Abstract
Recently, we showed that increased miR-181a expression was associated with improved outcomes in cytogenetically normal acute myeloid leukemia (CN-AML). Interestingly, miR-181a expression was increased in CN-AML patients harboring CEBPA mutations, which are usually biallelic and associate with better prognosis. CEBPA encodes the C/EBPα transcription factor. We demonstrate here that the presence of N-terminal CEBPA mutations and miR-181a expression are linked. Indeed, the truncated C/EBPα-p30 isoform, which is produced from the N-terminal mutant CEBPA gene or from the differential translation of wild-type CEBPA mRNA and is commonly believed to have no transactivation activity, binds to the miR-181a-1 promoter and up-regulates the microRNA expression. Furthermore, we show that lenalidomide, a drug approved for myelodysplastic syndromes and multiple myeloma, enhances translation of the C/EBPα-p30 isoform, resulting in higher miR-181a levels. In xenograft mouse models, ectopic miR-181a expression inhibits tumor growth. Similarly, lenalidomide exhibits antitumorigenic activity paralleled by increased miR-181a expression. This regulatory pathway may explain an increased sensitivity to apoptosis-inducing chemotherapy in subsets of AML patients. Altogether, our data provide a potential explanation for the improved clinical outcomes observed in CEBPA-mutated CN-AML patients, and suggest that lenalidomide treatment enhancing the C/EBPα-p30 protein levels and in turn miR-181a may sensitize AML blasts to chemotherapy.
Key Points
High miR-181a levels associate with CEBPA mutations and likely contribute to the favorable prognostic impact of these mutations in AML.
Lenalidomide induces CEBPA-dependent miR-181a expression and in turn increases sensitivity to chemotherapy in AML blasts.
Introduction
Cytogenetically normal acute myeloid leukemia (CN-AML) is the largest subset of AML, comprising up to 45% of cases.1 In CN-AML patients, several molecular markers have been identified as predictors of clinical outcome, including the mutational status of the CEBPA gene.2-5 CEBPA encodes the CCAAT enhancer-binding protein α (C/EBPα), a transcription factor critical for normal myeloid cell differentiation.6 In the hematopoietic system, C/EBPα is expressed early in myeloid precursors and up-regulated during their commitment to granulocytic pathway and maturation.6-8 Several reports demonstrated that diverse molecular mechanisms are responsible for C/EBPα inactivation of expression or function in various types of leukemia (AML and chronic myelogenous leukemia).9,10
The C/EBPα protein contains multiple N-terminal transactivation domains and 1 C-terminal basic-leucine zipper region, responsible for DNA binding and protein dimerization.11,12 As a result of differential translation initiation from a single CEBPA mRNA molecule, 2 protein isoforms are produced (full-length p42 and N-terminally truncated p30),13 which play different functions in gene regulation and proliferation.14-16 Approximately 15% of CN-AML patients carry mutations in the CEBPA gene that are clustered in 2 regions. The N-terminal frame-shift mutations, which occur in approximately 90% of CEBPA mutant patients, prevent expression of the full-length transcriptionally active C/EBPα-p42 protein, but allow translation of the truncated C/EBPα-p30 isoform.2,14,17,18 In contrast, the C-terminal mutations disrupt the basic-leucine zipper domain, and abolish C/EBPα DNA-binding.14 In most cases, the N-terminal mutations occur concurrently with a C-terminal mutation and are present on different alleles. Paradoxically, those biallelic double CEBPA mutations have a more favorable prognostic impact.3,19 However, the mechanisms through which CEBPA mutations lead to better response to treatment and improved outcomes have not been described.
MicroRNAs (miRNAs) are small noncoding RNAs that repress translation and/or initiate degradation of specific target messenger RNAs (mRNA) and have recently been shown to play a role in carcinogenesis.20 Genome-wide expression profiling of AML has revealed miRNA signatures associated with distinct cytogenetic or molecular subtypes of the disease5,21 and have proven useful to predict clinical outcome in AML patients.22
Recently, we and others reported an association of higher miR-181a expression and favorable outcomes especially in a molecular subset of high-risk CN-AML patients (FLT3-ITD positive and/or NPM1 wild-type) and in AML patients with cytogenetic abnormalities.22-24 In healthy cells, miR-181a regulates B-cell development, influences T-cell sensitivity to antigens by modulating T-cell receptor signaling intensity, and is involved in early steps of hematopoiesis.25 A tumor suppressor activity of miR-181a was reported in chronic lymphocytic leukemia,26 gliomas,27 and astrocytomas,28 and was attributed to direct targeting of BCL2 family members.28 Recently, it was shown that ectopic expression of miR-181a sensitizes AML cell lines to chemotherapy.29 In addition, we reported that higher miR-181a expression was associated with the presence of CEBPA mutations in CN-AML patients.5
In this study, we show that higher miR-181a expression in CN-AML patients is predominantly present in the patients harboring CEBPA N-terminal mutations, as opposed to those having single C-terminal mutations or wild-type CEBPA. We also demonstrate that miR-181a expression is directly modulated by the C/EBPα-p30 isoform. Furthermore, we show that the immunomodulatory agent lenalidomide enhances the expression of the N-truncated C/EBPα-p30 that is followed by increased miR-181a expression and augmented sensitivity of leukemia cells to chemotherapy. Finally, using murine models engrafted with human AML cells either expressing ectopic miR-181a, or treated in vivo with lenalidomide, we observed a strong inhibition of tumor growth. Thus, our findings support future investigations seeking novel pharmacologic therapies for AML patients with inadequate expression and/or function of the full-length C/EBPα-p42, with the intent to enhance the expression of the N-truncated C/EBPα-p30 isoform and thus, the expression of miR-181a. The therapeutic induction of C/EBPα-p30 and in turn of miR-181a are intended to recapitulate the favorable outcomes of AML patients with double CEBPA mutations after treatment with intensive chemotherapy.
Methods
Cell lines and AML patient samples
The cell lines THP-1 (TIB-202), MV4-11 (CRL-9591), HL60 (CCL-240), KG1a (CCL-246.1), and HEK-293T were from ATCC. CEBPA mutations were analyzed in adults < 60 years of age with untreated, primary molecular high-risk CN-AML enrolled onto the cancer and leukemia Group B (CALGB) treatment protocols 9621 and 19 808.5 RNA samples were analyzed for miRNA expression using a previously reported Ohio State University (OSU) customized miRNA chip.5 Images of the miRNA microarrays (E-MTAB-1320) were acquired, and calculations, normalizations, and filtering of signal intensity for each microarray spot were performed as previously reported.5 Expression levels of miR-181a were compared according to CEBPA mutation status using the Wilcoxon-Rank sum test. Estimated probabilities of survival were calculated using the Kaplan-Meier method, and the log-rank test evaluated differences between survival distributions. Written informed consent for these studies was obtained from all patients in accordance with the Declaration of Helsinki. The AML patient blasts used for ex vivo experiments were obtained from apheresis blood samples collected from patients treated at OSU and stored in the OSU Leukemia Tissue Bank. After thawing, the cells were placed in RPMI medium supplemented with 20% FBS and cytokine cocktail supporting proliferation of hematopoietic progenitors (StemSpan CC100, StemCell Technologies) and the adherent cells were removed. Informed consent to use the tissue for laboratory studies was obtained from each patient according to the OSU institutional guidelines.
Plasmids transfection and miRNA detection by nanoelectroporation
Nanoelectroporation (NEP) is a microchannel-nanochannel array with the 2 separate microchannels for cell and plasmid loading, that is capable of delivering the precise amount of transfection reagents into individual cells.30 C/EBPα expression vectors containing either wild-type, N-terminal (H24Afs), or C-terminal (R300L) mutants were loaded in one side of microchannels, whereas KG1a cell suspension (< 103/mL) was loaded into the other side of microchannels with a single cell in each microchannel being moved to the tip of the nanochannel by optical tweezers. Two electric pulses across the nanochannels (240V, 5ms each) were applied through platinum electrodes to conduct the transfection. Transfected cells were cultured in situ after the removal of the plasmid solution. To detect the miR-181a expression level inside individual cells, a locked nucleic acid (LNA) molecular beacon (MB; miR-181a LNA-MB sequence: 5′-FAM-accgcg-ActCacCgaCagCgtTgaAtgtt-cgcggt-BHQ1-3′ [upper cases represent LNA]; Sigma-Aldrich) was transfected 24 hours later through the same nanochannel. The images were acquired 4 in 4% PFA solution, at 20°C by Nicon Eclipse Ti microscope (Nikon Instruments) using Nikon Plan Apo VC 100X, N.A. = 1 and Evolve 512 EMCCD camera (Photometrics). NIS-Elements AR 3.2 software (Nikon Instruments) was used for image acquisition and processing. The average fluorescence of each cell (with background correction) was measured 45 minutes after the MB transfection. Three or 4 cells were transfected by NEP for each plasmid.
Xenograft models in NOD/SCID mice
THP-1 cells were injected (10 × 106 cells/mouse) subcutaneously (1 tumor per mouse) into 4- to 6-week-old NOD/SCID mice obtained from The Jackson Laboratory. Lenalidomide (50 mg/kg) or vehicle control (1 × PBS) were directly injected into the tumors twice a week for 2 weeks. Five tumors were treated with lenalidomide and 3 tumors were injected with vehicle. After 6 weeks the mice were killed and tumor sizes were assessed using caliper and by weight. To test the effect of miR-181a overexpression on tumor growth, THP-1 cells were transiently transfected with the Amaxa system (Solution V, Lonza) with pSUPERIOR-retro-puro vector overexpressing miR-181a, or empty vector. After a 6-hour culture, the cells were injected, and 6 weeks later tumor sizes were determined as described above. The in vivo animal studies were conducted with the approval of the OSU Institutional Lab Animal Care and Use Committee, and in accordance with the National Institutes of Health Guidelines for animal care.
Additional methods are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
N-terminal C/EBPα mutations are associated with higher miR-181a levels in CN-AML
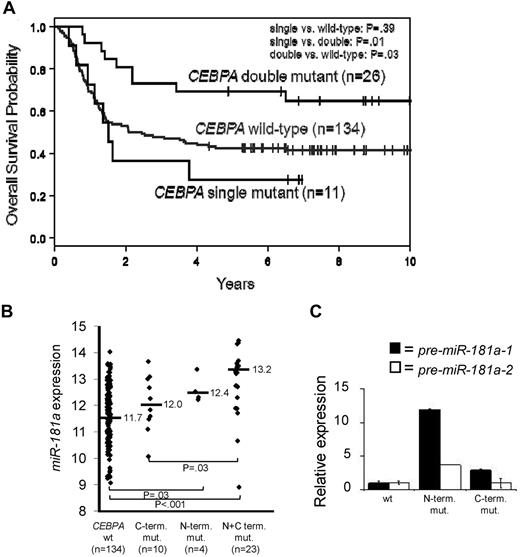
We demonstrated that CN-AML patients with either high miR-181a expression,22,24 or harboring CEBPA mutations5 have a more favorable prognosis. Mutations in CEBPA tend to cluster to 2 regions, N or C-terminal and are present mainly in CN-AML patients.5,13 The majority of these patients have mutations in both alleles (double mutants), which have better outcomes compared with the patients with monoallelic mutations or wild-type gene (Figure 1A). We also showed that patients with CEBPA mutations had higher miR-181a expression.5
C/EBPα-p30 expression is correlated with increased miR-181a expression. (A) Overall Survival of younger (18-59 years) CN-AML patients with CEBPA wild-type, single mutations (N or C-terminal), or double mutations (N and C-terminal mutations together). (B) miR-181a expression in younger (18-59 years) CN-AML patients with either CEBPA wild-type (CEBPA wt), monoallelic C-terminal mutations (C-term. mut.), monoallelic N-terminal mutations (N-term. mut.), or concurrent N and C-terminal mutations (N+C term. mut.). (C) Representative patient bone marrow samples with characterized CEBPA mutations effecting either the N-terminus or C-terminus of the C/EBPα protein. Quantitative real-time RT-PCR data showing the association of the CEBPA mutation status and expression of the precursor (pre) miR-181a-1 (encoded on chromosome 1 locus) or miR-181a-2 (encoded on chromosome 9 locus). Each bar represents the average of triplicate measurements and error bars denote SEM.
C/EBPα-p30 expression is correlated with increased miR-181a expression. (A) Overall Survival of younger (18-59 years) CN-AML patients with CEBPA wild-type, single mutations (N or C-terminal), or double mutations (N and C-terminal mutations together). (B) miR-181a expression in younger (18-59 years) CN-AML patients with either CEBPA wild-type (CEBPA wt), monoallelic C-terminal mutations (C-term. mut.), monoallelic N-terminal mutations (N-term. mut.), or concurrent N and C-terminal mutations (N+C term. mut.). (C) Representative patient bone marrow samples with characterized CEBPA mutations effecting either the N-terminus or C-terminus of the C/EBPα protein. Quantitative real-time RT-PCR data showing the association of the CEBPA mutation status and expression of the precursor (pre) miR-181a-1 (encoded on chromosome 1 locus) or miR-181a-2 (encoded on chromosome 9 locus). Each bar represents the average of triplicate measurements and error bars denote SEM.
We sought to determine whether the higher miR-181a expression was associated with a particular type of CEBPA mutation (N vs C-terminal). We analyzed the miR-181a expression among younger (< 60 years) CN-AML patients (n = 171) with wild-type CEBPA, monoallelic C-terminal, monoallelic N-terminal, or biallelic (concurrent N and C-terminal) mutated CEBPA (see supplemental Methods). Expectedly, very few patients presented with single N-mutations (n = 4). Patients with N-terminal mutations (single or double) tended to have a higher miR-181a expression than patients with a single C-terminal mutation or wild-type CEBPA (Figure 1B). These differences were statistically significant when patients with biallelic (N and C-terminal) mutations compared with patients with single C-terminal mutations (P = .03) or with wild-type alleles (P < .001). Given the very small number, single N-terminal mutation patients could not be compared with those in the remaining subgroups.
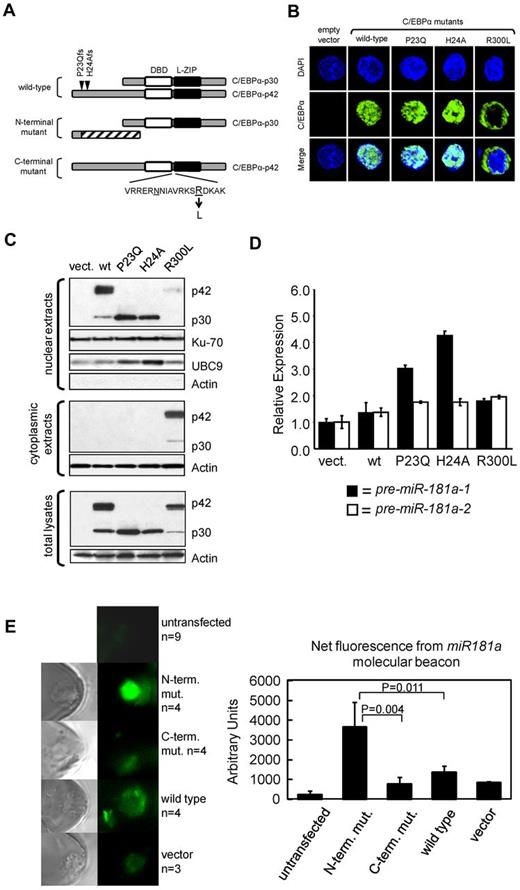
These results were confirmed by quantitative RT-PCR in 3 patients (Figure 1C), each representing a different CEBPA genotype (wild-type, N-terminal I68Lfs, and C-terminal E167GfsX3 mutants). The relative expression of C/EBPα-p42 and p30 polypeptides were determined (supplemental Figure 1). miR-181a is coded by 2 separate genes, miR-181a-1 and miR-181a-2, located on chromosomes 1q32.1 and 9q33.3, respectively. Thus, we tested if the expression of both genes were associated with the distinct C/EBPα mutants by measuring the levels of miR-181a precursors, chromosome 1-derived pre-miR-181a-1 and chromosome 9-derived pre-miR-181a-2. A 12-fold higher expression of pre-miR-181a-1 was observed in the patient harboring an N-terminal C/EBPα mutation compared with the patient expressing the wild-type C/EBPα (Figure 1C). Only a slight increase of pre-miR-181a-1 expression was seen in the sample containing C-terminal C/EBPα mutation. In contrast, the expression of precursor miR-181a-2 was not affected in the AML patient with C-terminal mutation, and up-regulated only 3.73-fold in the case with N-terminal mutation, compared with the patient with wild-type CEBPΑ (Figure 1C). These data suggest that miR-181a-1 gene expression derived from chromosome 1 is predominantly affected in patients harboring N-terminal CEBPA mutations. To further validate these results functionally and to avoid the interplay of various C/EBPα protein isoforms produced simultaneously from the wild-type and mutant alleles in individual AML specimens, we constructed vectors expressing either wild-type or mutant CEBPA genes and introduced them individually into the K562 cell line, which does not express endogenous C/EBPα.6,31 Schematic diagrams for the various C/EBPα wild-type and mutant proteins are shown in Figure 2A. Two N-terminal mutations (P23Qfs and H24Afs) led to frame-shifts precluding expression of the full-length p42 protein, but allowing for reinitiation of translation at the downstream in-frame translational start site; this resulted in the expression of N-terminally truncated C/EBPα-p30 isoform.9,14 The C-terminal mutation, R300L, has an amino acid substitution within the highly conserved nuclear localization sequence (NLS).32 Confocal microscopy of transiently transfected K562 cells (Figure 2B, supplemental Figure 2A) showed that the wild-type C/EBPα proteins were localized in the nuclei, as were the products from the N-terminally mutated genes (P23Q and H24A). In contrast, the C-terminal mutant protein (R300L) was found mainly in the cytoplasm. These results were confirmed by Western blot analyses using nuclear and cytoplasmic fractions prepared from the same cells (Figure 2C). In accord with the published data,14 the N-terminal mutants produced the C/EBPα-p30 protein isoform, which correlated with the increased expression of nuclear protein UBC9, previously described as being indirectly regulated by C/EBPα-p30 and not by the full-length p42 isoform.33
Forced expression of patient-derived N-terminal mutated CEBPA into K562 cells induces up-regulation of the endogenous miR-181a. (A) Schematic diagram of C/EBPα expressed from vectors containing the CEBPA gene isolated from AML patients with wild-type CEBPA or identified CEBPA mutations. N-terminal mutations locations, P23Qfs and H24Afs, are depicted by inverted triangles on the wild-type protein. N-terminal mutations produce the N-truncated C/EBPα-p30 isoform. C-terminal mutation contains an amino acid substitution R300L located within the nuclear localization sequence domain (V-K; underlined amino acids are believed to be required for nuclear localization). DNA-binding domain (DBD) and leucine-zipper (L-ZIP) domain are represented by white and black boxes, respectively. (B) Confocal microscopy images for K562 cells transiently expressing those C/EBPα isoforms described in panel A. All C/EBPα isoforms are identified with green labeling and DAPI staining performed as described,50 indicates location of nucleus shown in blue. The wild-type C/EBPα and N-terminal mutant C/EBPα were localized to the nucleus. In contrast, the C-terminal mutant C/EBPα (R300L) was localized in the cytoplasm. (C) Western blot analysis of cellular fractions after ectopic expression of C/EBPα isoforms described in panel A and expressed in the leukemia cell line K562. Notably, all isoforms are localized in the nucleus except for R300L which is predominately cytoplasmic. UBC9 serves as a positive-control for C/EBPα-p30 expression.33 Ku-70 and Actin serve as internal loading controls. (D) Quantitative real-time RT-PCR analysis for miR-181a expression in the K562 transiently transfected with the expression constructs described in panel A. Notably, miR-181-1 expression is highest in those cells transiently expressing the C/EBPα-p30 isoform. Each bar represents average of triplicate measurements and error bars denote SEM. (E) FAM fluorescence images of KG1a cells transfected with C/EBPα expression plasmids (N-terminal or C-terminal mutants, wild-type C/EBPA, or empty vector) and 1 day later with a fluorescent miR-181a LNA-MB (left), assessed for relative expression of endogenous miR-181a levels (right). FAM is released from its quencher and emitted fluorescence when MBs unfold and bind to miR-181a. Average of 3 to 9 (n) measurements is shown and error bars denote SEM.
Forced expression of patient-derived N-terminal mutated CEBPA into K562 cells induces up-regulation of the endogenous miR-181a. (A) Schematic diagram of C/EBPα expressed from vectors containing the CEBPA gene isolated from AML patients with wild-type CEBPA or identified CEBPA mutations. N-terminal mutations locations, P23Qfs and H24Afs, are depicted by inverted triangles on the wild-type protein. N-terminal mutations produce the N-truncated C/EBPα-p30 isoform. C-terminal mutation contains an amino acid substitution R300L located within the nuclear localization sequence domain (V-K; underlined amino acids are believed to be required for nuclear localization). DNA-binding domain (DBD) and leucine-zipper (L-ZIP) domain are represented by white and black boxes, respectively. (B) Confocal microscopy images for K562 cells transiently expressing those C/EBPα isoforms described in panel A. All C/EBPα isoforms are identified with green labeling and DAPI staining performed as described,50 indicates location of nucleus shown in blue. The wild-type C/EBPα and N-terminal mutant C/EBPα were localized to the nucleus. In contrast, the C-terminal mutant C/EBPα (R300L) was localized in the cytoplasm. (C) Western blot analysis of cellular fractions after ectopic expression of C/EBPα isoforms described in panel A and expressed in the leukemia cell line K562. Notably, all isoforms are localized in the nucleus except for R300L which is predominately cytoplasmic. UBC9 serves as a positive-control for C/EBPα-p30 expression.33 Ku-70 and Actin serve as internal loading controls. (D) Quantitative real-time RT-PCR analysis for miR-181a expression in the K562 transiently transfected with the expression constructs described in panel A. Notably, miR-181-1 expression is highest in those cells transiently expressing the C/EBPα-p30 isoform. Each bar represents average of triplicate measurements and error bars denote SEM. (E) FAM fluorescence images of KG1a cells transfected with C/EBPα expression plasmids (N-terminal or C-terminal mutants, wild-type C/EBPA, or empty vector) and 1 day later with a fluorescent miR-181a LNA-MB (left), assessed for relative expression of endogenous miR-181a levels (right). FAM is released from its quencher and emitted fluorescence when MBs unfold and bind to miR-181a. Average of 3 to 9 (n) measurements is shown and error bars denote SEM.
Next, we measured the expression of miR-181a precursors (pre-miR-181a-1 and pre-miR-181a-2) in K562 cells expressing ectopic C/EBPα mutant or wild-type proteins (Figure 2D). Consistent with our previous data, the highest expression of pre-miR-181a-1 was observed in the cells expressing the N-terminal mutants (3- to 4.5-fold), however only a slight increase in pre-miR-181a-1 expression was observed for the wild-type and C-terminally mutated isoforms, compared with the empty vector control. The slight increase of pre-miR-181a-1 expression seen among the C-terminally mutated isoform was probably because of the small but noticeable presence of C/EBPα protein in the nuclear fraction. The expression of H24Afs N-terminal mutant resulted in the highest levels of pre-miR-181a-1 (3.2-fold more than the levels induced by the wild-type C/EBPα), whereas the expression of the P23Qfs N-terminal mutant was associated with 2.2-fold higher pre-miR-181a-1 expression (Figure 2D). In general, the increase in miR-181a-1 expression was associated with the expression and function (as determined by UBC9 expression; Figure 2C) of the C/EBPα-p30. The expression of miR-181a-1 was also up-regulated in cells coexpressing N and C-terminal mutants of C/EBPα similarly to what we observed in patients with double CEBPA mutations (supplemental Figure 2). In agreement with the results obtained from AML patient samples (Figure 1B), the effect of each C/EBPα isoform on the chromosome 9-derived pre-miR-181a-2 expression was not as prominent (Figure 2D).
To demonstrate the differential impact of CEBPA mutants and wild-type on miR-181a expression, we transfected plasmids encoding wild-type, N-terminal and C-terminal mutated CEBPA in AML KG1a cells that do not express CEBPA (C.J.H., unpublished data, February 2013; also Radomska et al37 ) using a nanochannel electroporation (NEP) technique, which was developed by our group to deliver precise amounts of charged molecules or particles into individual cells with minimal cell damage.30 NEP relies on the selection of applied electric voltage, pulse duration and pulse number to achieve plasmid dosage control. Using the same poration conditions, a similar amount of plasmids was delivered to each transfected cell.30 A fluorescent LNA-MB was used to measure miR-181a at a single cell resolution. Figure 2E shows a significant increase in miR-181a expression in cells receiving the N-terminal mutated CEBPA compared with those receiving C-terminal mutated or wide-type CEBPA.
Taken together, our data suggest that N-terminal mutants are functionally more active than wild-type and C-terminal mutants in enhancing the expression of miR-181a-1.
The expression of the pre-miR-181a-1 is regulated by the N-terminally truncated C/EBPα-p30 protein
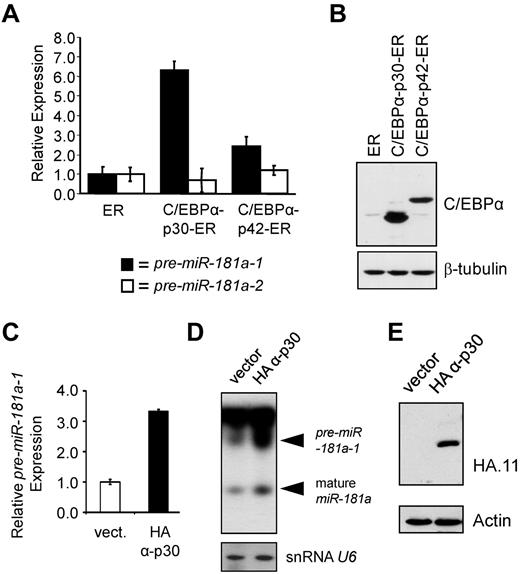
As noted in the introduction and diagrammed in Figure 2A, CEBPA mRNA can be translated from at least 2 in-frame translation start sites, leading to production of full-length C/EBPα-p42 and N-terminally truncated C/EBPα-p30 proteins.34 Because we observed a correlation between the expression of patient-derived N-terminal frame-shift mutants and miR-181a levels, we investigated the C/EBPα-p30/miR-181a interplay using inducible K562 stable cell lines expressing either C/EBPα-p42 or C/EBPα-p30 isoforms fused to human estrogen receptor (ER) ligand binding domain.31 On β-estradiol treatment, C/EBPα-ER fusion proteins translocate into nuclei, where they can exert their effects. Figure 3A shows miR-181a precursor levels assayed in K562 stable lines after a 48-hour treatment with β-estradiol and compared with the levels in the control cell line that was transduced with the vector expressing the ER portion of the fusion protein. In the presence of C/EBPα-p30-ER, pre-miR-181a-1 expression was increased 6.3-fold (Figure 3A). A somewhat lesser effect (2.42-fold) was observed in the cell line expressing C/EBPα-p42-ER (Figure 3A), similar to the data observed among cells expressing the full-length C/EBPα isoform (Figure 2D). Similar data were also obtained for THP-1 cells stably transduced with constitutively active HA-tagged C/EBPα-p3035 expression construct (Figures 3C-E). Viable THP-1 cells constitutively expressing HA-C/EBPα-p42 could not be established because of differentiation-inducing activity of C/EBPα-p42 protein (unpublished observations). However, compared with the empty vector control line, the cells expressing C/EBPα-p30 demonstrated a 3.5-fold increase in expression of pre-miR-181a-1 (Figure 3C). Increases in precursor miR-181a-1, as well as mature species were also apparent in Northern blot (Figure 3D).
Truncated C/EBPα-p30 isoform induces expression of pre-miR-181a-1. (A) K562 cells were stably transfected with β-estradiol inducible C/EBPα-p30 or p42 ER fusion constructs31,37 enabling nuclear translocation of ectopically expressed C/EBPα proteins. For negative control, vector expressing the ER domain alone was included. Total RNA was analyzed for the expression of miR-181a-1 (black bars) and miR-181a-2 (white bars) precursors by quantitative real-time PCR. (B) Total protein lysates from K562 stable lines described in panel A were analyzed for the relative expression of C/EBPα-ER fusion proteins by Western blot stained with C/EBPα antibody. To control for loading, staining with β-tubulin antibody was used. (C) Quantitative real-time RT-PCR data and (D) Northern blot data of THP-1 cells stably expressing HA-tagged C/EBPα-p30 isoforms or empty vector.35 Mature miR-181a expression was found to be highest in those THP-1 cells expressing the HA-tagged C/EBPα-p30 isoform. Northern blot shows an increase for both pre-miR-181a-1 and mature miR-181a expression for those cells expressing the HA-tagged C/EBPα-p30 (snRNA U6 was used as an internal loading control). (E) Western blot data showing the expression of HA-tagged C/EBPα-p30 and empty vector for those cells described in panels C and D. In panels A and C, average of triplicate measurements is shown and error bars denote SEM.
Truncated C/EBPα-p30 isoform induces expression of pre-miR-181a-1. (A) K562 cells were stably transfected with β-estradiol inducible C/EBPα-p30 or p42 ER fusion constructs31,37 enabling nuclear translocation of ectopically expressed C/EBPα proteins. For negative control, vector expressing the ER domain alone was included. Total RNA was analyzed for the expression of miR-181a-1 (black bars) and miR-181a-2 (white bars) precursors by quantitative real-time PCR. (B) Total protein lysates from K562 stable lines described in panel A were analyzed for the relative expression of C/EBPα-ER fusion proteins by Western blot stained with C/EBPα antibody. To control for loading, staining with β-tubulin antibody was used. (C) Quantitative real-time RT-PCR data and (D) Northern blot data of THP-1 cells stably expressing HA-tagged C/EBPα-p30 isoforms or empty vector.35 Mature miR-181a expression was found to be highest in those THP-1 cells expressing the HA-tagged C/EBPα-p30 isoform. Northern blot shows an increase for both pre-miR-181a-1 and mature miR-181a expression for those cells expressing the HA-tagged C/EBPα-p30 (snRNA U6 was used as an internal loading control). (E) Western blot data showing the expression of HA-tagged C/EBPα-p30 and empty vector for those cells described in panels C and D. In panels A and C, average of triplicate measurements is shown and error bars denote SEM.
In summary, using different in vitro systems we demonstrated that C/EBPα-p30, and to a lesser extent C/EBPα-p42, can induce chromosome 1-derived miR-181a-1 expression in AML cell line models. Because the N-terminal CEBPΑ mutations in AML are not capable of producing the wild-type full-length C/EBPα-p42 isoform, but allow for translation of the C/EBPα-p30 protein (Figure 2C),14 we conclude that C/EBPα-p30 is the main isoform responsible for up-regulation of miR-181a-1 in CEBPA mutated CN-AML patients.
C/EBPα-p30 physically interacts with the promoter of miR-181a-1 and regulates its expression
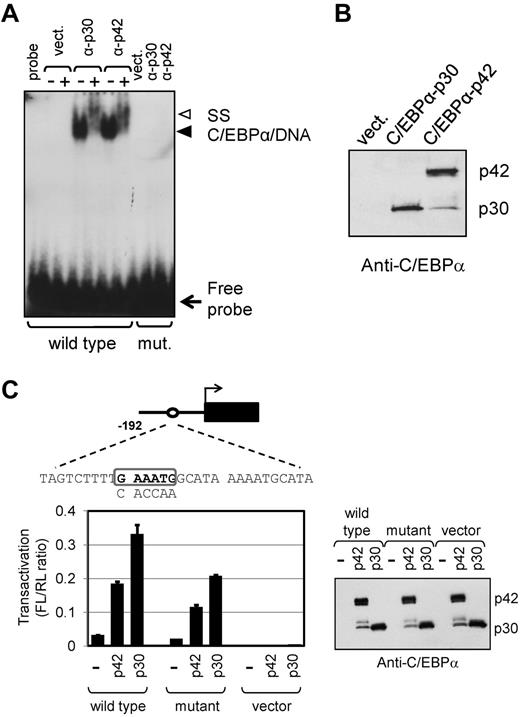
Given that our previous5 and current data showed a correlation between the expression of C/EBPα-p30 and increased expression of miR-181a-1 in AML patients, next we sought to identify the putative C/EBPα-responsive element within the miR-181a-1 promoter. It was recently reported that a region of approximately 600 base pairs (bp) upstream of the miR-181a-1 gene exhibits a promoter activity in immortalized human oral keratinocytes, but the molecular mechanism(s) regulating this activity were not investigated.36 Using several transcription factor binding site prediction engines (TFSearch, TESS, MatInspector), we identified 2 potential C/EBP-binding sites located between nucleotides −101/−96 and −159/−154 of the miR-181a-1 promoter. To test the binding of C/EBPα to those sites, we prepared nuclear extracts from HEK-293T cells transiently transfected with CEBPA expression vectors coding for either the full-length or N-truncated C/EBPα isoforms. As shown in Figure 4A, the more proximal site (nts −101/−96) bound both C/EBPα-p42 and p30 isoforms in a sequence-specific manner: mutation of the putative site abrogated the binding of both C/EBPα proteins. The addition of C/EBPα antibody (which recognizes both p42 and p30 isoforms) to the binding reactions led to formation of super-shifted complexes. Western blot data revealed that C/EBPα-p42 and C/EBPα-p30 isoforms were expressed at similar levels (Figure 4B). In contrast, the binding of C/EBPα proteins to the distal predicted C/EBP site (nts −159/−154) was not observed (data not shown).
Human miR-181a-1 promoter is regulated by C/EBPα. (A) C/EBPα binds specifically to a site within the human miR-181a-1 proximal promoter. Nuclear extracts from HEK-293T cells transiently transfected with pcDNA3-FLAG (vect.), pcDNA3-C/EBPα-p30-FLAG (α-p30), or pcDNA3-C/EBPα-p42-FLAG (α-p42) were used in electrophoretic mobility shift assay (EMSA). The radiolabeled probes contained either wild-type predicted C/EBP-binding site, or mutant (mut.) sequences (shown below). Lane labeled “probe” contains binding reaction in the absence of nuclear extract. Where indicated by “+” above the lanes, C/EBPα specific antibody was added to the binding reactions. Solid arrowhead shows protein/DNA complex, whereas open arrowhead indicates binding complex super-shifted with the antibody. Unbound probe is shown on the bottom of the gel (free probe). (B) Relative expression of C/EBPα proteins in nuclear extracts used in EMSA in panel A is demonstrated in Western blot stained with C/EBPα-specific antibody. The p42 and p30 C/EBPα polypeptides are indicated to the right. (C) C/EBPα-dependent transactivation of the miR-181a-1 promoter. Human miR-181a-1 192 bp promoter fragment containing wild-type C/EBP-binding site (boxed sequence) or mutated sequence (indicated below the box) were linked to firefly luciferase gene (black box; diagrammed on top) and transiently transfected to HEK-293T cells with either empty expression vector pcDNA3-FLAG (−), or pcDNA3-C/EBPα-FLAG vectors (p42 or p30). Cell lysates were analyzed for luciferase activity and normalized to cotransfected Renilla luciferase activity. For control, C/EBPα expression vectors were also tested with promoter-less luciferase vector, pGL4-11 (vector). Each bar represents average of 3 transfection experiments and SEM bars are shown. On the right, representative aliquots of cell lysates used for luciferase assay were also analyzed by Western blot to demonstrate comparable levels of C/EBPα protein.
Human miR-181a-1 promoter is regulated by C/EBPα. (A) C/EBPα binds specifically to a site within the human miR-181a-1 proximal promoter. Nuclear extracts from HEK-293T cells transiently transfected with pcDNA3-FLAG (vect.), pcDNA3-C/EBPα-p30-FLAG (α-p30), or pcDNA3-C/EBPα-p42-FLAG (α-p42) were used in electrophoretic mobility shift assay (EMSA). The radiolabeled probes contained either wild-type predicted C/EBP-binding site, or mutant (mut.) sequences (shown below). Lane labeled “probe” contains binding reaction in the absence of nuclear extract. Where indicated by “+” above the lanes, C/EBPα specific antibody was added to the binding reactions. Solid arrowhead shows protein/DNA complex, whereas open arrowhead indicates binding complex super-shifted with the antibody. Unbound probe is shown on the bottom of the gel (free probe). (B) Relative expression of C/EBPα proteins in nuclear extracts used in EMSA in panel A is demonstrated in Western blot stained with C/EBPα-specific antibody. The p42 and p30 C/EBPα polypeptides are indicated to the right. (C) C/EBPα-dependent transactivation of the miR-181a-1 promoter. Human miR-181a-1 192 bp promoter fragment containing wild-type C/EBP-binding site (boxed sequence) or mutated sequence (indicated below the box) were linked to firefly luciferase gene (black box; diagrammed on top) and transiently transfected to HEK-293T cells with either empty expression vector pcDNA3-FLAG (−), or pcDNA3-C/EBPα-FLAG vectors (p42 or p30). Cell lysates were analyzed for luciferase activity and normalized to cotransfected Renilla luciferase activity. For control, C/EBPα expression vectors were also tested with promoter-less luciferase vector, pGL4-11 (vector). Each bar represents average of 3 transfection experiments and SEM bars are shown. On the right, representative aliquots of cell lysates used for luciferase assay were also analyzed by Western blot to demonstrate comparable levels of C/EBPα protein.
To determine the transactivation potential of each C/EBPα isoform on the miR181a-1 promoter, we inserted a 192 bp proximal promoter fragment upstream of the firefly luciferase gene and tested the resulting luciferase activity in the absence and presence of each C/EBPα isoform. As demonstrated in HEK-293T cells and shown in Figure 4C, both isoforms transactivated the reporter gene, although C/EBPα-p30 exhibited a stronger activity than the C/EBPα-p42 isoform (6.5-fold vs 4.2-fold, respectively). Part of the promoter activity in C/EBPα-p42–transfected cells may be caused by the expression of the p30 isoform produced from the p42 expression vector (Figure 4A-C). The reporter construct containing a C/EBPα site mutation (which abrogated C/EBPα binding, Figure 4A) showed a decrease in transactivation by both C/EBPα-p42 and p30 (22% and 32%, respectively) but did not completely inhibit it. These data suggest that in addition to the direct binding to the miR181a-1 promoter, both C/EBPα isoforms, albeit p30 more efficiently than p42, may contribute to miR181a-1 regulation by an indirect mechanism via protein-protein interaction with other miR-181a-1 promoter-binding factors.
The immunomodulatory compound lenalidomide induces both C/EBPα-p30 and miR-181a expression
Thus far, our data showed that N-terminal CEBPA mutations (expressing C/EBPα-p30 protein) associated with higher miR-181a levels, which in turn predict better outcomes in CN-AML patients,22-24 and that C/EBPα-p30 was more potent in the up-regulation of miR-181a-1 than C/EBPα-p42. Therefore, we reasoned that the pharmacologic up-regulation of C/EBPα-p30 and in turn of miR-181a would be an attractive therapeutic option to improve the clinical outcomes of AML patients carrying wild-type CEBPA, but with the expression of a nonfunctional C/EBPα-p42 (such as C-terminal mutant, or serine 21 phosphorylated form).31,37 Thus, we focused our attention on compounds that could act as modulators of C/EBPα protein expression. The immunomodulatory compound, lenalidomide (Revlimid, Celgene) was selected based on its ability to up-regulate erythroid-specific genes and its clinical activity in AML.38-40 We also reported that a gene profile for CEBPA mutant AML was consistent with partial erythroid differentiation.5 Based on these observations, we hypothesized that lenalidomide was a probable candidate for inducing expression of miR-181a via modulation of C/EBPα isoforms.
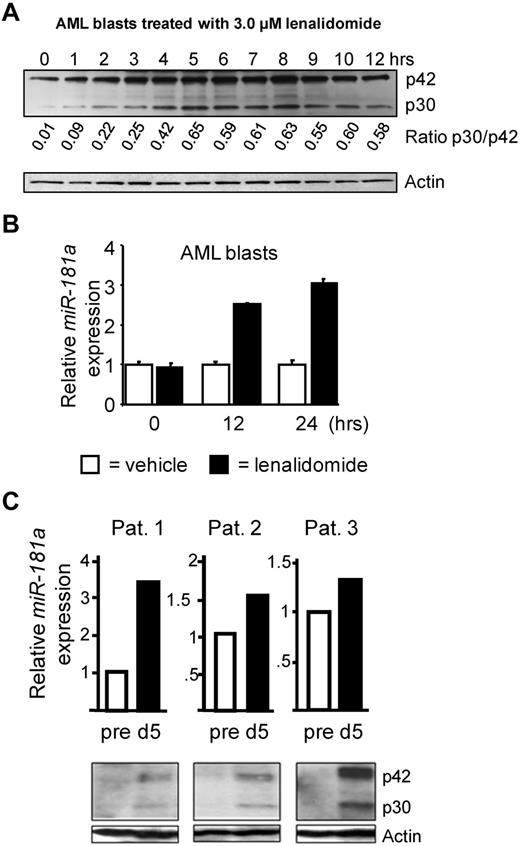
AML patient-derived blasts were cultured in vitro in the presence of 3.0μM lenalidomide and cells were collected hourly for Western blot analyses. The 3.0μM concentration of lenalidomide was previously shown to be clinically achievable in vivo.39 We observed an induction of both C/EBPα isoforms and an increase of C/EBPα-p30 expression compared with C/EBPα-p42 (Figure 5A). The induction of the C/EBPα-p30 isoform expression reached maximum between 5 and 8 hours of the treatment. As expected the miR-181a expression increased 2.5-fold after 12 hours and 3.0-fold after 24 hours of lenalidomide treatment (Figure 5B; same patient sample is shown in Figure 5A-B; also see supplemental Figure 3). Furthermore, AML primary samples treated with lenalidomide in vitro for 5 days showed a decrease in cell viability (supplemental Figure 3). Next, we performed analyses of bone marrow samples from relapse/refractory AML patient who participated in the OSU clinical trial (NCT01132586) receiving lenalidomide before induction chemotherapy. Bone marrow samples (Nos. 1-3) from 3 younger AML patients (< 60 years) were selected based on the availability of material for the analysis. As shown in Figure 5C, C/EBPα protein expression was induced by day 5 of lenalidomide treatment (before initiation of chemotherapy) compared with the pretreatment baseline and paralleled by the increase in miR-181a levels. Although an increase of both C/EBPα isoforms was observed in these samples, it was probable that C/EBPα-p30 induced miR-181a expression more effectively than C/EBPα-p42, as we demonstrated in our previous experiments (Figures 3 and 4). Consistently, the expression levels of miR-181a and C/EBPα expression were higher after lenalidomide treatment in bone marrow samples from 3 AML patients (Nos. 4-6) with available material and treated on a Washington University clinical trial (supplemental Figure 4).40 We conclude that lenalidomide treatment in vitro and in vivo leads to increase of C/EBPα expression and induction of miR-181a expression in AML patient blasts. Of note, the lenalidomide-mediated induction of miR-181a expression was found only in C/EBPα-expressing cells (THP-1 and HL60; Figure 6B and supplemental Figure 5, respectively), and not in cells lacking the C/EBPα expression (K562; supplemental Figure 5), supporting that C/EBPα mediates the lenalidomide-dependent mechanisms of miR-181a up-regulation.
Increased C/EBPα-p30 expression after treatment with the immunomodulatory compound, lenalidomide. (A) Western blot analysis of AML patient blasts treated in vitro with 3.0μM lenalidomide followed by hourly collections. The expression of C/EBPα (p30 and p42) was detected by C/EBPα antibody. The signal intensities were assessed and the p30/p42 ratios were calculated (shown below C/EBPα stained blot). Staining with Actin antibody served as an internal loading control. The data shown are representative for 3 patient samples. (B) Quantitative real-time RT-PCR data for miR-181a expression in the same AML blasts shown in panel A. The expression of miR-181a was increased at 12 hours and 24 hours after 3.0μM lenalidomide treatment (black bars) compared with the vehicle control (white bars) for the same time point. Average of triplicate measurements from a single patient sample is shown and error bars denote SEM. Similar results shown in panels A and B were observed in a total of 3 separate AML patient blasts used in the same experiment (not shown). Additional data from the long-term in vitro treatment with lenalidomide are shown in supplemental Figure 3. (C) Quantitative real-time RT-PCR data from 3 bone marrow samples from patients treated with lenalidomide induction therapy (top). Samples were analyzed before treatment (white bars) and 5 days after lenalidomide induction therapy (black bars). Data are shown as average of triplicate measurements and error bars denote SEM. Increased expression of miR-181a was observed on day 5 after lenalidomide induction therapy. Corresponding whole cell lysates for those patients were analyzed using Western blot (bottom). The expression of C/EBPα (p42 and p30) was increased on day 5 after lenalidomide induction therapy. Actin served as an internal loading control. Patients' cytogenetic and clinical characteristics before the therapy are summarized in supplemental Table 1.
Increased C/EBPα-p30 expression after treatment with the immunomodulatory compound, lenalidomide. (A) Western blot analysis of AML patient blasts treated in vitro with 3.0μM lenalidomide followed by hourly collections. The expression of C/EBPα (p30 and p42) was detected by C/EBPα antibody. The signal intensities were assessed and the p30/p42 ratios were calculated (shown below C/EBPα stained blot). Staining with Actin antibody served as an internal loading control. The data shown are representative for 3 patient samples. (B) Quantitative real-time RT-PCR data for miR-181a expression in the same AML blasts shown in panel A. The expression of miR-181a was increased at 12 hours and 24 hours after 3.0μM lenalidomide treatment (black bars) compared with the vehicle control (white bars) for the same time point. Average of triplicate measurements from a single patient sample is shown and error bars denote SEM. Similar results shown in panels A and B were observed in a total of 3 separate AML patient blasts used in the same experiment (not shown). Additional data from the long-term in vitro treatment with lenalidomide are shown in supplemental Figure 3. (C) Quantitative real-time RT-PCR data from 3 bone marrow samples from patients treated with lenalidomide induction therapy (top). Samples were analyzed before treatment (white bars) and 5 days after lenalidomide induction therapy (black bars). Data are shown as average of triplicate measurements and error bars denote SEM. Increased expression of miR-181a was observed on day 5 after lenalidomide induction therapy. Corresponding whole cell lysates for those patients were analyzed using Western blot (bottom). The expression of C/EBPα (p42 and p30) was increased on day 5 after lenalidomide induction therapy. Actin served as an internal loading control. Patients' cytogenetic and clinical characteristics before the therapy are summarized in supplemental Table 1.
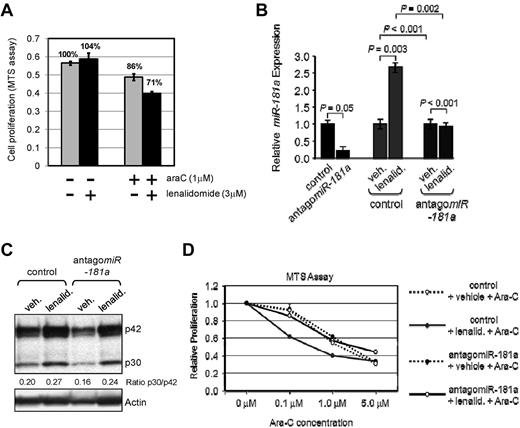
Lenalidomide induced miR-181a expression sensitizes leukemia cells to conventional chemotherapy. (A) Untransfected THP-1 cells were cultured for 6 days in the presence of 3μM lenalidomide alone, 1μM ara-C alone, both drugs together (as indicated by “+” or “−” below the graph), or vehicle control (PBS; the left-most bar) and MTS proliferation assay was performed. The bars represent averages from 3 to 4 readings. SEM and relative percentages of proliferation rate are shown. (B) Quantitative real-time RT-PCR analysis for the expression of miR-181a in THP-1 cells transiently transfected with nontargeting control (negative control) or antagomiR-181a. Endogenous expression of miR-181a was found to be lower in those cells transfected with antagomiR-181a before drug treatments (black bars). After the treatment with 3.0μM lenalidomide, miR-181a expression was increased among those cells previously transfected with nontargeting control (red bars). In contrast, the expression of miR-181a was relatively unchanged among those cells transfected with antagomiR-181a followed by 3.0μM lenalidomide treatment (blue bars). Data are shown as an average of measurements and error bars denote SEM. (C) Western blot for those cells described in panel A with the additional treatment with either 3.0μM lenalidomide or vehicle control for 3 days. The expression of C/EBPα (p42 and p30) was found to be higher in those cells treated with lenalidomide, regardless of earlier transfection status described in panel A. Actin served as an internal loading control. (D) THP-1 cells transfected with nontargeting control, or antagomiR-181a were cultured with various concentrations (0-5μM) of cytarabine (Ara-C) in the presence of 3.0μM lenalidomide (lenalid.; solid lines), or vehicle (broken lines) for 72 hours and cellular proliferation was measured by MTS assay. Each datapoint represents an average of 3 measurements.
Lenalidomide induced miR-181a expression sensitizes leukemia cells to conventional chemotherapy. (A) Untransfected THP-1 cells were cultured for 6 days in the presence of 3μM lenalidomide alone, 1μM ara-C alone, both drugs together (as indicated by “+” or “−” below the graph), or vehicle control (PBS; the left-most bar) and MTS proliferation assay was performed. The bars represent averages from 3 to 4 readings. SEM and relative percentages of proliferation rate are shown. (B) Quantitative real-time RT-PCR analysis for the expression of miR-181a in THP-1 cells transiently transfected with nontargeting control (negative control) or antagomiR-181a. Endogenous expression of miR-181a was found to be lower in those cells transfected with antagomiR-181a before drug treatments (black bars). After the treatment with 3.0μM lenalidomide, miR-181a expression was increased among those cells previously transfected with nontargeting control (red bars). In contrast, the expression of miR-181a was relatively unchanged among those cells transfected with antagomiR-181a followed by 3.0μM lenalidomide treatment (blue bars). Data are shown as an average of measurements and error bars denote SEM. (C) Western blot for those cells described in panel A with the additional treatment with either 3.0μM lenalidomide or vehicle control for 3 days. The expression of C/EBPα (p42 and p30) was found to be higher in those cells treated with lenalidomide, regardless of earlier transfection status described in panel A. Actin served as an internal loading control. (D) THP-1 cells transfected with nontargeting control, or antagomiR-181a were cultured with various concentrations (0-5μM) of cytarabine (Ara-C) in the presence of 3.0μM lenalidomide (lenalid.; solid lines), or vehicle (broken lines) for 72 hours and cellular proliferation was measured by MTS assay. Each datapoint represents an average of 3 measurements.
Lenalidomide treatment sensitizes leukemic cells to cytarabine chemotherapy
Cytarabine (Ara-C) is a pyrimidine antagonist, which interferes with DNA synthesis and is used in upfront and salvage regimens for AML.41,42 To improve the cytotoxic activity of Ara-C treatment, various novel drug combinations have been explored.43,44 Recently, it was demonstrated that miR-181a can sensitize a chemotherapy-resistant HL60 cell line to Ara-C treatment.29 Having found that miR-181a is up-regulated in response to lenalidomide, we asked whether pretreatment of AML cells with this compound can similarly sensitize the cells to Ara-C.
When THP-1 cells were treated with either 3μM lenalidomide, 1μM Ara-C, or both compounds simultaneously, an additive cytotoxic effect was observed (Figure 6A). Using THP-1 cells transiently transfected with antagomiR-181a, or a nonsilencing control oligoribonucleotide, we asked whether the lenalidomide effect was mediated by miR-181a expression. As shown in Figure 6B, antagomiR-181a efficiently down-regulated levels of the endogenous miR-181a and the lenalidomide-induced up-regulation of miR-181a was effectively blocked by antagomiR-181a. Regardless of the levels of miR-181a, lenalidomide treatment led to an increase in C/EBPα-p30 protein (Figure 6C), which indicates that the up-regulation of miR-181a is a downstream event with respect to regulation of C/EBPα.
After the transfection with antagomiR-181a or nontargeting control, THP-1 cells were treated with 3.0μM lenalidomide or vehicle every 24 hours for 3 days, followed by a single application of Ara-C at concentrations ranging between 0μM and 5.0μM and allowed to incubate for 72 hours. Cellular proliferation was determined by MTS assay. As shown in Figure 6D, Ara-C alone inhibited cell proliferation in a dose-dependent fashion; however, the addition of lenalidomide had a stronger inhibitory effect. Furthermore, down-regulation of miR-181a levels by the transfected antagomiR-181a prevented the antiproliferation effect associated with the lenalidomide treatment (Figure 6D solid blue), whereas antagomiR-181a had no effect on the cell response to Ara-C alone (Figure 6D). From these data, we concluded that lenalidomide sensitizes leukemic cells to conventional Ara-C therapy via miR-181a.
Lenalidomide treatment in vivo causes a decrease in xenograft tumor size
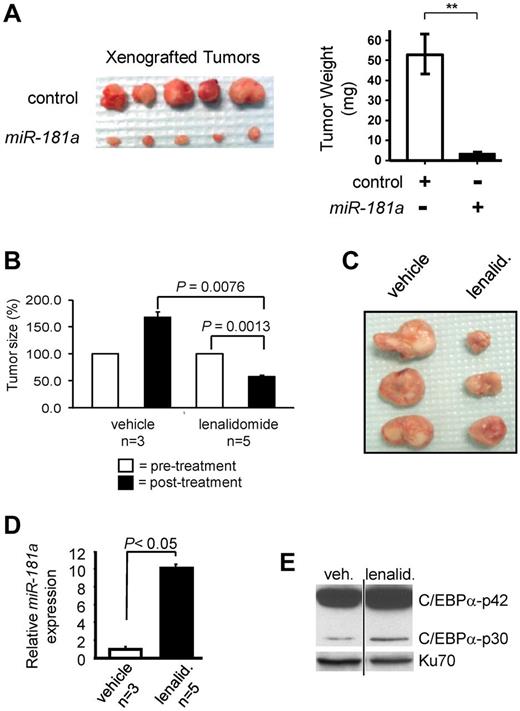
Patients responding to lenalidomide were found to have increased miR-181a expression levels (Figure 5). Moreover, forced expression of miR-181a resulted in reduced cell growth (supplemental Figure 6A) and colony-forming ability (supplemental Figure 6B), as well as increased spontaneous death (increased number of cells in sub-G1 phase; supplemental Figure 6C). In addition, transient expression of miR-181a in AML patient blasts led to a 2-fold increase of annexin V labeling, suggesting that blasts expressing increased levels of miR-181a had a lower apoptosis threshold (supplemental Figure 6D). To further prove the causal role of miR-181a in in vivo tumor growth inhibition, mice were injected with THP-1 cells transiently transfected with miR-181a expression vector (n = 5) or with empty control vector (n = 5). After 6 weeks, mice were killed and tumors measured. A tumor-suppressing activity of miR-181a was revealed by decreased tumor sizes and weights, in mice engrafted with cells forced to express miR-181a (P < .01; Figure 7A).
Lenalidomide treatment in vivo induces miR-181a-mediated inhibition of xenograft AML tumor growth. (A) THP-1 cells were transiently transfected with empty expression vector (control), or construct expressing the ectopic miR-181a and injected subcutaneously into NOD/SCID mice (5 mice per construct). Six weeks later the tumors were excised (left) and their sizes were determined (right). (B-E) THP-1 cells were xenografted subcutaneously to NOD/SCID mice (1 tumor per mouse). Four weeks later, tumors were directly injected with lenalidomide (lenalid.; 50 mg/kg; n = 5) or vehicle control (n = 3), twice a week for 2 weeks. Six weeks after transplantation mice were killed and tumors excised. (B) Xenograft tumors assessed at the onset (white bars) and the end (black bars) of the treatment. Measurements were plotted as the relative percentages of the tumors at the beginning of the treatment. (C) Dissected tumors after the lenalidomide treatment (lenalid.; on the right) or vehicle control (on the left). (D) Quantitative real-time RT-PCR assessment of miR-181a expression for the xenografts after treatment with either lenalidomide (lenalid.; black bar) or vehicle (white bar). (E) The relative expression of C/EBPα-p42 and C/EBPα-p30 in lenalidomide (lenalid.) or vehicle-treated xenograft tumors was evaluated by Western blot of nuclear extracts prepared from the tumors. Blot was stained with C/EBPα antibody and staining with Ku70 served as a loading control. Both lanes were from the same blot and same exposure. Vertical line has been inserted to indicate repositioned gel lanes.
Lenalidomide treatment in vivo induces miR-181a-mediated inhibition of xenograft AML tumor growth. (A) THP-1 cells were transiently transfected with empty expression vector (control), or construct expressing the ectopic miR-181a and injected subcutaneously into NOD/SCID mice (5 mice per construct). Six weeks later the tumors were excised (left) and their sizes were determined (right). (B-E) THP-1 cells were xenografted subcutaneously to NOD/SCID mice (1 tumor per mouse). Four weeks later, tumors were directly injected with lenalidomide (lenalid.; 50 mg/kg; n = 5) or vehicle control (n = 3), twice a week for 2 weeks. Six weeks after transplantation mice were killed and tumors excised. (B) Xenograft tumors assessed at the onset (white bars) and the end (black bars) of the treatment. Measurements were plotted as the relative percentages of the tumors at the beginning of the treatment. (C) Dissected tumors after the lenalidomide treatment (lenalid.; on the right) or vehicle control (on the left). (D) Quantitative real-time RT-PCR assessment of miR-181a expression for the xenografts after treatment with either lenalidomide (lenalid.; black bar) or vehicle (white bar). (E) The relative expression of C/EBPα-p42 and C/EBPα-p30 in lenalidomide (lenalid.) or vehicle-treated xenograft tumors was evaluated by Western blot of nuclear extracts prepared from the tumors. Blot was stained with C/EBPα antibody and staining with Ku70 served as a loading control. Both lanes were from the same blot and same exposure. Vertical line has been inserted to indicate repositioned gel lanes.
Having established that lenalidomide increases miR-181a expression, we asked whether, similar to miR-181a, this compound could also suppress tumor growth in vivo. NOD/SCID mice were injected subcutaneously with untransfected THP-1 cells (10 × 106 cells). Four weeks after engraftment, lenalidomide (50 mg/kg; n = 5) or vehicle (n = 3) were injected directly into the tumors twice a week for 2 weeks. A relatively higher dose of lenalidomide was used in this experiment to compensate for a short half-life of the drug and to ensure a promptly evaluable pharmacologic effect in the leukemia tumors. The mice were killed and tumors excised to evaluate the effects of lenalidomide. The average size of the lenalidomide-treated tumors (57.5 ± 0.06 mm) was significantly decreased compared with the vehicle-treated tumors (166.8 ± 1.08 mm; P = .008; paired t test; Figure 7B-C). The tumor sizes after lenalidomide administration were decreased by approximately 3-fold. Consistent with our previous results, lenalidomide-treated tumors displayed increased miR-181a expression (Figure 7D). The expression of miR-181a was found to be nearly 10-fold higher (P < .05) in the tumors treated with lenalidomide compared with the vehicle-treated tumors.
In summary, the tumor-suppressing effect of lenalidomide, was accompanied by an increase of miR-181a expression, which is analogous to tumor growth suppression by constitutive ectopic expression of miR-181a, thereby suggesting antitumorigenic properties for this miRNA.
Discussion
We and others reported that CN-AML patients carrying mutations in CEBPA have better clinical outcomes compared with patients with wild-type CEBPA.2-5 We also showed a strong association of CEBPA mutations and high miR-181a levels5 and better outcomes of CN-AML patients with higher miR-181a levels.22-24 The favorable impact of CEBPA mutations is restricted to only those patients with double mutations (usually concurrent N and C-terminal mutations) and not observed in patients with single mutations, among which the majority are found only within the C-terminal region.3,4 A hallmark of AML with CEBPA N-terminal mutations present in > 90% of CEBPA mutated patients is that these mutations prevent the expression of the full-length C/EBPα-p42 isoform, whereas the expression of the truncated C/EBPα-p30 isoform is undisturbed, or even enhanced.14 In general, C/EBPα-p30 acts as a dominant-negative isoform of C/EBPα-p42 function,9,14,45 but proteomic approaches identified exclusive targets of C/EBPα-p30 (UBC9 and PIN1).33,46 Thus, we questioned whether C/EBPα-p30 could exert any positive activity on the regulation of miR-181a expression. Indeed, we found that although both C/EBPα isoforms can directly bind to the miR-181a-1 promoter, the N-terminally truncated C/EBPα is more potent in its transactivation. To the best of our knowledge, in contrast to UBC9 and PIN1, the miR-181a-1 gene located on chromosome 1 is the first reported direct target of C/EBPα-p30.
Because higher miR-181a expression is associated with a favorable response to treatment and better outcomes,22-24 we decided to pursue a strategy that increases the endogenous expression of this miRNA in myeloid blasts. Giving the similarities between the gene expression profile predictive of response to lenalidomide and the gene profile associated with CEBPA mutational status, lenalidomide was selected as a potential modulator of miR-181a expression via C/EBPα-dependent mechanisms. Indeed, in both preclinical models and treated AML patients, we showed that lenalidomide treatment of AML blasts harboring wild-type CEBPA led to induction of both C/EBPα-p30, C/EBPα-p42, and miR-181a. Lenalidomide has clinical activity in AML.39,40 Thus, in addition to its immunomodulatory properties, it is also possible that this activity occurs via 2 mechanisms that implicate C/EBPα. One mechanism involves the C/EBPα-p30–mediated increased expression of miR-181a, which contributes to better chemotherapy response29 (Figure 6), thereby improving the outcomes of AML patients with wild-type CEBPA expressing either, nonfunctional (phosphorylated on serine 21),31,37 or insufficient levels of C/EBPα-p42 to allow for granulocytic maturation.14 Because lenalidomide also increased C/EBPα-p42 levels, it is probable that a second antileukemic mechanism may involve up-regulation of the differentiation-promoting C/EBPα-p42 polypeptide. Of course, lenalidomide's immunomodulatory activity may also mediate clinical response in AML patients, especially in those patients who previously have undergone allogeneic stem cell transplantation.39 The mechanism of C/EBPα up-regulation by lenalidomide remains to be elucidated, but preliminary results suggest that lenalidomide modulates the activity of translation elongation factors responsible for C/EBPα-p30 expression (C. Hickey, unpublished data).
Different biologic functions of C/EBPα isoforms have been reported relating to myeloid proliferation and differentiation.14,45,47 C/EBPα-p30 lacks a differentiation-inducing and antimitotic activities15,48 Paradoxically, the lack of a cell-cycle inhibitory function of C/EBPα-p30 serves as an attractive feature when treating AML with chemotherapeutic agents, such as Ara-C that are most effective in dividing cells. Our data argue that the favorable impact of C/EBPα-p30 may also related to its ability to induce miR-181a, which in turn can sensitize AML cells to chemotherapy.29 In agreement, we showed here that lenalidomide can also sensitize THP-1 cells to Ara-C. This lenalidomide activity is inhibited in the presence of miR-181a antagomiR, thereby supporting that lenalidomide-induced sensitivity to Ara-C is dependent on its ability to increase miR-181a. To prove this principle, a phase 1 trial of lenalidomide followed by the Ara-C induction chemotherapy is currently ongoing at our institution (NCT01132586).
In summary, we presented a model for C/EBPα-p30–dependent miR-181a up-regulation. The absence of the C/EBPα-p30 antiproliferative property15,48 and higher levels of miR-181a,22,24 that have been shown to sensitize leukemia cells to chemotherapy (this study),29 may partly explain the improved outcomes in CEBPA mutated CN-AML patients, who usually present with N-mutations predictive of C/EBPα-p30 expression. One may raise the question as to why patients with wild-type CEBPA have lower levels of miR-181a than those with mutations affecting the N-terminal region of the protein. Indeed, the C/EBPα-p42 isoform, which is linked to differentiation, was found to be expressed concurrently with the truncated C/EBPα-p30 isoform among lenalidomide-treated AML patients (Figure 5, supplemental Figures 3 and 4). This polypeptide also contributes to miR-181a regulation in vitro (Figure 4). It is possible that in patients with wild-type CEBPA, lower miR-181a levels are because of a silenced CEBPA49 or nonfunctioning C/EBPα protein.31,37 Thus, these patients may benefit from pretreatment with agents such as lenalidomide alone, or in combination with hypomethylating agents (for epigenetically silenced CEBPA),49 or kinase inhibitors (for nonfunctioning serine 21 phosphorylated C/EBPα-p42).33 These therapies would increase C/EBPα-p30 expression and in turn miR-181a, thereby leading to increase sensitivity to chemotherapy and better clinical outcome.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Donna Bucci, manager of the OSU and Alliance (formerly Cancer and Leukemia Group B, CALGB) Leukemia Tissue Banks, for sample processing and storage services, and Lisa J. Sterling and Colin G. Edwards for data management. FLAG-tagged C/EBPα expression vectors were kindly provided by Daniel G. Tenen (Harvard Medical School, Boston, MA).
This work was supported in part by National Cancer Institute grants CA101140, CA114725, CA31946, CA33601, CA16058, CA77658, CA35279, CA03927, and CA41287; the Harry T. Mangurian Jr Foundation; the Coleman Leukemia Research Foundation; the Leukemia & Lymphoma Society (SCOR grant); and the National Science Foundation (EEC-0425626). H.B. was supported by the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation.
National Institutes of Health
Authorship
Contribution: C.J.H., H.S.R., S.S., and G.M. participated in designing the research and drafting the paper; C.H., S.S., H.S.R., A.M.D., A.M.E., S.R., A.M., Y.-Z.W., X.Z., and H.B. performed experiments; K.M. and D.N. performed the statistical analyses; H.A., K.M., S.W.P., L.-C.W., D.P., R.B., M.A.C., J.C.B., J.L., C.M.C., C.D.B., and R.G. provided technical insight and critically reviewed the paper; and W.B.,T.A.F., and R.V. participated in clinical trials, provided clinical samples, and critically reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guido Marcucci, The Ohio State University, Comprehensive Cancer Center, Biomedical Research Tower 410, 460 W 12th Ave, Columbus, OH 43210; e-mail: guido.marcucci@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal