Abstract
RUNX1 encodes a DNA-binding α subunit of the core-binding factor, a heterodimeric transcription factor. RUNX1 is a master regulatory gene in hematopoiesis and its disruption is one of the most common aberrations in acute leukemia. Inactivating or dominant-negative mutations in the RUNX1 gene have been also identified in pedigrees of familial platelet disorders with a variable propensity to develop acute myeloid leukemia (FPD/AML). We performed analysis of hematopoiesis from 2 FPD/AML pedigrees with 2 distinct RUNX1 germline mutations, that is, the R139X in a pedigree without AML and the R174Q mutation in a pedigree with AML. Both mutations induced a marked increase in the clonogenic potential of immature CD34+CD38− progenitors, with some self-renewal capacities observed only for R174Q mutation. This increased proliferation correlated with reduction in the expression of NR4A3, a gene previously implicated in leukemia development. We demonstrated that NR4A3 was a direct target of RUNX1 and that restoration of NR4A3 expression partially reduced the clonogenic potential of patient progenitors. We propose that the down-regulation of NR4A3 in RUNX1-mutated hematopoietic progenitors leads to an increase in the pool of cells susceptible to be hit by secondary leukemic genetic events.
Introduction
The importance of RUNX1 has been stressed by the finding that chromosomal translocations and gene mutations disrupting the protein function were observed in patients with acute leukemia. Fusion proteins and mutated RUNX1 proteins are thought to mediate their oncogenic activity by dominantly repressing RUNX1-target genes. Several lines of experimental evidence corroborate the hypothesis that RUNX1 alterations are the initiating event in the hematopoietic diseases in which they are found. These aberrations alter the differentiation and proliferation of myeloid or lymphoid progenitor cells, establishing a preleukemic state that requires additional hits for disease penetration.1
Familial plate disorders/acute myeloid leukemia (FPD/AML) is a familial thrombocytopenia with predisposition to AML with germline RUNX1 mutations, which represents a unique model to study how this mutation alters hematopoiesis before full-blown leukemic development. Analysis of 32 FPD/AML (OMIM 601399) pedigrees has identified various germline heterozygous alterations in the RUNX1 gene.2-16 Missense and nonsense mutations in the conserved N-terminal Runt homology domain (RHD) generate RUNX1 proteins with impaired DNA binding that behave as dominant-negative (DN) mutants whereas large deletions or frameshift mutations rather induce RUNX1 haploinsufficiency. RUNX1 DN mutants may induce a higher probability of acquiring additional mutations in RUNX1 or other genomic alterations that lead to leukemic development10,13 than “haploinsufficiency” mutations.4 In the present work, our aim was to obtain more insight into the mechanism involved in the first steps of hematopoietic deregulation leading to leukemic development. We have setup an exhaustive study of the biologic properties of hematopoietic progenitor cells harboring 2 different mutation types: an unreported mutation, R139X, in a pedigree with thrombocytopenia alone and a R174Q mutation in a pedigree with a thrombocytopenia and a strong predisposition to leukemia leading to the occurrence of AML in several members of the pedigree. To obtain further insight in the molecular basis of this hematopoietic deregulation, we performed gene profiling of CD34+ cells from controls and patients with R174Q or R139X RUNX1 mutations. We showed a correlation between the increased clonogenic potential of patient hematopoietic progenitors and NR4A3 expression. We demonstrate that NR4A3 is a direct target of RUNX1 and that its expression restoration partially reduces the clonogenic potential of progenitors in patients with both R174Q and R139X mutations.
Methods
Blood cells
Blood samples from FPD/AML patients, healthy subjects, patients with a primitive hemochromatosis undergoing phlebotomy and individuals after mobilization were collected after informed consent was obtained. The study was approved by the Local Research Ethics Committee from the Assistance Publique–Hôpitaux de Paris (AP-HP) and informed consent was obtained from each FPD/AML patient in accordance with the Declaration of Helsinki.
Granulocytes, platelets, and mononuclear cells were separated by standard techniques. CD34+ cells were separated by double-positive selection using a magnetic cell-sorting system (AutoMACS; Miltenyi Biotec). CD3+ mononuclear cells were purified from CD34− fraction by the same method. Fibroblast cells were derived from skin biopsies of patients obtained with their informed consent and were grown in HAM F10 medium (Invitrogen) with 20% FCS and antibiotics at 5% CO2, 37°C.
Gene mutations screening
Exons 3 to 8 of RUNX1 gene were sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and analyzed on the Applied Biosystems 3130xl Genetic Analyzer.
Cell transduction
CD34+ cells from mobilized peripheral blood (105/mL) were prestimulated for 24 hours with IL-3, IL-6, SCF, and FLT3-L.
shRUNX1 and control scramble sequence (SCR) containing lentiviruses were previously described.17 A lentivirus containing 2 promoters pRRL-EF1α_MCS-PGK-GFP was constructed as follows: the minimal EF1α promoter with multicloning site was amplified by PCR from the TRIPΔU3-EF1α-GFP lentivirus and cloned into the XhoI site upstream the PGK-GFP cassette of sinpRRL-PGK-GFP lentivirus. A NR4A3 cDNA was amplified by PCR from the pSV7d-NR4A3 plasmid and cloned downstream the EF1α promoter (supplemental Figure 1). Lentiviral production and transduction were performed as previously described.17 CD34+GFP+ cells transduced by shRUNX1 and SCR were sorted at day 4 of culture in presence of IL-3, IL-6, SCF, and FLT3-L and submitted to Q-PCR analysis. CD34+GFP+ cells transduced with empty or NR4A3-containing lentivirus were sorted 2 days after transduction. Clones with a B/NK/M phenotype were assessed as described in the next paragraph.
Assessment of simultaneous B, NK, and granulomonocytic differentiation
B, M, and NK potential was assessed as previously described.18 Briefly, CD34+ cells isolated from peripheral blood of 3 healthy donors or FPD/AML patients were stained by anti-CD34–PE and anti-CD38–FITC Ab (BD Pharmingen). CD34+CD38− cells were sorted using the FACS-DIVA cell sorter (BD Biosciences) at 1 cell per well in 96-well plates on MS-5 cells in presence of 10 ng/mL IL-3, 50 ng/mL SCF, 50 ng/mL FLT3-L (generous gift from Immunex), 10 ng/mL TPO (generous gift from Kirin Brewery), 20 ng/mL IL-7, 10 ng/mL IL-15 (PeproTech), and 5 ng/mL IL-2 (generous gift from Chiron Laboratories). Wells were collected after 5 weeks and cell phenotype was determined by flow cytometry using anti-CD15–FITC, anti-CD19–PE (BD Biosciences), and anti-CD56–APC (Beckman Coulter) Abs. Lentiviral-transduced CD34+GFP+ cells were sorted 4 days after transduction at 1 cell per well and cultured as CD34+ CD38 cells. The immunophenotypic analysis was performed by using anti-CD15–PE, anti-CD19–APC (BD Biosciences), and anti-CD56–PC7 (Beckman Coulter) Abs.
Assessment of long-term culture-initiating cell potential
CD34+CD38− cells were sorted as above at one cell per well in 96-well plates and incubated on MS-5 cells in long-term culture (LTC) medium (αMEM supplemented with 12.5% FCS, 12.5% horse serum (Hyclone Laboratories, 10-4 mol/L 2-β mercaptoethanol). The progenitor content of each well was assessed by sacrificing wells after 5 weeks of culture and plating cells in methycellulose colony assays (see next paragraph).
Assessment of colony-forming cell potential
Sorted CD34+CD38+ and CD34+CD38− cells were plated at a concentration of 0.5 to 1 × 103 cells/mL of complete methylcellulose medium (StemCell Technologies) supplemented with EPO, IL-3, SCF, G-CSF. Erythroid (BFU-E), granulocytic (CFU-GM) and mixed (CFU-GEMM) progenitors were quantified at day 15 of culture.19 Colony number was calculated per 1 × 103 CD34+ plated cells. In 2 independent experiments, 15 and 30 CFU-GM and CFU-GEMM colonies were replated in methylcellulose and percentage of secondary colonies was estimated 2 weeks later.
Flow cytometric analysis
CD34+CD38− cells were grown in serum-free liquid medium20 in the presence of recombinant human cytokines: SCF, G-CSF, GM-CSF, TPO, EPO, FLT3-L, IL-3, and IL-6. Anti-CD11b, anti-CD15, anti-CD14, and anti-CD36 Abs (Pharmingen) were used to determine the cell phenotype using a FACSort cytometer and the CellQuest software package (BD Biosciences).
Cytokine concentrations
If not specified, the cytokine were used at the following concentrations: SCF 25 ng/mL, G-CSF 10 ng/mL, GM-CSF 10 ng/mL, TPO 10 ng/mL, EPO 3 IU/mL, FLT3-L 1 ng/mL, IL-3 100 IU/mL, and IL-6 10 ng/mL.
NOG mice repopulation assays
Twenty-four hours before transplantation, 8-week-old NOG mice were sublethally irradiated (3 Gy). CD34+ cells (2 × 105; 90% purity) isolated from peripheral blood of donors with a primitive hemochromatosis undergoing phlebotomy or FPD/AML patients were intravenously injected into NOG mice. BM was collected from the mouse femur at 3, 6, and 12 weeks after transplantation. Mice were killed at week 15 and secondary transplantations were performed. At 3 and 6 weeks after secondary transplantation, cell phenotype was analyzed. Human cell engraftment was evaluated by flow cytometry by measuring human leukocytes (CD45+) and myeloid progenitors (CD33+CD45+) in BM.
Mutagenesis
MPI_HAa vector containing RUNX1 c isoform17 and NR4A3_Luc plasmids were used for directed mutagenesis performed using mutagenesis kit (Stratagene).
Western blot assay
Western blot analyses were performed as previously reported.21 The following Abs were used: anti-AML1/RHD (Domain Ab-2; Calbiochem), anti-RUNX1 (H-65), anti-HA (F-7; Santa Cruz Biotechnology), anti-V5 (Invitrogen), and anti-HSC70 (Stressgen). In addition to HSC70 staining, coloration with Ponceau S solution (Sigma-Aldrich) after transfer was used to control the quantity of proteins.
Protein immunoprecipitation assay
Plasmids driving expression of HA-RUNX1_wt, HA-RUNX1_R174Q, or HA-RUNX1_R139X and V5-tagged CBFβ17 were transfected into HEK293T cells. After 48 hours, cells were lysed (100mM Tris, 150mM NaCl, 5mM EDTA, 0.5% SDS, 0.5% NP-40, 0.5% sodium deoxycholate, and complete protease inhibitor ([Roche], pH 8) and immunoprecipitations were performed using the μMACS HA Isolation Kit (Miltenyi Biotec).
Immunohistochemistry cytologic analysis
HA-RUNX1_wt, HA-RUNX1_R174Q, or HA-RUNX1_R139X transfected HEK 293T cells were seeded on polylisine-coated slides (CML) for 2 hours at 37°C (5% CO2 in air). Cells were fixed in 2% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton X-100 for 5 minutes and washed with 1× PBS for 5 minutes. Primary anti-HA (F-7; Santa Cruz Biotechnology) Ab was applied to the slides at a concentration of 1 μg/mL and incubated for 1 hour at room temperature. Slides were then washed 3 times with 1× PBS for 5 minutes before and after application of the secondary Ab anti–rabbit TRITC (Jackson ImmunoResearch, 20 μg/mL for 30 minutes at room temperature) and observed under a Leica DMI 4000 SPE laser scanning microscope (Leica Microsystems) with a 63×/1.4 numeric aperture (NA) oil objective. Images were processed using Adobe Photoshop 6.0 software. Fifty cells transfected with each plasmid were analyzed. For cytologic study, cells were collected at week 10 of culture, cytocentrifuged at 50g for 5 minutes onto glass slides, stained with May-Grümwald-Giemsa or stained for myeloperoxydase using a benzidine dihydrochloryde-containing incubation mixture22 and analyzed at a magnification of ×1000.
Array expression analysis and data availability
RNAs were extracted from sorted CD34+ cells using the RNeasy Micro protocol (QIAGEN). For RNA linear amplification, we applied a strategy of 2 rounds of reverse transcription followed by T7 promoter dependent in vitro transcription. We used a modified version of the Agilent Low input amplification Kit protocol (ref. 5188-5340; Agilent). Raw data files from the Agilent Feature Extraction software for image analysis were imported into ResolverTM system (Rosetta) for gene expression data analysis. Then combined experiments were generated to obtain average values from the replicates experiments to avoid dye incorporation bias. An ANOVA test was performed with a P value threshold P < 10−3.
All data obtained by microarray analyses and protocols have been submitted to ArrayExpress at the European Bioinformatics Institute 8 with the accession number E-TABM-792.
Quantitative real-time RT-PCR
RNA was isolated with the RNeasy mini or micro kit (QIAGEN) and cDNA was generated by reverse transcription (Invitrogen). Array data confirmation was performed using CellsDirect One-Step RT-PCR Kits beginning from 200 to 500 sorted CD34+ cells (Invitrogen). Quantitative real-time RT-PCR (Q-PCR) for all genes were performed with probes or SYBR green (Applied Biosystems) and a PRISM 7700 (Applied Biosystems). Levels of gene expression were calculated relatively to housekeeping genes as HPRT, RPL0, 18S, or PPIA (for primer sequences see supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
ChIP and promoter activity assay
ChIP assays were performed with a ChIP assay kit (Millipore, Upstate Biotechnology) using the anti-RUNX1 Ab (H-65; Santa Cruz Biotechnology). Chromatin was prepared from human CD34+ cells isolated from peripheral blood after G-CSF mobilization and cultured in presence of IL-3, SCF, FLT3-L, and IL-6 for 36 hours. Quantification of precipitated DNA fragments was carried out on a PRISM 7700 sequence detection system using SYBR green (Applied Biosystems) in duplicate (for primers, see supplemental Table 1). The relative proportions of coimmunoprecipitated promoter fragments were determined based on the threshold cycle (CT) value for each PCR (see supplemental Methods). Promoter activity assay was performed as previously described.17
Statistical analyses
Data are presented as means (± SD). Statistical significance was determined by Student t test. Statistical significance for B/M/NK and LTC-IC experiments was determined by Pairwise t test with Holm correction. For both tests, a P value < 0.05 was considered as statistically significant.
Results
FPD/AML pedigrees and molecular characterization of mutations
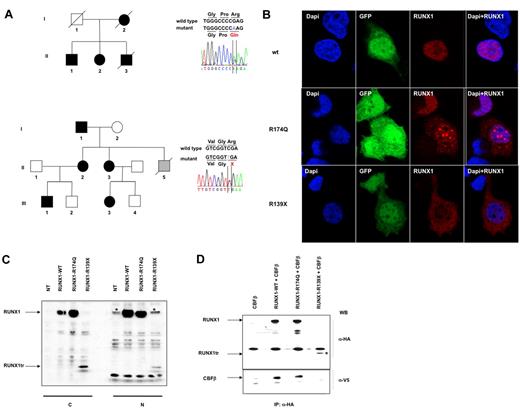
Hematopoiesis of 2 different pedigrees was studied. All the patients we investigated suffered from thrombocytopenia in the absence of any sign of myelodysplastic syndrome (MDS) or AML. In the first pedigree, we explored 2 patients (AII-1 and AII-2), who carried the previously described DN-like mutation R174Q (NM_001001890). Two members of their family previously died from a MDS/AML or an AML-M1 (Figure 1A top, Table 1). In the second pedigree, we studied 4 patients (BI-1, BII-2, BII-3, and BIII-1), who carried a previously unreported stop codon mutation, R139X (NM_001001890), generating a truncated RUNX1 protein devoid of nucleus localization sites (NLS). None in this family had developed AML or MDS (Figure 1A bottom; Table 1). The normal leukocyte count and the slightly decreased platelet count observed in all patients did not change during the experimental period (supplemental Figure 2A). In addition, the percentage of peripheral blood CD34+ cells and the proportion of CD34+CD38− cells of patients used in the experiments did not differ from controls (supplemental Figure 2B). Moreover, no blast cells were detected in BM. R139X mutant is devoid of NLS and as shown in Figure 1B and C, it is principally localized in the cytoplasm. A low quantity of the mutant was observed in the nucleus after transfection and this could be explained by a passive transport. Moreover, this truncated mutant lacks 2 of the 3 loop-containing regions responsible for DNA binding βE′ loop and βP2 loop,23 suggesting that this mutant does not bind DNA. Another already described mutation at the same position, R139Q, gives rise to the full-length protein, which does not dimerize with CBFβ4 and here we demonstrated that it is the same for R139X (Figure 1D; supplemental Figure 3). Only a small total quantity of this truncated protein was observed in the cells after transfection (Figure 1C) and this may be explained by the rapid degradation of protein because of the absence of RUNX1/CBFβ complex. This observation was strongly enhanced by the fact that, in primary monocytes isolated from peripheral blood of 2 patients BII-2 and BIII-1 from pedigree B (R139X mutation), the truncated protein was detected neither in nucleus nor in cytoplasm. In contrast, a small quantity of wt RUNX1 was present in the nucleus suggesting that this mutant induces a haploinsuficiency state (supplemental Figure 4). As previously described, R174Q mutant did not bind DNA, was only partially localized in the cytoplasm of transfected cells, and was still able to dimerize with CBFβ (Figure 1B-D).4 However, we were unable to detect wt RUNX1 and R174Q RUNX1 in patient (AII-1 and AII-2) peripheral blood monocytes (supplemental Figure 4). One possibility is that the R174Q mutant, which dimerizes with CBFβ with a stronger affinity than the wt protein but does not bind DNA, is degraded very quickly. The wt protein becomes unstable because of the absence of dimerization with CBFβ. Thus, the R174Q mutant leads to an almost complete absence of RUNX1.
Two germline mutations in the RUNX1 gene in a familial thrombocytopenia with a propensity to develop acute myeloid leukemia (FPD/AML). (A) Left panels: pedigree A (top) harbors the R174Q; pedigree B (bottom), the R139X mutations. Circles indicate females; squares, males; black filled symbols, affected individuals; gray filled symbol, individual with non hematologic disease; white symbols, nonaffected individuals; and symbols with a diagonal line, deceased individuals. Right panels: analysis of RUNX1 (NM_001001890.2) exon 5 (top panel) showing the heterozygous germline R174Q mutation and exon 4 (bottom panel) showing the heterozygous R139X mutation. (B) Localization of RUNX1 in HEK293T cells after transfection with the lentiviruses encoding for wtRUNX1, or R174Q RUNX1 or R139X RUNX1 and GFP. Immunofluorescence staining of RUNX1 was performed using an anti-HA Ab (red); the nucleus is stained with DAPI (blue). wtRUNX1 protein is localized exclusively in the nucleus with a diffuse pattern. R139X RUNX1 protein is mainly diffused in cytoplasm. R174Q RUNX1 protein is mainly localized in nucleus. In 65% of cells it is localized in dense foci and in 35% of cells, a diffuse pattern is observed. (C) Western blot (WB) analysis of RUNX1, expression in cytoplasmic (C) and nuclear (N) extracts from HEK293T cells overexpressing wild-type (wt), mutant R174Q or mutant R139X RUNX1 (RUNX1tr). RUNX1 forms were tagged with a HA epitope. Western blots (WB) were performed using anti-HA Ab. RUNX1tr: R139X RUNX1 truncated protein. *The nonspecific bands appearing using the anti-HA Ab in nuclear extracts. (D) Interactions between CBFβ and wt or mutant RUNX1 proteins in nuclear extracts from HEK293T cells transfected with CBFβ alone (lane 1) or together with wt RUNX1 (lane 2) or R174Q RUNX1 (lane 3) or R139X RUNX1 (lane 4). CBFβ was tagged with the V5 epitope and RUNX1 forms with a HA epitope. Immunoprecipitation (IP) was performed using an anti-HA Ab and Western blots (WB) using anti-HA and anti-V5 Abs. *Presence of RUNX1tr which is R139X RUNX1 truncated protein.
Two germline mutations in the RUNX1 gene in a familial thrombocytopenia with a propensity to develop acute myeloid leukemia (FPD/AML). (A) Left panels: pedigree A (top) harbors the R174Q; pedigree B (bottom), the R139X mutations. Circles indicate females; squares, males; black filled symbols, affected individuals; gray filled symbol, individual with non hematologic disease; white symbols, nonaffected individuals; and symbols with a diagonal line, deceased individuals. Right panels: analysis of RUNX1 (NM_001001890.2) exon 5 (top panel) showing the heterozygous germline R174Q mutation and exon 4 (bottom panel) showing the heterozygous R139X mutation. (B) Localization of RUNX1 in HEK293T cells after transfection with the lentiviruses encoding for wtRUNX1, or R174Q RUNX1 or R139X RUNX1 and GFP. Immunofluorescence staining of RUNX1 was performed using an anti-HA Ab (red); the nucleus is stained with DAPI (blue). wtRUNX1 protein is localized exclusively in the nucleus with a diffuse pattern. R139X RUNX1 protein is mainly diffused in cytoplasm. R174Q RUNX1 protein is mainly localized in nucleus. In 65% of cells it is localized in dense foci and in 35% of cells, a diffuse pattern is observed. (C) Western blot (WB) analysis of RUNX1, expression in cytoplasmic (C) and nuclear (N) extracts from HEK293T cells overexpressing wild-type (wt), mutant R174Q or mutant R139X RUNX1 (RUNX1tr). RUNX1 forms were tagged with a HA epitope. Western blots (WB) were performed using anti-HA Ab. RUNX1tr: R139X RUNX1 truncated protein. *The nonspecific bands appearing using the anti-HA Ab in nuclear extracts. (D) Interactions between CBFβ and wt or mutant RUNX1 proteins in nuclear extracts from HEK293T cells transfected with CBFβ alone (lane 1) or together with wt RUNX1 (lane 2) or R174Q RUNX1 (lane 3) or R139X RUNX1 (lane 4). CBFβ was tagged with the V5 epitope and RUNX1 forms with a HA epitope. Immunoprecipitation (IP) was performed using an anti-HA Ab and Western blots (WB) using anti-HA and anti-V5 Abs. *Presence of RUNX1tr which is R139X RUNX1 truncated protein.
Clinical characteristics of members of two FPD/AML pedigrees
| Pedigree/patient . | Age at diagnosis, y . | Actual age, y . | Platelet count, 109/L . | Platelet function . | Medular MK morphology . | Bleeding history malignancy . | Germline mutation . | Actual status . |
|---|---|---|---|---|---|---|---|---|
| AI-2 | 56 | 84 | Abnormal | dysMKpoiesis | MDS/AML | R174Q | Deceased | |
| AII-1* | 19 | 40 | 109 | Abnormal | dysMKpoiesis | Easy bruising | R174Q | Alive |
| AII-2* | 18 | 39 | 134 | Abnormal | dysMKpoiesis | Easy bruising | R174Q | Alive |
| AII-3 | 15 | 50 | nk | nk | Medullar aplasia and AML-M1 | R174Q | Deceased | |
| BI-1 | 58 | 130 | nk | nk | Easy bruising | R139X | Alive | |
| BII-2* | nk | 41 | 129 | Abnormal | dysMKpoiesis | Hematomas | R139X | Alive |
| BII-3* | 41 | 160 | Abnormal | nk | Easy bruising | R139X | Alive | |
| BII-5 | 22 | nk | nk | nk | Posttraumatic bleeding inducing death | nk | Deceased | |
| BIII-1* | 1 | 4 | 96 | Abnormal | dysMKpoiesis | Easy bruising | R139X | Alive |
| Pedigree/patient . | Age at diagnosis, y . | Actual age, y . | Platelet count, 109/L . | Platelet function . | Medular MK morphology . | Bleeding history malignancy . | Germline mutation . | Actual status . |
|---|---|---|---|---|---|---|---|---|
| AI-2 | 56 | 84 | Abnormal | dysMKpoiesis | MDS/AML | R174Q | Deceased | |
| AII-1* | 19 | 40 | 109 | Abnormal | dysMKpoiesis | Easy bruising | R174Q | Alive |
| AII-2* | 18 | 39 | 134 | Abnormal | dysMKpoiesis | Easy bruising | R174Q | Alive |
| AII-3 | 15 | 50 | nk | nk | Medullar aplasia and AML-M1 | R174Q | Deceased | |
| BI-1 | 58 | 130 | nk | nk | Easy bruising | R139X | Alive | |
| BII-2* | nk | 41 | 129 | Abnormal | dysMKpoiesis | Hematomas | R139X | Alive |
| BII-3* | 41 | 160 | Abnormal | nk | Easy bruising | R139X | Alive | |
| BII-5 | 22 | nk | nk | nk | Posttraumatic bleeding inducing death | nk | Deceased | |
| BIII-1* | 1 | 4 | 96 | Abnormal | dysMKpoiesis | Easy bruising | R139X | Alive |
FPD indicates familial platelet disorder; AML, acute myeloid leukemia; MK, megakaryocyte; MDS, myelodysplasia syndrome; and nk, not known.
Patients samples used in experiments.
Effect of R174Q and R139X RUNX1 mutations on HSC and progenitor capacities
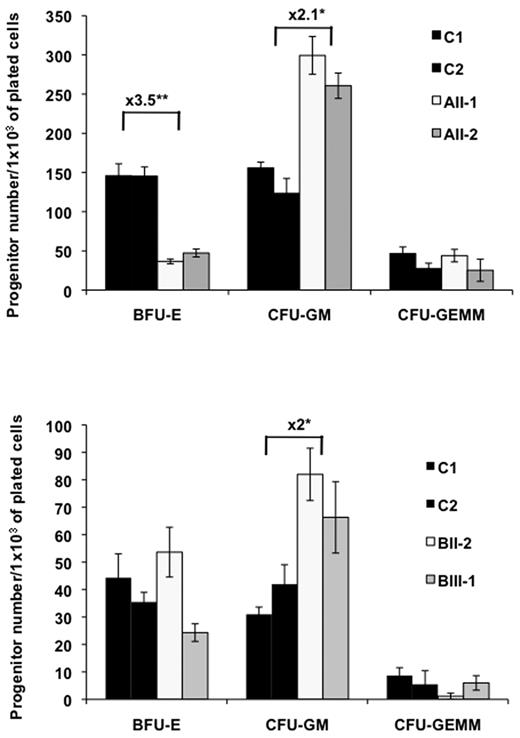
To characterize the abnormalities of hematopoiesis associated with this predisposition to leukemia, we studied the biologic properties of hematopoietic progenitor cells in patients belonging to 2 FPD/AML pedigrees. Short-term methylcellulose assays were used to compare the capacity of patient and control peripheral blood CD34+CD38+ and more primitive CD34+CD38− cells to give rise to hematopoietic colonies. Each experiment was performed twice for each patient and P value was calculated as a function of the mutation. In both pedigrees, the number of granulo-macrophage colony-forming unit (CFU-GM) present in CD34+CD38+ cells was increased by 2- to 2.1-fold (n = 2, P < .05). In contrast a 3.5-fold decrease in the number of erythroid-burst forming units (BFU-E) was observed, but only in pedigree with the R174Q mutation (n = 2, P < .005). No difference was observed for CFU-GEMM–derived colonies suggesting that RUNX1 may regulate the commitment between the erythroid and the granulomonocytic lineages (Figure 2). In the CD34+CD38− cell fraction containing more primitive progenitor cells, the increase in CFU-GM number was more pronounced for the R174Q mutation (3-fold, n = 2, P < .05; Figure 3 top) than for the R139X mutation (1.7-fold, n = 2, P < .05, Figure 3A bottom). A profound decrease in BFU-E number was also detected for pedigree with the R174Q mutation (21-fold, n = 2, P < .05, Figure 3A top). Remarkably, the CFU-GM derived from CD34+CD38− cells with R174Q mutation showed an increase in replating capacities and could generate a 2-fold increased number in secondary colonies compared with control cells using methylcellulose assays (n = 2, P < .05). However, no tertiary colonies could be observed. This observation contrasts with the R139X mutant, which was not associated with an increased replating ability.
Assessment of colony forming cells (CFC) in control and FPD/AML patient CD34+CD38+ cells. CD34+CD38+ cells from peripheral blood of FPD/AML patients with R174Q (AII-1, AII-2), or R139X (BII-2, BIII-1) RUNX1 mutation, and from peripheral blood of different healthy individuals (C) were plated in methylcellulose. Progenitor content (BFU-E, CFU-GM, and CFU-GEMM) was determined at 2 weeks of culture. The number of colonies was expressed per 1 × 103 of plated CD34+ cells.
Assessment of colony forming cells (CFC) in control and FPD/AML patient CD34+CD38+ cells. CD34+CD38+ cells from peripheral blood of FPD/AML patients with R174Q (AII-1, AII-2), or R139X (BII-2, BIII-1) RUNX1 mutation, and from peripheral blood of different healthy individuals (C) were plated in methylcellulose. Progenitor content (BFU-E, CFU-GM, and CFU-GEMM) was determined at 2 weeks of culture. The number of colonies was expressed per 1 × 103 of plated CD34+ cells.
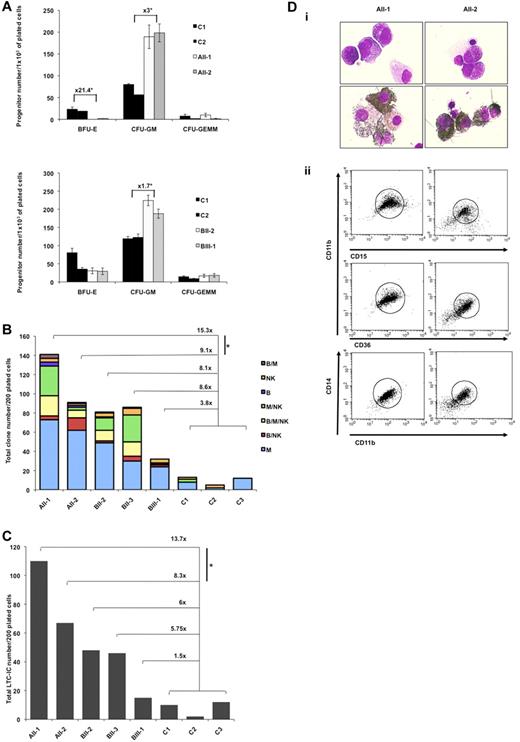
Effect of R174Q and R139X RUNX1 mutations on HSC and progenitor capacities. (A-C) CD34+CD38− peripheral blood cells from FPD/AML patients with R174Q (AII-1, AII-2) or R139X (BII-2, BII-3, BIII-1) RUNX1 mutation, and from healthy individuals (C) were analyzed. (A) Assessment of colony forming cells (CFC). CD34+CD38− cells were plated in methylcellulose. The number of colonies (BFU-E, CFU-GM, and CFU-GEMM) was expressed per 1 × 103 of plated CD34+ cells, pedigree A (top), and pedigree B (bottom). (B) Assessment of B/NK/myeloid potentialities. Analysis of the progeny of single CD34+CD38− cells. CD34+CD38− cells were plated at one cell per well in 96-well plates on MS-5 cells in presence of IL-3, SCF, FLT3-L, TPO, IL-7, IL-15, and IL-2. The experiment was performed once for each patient or control sample. Histograms indicate the number of positive wells in 200 plated wells containing 1 (B, NK, M), 2 (B/M, B/NK, M/NK), or 3 (B/NK/M) lineages. Myeloid cells correspond to CD15+ cells, B cells to CD19+ cells, and NK cells to CD56+ cells. *P < .02 (calculated as described in “Statistical analyses”). (C) Assessment of LTC-IC frequency. Analysis of the progeny of single CD34+CD38− cells. (C, Mean ± SD). Histograms: number of positive wells in 200 plated wells. *P < .02 (calculated as described in “Statistical analyses”). (D) FPD/AML CD34+ cell ability to grow in liquid medium. CD34+CD38− peripheral blood cells from AII-1, AII-2 (R174Q mutation) patients were grown in liquid medium containing cytokine cocktail. May-Grünwald-Giemsa (top panels) and myeloperoxidase (bottom panels) staining were performed at 10 weeks of culture (i). Flow cytometric analysis was performed at 4 months of culture (ii).
Effect of R174Q and R139X RUNX1 mutations on HSC and progenitor capacities. (A-C) CD34+CD38− peripheral blood cells from FPD/AML patients with R174Q (AII-1, AII-2) or R139X (BII-2, BII-3, BIII-1) RUNX1 mutation, and from healthy individuals (C) were analyzed. (A) Assessment of colony forming cells (CFC). CD34+CD38− cells were plated in methylcellulose. The number of colonies (BFU-E, CFU-GM, and CFU-GEMM) was expressed per 1 × 103 of plated CD34+ cells, pedigree A (top), and pedigree B (bottom). (B) Assessment of B/NK/myeloid potentialities. Analysis of the progeny of single CD34+CD38− cells. CD34+CD38− cells were plated at one cell per well in 96-well plates on MS-5 cells in presence of IL-3, SCF, FLT3-L, TPO, IL-7, IL-15, and IL-2. The experiment was performed once for each patient or control sample. Histograms indicate the number of positive wells in 200 plated wells containing 1 (B, NK, M), 2 (B/M, B/NK, M/NK), or 3 (B/NK/M) lineages. Myeloid cells correspond to CD15+ cells, B cells to CD19+ cells, and NK cells to CD56+ cells. *P < .02 (calculated as described in “Statistical analyses”). (C) Assessment of LTC-IC frequency. Analysis of the progeny of single CD34+CD38− cells. (C, Mean ± SD). Histograms: number of positive wells in 200 plated wells. *P < .02 (calculated as described in “Statistical analyses”). (D) FPD/AML CD34+ cell ability to grow in liquid medium. CD34+CD38− peripheral blood cells from AII-1, AII-2 (R174Q mutation) patients were grown in liquid medium containing cytokine cocktail. May-Grünwald-Giemsa (top panels) and myeloperoxidase (bottom panels) staining were performed at 10 weeks of culture (i). Flow cytometric analysis was performed at 4 months of culture (ii).
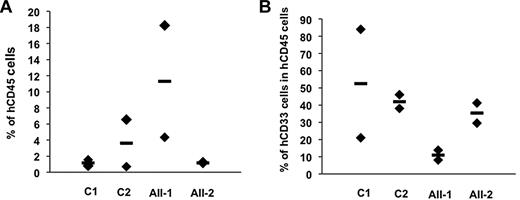
To investigate the role of RUNX1 on early progenitor compartment, we studied the clonogenic capacities of multipotent progenitors (lymphomyeloid). In comparison to the 3 controls, a 9- to 15-fold increase in their cloning efficiency was detected for 2 patients with the R174Q mutation (P < .05). This increase was even higher when taking into account only multipotent clones giving rise to 3 lineages that is, myeloid, B, and NK cells. A lower 4- to 8-fold increase was observed in the R139X pedigree (Figure 3B). To confirm the expansion of the lymphomyeloid compartment, in particular in the R174Q pedigree, we studied the most primitive progenitor assay assessable in vitro, LTC-initiating cells (LTC-ICs). The number of LTC-ICs was increased by 8- to 14-fold in the R174Q pedigree (P < .05) compared with 1.5- to 6.0-fold in the R139X pedigree (Figure 3C). To further investigate this gain of function, CD34+CD38− cells were cultured in liquid medium. Neither cells from control donors nor those from R139X patients could be kept in culture longer than 4 weeks. In contrast, for 2 R174Q patients, 80% of peroxidase-positive cells were detected 10 weeks after the initiation of culture (Figure 3Di). Six weeks later, cytokine-dependent cell lines negative for the CD34 marker, but expressing myelo/monocytic markers CD11b, CD36, CD14, and CD15 were established (Figure 3Dii). Their proliferation rate decreased after 8 months and the cell viability was lost. These results suggest that the R174Q mutant increases the compartment of primitive progenitors and amplifies the pool of early CFU-GM better than R139X. In addition, in contrast to R139X, it induces some self-renewal capacities to myeloid committed progenitors. However, xenotransplantation of CD34+ cells isolated from peripheral blood of patients AII-1 and AII-2 with the R174Q mutation into irradiated NOG did not demonstrate any advantage on cells isolated from 2 control donors (Figure 4). In addition, neither patient nor control cells were able to give secondary transplants. Altogether, our results show that mutations in the RUNX1 gene are associated with the expansion of multipotent and myeloid progenitors without significant effect on HSC. Furthermore, the severity of this expansion appears to depend on the mutation type (Figure 3A-C).
Xenotransplantation of RUNX1 R174Q CD34+ cells into irradiated NOG mice. FPD/AML CD34+ cell capacity for hematopoietic reconstitution in a xenotransplantation model. Human cell engraftment for 2 normal donors (C) and for 2 patients with R174Q mutation (AII-1 and AII-2) was estimated at week 15 by flow cytometry as percentage of human CD45+ (A) or CD33+ (myeloid) among CD45+ cells (B) in the BM. Mean ± SD of 2 mice per donor and patient.
Xenotransplantation of RUNX1 R174Q CD34+ cells into irradiated NOG mice. FPD/AML CD34+ cell capacity for hematopoietic reconstitution in a xenotransplantation model. Human cell engraftment for 2 normal donors (C) and for 2 patients with R174Q mutation (AII-1 and AII-2) was estimated at week 15 by flow cytometry as percentage of human CD45+ (A) or CD33+ (myeloid) among CD45+ cells (B) in the BM. Mean ± SD of 2 mice per donor and patient.
Expression profiling in R174Q and R139X RUNX1 mutated patients' CD34+ cells
To obtain further insight in the molecular basis of this hematopoietic deregulation, we performed gene profiling of CD34+ cells from controls and R174Q or R139X patients (supplemental Table 2). Of the 44 000 tested probe sets, 3950 probes sets were deregulated at least 2-fold in R139X cells. They correspond to 1222 and 1058 genes or expressed sequence tags (ESTs) down or up-regulated, respectively. Probe sets (2647) were deregulated at least 2-fold in R174Q cells corresponding to 1015 and 529 genes or ESTs down or up-regulated, respectively. Most genes identified by this approach have molecular or cellular function involved in cell death, cell cycle or cellular growth and proliferation (Ingenuity Systems software analysis).
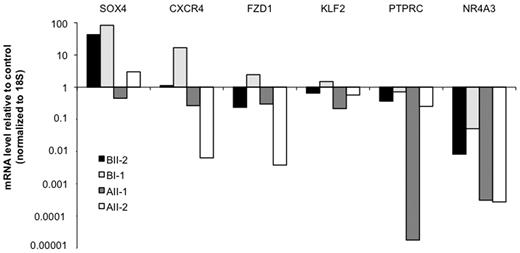
Because R139X and R174Q mutants have not exactly the same molecular and biologic properties (Figures 1,Figure 2–3), we focused on differentially deregulated genes. First we validated the microarray analysis. Changes in the expression of some selected genes were confirmed by Q-PCR in an independent experiment including 2 patient samples for each pedigree (Figure 5). Among the most differentially expressed genes, we focused on NR4A3, because its down-regulation is a common feature in leukemic blast cells and that mice deficient in both Nr4a3 and Nr4a1 have a high predisposition to AML.24 NR4A3 expression level was strongly reduced, but clearly detectable in R139X progenitors while it was nearly undetectable in those with the R174Q mutation (Figure 5; S1 and S2 in supplemental Table 2). A NR4A1 down-regulation was also detected, but less pronounced than that of NR4A3 (S1 and S2 in supplemental Table 2), suggesting that the level of NR4A3 expression could be an important parameter in the leukemic predisposition induced by RUNX1 mutations. This hypothesis was strengthened by the fact that in control cells, NR4A3 was highly expressed in the CD34+CD38− cell population, its expression decreased in CD34+CD38+ population to highly increase during monocytic differentiation. NRA41 was only weakly expressed in the same cell populations (supplemental Figure 5). This high expression of NR4A3 in the hematopoietic CD34+CD38− progenitors suggests that it may play an important role in early stages of hematopoiesis and that its decrease in RUNX1 mutant patients may explain the altered properties of their immature progenitors (Figure 3B-C).
Expression profiling in R174Q and R139X RUNX1 mutated patients CD34+ cells. Expression fold-change analysis of genes selected from array data (supplemental Table 2) in independent samples by Q-PCR. RNA was directly extracted and reverse-transcribed from sorted CD34+ cells obtained from AII-1, AII-2 (R174Q mutation) and BI-1, BII-2 (R139X mutation) patients and healthy donors peripheral blood. Gene expression was normalized to 18S and results are expressed as fold changes related to healthy control.
Expression profiling in R174Q and R139X RUNX1 mutated patients CD34+ cells. Expression fold-change analysis of genes selected from array data (supplemental Table 2) in independent samples by Q-PCR. RNA was directly extracted and reverse-transcribed from sorted CD34+ cells obtained from AII-1, AII-2 (R174Q mutation) and BI-1, BII-2 (R139X mutation) patients and healthy donors peripheral blood. Gene expression was normalized to 18S and results are expressed as fold changes related to healthy control.
NR4A3 is a direct target gene of RUNX1 in hematopoietic progenitors
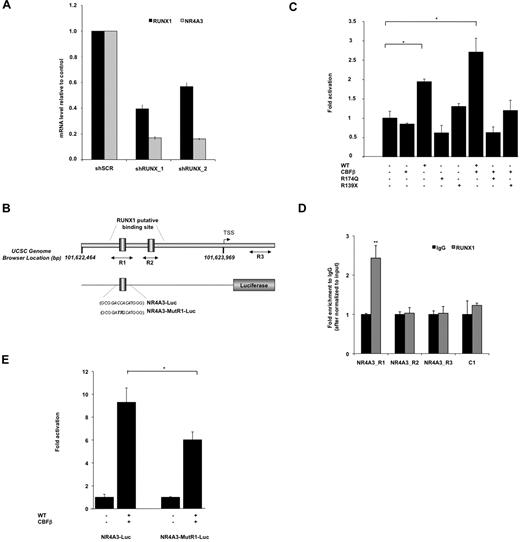
We explored the molecular mechanisms underlying the ability of RUNX1 to regulate NR4A3 expression. By using an RNA interference strategy using 2 lentiviruses encoding for 2 different shRNA of RUNX1 (shRUNX1_117 and shRUNX1_2), we confirmed that the inhibition of RUNX1 expression in primary CD34+ cells leads to the down-regulation of NR4A3 (Figure 6A). To address whether NR4A3 is a direct RUNX1 transcriptional target, we performed an in silico conserved sequence analysis25 of its promoter region and identified 2 potential RUNX1-binding sites. To test the functional relevance of these RUNX1 sites, we cloned this promoter region upstream the luciferase gene (Figure 6B) and performed gene reporter assays. Overexpression of wt RUNX1 with its cofactor CBFβ increased luciferase activity in transient transfection assays in HEL cells; in contrast, the R174Q or R139X mutants did not exhibit this transactivation capacity (Figure 6C). Finally, ChIP assays performed in primary CD34+ cells showed that RUNX1 binds to the R1 and not to the R2 site with a significant enrichment compare with R3 region, which is a downstream promoter region without RUNX1 putative-binding site (Figure 6E). To demonstrate its functionality, we mutated the R1 site of the NR4A3 promoter luciferase vector (Figure 6B) and showed a significant decrease in RUNX1/CBFβ induced luciferase activity (Figure 6E). Moreover, gel shift assays confirmed that RUNX1 binds to the R1 site in the NR4A3 promoter (supplemental Figure 6). These data strongly support that NR4A3 is a direct, positively regulated transcriptional target of RUNX1 in hematopoietic progenitor cells.
NR4A3 is a target gene of RUNX1 in hematopoietic progenitors. (A) Effect of RUNX1 knockdown on NR4A3 expression. Q-PCR analysis of CD34+GFP+ cells transduced with the lentivirus expressing shRUNX1_1, shRUNX1_2, or a shSCR (scramble control sequence). The mRNA level of RUNX1 and NR4A3 was normalized to HPRT mRNA level. The histograms show 1 representative experiment of 2, each in triplicate. Error bars represent ± SD of triplicate. (B) Schematic representation of the NR4A3 human promoter region. The arrowhead represents the putative transcription start site. 101,622,464_101,623,969 designates Chr9 genomic positions of the region cloned in luciferase analysis (UCSC hg18 assembly, http://genome.ucsc.edu/). Two-side black arrows designate genomic positions of the amplicons used in ChIP analysis. NR4A3-Luc and NR4A3-MutR1-Luc designate luciferase reporter vector with the region of NR4A3 promoter with or without the mutated R1 RUNX1 binding site in italic bold. (C) Luciferase levels are shown as fold increase relative to cells transfected with promoter construct alone. Error bars represent ± SD of 2 experiments, each in triplicate. *P < .05. (D) ChIP assay was performed on CD34+ cells and shows the fold difference in chromatin immunoprecipitated between anti-RUNX1 and irrelevant IgG after normalization to input chromatin (means ± SD, n = 2). Amplified NR4A3 promoter regions R1, R2, and R3 illustrated in panel B were tested. A nearby region without RUNX1 binding site (C1) was amplified and used as a negative control. (E) Luciferase levels are shown as the fold increase relative to cells transfected with normal promoter construct (NR4A3-Luc) alone (on left) and with mutated promoter construct (NR4A3-MutR1-Luc) alone (on right). Error bars represent ± SD of 2 experiments, each in triplicate. *P < .05.
NR4A3 is a target gene of RUNX1 in hematopoietic progenitors. (A) Effect of RUNX1 knockdown on NR4A3 expression. Q-PCR analysis of CD34+GFP+ cells transduced with the lentivirus expressing shRUNX1_1, shRUNX1_2, or a shSCR (scramble control sequence). The mRNA level of RUNX1 and NR4A3 was normalized to HPRT mRNA level. The histograms show 1 representative experiment of 2, each in triplicate. Error bars represent ± SD of triplicate. (B) Schematic representation of the NR4A3 human promoter region. The arrowhead represents the putative transcription start site. 101,622,464_101,623,969 designates Chr9 genomic positions of the region cloned in luciferase analysis (UCSC hg18 assembly, http://genome.ucsc.edu/). Two-side black arrows designate genomic positions of the amplicons used in ChIP analysis. NR4A3-Luc and NR4A3-MutR1-Luc designate luciferase reporter vector with the region of NR4A3 promoter with or without the mutated R1 RUNX1 binding site in italic bold. (C) Luciferase levels are shown as fold increase relative to cells transfected with promoter construct alone. Error bars represent ± SD of 2 experiments, each in triplicate. *P < .05. (D) ChIP assay was performed on CD34+ cells and shows the fold difference in chromatin immunoprecipitated between anti-RUNX1 and irrelevant IgG after normalization to input chromatin (means ± SD, n = 2). Amplified NR4A3 promoter regions R1, R2, and R3 illustrated in panel B were tested. A nearby region without RUNX1 binding site (C1) was amplified and used as a negative control. (E) Luciferase levels are shown as the fold increase relative to cells transfected with normal promoter construct (NR4A3-Luc) alone (on left) and with mutated promoter construct (NR4A3-MutR1-Luc) alone (on right). Error bars represent ± SD of 2 experiments, each in triplicate. *P < .05.
NR4A3 regulates the clonogenic potential of hematopoietic progenitors from patients and controls
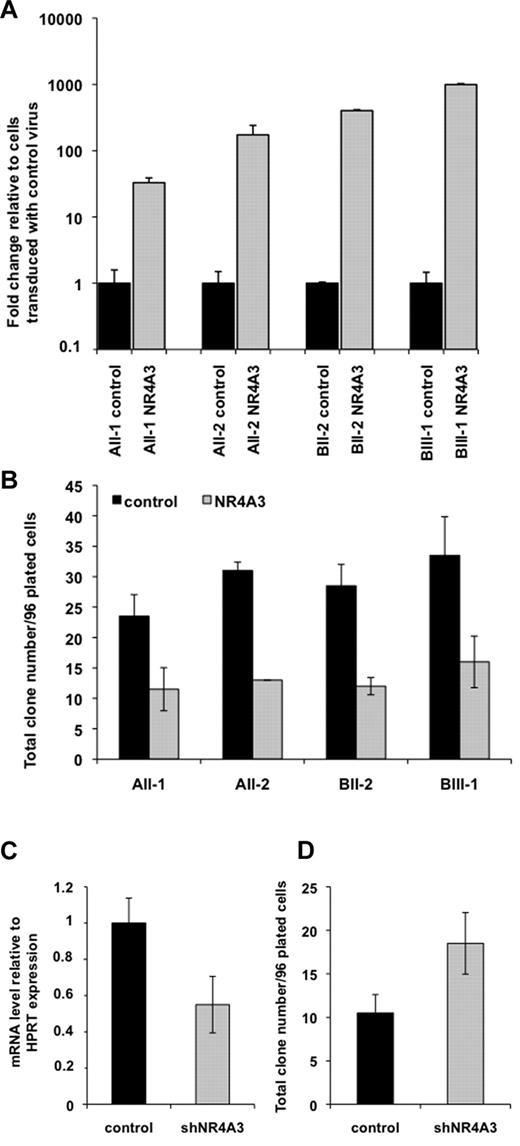
To determine whether down-regulation of NR4A3 expression in FPD/AML patient CD34+ cells played a role in the abnormal expansion of multipotent progenitors, we transduced CD34+ cells from the R174Q and R139X patients with a lentivirus encoding GFP alone or together with NR4A3 (Figure 7A, supplemental Figure 7a) and tested their capacity to give rise to lymphoid and myeloid clones. For AII-1, AII-2 (R174Q mutation) and BII-2, BIII-1 (R139X mutation) patients, we demonstrated that NR4A3 overexpression induced a significant 2-fold (n = 2, P < .01) decrease in the number of total clones as well as in those containing lymphoid and myeloid cells (Figure 7A-B). In this experiment, 90% of clones were myeloid for both conditions. However, a possible bias in these experiments lie in the fact that NR4A3 might be more expressed after transduction than in the control cells (supplemental Figure 7B). Thus, we performed a knockdown strategy to further demonstrate the role of the NR4A3 down-regulation in the regulation of immature hematopoietic progenitors. The transduction of normal CD34+ cells with a shRNA against NR4A3 (supplemental Figure 7A) induced a significant 1.7-fold increase in colony numbers associated with a 45% inhibition of NR4A3 mRNA level (Figure 7C-D, n = 2, P < .05). This result strongly suggests that NR4A3 down-regulation in FPD/AML patients is responsible for the expansion of primitive hematopoietic progenitors.
NR4A3 regulates the clonogenic potential of hematopoietic progenitors from patients and controls. (A-B) Overexpression of NR4A3 in CD34+ FPD/AML cells. CD34+ cells were transduced with an empty lentivirus (control) and a lentivirus expressing NR4A3 (NR4A3), and GFP+ cells were sorted at day 4 of culture. (A) NR4A3 expression was performed by Q-PCR and normalized to RPL0 expression. (B) B/NK/myeloid clonogenic potential of CD34+GFP+ cells. The histograms represent number of positive wells in 96-plated wells. Error bars represent ± SD of 1 representative experiment performed in duplicate. One representative of 2 independent experiments giving similar results is shown (P < .01). (C-D) Effect of NR4A3 knockdown expression on clonogenic progenitors. (C) Q-PCR analysis of CD34+GFP+ cells transduced with the lentivirus expressing shRNA of NR4A3 or a scramble sequence (control). The expression of NR4A3 was normalized to HPRT expression. Error bars represent ± SD of triplicate. (D) B/NK/myeloid clonogenic potential of CD34+GFP+ cells transduced by shNR4A3 or scramble sequence (control). The histograms represent number of positive wells in 96-plated wells. Error bars represent ± SD of 1 representative experiment performed in duplicate. One representative of 2 independent experiments giving similar results is shown (P < .05).
NR4A3 regulates the clonogenic potential of hematopoietic progenitors from patients and controls. (A-B) Overexpression of NR4A3 in CD34+ FPD/AML cells. CD34+ cells were transduced with an empty lentivirus (control) and a lentivirus expressing NR4A3 (NR4A3), and GFP+ cells were sorted at day 4 of culture. (A) NR4A3 expression was performed by Q-PCR and normalized to RPL0 expression. (B) B/NK/myeloid clonogenic potential of CD34+GFP+ cells. The histograms represent number of positive wells in 96-plated wells. Error bars represent ± SD of 1 representative experiment performed in duplicate. One representative of 2 independent experiments giving similar results is shown (P < .01). (C-D) Effect of NR4A3 knockdown expression on clonogenic progenitors. (C) Q-PCR analysis of CD34+GFP+ cells transduced with the lentivirus expressing shRNA of NR4A3 or a scramble sequence (control). The expression of NR4A3 was normalized to HPRT expression. Error bars represent ± SD of triplicate. (D) B/NK/myeloid clonogenic potential of CD34+GFP+ cells transduced by shNR4A3 or scramble sequence (control). The histograms represent number of positive wells in 96-plated wells. Error bars represent ± SD of 1 representative experiment performed in duplicate. One representative of 2 independent experiments giving similar results is shown (P < .05).
Discussion
RUNX1 was first identified as a target of chromosomal translocation in the t(8;21)(q22;q22) found in approximately 12% of AML. Other chromosomal aberrations, different translocations, acquired mutations and duplications of RUNX1 gene are frequently found in MDS and acute leukemia. The chromosomal inversion (inv(16)) affecting CBFβ is associated with the AML-M4Eo form of AML. Sporadic point mutations found in AML-M0 are often biallelic, additional karyotypic abnormalities are found in MDS-AML and secondary MDS/AML. Heterozygous germline alterations in RUNX1 leading to FPD/AML per se are not sufficient to induce full-blown leukemia, one or more additional events are required. Therefore, this pathology seems to be an ideal model to understand how RUNX1 germline mutations contribute to an early hematopoietic deregulation, which will predispose to leukemia.
A previous study in FPD/AML showed that missense and nonsense mutations retained the capacity to dimerize with CBFβ and suggested that by inhibiting the transactivating capacities of wild-type RUNX1, they may create a higher propensity to develop leukemia than RUNX1 deletions and frameshifts that lead to haploinsufficiency.4 However, this study was performed only at the molecular level and the correlation between the type of mutations and the proportion of patients who developed leukemia was limited taking in consideration the patient number with available clinical data. In the present work, we performed a functional analysis of hematopoiesis in patients harboring a previously described DN-like R174Q mutation and in patients harboring a R139X mutation leading to a truncated RUNX1 protein, which is unable to dimerize with CBFβ subunit. It should be noted that all the patients from the 2 pedigrees included in our study present only a thrombocytopenia though 2 other patients of the pedigree with R174Q mutation died from a MDS/AML or an AML-M1, respectively. Our results demonstrate that in the R174Q mutant context, RUNX1 is not detectable in the nucleus of mononuclear cells whereas the wt protein, even at a lower level, remains present in cells from patient carrying the R193X mutant. Thus the R174Q pedigree appears to be close to a RUNX1 knockout while the R139X pedigree is closer to a haploinsufficiency. The R174Q mutant was associated with a marked increase in the clonogenic potential and the proliferative rate of immature CD34+CD38− progenitors. However, the progenitors of patients with the R139X mutation also displayed an increase in clonogenic potential. This parallels the results observed in AML1+/− mice where the haploinsufficiency induces an increase in mutlilinegae and lineage-restricted progenitors.26 R139X mutant, which does not interact with CBFβ, can be thus considered as a “dead” mutant. However, it may be also associated with a predisposition to leukemia, but with a lower penetrance than the R174Q RUNX1 mutant. Interestingly, an increase in CFU-GM accompanied by a decrease in BFU-E colony number was observed in one patient of each pedigree (as previously described by Song et al2 ), suggesting that RUNX1 could be also involved in the commitment of a common myeloid progenitor.
To understand the mechanism by which different point mutations in RUNX1 can induce different pathologies, the mutations in RHD domain (K83N, R135G, and D171N) or in the C-terminal domain (S291fsX300) were introduced in RUNX1 cDNA and vectorized in retroviruses.27,28 The overexpression in mice of Runx1 DNA-binding mutants K83N and R135G (initially found in patients with AML and/or FPD/AML) did not completely superimpose with our results described with the R174Q mutant. Indeed K83N and R135G mutants have not been found in vivo as dominant-negative suppressors during hematopoiesis. The mice exhibited normal level of myeloid progenitors and no defect in megakaryocyte differentiation or in the lymphoid compartment. However, in agreement with our results, an increased replating capacity of myeloid progenitors and an impaired erythropoiesis were found.27 In the approach using retroviruses, the integration site may influence the phenotype obtained. During the retroviral overexpression of D171N and S291fs mutants, retroviruses integrated, respectively, near the Evi1 or into the Mn1 gene and the induction of MDS/leukemia phenotype probably resulted from oncogenic collaboration.28 In addition, retroviral transduction led to a sustained overexpression of mutant proteins in mouse bone marrow cells, while both R139X and R174Q mutants were not detected in mature patient mononuclear cells.
Xenotransplantation of RUNX1 R174Q CD34+ cells into irradiated NOG mice showed that this DN-like mutation does not affect significantly the human HSC compartment. Altogether our results obtained with FPD/AML patient cells corroborate those found in mice deficient for Runx1. In both cases, an increase in hematopoietic multipotent and committed progenitors29,30 without increasing long-term myeloid-repopulating capacity in the competitive transplantation assays30 was observed.
However, we could not rule out that the differences between the R174Q and R139X mutants were related to the acquisition of a second genetic event in the R174Q pedigree, which would lead to a clonal dominance although a similar phenotype was found in 2 members of the pedigree. We performed clonality studies, as previously described,31 in one female patient of R174Q pedigree and one female patients of R139X pedigree using polymorphic genes located on the X chromososme (data not shown). This study showed that hematopoiesis was polyclonal for the R174Q patient. Surprisingly, in the female with the R139X mutant, the entire hematopoiesis including T cells was clonal. This type of clonality is extremely unusual in malignant hematopoietic disorder hitting the HSC where clonality only involves myeloid cells and eventually B cells but not T cells. Thus, this X skewing in the hematopoietic cells might be constitutive; such an extreme skewing has been calculated to occur in 1 of 1200 subjects based on the concept that X inactivation occurs when 8 HSC have emerged. However, we cannot exclude that the RUNX1 mutation has modified the kinetics of HSC emergence and thus that X inactivation has occurred at a time where less than 8 HSC were present increasing the probability to have an extreme skewing. Whatever the explanation, this result demonstrates that in the R174Q patients the deregulation of hematopoiesis is related to the RUNX1 mutation, which leads to an amplification of multipotent and myeloid compartment fulfilling the necessary conditions of leukemia development.
We performed a transcriptional analysis in the hematopoietic CD34+ progenitors of both pedigrees to better understand the molecular mechanism of this hematopoietic deregulation. Gene profiling showed an almost complete loss in NR4A3 expression in R174Q hematopoietic progenitors and a less pronounced decrease in R139X cells. NR4A3 encodes an orphan nuclear receptor, the transcriptional activity of which depends on serum growth factors, mitogenic, apoptotic, or inflammatory stimuli and is thus involved in many different cellular processes such as anti-inflammatory response, apoptosis and cell proliferation.32-34 The role of Nr4a3 alone in hematopoiesis is not described; however, the reduced gene dosage of Nr4a1 and Nr4a3 in hypoallelic mice below a critical level induces myelodysplastic/myeloproliferative neoplasms35 and mice deficient for both Nr4a3 and Nr4a1 demonstrate abnormal expansion of HSCs and myeloid progenitors, decreased expression of AP-1 transcription factors, and defective apoptosis, altogether leading to AML development.24 Although an epigenetic silencing of NR4A3 has been previously reported in AML patients,36 here we demonstrate for the first time that NR4A3 is a direct target gene of RUNX1 transcription factor in normal hematopoietic progenitors. The NR4A3 decrease contributes to the increased clonogenic potential of both multipotent progenitors and CFU-GM as well as the proliferative capacity of myeloid progenitors. Indeed, restoration of NR4A3 expression partially reduced the clonogenic potential of patient progenitors with both R174Q and R139X mutations and conversely, NRA4A3 inhibition increased the clonogenic potential of normal hematopoietic progenitors. The slight decrease in NR4A1 expression comparable in the 2 pedigrees could synergically contribute to the deregulation of hematopoiesis, especially in the cases with an almost complete loss of NR4A3 expression.
Altogether, we propose that the NR4A3 down-regulation below a critical threshold depending on the type of RUNX1 mutation in FPD/AML patients is involved in the deregulation of the first steps of hematopoiesis. This deregulation may either directly lead to the development of a myelodysplastic/myeloproliferative neoplasm or by amplification of the hematopoietic progenitor compartment may favor the occurrence of secondary genetic events. Therefore, NR4A3 may represent a potential target for therapeutic strategies aiming to prevent leukemic development in FPD/AML patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for participation in this study, as well as Dr F. Comte (Bastia, France) and Miss S. Cadic (Pasteur Cerba laboratory, Paris, France). They are grateful to O. Bluteau, E. Solary (Inserm U1009, Paris, France), and F. Wendling (Ligue National contre le Cancer, Paris, France) for helpful suggestions on the manuscript; Genethon (Evry, France) for sinpRRL-PGK-GFP lentivirus vector; A. Dubart-Kupperschmitt (Institut Cochin, Paris, France) for TRIPΔU3-EF1a-GFP lentivirus vector; C. J. M. de Vries (Academic Medical Center, Amsterdam, The Netherlands) for pSV7 d-NR4A3 plasmid; F. Mazurier (Inserm U876, Bordeaux, France) for the NOG experiments and to genomic platform of IGR for microarrays; and P. Nusbaum and A. Hubas for patient fibroblast cultures (Hôpital Cochin, Banque de cellules, Paris, France).
This work was supported by grants from the Agence Nationale de la Recherche (ANR-GIS maladies rares), the Ligue contre le Cancer (équipe labellisée 2009), the Association de Recherche Contre le Cancer (ARC), and the Center de Référence des pathologies plaquettaires. D.B. was supported by a postdoctoral fellowship from ANR, and L.G. was partially supported by ARC.
Authorship
Contribution: D.B. designed and performed experiments, analyzed data, and wrote the manuscript; L.G., M.H., and V.C.-L. performed experiments; I.A.-D. performed results; C.J., N.D., V.C.-C., and O.W.-B. performed experiments and analyzed data; T.R. performed arrays experiments; H. Ripoche performed arrays analysis; P.G. performed statistical analysis; S.S. performed clonality experiments; J.P. directed clonality experiments and discussed results; W.V. designed the work, discussed the results, and wrote the manuscript; R.F. followed up with patients, performed experiments, and discussed the results; and H. Raslova designed the work, performed and supervised experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hana Raslova, Inserm, U1009, 114 rue Edouard Vaillant, Villejuif, France; e-mail: hraslova@igr.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal