,11,13 implying the use of KIT mutation status for risk stratification in this AML subgroup.
In addition, secondary chromosomal aberrations are present in 35% to 40% of inv(16) AML cases, with trisomy 22 representing the most frequent abnormality followed by trisomy 8.14-16 As reported by our group, 7q deletions occur in approximately 10% of CBF-AML, with one-half of them being microdeletions that are only detectable by high-resolution genetic profiling.17 Among the secondary chromosomal aberrations, only trisomy 22 has so far been reported to affect outcome in inv(16) AML.14-16
In this study, we assessed the incidence and prognostic impact of secondary genetic lesions, that is, additional chromosomal aberrations and gene mutations affecting NRAS, KRAS, KIT, FLT3, and JAK2, in a large cohort of adult patients with inv(16)–positive AML who were intensively treated on German-Austrian AML Study Group (AMLSG) protocols.
Methods
Patients
Only patients enrolled on AMLSG treatment trials were considered for this study. The inclusion criteria comprised (1) presence of inv(16) abnormality at diagnosis detected by cytogenetic analysis, fluorescence in situ hybridization (FISH), or molecular analysis using reverse transcriptase–polymerase chain reaction (RT-PCR); (2) availability of pretreatment bone marrow samples, blood samples, or both for mutational analyses; and (3) availability of clinical follow-up data. In total, 176 patients from 7 AMLSG treatment trials were identified for this study (AML HD93 [n = 20]),18 AML-SHG 02/95 [n = 12],19 AML HD98A [clinicaltrials.gov Identifier NCT00146120; n = 56], AML HD98B [n = 6],20 AML-SHG 01/99 [NCT00209833; n = 23], AMLSG 06-04 [NCT00151255; n = 9], and AMLSG 07-04 [NCT00151242; n = 50]). The treatment protocols on all trials included intensive anthracycline-/cytarabine-based induction therapy and intensive postremission therapy. All patients gave informed consent for both treatment and genetic analysis according to the Declaration of Helsinki. Approval was obtained from the institutional review boards of the participating AMLSG institutions. All cytogenetic and molecular analyses were carried out centrally in the 2 AMLSG reference laboratories at Ulm University and Hannover Medical School.
Cytogenetic and molecular analyses
Cytogenetic studies on pretreatment samples from all patients were performed centrally, and chromosomal aberrations were described according to the International System for Human Cytogenetic Nomenclature.21 For gene mutation analyses, genomic DNA was used as template for PCR-based amplification of DNA fragments spanning mutational “hot spots” in the distinct genes. The amplicons were subsequently assessed for mutations by established methods. The primer sequences used for PCR amplification of NRAS, KRAS, and KIT are described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The amplified fragments of NRAS (exons 1 and 2), KRAS (exons 1 and 2), and KIT (exons 8, 10, 11, and 17) were analyzed for the presence of heterozygous DNA sequence variations by denaturing high-performance liquid chromatography (DHPLC) on a WAVE 3500HT DNA Fragment Analysis System (Transgenomic). The conditions for a successful resolution of heteroduplex molecules on the WAVE-system were determined using the Navigator Version 2.0.0 software (Transgenomic). The samples with a chromatogram that differed from a wild-type reference were further analyzed by sequencing. To test for the presence of homozygous mutations, the amplicons were pooled with corresponding wild-type amplicons, denatured for 10 minutes at 95°C, cooled down slowly to room temperature, and kept at 4°C until the DHPLC analysis. The reannealed DNA duplexes were reanalyzed for mutations in a second DHPLC round. The screening of the FLT3 gene for the presence of internal tandem duplications (ITDs) and tyrosine kinase domain (TKD) mutations was performed as described previously.22 JAK2 was assessed for the presence of JAK2V617F mutation using an allele-specific PCR assay.23
Definition of clinical end points and statistical analyses
The median follow-up for survival was calculated according to the method of Korn.24 The definition of complete remission (CR), relapse-free survival (RFS), and overall survival (OS) followed recommended criteria.25 All univariable and multivariable survival analyses were stratified according to age, with a cut-point of 60 years, because treatment intensity in older patients was lower compared with that in younger patients. Pairwise comparisons between patient characteristics (covariates) were performed by Mann-Whitney or Kruskal-Wallis test for continuous variables and by Fisher exact test for categorical variables. The Kaplan-Meier method was used to estimate the distribution of RFS and OS.26 For RFS analysis, patients receiving an allogeneic hematopoietic stem cell transplantation (HSCT) during first CR (n = 15) were censored at the date of transplantation. Confidence interval (CI) estimation for the survival curves was based on the cumulative hazard function using the Greenwood formula for the SE estimation.27 Cox models were used to identify the prognostic value of gene mutations and chromosome aberrations on RFS and OS and also to identify other prognostic factors.28 In all models, the proportional hazard assumptions were tested.29 A limited backward-selection procedure was used to exclude redundant or unnecessary variables.30 For multivariable analyses, missing data were estimated for covariates using 50 multiple imputations in chained equations incorporating predictive mean matching.30 Statistical analyses were performed with the use of the R packages rms (Version 3.3-1) of the R statistical software platform (Version 2.14.0).31
Results
Demographics, clinical baseline characteristics, and outcomes of the entire study population
The median age of all patients at diagnosis was 41 years (range, 18-74 years), with 92% of the patients being under the age of 61 years. The majority of the patients (164 of 176, 93%) had de novo AML, and 12 (7%) patients had therapy-related AML. Patient baseline characteristics are summarized in Table 1. Complete remission rate was 90% (159 of 176 patients). For postremission therapy, 109 (67%) of the 159 patients received chemotherapy with high-dose cytarabine, 32 (20%) patients had autologous HSCT, and 15 (9%) patients allogeneic HSCT in first CR. Three patients received no further consolidation therapy; 2 patients died during induction therapy and 1 patient declined further treatment. With a median follow-up of 6.04 years (95% CI, 5.3-6.5 years), the estimated RFS and OS rates at 6 years in the entire study cohort were 52% and 66%, respectively.
Clinical characteristics of the entire study cohort
| Characteristic . | All patients, n = 176 . |
|---|---|
| Median age, y (range) | 41 (18-74) |
| % of patients younger than 61 y | 92 |
| Male sex, n (%) | 94 (53) |
| AML history, n (%) | |
| De novo | 164 (93) |
| Therapy-related | 12 (7) |
| Median WBC count, × 109/L (range) | 38.8 (1.1-294.9) |
| Missing data, n | 3 |
| Median platelet count, × 109/L (range) | 34 (7-529) |
| Missing data, n | 3 |
| Median hemoglobin, g/dL (range) | 9.2 (2.5-14.5) |
| Missing data, n | 3 |
| Median % blood blasts (range) | 44 (0-97) |
| Missing data, n | 9 |
| Median % bone marrow blasts (range) | 80 (4-99) |
| Missing data, n | 19 |
| Extramedullary involvement, n (%) | |
| Present | 56 (32) |
| Absent | 117 (68) |
| Missing data | 3 |
| Characteristic . | All patients, n = 176 . |
|---|---|
| Median age, y (range) | 41 (18-74) |
| % of patients younger than 61 y | 92 |
| Male sex, n (%) | 94 (53) |
| AML history, n (%) | |
| De novo | 164 (93) |
| Therapy-related | 12 (7) |
| Median WBC count, × 109/L (range) | 38.8 (1.1-294.9) |
| Missing data, n | 3 |
| Median platelet count, × 109/L (range) | 34 (7-529) |
| Missing data, n | 3 |
| Median hemoglobin, g/dL (range) | 9.2 (2.5-14.5) |
| Missing data, n | 3 |
| Median % blood blasts (range) | 44 (0-97) |
| Missing data, n | 9 |
| Median % bone marrow blasts (range) | 80 (4-99) |
| Missing data, n | 19 |
| Extramedullary involvement, n (%) | |
| Present | 56 (32) |
| Absent | 117 (68) |
| Missing data | 3 |
Frequency and types of secondary genetic lesions
The frequencies and types of secondary genetic lesions are summarized in Table 2. Results of pretreatment cytogenetics were available in 168 (95%) of 176 patients. Overall, secondary chromosome aberrations were identified in 65 (39%) patients. The most frequent secondary chromosome aberration was trisomy 22 identified in 31 (18%) patients, followed by trisomy 8 (n = 27; 16%) and deletions of 7q (n = 9; 5%).
Frequency and types of secondary genetic abnormalities in 176 patients with inv(16) AML
| Secondary genetic abnormality . | Patients with aberration, n (%) . | Patients with missing values, n . |
|---|---|---|
| Chromosomal aberration | 8 | |
| Absent | 103 (61) | |
| Present* | 65 (39) | |
| Trisomy 22 | 31 (18) | |
| Trisomy 8 | 27 (16) | |
| Del(7q) | 9 (5) | |
| Other | 10 (6) | |
| RAS mutation† | 91 (53) | 3 |
| NRAS mutation‡ | 78 (45) | 1 |
| Exon 1 | 45 (26) | 1 |
| Exon 2 | 42 (24) | 1 |
| KRAS mutation | 22 (13) | 3 |
| Exon 1 | 19 (11) | 3 |
| Exon 2 | 3 (2) | 1 |
| KIT mutation§ | 65 (37) | 1 |
| Exon 8‖ | 44 (25) | 1 |
| Exon 10 | 0 (0) | 0 |
| Exon 11 | 3 (2) | 0 |
| Exon 17 | 24 (14) | 0 |
| FLT3 mutation¶ | 30 (17) | 1 |
| FLT3-ITD | 8 (5) | 1 |
| FLT3-TKD | 25 (14) | 1 |
| JAK2V617F mutation | 0 | 2 |
| Secondary genetic abnormality . | Patients with aberration, n (%) . | Patients with missing values, n . |
|---|---|---|
| Chromosomal aberration | 8 | |
| Absent | 103 (61) | |
| Present* | 65 (39) | |
| Trisomy 22 | 31 (18) | |
| Trisomy 8 | 27 (16) | |
| Del(7q) | 9 (5) | |
| Other | 10 (6) | |
| RAS mutation† | 91 (53) | 3 |
| NRAS mutation‡ | 78 (45) | 1 |
| Exon 1 | 45 (26) | 1 |
| Exon 2 | 42 (24) | 1 |
| KRAS mutation | 22 (13) | 3 |
| Exon 1 | 19 (11) | 3 |
| Exon 2 | 3 (2) | 1 |
| KIT mutation§ | 65 (37) | 1 |
| Exon 8‖ | 44 (25) | 1 |
| Exon 10 | 0 (0) | 0 |
| Exon 11 | 3 (2) | 0 |
| Exon 17 | 24 (14) | 0 |
| FLT3 mutation¶ | 30 (17) | 1 |
| FLT3-ITD | 8 (5) | 1 |
| FLT3-TKD | 25 (14) | 1 |
| JAK2V617F mutation | 0 | 2 |
To estimate the frequency of a distinct genetic lesion in the entire cohort data on missing cases is considered in the denominator.
There were patients with more than 1 additional chromosome aberration; thus, the numbers for the distinct additional chromosome aberrations add to more than 65.
Nine cases with a mutation in both NRAS and KRAS.
Nine cases with a NRAS mutation in both exon 1 and 2.
Six cases with a KIT mutation in both exon 8 and 17.
One case scored as mutated by DHPLC but could not be resolved by sequencing due to the presence of low number of mutated cells in the sample.
Three cases with concurrent FLT3-ITD and FLT3-TKD mutation.
NRAS, KRAS, KIT, FLT3, and JAK2 were all assessed for mutations in 171 cases; the number of cases with missing mutation status is given for each gene in Table 2. At least 1 gene mutation was found in 143 (84%) of the 171 cases. Most frequent were NRAS mutations (n = 78 of 175, 45%), followed by mutations of KIT (n = 65 of 175, 37%), FLT3 (n = 30 of 175, 17%), and KRAS (n = 22 of 173, 13%); none of the patients had a JAK2V617F mutation.
Overall, RAS mutations were found in 91 (53%) of 173 patients, including 9 patients having mutations in both NRAS and KRAS. All but 2 RAS mutations were point mutations affecting the known hot spots, that is, glycine 12 and/or Gly13 (exon 1), glutamine 61 (exon 2); in 1 patient with a KRAS mutation, an insertion of proline between alanine 11 and Gly12 was identified, and in a second KRAS mutated case a duplication of Gly10 was found. All but 1 RAS mutation (NRASGly12Cys) were heterozygous.
KIT mutations were found in 65 (37%) of 175 patients with complete analysis on all 4 KIT exons, and all of them were heterozygous. KIT exon 8 was involved most frequently (44 [68%] of 65 mutated cases corresponding to 25% of the 175 patients analyzed in the entire cohort). KIT exon 8 mutations were either small deletions or insertions of variable size, or combinations of deletions and insertions that clustered between leucine 416 and valine 422. A common feature of KIT exon 8 mutations was the loss or replacement of aspartic acid 419. KIT exon 17 mutations comprised 24 (37%) of 65 mutated cases (24 [14%] of the 176 patients analyzed in the entire cohort), and all were point mutations within the activation loop of the kinase domain. In 19 patients, Asp816 was involved, 4 patients showed mutations of asparagine 822, 1 patient of Asp820, and 1 patient had a mutation in both Asp816 and Asn822. KIT exon 11 mutations were only detected in 3 (2%) of 176 patients, including 2 cases with internal duplications and 1 case with a point mutation p.Val560Asp. No mutation was detected in KIT exon 10. Among patients with mutated KIT, 6 (3%) of 175 patients had concurrent mutations in exons 8 and 17.
FLT3 mutations were identified in 30 (17%) of the 175 patients, with FLT3-TKD (n = 25; 14%) being more frequent than FLT3-ITD (n = 8; 5%) mutations. Three of the 175 patients (2%) had both FLT3-ITD and FLT3-TKD mutation.
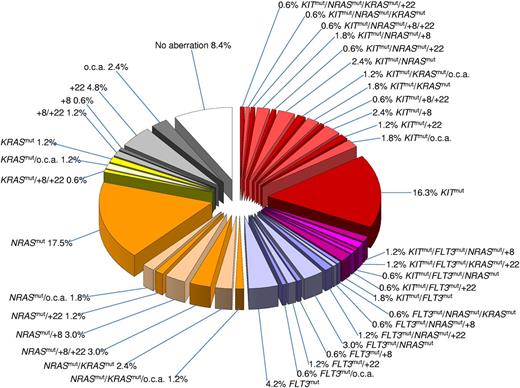
Notably, 25% of patients in our study had more than 1 gene mutation by considering NRAS, KRAS, KIT, FLT3-ITD, and FLT3-TKD as separate mutations. The genetic heterogeneity in inv(16) AML and coexistence of the distinct secondary genetic abnormalities is illustrated in Figure 1. Trisomy 8 frequently co-occurred with NRAS mutations (P = .03). In contrast, concurrent KIT and RAS mutations were less likely (P = .003) than expected based on their frequencies as single molecular alterations. Mutations affecting KIT and FLT3, which both encode class III receptor tyrosine kinases, were found concurrently in 10 patients. No other significant interaction between the distinct secondary genetic abnormalities was observed. None of the secondary genetic alterations was more likely to occur in therapy-related AML compared with cases with de novo AML.
Pie chart illustrating the genetic heterogeneity and coexistence of the distinct secondary genetic abnormalities in AML with inv (16). The chart is based on 166 patients with complete cytogenetic data and complete mutation status on KIT, FLT3, NRAS, and KRAS. Among the secondary chromosome aberrations, trisomy 22 (+22) and trisomy 8 (+8) are indicated; all other secondary chromosome aberrations constitute 1 group abbreviated in the chart as “o.c.a.” Because of the rounding error, all values do not add up to exactly 100%.
Pie chart illustrating the genetic heterogeneity and coexistence of the distinct secondary genetic abnormalities in AML with inv (16). The chart is based on 166 patients with complete cytogenetic data and complete mutation status on KIT, FLT3, NRAS, and KRAS. Among the secondary chromosome aberrations, trisomy 22 (+22) and trisomy 8 (+8) are indicated; all other secondary chromosome aberrations constitute 1 group abbreviated in the chart as “o.c.a.” Because of the rounding error, all values do not add up to exactly 100%.
Clinical characteristics associated with KIT and FLT3 mutations
Differences in clinical characteristics were assessed with respect to mutation status of the 2 genes that had significant impact on outcome, that is, KIT and FLT3. Four groups were included into the comparison: patients with mutated KIT and FLT3, mutated KIT only, mutated FLT3 only, and those with wild type of both genes (Table 3). Between these 4 groups, significant differences were observed in white blood cell (WBC) counts (P = .033) and proportion of circulating blasts (P = .009); the highest values were noted in patients with both KIT and FLT3 mutation, and patients with either KIT or FLT3 mutation also had higher values than patients lacking any KIT and FLT3 mutation.
Clinical characteristics according to mutation status of KIT and FLT3
| Clinical characteristic . | Mutation status . | P . | |||
|---|---|---|---|---|---|
| KIT and FLT3 mutated, n = 10 . | KIT mutated, n = 55 . | FLT3 mutated, n = 20 . | KIT/FLT3 wild type, n = 90 . | ||
| Median age, y (range) | 33 (19-50) | 42 (18-72) | 41 (19-74) | 42 (19-74) | .081 |
| Patients > 61 y, n (%) | 0 | 6 (11) | 2 (10) | 7 (8) | .76 |
| Male sex, n (%) | 8 (80) | 33 (60) | 9 (45) | 43 (48) | .14 |
| AML history, n (%) | .96 | ||||
| De novo | 10 (100) | 50 (91) | 19 (95) | 84 (93) | |
| Therapy-related | 0 | 5 (9) | 1 (5) | 6 (7) | |
| Median WBC count, × 109/L (range) | 86.3 (18.0-243.6) | 47.4 (1.2-271.0) | 43.4 (2.7-284.0) | 32.7 (1.1-294.9) | .033 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median platelet count, × 109/L (range) | 34.5 (17-55) | 34 (7-529) | 37 (7-117) | 35 (8-380) | .92 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median hemoglobin, g/dL (range) | 8.4 (2.5-12.3) | 9.3 (3-14.19) | 9 (5.5-12.69) | 9.2 (3.5-14.5) | .59 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median % blood blasts (range) | 66 (44-94) | 55 (3-97) | 50 (7-90) | 35.5 (0-95) | .009 |
| Missing data, n | 0 | 4 | 1 | 4 | |
| Median % bone marrow blasts (range) | 80 (75-94) | 90 (80-99) | 90 (78.5-91) | 90 (78.5-99) | .72 |
| Missing data, n | 1 | 6 | 2 | 10 | |
| Extramedullary involvement, n (%) | .98 | ||||
| Present | 3 (30) | 16 (31) | 7 (35) | 30 (33) | |
| Absent | 7 (70) | 36 (69) | 13 (75) | 60 (67) | |
| Missing data | 0 | 3 | 0 | 0 | |
| Secondary chromosome aberrations, n (%) | |||||
| None | 5 (50) | 35 (66) | 13 (65) | 50 (59) | .72 |
| Trisomy 22 | 3 (30) | 6 (11) | 4 (20) | 18 (21) | .30 |
| Trisomy 8 | 2 (20) | 9 (17) | 2 (10) | 14 (17) | .88 |
| Other | 0 | 5 (9) | 1 (5) | 11 (13) | .65 |
| Missing data | 0 | 2 | 0 | 6 | |
| Clinical characteristic . | Mutation status . | P . | |||
|---|---|---|---|---|---|
| KIT and FLT3 mutated, n = 10 . | KIT mutated, n = 55 . | FLT3 mutated, n = 20 . | KIT/FLT3 wild type, n = 90 . | ||
| Median age, y (range) | 33 (19-50) | 42 (18-72) | 41 (19-74) | 42 (19-74) | .081 |
| Patients > 61 y, n (%) | 0 | 6 (11) | 2 (10) | 7 (8) | .76 |
| Male sex, n (%) | 8 (80) | 33 (60) | 9 (45) | 43 (48) | .14 |
| AML history, n (%) | .96 | ||||
| De novo | 10 (100) | 50 (91) | 19 (95) | 84 (93) | |
| Therapy-related | 0 | 5 (9) | 1 (5) | 6 (7) | |
| Median WBC count, × 109/L (range) | 86.3 (18.0-243.6) | 47.4 (1.2-271.0) | 43.4 (2.7-284.0) | 32.7 (1.1-294.9) | .033 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median platelet count, × 109/L (range) | 34.5 (17-55) | 34 (7-529) | 37 (7-117) | 35 (8-380) | .92 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median hemoglobin, g/dL (range) | 8.4 (2.5-12.3) | 9.3 (3-14.19) | 9 (5.5-12.69) | 9.2 (3.5-14.5) | .59 |
| Missing data, n | 0 | 2 | 1 | 0 | |
| Median % blood blasts (range) | 66 (44-94) | 55 (3-97) | 50 (7-90) | 35.5 (0-95) | .009 |
| Missing data, n | 0 | 4 | 1 | 4 | |
| Median % bone marrow blasts (range) | 80 (75-94) | 90 (80-99) | 90 (78.5-91) | 90 (78.5-99) | .72 |
| Missing data, n | 1 | 6 | 2 | 10 | |
| Extramedullary involvement, n (%) | .98 | ||||
| Present | 3 (30) | 16 (31) | 7 (35) | 30 (33) | |
| Absent | 7 (70) | 36 (69) | 13 (75) | 60 (67) | |
| Missing data | 0 | 3 | 0 | 0 | |
| Secondary chromosome aberrations, n (%) | |||||
| None | 5 (50) | 35 (66) | 13 (65) | 50 (59) | .72 |
| Trisomy 22 | 3 (30) | 6 (11) | 4 (20) | 18 (21) | .30 |
| Trisomy 8 | 2 (20) | 9 (17) | 2 (10) | 14 (17) | .88 |
| Other | 0 | 5 (9) | 1 (5) | 11 (13) | .65 |
| Missing data | 0 | 2 | 0 | 6 | |
Response to induction therapy
The impact on response to induction therapy was assessed for individual genetic lesions (Table 4). The 2 most frequent secondary chromosome aberrations, namely, trisomy 22 (P = .31) and trisomy 8 (P = .72), were not associated with achievement of CR. Likewise, none of the gene mutations, that is, all RAS (P = .99), NRAS (P = .99), KRAS (P = .51), all KIT (P = .29), KIT exon 8 (P = .57), KIT exon 17 (P = .47), all FLT3 (P = .18), FLT3-ITD (P = .18), and FLT3-TKD (P = .71) influenced achievement of CR (Table 4).
Univariable outcome analyses according to secondary genetic abnormalities
| Clinical end point . | Genetic abnormality . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trisomy 8 . | Trisomy 22 . | RAS . | KIT . | FLT3 . | |||||||||||
| Present, n = 27 . | Absent, n = 141 . | P . | Present, n = 31 . | Absent, n = 137 . | P . | Mutation, n = 91 . | Wild type, n = 82 . | P . | Mutation, n = 65 . | Wild type, n = 110 . | P . | Mutation, n = 30 . | Wild type, n = 145 . | P . | |
| CR rate, % | 89 | 91 | .72 | 97 | 89 | .31 | 90 | 90 | .99 | 94 | 88 | .29 | 83 | 92 | .18 |
| RFS | .89 | .02 | .83 | .01 | .71 | ||||||||||
| Median, y | 1.56 | 3.45 | 8.97 | 1.29 | 3.03 | 8.97 | 1.26 | 8.97 | 2.84 | 3.65 | |||||
| 6-y RFS, % | 50 | 49 | 70 | 43 | 49 | 50 | 38 | 60 | 49 | 50 | |||||
| 95% CI | 33-76 | 40-59 | 55-88 | 35-54 | 39-62 | 40-64 | 27-54 | 47-68 | 32-74 | 41-60 | |||||
| OS | .04 | .12 | .41 | .49 | .04 | ||||||||||
| Median, y | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |||||
| 6-y OS, % | 53 | 69 | 79 | 64 | 64 | 69 | 61 | 70 | 51 | 70 | |||||
| 95% CI | 36-76 | 61-78 | 65-96 | 55-73 | 55-76 | 59-82 | 49-76 | 61-80 | 34-76 | 62-78 | |||||
| Clinical end point . | Genetic abnormality . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trisomy 8 . | Trisomy 22 . | RAS . | KIT . | FLT3 . | |||||||||||
| Present, n = 27 . | Absent, n = 141 . | P . | Present, n = 31 . | Absent, n = 137 . | P . | Mutation, n = 91 . | Wild type, n = 82 . | P . | Mutation, n = 65 . | Wild type, n = 110 . | P . | Mutation, n = 30 . | Wild type, n = 145 . | P . | |
| CR rate, % | 89 | 91 | .72 | 97 | 89 | .31 | 90 | 90 | .99 | 94 | 88 | .29 | 83 | 92 | .18 |
| RFS | .89 | .02 | .83 | .01 | .71 | ||||||||||
| Median, y | 1.56 | 3.45 | 8.97 | 1.29 | 3.03 | 8.97 | 1.26 | 8.97 | 2.84 | 3.65 | |||||
| 6-y RFS, % | 50 | 49 | 70 | 43 | 49 | 50 | 38 | 60 | 49 | 50 | |||||
| 95% CI | 33-76 | 40-59 | 55-88 | 35-54 | 39-62 | 40-64 | 27-54 | 47-68 | 32-74 | 41-60 | |||||
| OS | .04 | .12 | .41 | .49 | .04 | ||||||||||
| Median, y | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |||||
| 6-y OS, % | 53 | 69 | 79 | 64 | 64 | 69 | 61 | 70 | 51 | 70 | |||||
| 95% CI | 36-76 | 61-78 | 65-96 | 55-73 | 55-76 | 59-82 | 49-76 | 61-80 | 34-76 | 62-78 | |||||
NA indicates not applicable.
Survival analyses
Secondary chromosomal aberrations.
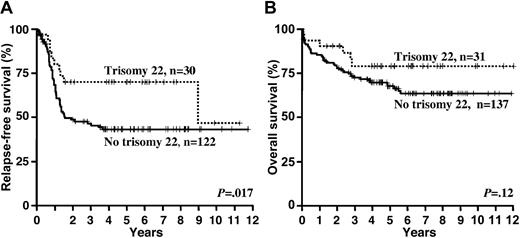
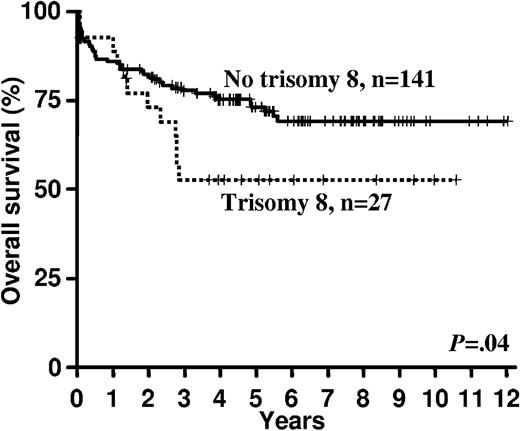
In univariable analyses the presence of trisomy 22 (Table 4 and Figure 2) was associated with a superior RFS (P = .02) and in trend also better OS (P = .12). Trisomy 8 had no impact on RFS (P = .89), but it was associated with an inferior OS (P = .04; Table 4 and Figure 3).
Gene mutations.
In univariable analyses, RAS mutations did not affect RFS (P = .83) and OS (P = .41); the same was true when NRAS and KRAS mutations were analyzed as separate variables (data not shown). In contrast, patients with KIT mutations had an inferior RFS (P = .01; Table 4 and Figure 4); however, the adverse impact of KIT mutations on RFS did not translate into an inferior OS (P = .49). Subset analyses revealed that the adverse impact of mutated KIT on RFS was mainly attributed to mutations affecting KIT exon 8 (P = .006), but not KIT exon 17 (P = .81). Again, the adverse impact of KIT exon 8 mutations on RFS did not translate into an inferior OS (P = .14). There was no effect of FLT3 mutations on RFS (P = .71). In subset analyses, neither FLT3-ITD (P = .26) nor FLT3-TKD (P = .30) mutations affected RFS. However, patients with FLT3 mutations had inferior OS (P = .04; Table 4 and Figure 5) than those without FLT3 mutations which seemed to be because of FLT3-TKD (P = .06) rather than FLT3-ITD (P = .45) mutations.
Evaluation of prognostic variables.
In multivariable analysis on RFS (Table 5), the presence of a KIT mutation was an unfavorable prognostic factor (hazard ratio [HR] = 1.67, P = .04); other variables remaining in the model were log10(WBC) (HR = 1.33; P = .02), and trisomy 22 (HR = 0.54; P = .08). Multivariable analysis on OS (Table 5) revealed the presence of a FLT3 mutation (HR = 2.56; P = .006) as an unfavorable factor; other relevant variables were trisomy 22 (HR = 0.45; P = .07), trisomy 8 (HR = 2.26; P = .02), age (for difference of 10 years HR = 1.46; P = .01), and the presence of therapy-related AML (HR = 2.13; P = .14).
Multivariable analysis for RFS and OS
| End point . | Variable . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| RFS | KIT mutation | 1.67 | 0.99-2.69 | .04 |
| Log10(WBC) | 1.33 | 1.08-1.73 | .02 | |
| Trisomy 22 | 0.54 | 0.27-1.07 | .08 | |
| OS | FLT3 mutation | 2.56 | 1.15-4.85 | .006 |
| Trisomy 22 | 0.45 | 0.19-1.09 | .07 | |
| Trisomy 8 | 2.26 | 1.26-5.00 | .02 | |
| Age | 1.46* | 1.02-1.94 | .01 | |
| t-AML | 2.13 | 0.86-6.61 | .14 |
| End point . | Variable . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| RFS | KIT mutation | 1.67 | 0.99-2.69 | .04 |
| Log10(WBC) | 1.33 | 1.08-1.73 | .02 | |
| Trisomy 22 | 0.54 | 0.27-1.07 | .08 | |
| OS | FLT3 mutation | 2.56 | 1.15-4.85 | .006 |
| Trisomy 22 | 0.45 | 0.19-1.09 | .07 | |
| Trisomy 8 | 2.26 | 1.26-5.00 | .02 | |
| Age | 1.46* | 1.02-1.94 | .01 | |
| t-AML | 2.13 | 0.86-6.61 | .14 |
HR greater than (less than) 1 indicates an increased (decreased) risk for the category “present” for a dichotomous variable and for a higher value of a continuous variable.
t-AML indicates therapy-related acute myeloid leukemia.
HR for difference of 10 y.
Discussion
In this study, 84% of patients with inv(16) AML had a mutation affecting at least 1 of the genes analyzed, that is, NRAS, KRAS, KIT, and FLT3. The high frequency of secondary gene mutations in our study is in line with previous studies in inv(16) AML.12,32,33 Thus, inv(16) AML constitutes a paradigm for the model explaining AML leukemogenesis, where second hits causing survival/proliferation advantage (RAS, KIT, or FLT3 mutations) cooperate with a primary hit (CBFB/MYH11 rearrangement) that confers a block in hematopoietic differentiation.4 This model is supported by animal studies, where the coexpression of Cbfb/MYH11 with mutant KIT34 or CBFB/MYH11 with FLT3-ITD35 induced or accelerated the development of leukemia. Although intensive postremission therapy, such as repetitive cycles of higher doses of cytarabine, has substantially improved outcome of patients with inv(16) AML,15 approximately half of the patients in this cytogenetic AML subgroup are still not cured.14,15 Thus, there is a need for markers to refine risk stratification of patients at diagnosis. Ideally, such markers also could serve as therapeutic targets.
In our study, KIT mutations were found in 37% of the patients, a value in line with other data published for inv(16) AML.7-11,13 The presence of a KIT mutation was a significant factor for shorter RFS, but not for OS. One explanation why the adverse impact on RFS did not translate into shorter OS might be the high sensitivity to salvage therapy.14,15 Indeed, in our study a second CR was achieved in 76% (25 of 33) of KIT-mutated cases. The prognostic impact of KIT mutations in adult inv(16) AML was evaluated in several smaller studies yielding somewhat controversial results. In analogy to our study, Care et al showed in 63 adult patients with inv(16) AML that KIT exon 8 mutations were associated with a higher relapse rate, but not with inferior OS.11 In the study by Cairoli et al, 50 adult AML with inv(16) were studied for the prognostic relevance of KIT mutations located within exons 8, 10, and 17. In their study, KIT mutations as 1 group were associated with a higher relapse rate, whereas no difference was observed in OS.8 However, in contrast to our study in none of the 2 studies the prognostic impact of KIT mutations was assessed in a multivariable setting, in particular within the context of other relevant factors and molecular markers.8,11 In the study from the Cancer and Leukemia Group B (CALGB) on 61 adult inv(16) AML, KIT mutations conferred a higher cumulative incidence of relapse, but this negative impact on cumulative incidence of relapse was attributed to KIT exon 17 mutations.13 In this CALGB study, KIT mutations also were found as an adverse factor for OS on multivariable analysis, whereas no effect on OS was observed on univariable analysis.13 In contrast, several other smaller studies did not identify KIT mutations as a relevant prognostic factor in both adult as well as pediatric inv(16) AML.7,9,10,36 The discrepancies among the studies might be in part related to differences in treatment regimens, including salvage therapy, selection of the study cohorts, and the relatively small numbers of patients analyzed. However, because of the high frequency of KIT mutations in inv(16) AML and their “gain-of-function“ nature that leads to constitutive kinase activity (exon 17) or KIT receptor hyperactivation in response to stem cell factor stimulation (exon 8),32,37,38 the mutated KIT protein offers an attractive target for tyrosine kinase inhibitors. Trials targeting mutant or overexpressed KIT are currently ongoing in patients with CBF-AML (NCT01238211, NCT00850382).
In our study, we detected FLT3 mutations in 17% of the patients with a 3-fold higher frequency of TKD mutations compared with ITDs. In previous studies, FLT3-ITD has been reported as a relatively uncommon molecular alteration in inv(16) AML occurring in 0% to 7% of patients.39-42 Fourteen percent of patients in our study carried an FLT3-TKD mutation. The reported frequencies for FLT3-TKD mutations in inv(16) AML in previous smaller studies ranged between 3.7% and 8.6%7,11,33,42,43 ; only a study by the United Kingdom Medical Research Council (MRC) reported a high incidence of FLT3-TKD mutations of approximately 24%.40 The prognostic relevance of FLT3 mutations in inv(16)–positive AML is still not well established. Boissel et al analyzed 47 patients and found 3 patients with a FLT3-TKD mutation; all 3 patients had a dismal outcome.7 In a more recent study from MD Anderson Cancer Center, FLT3-ITD and FLT3-TKD mutations as 1 group conferred inferior progression-free survival in inv(16) AML.36 However, lack of multivariable analysis in the context of other relevant prognostic factors hampers the interpretation of the results in this study.36 In contrast, a favorable effect of FLT3-TKD mutations on OS was reported in one MRC study in unselected AML40 ; this favorable effect of FLT3-TKD mutations on OS also was retained in a subset analysis on 55 patients with inv(16) AML.40 To our knowledge, our large study is the first one showing the independent adverse impact of FLT3 mutations on OS in inv(16) AML within the context of other clinically and genetically relevant factors. Importantly, this adverse effect appeared to be mainly conferred by FLT3-TKD mutations.
Compared with other cytogenetic AML subgroups, RAS mutations represent a particularly frequent molecular abnormality in inv(16) AML,7,44,45 with an incidence of up to 50%.7,33,44,45 We found NRAS mutations, KRAS mutations, or both in 53% of the patients. NRAS mutations were almost 4 times more frequent than mutations affecting KRAS. In analogy to previous studies,7,44 we did not find RAS mutations to be associated with outcome in inv(16)–positive AML. We did not identify any significant interaction between the distinct gene mutations and none of the mutated genes were mutually exclusive.
Notably, among the additional genetic lesions observed also secondary chromosomal aberrations significantly impacted outcome of patients. In a previous study, we identified trisomy 22 as a favorable prognostic factor for RFS in AML with inv(16).14 This finding was confirmed in a subsequent CALGB study.15 More recently, an MRC study reported that the presence of trisomy 22 is associated with a better survival in inv(16) AML.16 In the present study, we could confirm the favorable impact of trisomy 22 on RFS. In addition and in line with the MRC study,16 we found trisomy 22 to be a favorable factor for OS. Moreover, with trisomy 8 we have identified a second chromosomal aberration that impacts outcome in inv(16) AML. In both univariable and multivariable analysis, the presence of trisomy 8 was a relevant factor for inferior OS. In our study, 7q deletions were found using standard cytogenetic methods in 9 (5%) patients. In a recent study from our group, 7q deletions represented a more frequent genetic alteration occurring in approximately 10% of CBF-AML cases.17 However, one- half of 7q deletions in that study were only detectable using high-resolution genetic profiling.17 In the present study in which diagnosis of 7q deletion was based on conventional cytogenetics only, none of the clinical end points, that is, CR, RFS, and OS, was adversely affected by the presence of the deletions (data not shown).
Among the clinical characteristics, we confirmed the unfavorable impact of age on the risk of relapse in adult inv(16) AML,15,46 and we showed that higher WBC at diagnosis independently predicts for an inferior RFS in this AML subgroup. The presence of extramedullary involvement was not a significant prognostic factor in our study. On multivariable analysis, the presence of therapy-related AML was in trend a relevant factor for a shorter survival. This supports the findings of one of our recent studies showing that patients with therapy-related inv(16) AML have inferior outcome compared with those with de novo disease.47
Our large study highlights the importance of both secondary chromosomal aberrations as well as gene mutations for the clinical heterogeneity of AML with inv(16). Based on our findings, trisomy 8 as well as KIT mutations, FLT3 mutations, or both represent adverse prognostic markers in this favorable AML entity and therefore allow the identification of patients at higher risk for relapse and with inferior OS, whereas the presence of trisomy 22 is associated with a very low relapse probability and superior survival. These findings are of clinical relevance, particularly in light of current approaches aiming for the development of small molecule inhibitors that target mutant KIT and FLT3 as well as for intensification of therapy using, for example, allogeneic HSCT.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the AMLSG institutions and investigators who contributed to this study. They also thank Patricia Erdmann and Susanne Kuhn for technical support with molecular analyses.
This work was supported in part by grants 01GI9981 (Network of Competence Acute and Chronic Leukemias) and 01KG0605 (IPD-Meta-Analysis: A Model-based Hierarchical Prognostic System for Adult Patients with AML) from the Bundesministerium für Bildung und Forschung, Germany. L.B. was supported by the Deutsche Forschungsgemeinschaft (Heisenberg-Stipendium BU 1339/3-1).
Authorship
Contribution: P.P., J.D., R.F.S., H.D., and K.D. designed research; K.D. contributed reagents and analytical tools to this study; P.P. and J.D. carried out laboratory-based research; P.P., J.D., V.I.G., L.B., A.C., D.S., R.F.S., H.D., and K.D. contributed to data collection and interpretation; R.F.S., H.D., and K.D. provided administrative support; B.S. and S.K. participated in cytogenetic review and interpretation of the results; R.F.S. performed statistical analyses; A.G., A.K., C.-H.K., G.H., H.-A.H., H.K., H.R.S., K.G., J.K., M.L., v.M.L.-T., M.Ri., M.Ru., M.W., R.F.S., H.D., and K.D. were involved directly or indirectly in care of patients, sample procurement, or both; P.P., R.F.S., H.D., and K.D. drafted the manuscript; and all authors agreed on the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the AMLSG institutions and investigators who contributed to this study appears in the online supplemental Appendix.
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, D-89081 Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.
References
Author notes
P.P. and J.D. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal