Abstract
Both monoallelic and biallelic oncogenic NRAS mutations are identified in human leukemias, suggesting a dose-dependent role of oncogenic NRAS in leukemogenesis. Here, we use a hypomorphic oncogenic Nras allele and a normal oncogenic Nras allele (Nras G12Dhypo and Nras G12D, respectively) to create a gene dose gradient ranging from 25% to 200% of endogenous Nras G12D/+. Mice expressing Nras G12Dhypo/G12Dhypo develop normally and are tumor-free, whereas early embryonic expression of Nras G12D/+ is lethal. Somatic expression of Nras G12D/G12D but not Nras G12D/+ leads to hyperactivation of ERK, excessive proliferation of myeloid progenitors, and consequently an acute myeloproliferative disease. Using a bone marrow transplant model, we previously showed that ∼ 95% of animals receiving Nras G12D/+ bone marrow cells develop chronic myelomonocytic leukemia (CMML), while ∼ 8% of recipients develop acute T-cell lymphoblastic leukemia/lymphoma [TALL] (TALL-het). Here we demonstrate that 100% of recipients transplanted with Nras G12D/G12D bone marrow cells develop TALL (TALL-homo). Although both TALL-het and -homo tumors acquire Notch1 mutations and are sensitive to a γ-secretase inhibitor, endogenous Nras G12D/+ signaling promotes TALL through distinct genetic mechanism(s) from Nras G12D/G12D. Our data indicate that the tumor transformation potential of endogenous oncogenic Nras is both dose- and cell type-dependent.
Introduction
In mammals 3 different ras gene loci encode 4 highly homologous 21-KD proteins: Hras, Nras, Kras.4A and Kras.4B.1 Ras proteins belong to the super family of small GTPases. They cycle between the active GTP-bound form and the inactive GDP-bound form.2,3 Once Ras proteins are activated, they subsequently activate multiple downstream signaling pathways (including the Raf/MEK/ERK and PI3K/Akt pathways) and regulate cell survival, proliferation, and differentiation.
Oncogenic mutations in the 3 RAS genes have been identified in virtually all types of human cancers, with characteristic incidences and RAS gene associations.4 In particular, mutations in the KRAS and NRAS genes (but rarely in the HRAS gene) are frequently identified in myeloid disorders (15%-60%), including acute myeloid leukemia (AML),5,6 atypical chronic myeloid leukemia,7 chronic myelomonocytic leukemia (CMML),8-10 and juvenile myelomonocytic leukemia (JMML).8-10 In contrast, although hyperactivated Ras signaling is identified in 50% of patients with acute T-cell lymphoblastic leukemia/lymphoma (TALL), oncogenic NRAS mutations only occur in < 5% of these patients.4,11-13
Uniparental disomy (UPD) of an oncogenic RAS allele is reported in both primary human tumor samples and tumor cell lines,14,15 suggesting a dose-dependent role of oncogenic RAS in tumor development. Results obtained from several mouse models under conditions resulting in overexpression of oncogenic Nras support this hypothesis; increasing levels of oncogenic Nras leads to stronger phenotypes and significantly shorter disease latency.16,17 However, whether or not this conclusion would be true with endogenous transcriptional regulation of the Nras locus was, until now, uncertain.
Haigis et al generated a conditional oncogenic Nras allele,18 in which the oncogenic mutation G12D was introduced into the endogenous Nras locus and expression of oncogenic Nras is blocked by a floxed STOP cassette (LSL Nras G12D). In the presence of Cre recombinase, the STOP cassette is removed and oncogenic Nras is expressed at a level comparable with the wild-type (WT) Nras allele.19 Using this allele, we established a mouse bone marrow transplantation model in which monoallelic oncogenic Nras is expressed in bone marrow cells alone.19 Approximately 95% of recipient mice developed a CMML-like disease after a prolonged latency. Moreover, CMML development is associated with aberrant GM-CSF signaling and UPD of the oncogenic Nras allele, both of which are reported in human CMML specimens.14,20 These results highlight the relevance of our model to the human disease.
The utility of genetically engineered alleles to study gene dosages has been reported extensively in mouse.21 Here, we report that using a sophisticated mouse genetic approach, we created a gradient of gain-of Nras signaling ranging from 25%-200% of endogenous monoallelic expression of oncogenic Nras. We studied the effects of this signaling gradient on leukemogenesis.
Methods
Mice
The hypomorphic LSL Nras G12D (LSL Nras G12Dhypo) mice were constructed as following: BAC clones covering the Nras locus were purchased from BACPAC Resources. A 6.6 Kb fragment double-digested with EcoRI and HindIII was cloned into the vector pGEM-3Z (Promega). G12D mutation and intron 1 mutations (changing WT sequence from gtaattgctgcttttctacag to gtaattgctgcgtttaaacag) were introduced via a site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). A PmeI site was created in intron 1 to insert the LoxP-stop casette-LoxP. The targeting construct was electroporated into V6.5 embryonic stem (ES) cells and G418 resistant colonies were screened by genomic Southern blot analysis. The germ line-transmitted LSL mice were generated by tetraploid injection of correct ES cells. ES cells were screened using genomic Southern analysis. The mice were backcrossed into C57BL/6 genetic background for more than 10 generations. The embryos and adult mice were genotyped using primers 5′ WT (5′-CCACGTGTATCGAATGGGTGCCTTAG-3′), 3′ WT (5′-GGGATCATATTCATCCACAAAGTGG-3′), and 3′ mt (5′-GCGGCCAAACGCAGCATTAC-3′).
All mouse lines were maintained on a pure C57BL/6 genetic background (N > 10). The conditional Nras G12D allele with normal expression level comparable with WT allele is described in Haigis et al.18 Mice bearing this allele were crossed to Mx1-Cre mice to generate mice carrying both alleles (LSL Nras/+; Mx1-Cre). LSL Nras G12D/+; Mx1-Cre males were crossed to LSL Nras G12D/+ females to generate LSL Nras G12D/G12D; Mx1-Cre mice. Genotyping of the adult mice was performed as described in Haigis et al.18
Mox2 Cre mice were purchased from Jackson Laboratories. CD45.1-positive congenic C57BL/6 recipient mice were purchased from the National Cancer Institute.
To induce Mx1-Cre expression, 5-6 week old mice were injected intraperitoneally with 250 μg of polyinosinic-polycytidylic acid (pI-pC; Sigma Aldrich) every other day for 2 doses. All the experiments were performed 2 days after the second injection of pI-pC unless specified. The injected mice were monitored daily for evidence of disease. All experiments were conducted with the ethical approval of International Association for Assessment and Accreditation of Laboratory Animal Care at the University of Wisconsin-Madison.
Sequence analysis of Nras G12 codon
Total RNAs and genomic DNAs were extracted and analyzed for Nras G12 codon as described.19
Western blot analysis
TER119-negative fetal liver cells (enriched for erythroid progenitors) were purified from E14.5 individual embryos using the StemSep magnetic bead system (StemCell Technologies Inc)22 and Western blot analysis was performed essentially as described.23 To detect the levels of total Nras proteins and TdT in tumor samples, 40 and 20 μg of cell lysates were loaded in each lane, respectively. Anti-Nras (F155) and anti-TdT (N-20) antibodies were purchased from Santa Cruz Biotechnology, whereas anti–mouse β-actin antibodies were from Sigma-Aldrich.
Flow cytometric analysis of hematopoietic tissues
For lineage analysis in peripheral blood, bone marrow, spleen, and thymus tissues, flow cytometric analyses were performed as previously described.24 Myeloid progenitors in bone marrow and spleen were analyzed as previously described.25,26 The stained cells were analyzed on a FACSCalibur or LSRII (BD Biosciences).
Directly conjugated antibodies specific for the following surface antigens were purchased from BD Biosciences: CD45.1 (A20), CD45.2, (104) B220 (6B2), CD19 (1D3), Thy1.2 (53-2.1), Mac-1 (M1/70), Gr-1 (RB6-8C5), CD4 (RM4-5), CD8 (53-6.7), CD3 (145-2C11), IgM (II/41), IL7Rα (B12-1), Sca-1 (E13-161.7), TER119, CD34 (RAM34), FcγRII/III (2.4G2), CD25 (3C7) and CD44 (IM7). Anti–c-Kit (2B8) antibodies were purchased from eBiosciences.
Cell-cycle analysis
Cell-cycle analysis was performed essentially as described.27 Lineage markers (CD3, CD4, CD8, CD19, B220, TER119, Gr1, and IgM) and IL-7Rα were stained with biotin conjugated antibodies followed by PECy7 conjugated streptavidin. Cells were also simultaneously stained for FITC-Ki67 (BD Biosciences), PE-CD45.1, APC-cKit, PerCP-Sca1, and DAPI (Invitrogen). The stained cells were analyzed on a LSRII (BD Biosciences).
Flow cytometric analysis of phospho-ERK1/2
Phosphorylated ERK1/2 in defined Lin− c-Kit+ cells were analyzed essentially as previously described.19 U0126 (Cell Signaling Technology) was mixed with cells for 30 minutes before GM-CSF stimulation.
Murine bone marrow transplantation
Bone marrow transplantation was performed as described.19 For secondary transplantation, recipient mice (CD45.1+) were sublethally irradiated (650 rads). Subsets of primary bone marrow tumor cells were sorted using FACSAria II (BD Biosciences). Different doses of tumor cells (CD45.2+) were mixed with 2.5 × 105 helper cells (CD45.1+ splenocytes) and transplanted into recipient mice.
Histopathology
Mouse organs were fixed in 10% neutral buffered formalin (Sigma-Aldrich) and further processed at the Histology Lab of the University of Wisconsin Carbone Cancer Center.
TCR-β genomic Southern
Genomic DNA samples were prepared using the Puregene Genomic DNA Purification Kit (QIAGEN) per manufacturer's instructions. Rearrangement at the T-cell receptor β locus was detected as previously described.28
DNA sequencing of Notch1
Genomic DNA was isolated from tumor cells using the Puregene Genomic DNA Purification Kit (QIAGEN). Amplification of Notch1 exons 26, 27, and 34 was performed as described previously.29 PCR products were subcloned into pGEM-T Easy vector (Promega). Ten to 15 clones were randomly selected and sequenced at the Biotechnology Center (University of Wisconsin-Madison). The sequencing data were analyzed using the Lasergene Version 8.0 software (DNASTAR).
Results
Mice expressing Nras G12Dhypo/G12Dhypo develop normally and are tumor-free
To study the dose-dependent role of endogenous oncogenic Nras in leukemogenesis, we generated a conditional hypomorphic oncogenic Nras allele (LSL Nras G12Dhypo; Figure 1A). Through a computational modeling design,30 mutations in Nras intron 1 were created in sites adjacent to the splicing acceptor to lower the splicing efficiency of the transcript and to insert the floxed STOP cassette (see “Mice” for details). Thus, the floxed STOP cassette in our hypomorphic allele is at a different position from that in the allele created by Haigis et al.18 Correct targeting in ES cells was confirmed using Southern blot analysis with a 5′ end and a 3′ end probe (Figure 1B). Germ line transmission of the conditional allele was confirmed by genotyping PCR (data not shown). The presence of the G12D point mutation in germ line transmitted mice was confirmed by genomic sequencing (Figure 1C).
Construction of a conditional hypomorphic Nras G12D allele. (A) Schematic diagram of WT Nras allele, targeting vector, and constructed LSL (LoxP-STOP cassette-LoxP) Nras G12Dhypo allele. The asterisk shows the substitution of amino acid aspartic acid for glycine by mutation of GGT to GAT at the codon 12. Please see “Mice” for the details of intron 1 mutations. The positions of the probes for Southern blotting are shown. (B) Southern blot analysis of BglII digested genomic DNA isolated from different ES cell clones to confirm correct targeting at the endogenous Nras locus. The WT allele is denoted by the 10.7 KB fragment. The correctly targeted LSL allele is indicated by the 7.4 KB fragment using the 5′ internal probe and by the 5.6 Kb fragment using the 3′ external probe. (C) Direct sequencing with a reverse primer of genomic DNA isolated from WT and germ line transmitted LSL Nras G12Dhypo mice demonstrates the presence of G12D mutation at the Nras locus. Arrows indicate the WT and mutated nucleotides at the codon 12. (D) Evaluation of recombination efficiency of Mox2-Cre and expression level of Nras G12D hypo allele in E14.5 fetal liver erythoid progenitors. Because Mox2-Cre recombines the conditional LSL cassette and leads to Nras G12Dhypo expression, we refer the compound mice harboring both LSL and Mox2-Cre alleles as Nras G12Dhypo/+; Mox2-Cre/+. We further crossed Nras G12Dhypo/+; Mox2-Cre/+ mice to LSL Nras G12Dhypo/+ (LSL/+) mice and generate Nras G12Dhypo/LSL progenies inherit a recombined Nras G12Dhypo allele and a nonrecombined LSL allele from parents but does not carry Mox2-Cre allele. To simplify the genotyping results, we omit the Mox2-Cre status. Southern blot analysis of SpeI digested genomic DNA using the 3′ external probe confirmed the recombination efficiency at the endogenous Nras locus (top panel). Expression levels of total Nras were measured by Western blotting and normalized against actin using the Molecular Analyst Version 1.4 software (Bio-Rad).
Construction of a conditional hypomorphic Nras G12D allele. (A) Schematic diagram of WT Nras allele, targeting vector, and constructed LSL (LoxP-STOP cassette-LoxP) Nras G12Dhypo allele. The asterisk shows the substitution of amino acid aspartic acid for glycine by mutation of GGT to GAT at the codon 12. Please see “Mice” for the details of intron 1 mutations. The positions of the probes for Southern blotting are shown. (B) Southern blot analysis of BglII digested genomic DNA isolated from different ES cell clones to confirm correct targeting at the endogenous Nras locus. The WT allele is denoted by the 10.7 KB fragment. The correctly targeted LSL allele is indicated by the 7.4 KB fragment using the 5′ internal probe and by the 5.6 Kb fragment using the 3′ external probe. (C) Direct sequencing with a reverse primer of genomic DNA isolated from WT and germ line transmitted LSL Nras G12Dhypo mice demonstrates the presence of G12D mutation at the Nras locus. Arrows indicate the WT and mutated nucleotides at the codon 12. (D) Evaluation of recombination efficiency of Mox2-Cre and expression level of Nras G12D hypo allele in E14.5 fetal liver erythoid progenitors. Because Mox2-Cre recombines the conditional LSL cassette and leads to Nras G12Dhypo expression, we refer the compound mice harboring both LSL and Mox2-Cre alleles as Nras G12Dhypo/+; Mox2-Cre/+. We further crossed Nras G12Dhypo/+; Mox2-Cre/+ mice to LSL Nras G12Dhypo/+ (LSL/+) mice and generate Nras G12Dhypo/LSL progenies inherit a recombined Nras G12Dhypo allele and a nonrecombined LSL allele from parents but does not carry Mox2-Cre allele. To simplify the genotyping results, we omit the Mox2-Cre status. Southern blot analysis of SpeI digested genomic DNA using the 3′ external probe confirmed the recombination efficiency at the endogenous Nras locus (top panel). Expression levels of total Nras were measured by Western blotting and normalized against actin using the Molecular Analyst Version 1.4 software (Bio-Rad).
We examined the expression level of oncogenic Nras at the protein level. We crossed LSL Nras G12Dhypo/+ mice to Mox2-Cre/+ mice to generate compound mice carrying both alleles. Mox2-Cre efficiently activates oncogenic Nras expression in epiblasts beginning at E5, as judged by Southern blot analysis (Figure 1D top panel).31 Analysis of E14.5 fetal liver erythroid progenitors showed that Nras G12Dhypo/+ expresses oncogenic Nras at a level equivalent to 25%-40% of single copy of the Nras WT allele (Figure 1D bottom panel). Thus, Nras G12Dhypo/G12Dhypo expresses 50%-80% of single copy of the Nras WT allele. To our surprise, despite efficient recombination at the Nras locus, both LSL Nras G12Dhypo/+; Mox2-Cre/+ mice and LSL Nras G12Dhypo/G12Dhypo; Mox2-Cre/+ mice were viable and generated with the expected percentages (supplemental Table 1, available on Blood Web site; see the Supplemental Materials link at the top of the online article). None of these mice display any morphologic abnormalities. We killed 5 LSL Nras G12Dhypo/G12Dhypo;Mox2-Cre/+ mice at 12 months and 10 at 18-24 months, and examined the thymus, spleen, peripheral blood, liver, lung, kidney, and intestine. We did not observe any hyperplasia phenotypes (data not shown). Our results indicate that expression of Nras G12Dhypo/G12Dhypo is insufficient to cause developmental abnormalities and cancers.
We obtained another conditional oncogenic Nras allele (LSL Nras G12D) independently generated by Haigis et al.18 This allele expresses oncogenic Nras at a comparable level to the WT Nras allele.19 We crossed LSL Nras G12D/+ mice to Mox2-Cre. None of LSL Nras G12D/+; Mox2-Cre/+ mice were born alive (supplemental Table 2), suggesting that early embryonic expression of Nras G12D/+ leads to embryonic lethality. Our data are consistent with a recent report from Dr Martin Zenker's group.32 In this report, they failed to identify a germ line oncogenic NRAS mutation in 917 Noonan Syndrome patients who are negative for previously known mutations in the Ras pathway, suggesting that germ line oncogenic NRAS leads to embryonic/fetal lethality.
Somatic activation of Nras G12D/G12D but not Nras G12D/+ leads to an acute myeloproliferative disease
We further studied oncogenic Nras-initiated hematopoietic malignancies using the normally expressed LSL Nras G12D allele.18 We crossed LSL Nras G12D/+ mice to Mx1-Cre transgenic mice to generate compound mice (LSL Nras G12D/+; Mx1-Cre) and further crossed LSL Nras G12D/+; Mx1-Cre mice to LSL Nras G12D/+ mice to generate compound mice (LSL Nras G12D/G12D; Mx1-Cre). Administration of pI-pC in compound mice stimulates endogenous IFN production and thus induces Cre expression from the IFN-α/β–inducible Mx1 promoter, which in turn leads to the expression of oncogenic Nras from its endogenous locus.33 We refer to pI-pC–treated compound mice as Nras G12D/+ and Nras G12D/G12D mice, respectively, and pI-pC–treated Mx1-Cre mice as WT control mice throughout this manuscript.
pI-pC treatment of LSL Nras G12D/+; Mx1-Cre and LSL Nras G12D/G12D; Mx1-Cre mice efficiently induced oncogenic Nras expression in bone marrow (Figure 2A-B). Two days after the second injection of pI-pC, genotypic analysis of genomic DNA showed that the recombined allele was readily detectable in bone marrow and gain of the recombined allele was associated with the loss of the LSL allele (Figure 2A). Sequencing of RT-PCR amplification products confirmed that oncogenic Nras was efficiently transcribed at the mRNA level (Figure 2B).
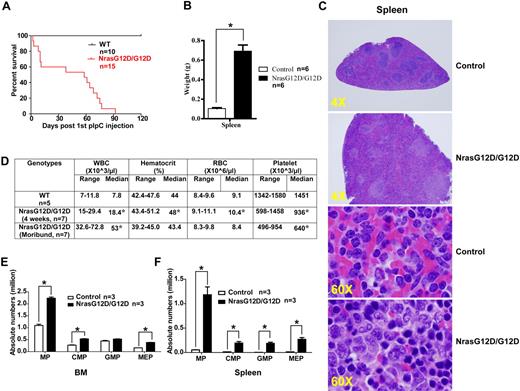
Somatic activation of Nras G12D/G12D but not Nras G12D/+ leads to acute myeloproliferative disease. Five- to 6-week-old mice were injected with pI-pC as described in “Mice.” Two days after the second pI-pC injection, different tissues were isolated and analyzed. Nras G12D/+ and Nras G12D/G12D refer to pI-pC treated compound mice expressing monoallelic and biallelic oncogenic Nras, respectively, as described in “Somatic activation of Nras G12D/G12D but not Nras G12D/+ leads to an acute myeloproliferative disease.” (A) Genotyping analysis of genomic DNA to detect WT allele, LSL allele, and recombined LSL allele (1 LoxP allele). (B) Total RNA was extracted from bone marrow cells. Direct sequencing of RT-PCR amplified Nras gene using a reverse primer to confirm the sequences at the codon 12. Arrows indicate the WT and mutated nucleotides at the codon 12. (C) Flow cytometric analysis of peripheral blood (PB), spleen and bone marrow (BM) cells isolated from control (n = 5) and Nras G12D/G12D (n = 5) mice using myeloid lineage specific markers. Debris and unlysed red blood cells (low forward scatter) and dead cells (propidium iodide positive) were excluded from analysis. Data are presented as averages + SDs. (D) Splenomegaly in Nras G12/G12D mice. Results are presented as the average of spleen weights + SD. *P < .01. (E, F) 5 × 104 bone marrow cells isolated from control, Nras G12D/+, and Nras G12D/G12D mice were plated in duplicate in semisolid medium with or without GM-CSF (E) or IL-3 (F). The data are presented as average percentages (from multiple mice of each group) of maximum number of colonies formed in culture with 0.2 ng/mL of GM-CSF or 10 ng/mL of IL-3. Student t test was performed. Error bars show SD. (E) Crosses indicate P < .01. (F) *P < .05.
Somatic activation of Nras G12D/G12D but not Nras G12D/+ leads to acute myeloproliferative disease. Five- to 6-week-old mice were injected with pI-pC as described in “Mice.” Two days after the second pI-pC injection, different tissues were isolated and analyzed. Nras G12D/+ and Nras G12D/G12D refer to pI-pC treated compound mice expressing monoallelic and biallelic oncogenic Nras, respectively, as described in “Somatic activation of Nras G12D/G12D but not Nras G12D/+ leads to an acute myeloproliferative disease.” (A) Genotyping analysis of genomic DNA to detect WT allele, LSL allele, and recombined LSL allele (1 LoxP allele). (B) Total RNA was extracted from bone marrow cells. Direct sequencing of RT-PCR amplified Nras gene using a reverse primer to confirm the sequences at the codon 12. Arrows indicate the WT and mutated nucleotides at the codon 12. (C) Flow cytometric analysis of peripheral blood (PB), spleen and bone marrow (BM) cells isolated from control (n = 5) and Nras G12D/G12D (n = 5) mice using myeloid lineage specific markers. Debris and unlysed red blood cells (low forward scatter) and dead cells (propidium iodide positive) were excluded from analysis. Data are presented as averages + SDs. (D) Splenomegaly in Nras G12/G12D mice. Results are presented as the average of spleen weights + SD. *P < .01. (E, F) 5 × 104 bone marrow cells isolated from control, Nras G12D/+, and Nras G12D/G12D mice were plated in duplicate in semisolid medium with or without GM-CSF (E) or IL-3 (F). The data are presented as average percentages (from multiple mice of each group) of maximum number of colonies formed in culture with 0.2 ng/mL of GM-CSF or 10 ng/mL of IL-3. Student t test was performed. Error bars show SD. (E) Crosses indicate P < .01. (F) *P < .05.
Two days after the second injection of pI-pC, all of the Nras G12D/+ mice were grossly unremarkable; both white blood cell counts and differentials were normal.19 In contrast, myeloproliferative phenotypes were prominent in all of the Nras G12D/G12D mice (n = 20). Diseased animals showed marked splenomegaly (Figure 2D). Flow cytometric analysis using myeloid cell–specific markers demonstrated a predominantly granulocytic/monocytic myeloid hyperplasia in the peripheral blood, spleen, and bone marrow of diseased mice (Figure 2C). These phenotypes closely resembled the acute myeloproliferative disease (MPD) developed in Kras G12D mice in a similar experimental setting.24,34,35 As in Kras G12D mice, MPD develops in Nras G12D/G12D mice without pI-pC injections (n = 5).
To determine whether Nras G12D/G12D myeloid progenitors display any abnormal growth patterns, bone marrow cells isolated from control, Nras G12D/+, or Nras G12D/G12D mice were plated in semisolid cultures in the presence of various concentrations of mGM-CSF (Figure 2E) or IL-3 (Figure 2F). Bone marrow cells from Nras G12D/+ and Nras G12D/G12D mice, but not controls, formed significant numbers of CFU-GM colonies in the absence of exogenous cytokines. In the presence of GM-CSF or IL-3, Nras G12D/+ and Nras G12D/G12D colonies were significantly enlarged compared with that of controls in a dose-dependent manner. Nras G12D/+ cells did not show characteristic hypersensitivity to both cytokines, whereas Nras G12D/G12D cells displayed hypersensitivity to IL-3 but not to GM-CSF. These results are in sharp contrast to those of mice expressing oncogenic Kras in a similar experimental system34,35 ; their bone marrow cells show hypersensitivity to both IL-3 and GM-CSF.
Oncogenic Nras hyperactivates ERK and promotes cell proliferation in myeloid progenitors in a dose-dependent manner
We analyzed the myeloid progenitors (MPs, Lin− IL7Rα− Sca1− c-Kit+) in the bone marrow and spleen of control, Nras G12D/+ and Nras G12D/G12D mice. In bone marrow, the percentages of MPs in control and Nras G12D/G12D mice were comparable with each other (P = .11) but significantly lower than that of Nras G12D/+ mice (P < .005) (supplemental Figure 1). However, because of the hypercellularity in Nras G12D/G12D bone marrow, the absolute numbers of MPs in both Nras G12D/+ and Nras G12D/G12D mice were comparable with each other but ∼ 2-fold over that of control mice (Figure 3A). The increased numbers of MPs in Nras G12D/+ and Nras G12D/G12D mice were mainly because of the expansion of common myeloid progenitor (CMP) and granulocyte-monocyte progenitor (GMP) compartments. In spleen, the percentages of MPs in control and Nras G12D/+ mice were comparable with each other (P = .43) but lower than that of Nras G12D/G12D mice (supplemental Figure 1). Because of the marked splenomegaly in Nras G12D/G12D mice, the absolute numbers of MPs, CMPs, GMPs, and MEPs (megakaryocyte- erythroid progenitors) in these animals were all much higher than those of control and Nras G12D/+ mice (Figure 3A).
Oncogenic Nras signaling engages myeloid progenitors into cell cycle and leads to their expansion in a dose-dependent manner. Different tissues were isolated and analyzed 2 days after pI-pC injections. (A) Quantitative analysis of myeloid progenitor (MP) compartment in bone marrow and spleen of control, Nras G12D/+, and Nras G12D/G12D mice. CMP indicates common myeloid progenitor; GMP, granulocyte-monocyte progenitor; and MEP, megakaryocyte-erythroid progenitor. Results are presented as averages + SDs. Student t test was performed: *P < .05, and **P < .01. (B) Cell cycle analysis of MPs in bone marrow of control, Nras G12D/+, and Nras G12D/G12D mice. Cell-cycle phases are defined as G0 (Ki67−, DAPIlo), G1 (Ki67+, DAPIlo), and S/G2/M (Ki67+, DAPIhi). The percentages of MPs in individual cell-cycle phases are indicated on the density plots. Average values + SDs are shown in the right graph. Student t test was performed: *P < .05. (C,D) Phospho-flow analysis of p-ERK1/2 in Lin−/low c-Kit+ bone marrow cells of control, Nras G12D/+, and Nras G12D/G12D mice 2 days after the second pI-pC injection. (C) Total bone marrow cells were freshly isolated and stimulated with or without 2ng/mL of GM-CSF at 37°C for 10 minutes. Basal condition is defined as without GM-CSF stimulation. U0126 was mixed with cells for 30 minutes before fixation or GM-CSF stimulation. Levels of p-ERK1/2 were measured using phospho-specific flow cytometry. Nonneutrophil Lin−/low c-Kit+ cells were gated for data analysis. Results obtained from one representative experiment are shown (left panel). Quantification of 6 independent experiments is shown as average values + SDs (middle panel). Solid lines indicate the median intensity of p-ERK1/2 in control cells without GM-CSF stimulation (right panels). (D) Total bone marrow cells were serum- and cytokine-starved for 1 hour and stimulated with various concentrations of GM-CSF (0, 0.16 and 2 ng/mL) at 37°C for 10 minutes. Gating strategy and plots of p-ERK1/2 are representative of 4 independent experiments. To quantify the activation of ERK1/2, median intensities of p-ERK1/2 at different GM-CSF concentrations in different animals are compared with control cells at 0 ng/mL, which is arbitrarily set at 1. Average values ± SDs are shown in the right graph. Student t test was performed: *P < .05.
Oncogenic Nras signaling engages myeloid progenitors into cell cycle and leads to their expansion in a dose-dependent manner. Different tissues were isolated and analyzed 2 days after pI-pC injections. (A) Quantitative analysis of myeloid progenitor (MP) compartment in bone marrow and spleen of control, Nras G12D/+, and Nras G12D/G12D mice. CMP indicates common myeloid progenitor; GMP, granulocyte-monocyte progenitor; and MEP, megakaryocyte-erythroid progenitor. Results are presented as averages + SDs. Student t test was performed: *P < .05, and **P < .01. (B) Cell cycle analysis of MPs in bone marrow of control, Nras G12D/+, and Nras G12D/G12D mice. Cell-cycle phases are defined as G0 (Ki67−, DAPIlo), G1 (Ki67+, DAPIlo), and S/G2/M (Ki67+, DAPIhi). The percentages of MPs in individual cell-cycle phases are indicated on the density plots. Average values + SDs are shown in the right graph. Student t test was performed: *P < .05. (C,D) Phospho-flow analysis of p-ERK1/2 in Lin−/low c-Kit+ bone marrow cells of control, Nras G12D/+, and Nras G12D/G12D mice 2 days after the second pI-pC injection. (C) Total bone marrow cells were freshly isolated and stimulated with or without 2ng/mL of GM-CSF at 37°C for 10 minutes. Basal condition is defined as without GM-CSF stimulation. U0126 was mixed with cells for 30 minutes before fixation or GM-CSF stimulation. Levels of p-ERK1/2 were measured using phospho-specific flow cytometry. Nonneutrophil Lin−/low c-Kit+ cells were gated for data analysis. Results obtained from one representative experiment are shown (left panel). Quantification of 6 independent experiments is shown as average values + SDs (middle panel). Solid lines indicate the median intensity of p-ERK1/2 in control cells without GM-CSF stimulation (right panels). (D) Total bone marrow cells were serum- and cytokine-starved for 1 hour and stimulated with various concentrations of GM-CSF (0, 0.16 and 2 ng/mL) at 37°C for 10 minutes. Gating strategy and plots of p-ERK1/2 are representative of 4 independent experiments. To quantify the activation of ERK1/2, median intensities of p-ERK1/2 at different GM-CSF concentrations in different animals are compared with control cells at 0 ng/mL, which is arbitrarily set at 1. Average values ± SDs are shown in the right graph. Student t test was performed: *P < .05.
To determine whether the expansion of the MP compartment is associated with the hyperproliferation of these cells, we performed flow cytometry-based cell-cycle analysis of MPs in control, Nras G12D/+ and Nras G12D/G12D bone marrow (Figure 3B).27 Through intracellular staining for Ki67 and DNA content (using DAPI), we could define G0 (Ki67−, 2n DNA content), G1 (Ki67+, 2n), and S/G2/M (Ki67+, > 2n) subpopulations within the MP population. Our results demonstrated an oncogenic Nras dose-dependent decrease in the proportion of MPs in G0 with a corresponding increase in the proportions in G1 and S/G2/M.
We studied whether the hyperproliferation of MPs corresponds to the hyperactivation of ERK, the major signaling component downstream of oncogenic Nras (Figure 3C-D). First, total bone marrow cells were freshly isolated and immediately fixed and permeabilized (basal condition) to measure phospho-ERK1/2 (p-ERK1/2) levels in vivo (Figure 3C). P-ERK1/2 in Lin–/low c-Kit+ cells (enriched for MPs) were analyzed using multiparameter flow cytometry.19 As expected, the ERK pathway was hyperactivated in Nras G12D/G12D cells over control and Nras G12D/+ cells. To determine whether the higher intensity of p-ERK1/2 staining in Nras G12D/G12D cells truthfully represents elevated p-ERK1/2 level, we added U0126, a MEK inhibitor, before fixation or GM-CSF stimulation (Figure 3C). Our data show that in the presence of U0126, the p-ERK1/2 level in Nras G12D/G12D cells was shifted back to a level comparable with that in control cells, indicating an elevated ERK1/2 activation in Nras G12D/G12D cells in vivo. Second, total bone marrow cells were deprived of serum and cytokines and stimulated with various concentrations of GM-CSF (Figure 3D). The cells were fixed and permeabilized after cytokine stimulation. P-ERK1/2 was analyzed in Lin–/low c-Kit+ cells. We found that Nras G12D/G12D does not constitutively activate ERK. Rather, it significantly hyperactivates ERK at a low concentration of GM-CSF. The magnitude of ERK hyperactivation is similar to that of basal in vivo levels shown in Figure 3C.
We also examined the activation of Stat5 in Nras G12D/G12D cells (supplemental Figure 2). Our results demonstrate that activation of Stat5 in Nras G12D/G12D cells is indistinguishable from that in control or Nras G12D/+ cells. This result is similar to that obtained from Kras G12D cells.36
Nras G12D/G12D mice die with a myeloproliferative disease
After pI-pC injections, control, Nras G12D/+, and Nras G12D/G12D mice were kept for an extended period of time. Approximately 50% of Nras G12D/+ mice died with either histiocytic sarcoma (predominant) or a chronic MPD (occasional) closely resembling CMML at 12 months,19 whereas all of the Nras G12D/G12D mice died with an acute MPD (Figure 4A). At a moribund stage, Nras G12D/G12D mice displayed a ∼ 7-fold increase in their average spleen weights (Figure 4B). Extramedullary hematopoiesis was evident with various proportions of granulocytic, monocytic, erythroid, and megakaryocytic lineage cells (Figure 4C). Flow cytometric analysis using myeloid cell specific markers revealed a predominantly granulocytic/monocytic myeloid hyperplasia in the peripheral blood, spleen, and bone marrow of diseased mice (supplemental Figure 3). Complete blood counts were performed on peripheral blood samples obtained from control and Nras G12D/G12D mice (Figure 4D). The median white blood cell count in Nras G12D/G12D mice was significantly elevated (18.4 × 103/μL at 4 weeks after pI-pC injections and 53 × 103/μL at a moribund stage versus 7.8 × 103/μL in controls). The median platelet count in Nras G12D/G12D mice was significantly lower than that in controls (936 × 103/μL at 4 weeks after pI-pC injections and 640 × 103/μL at a moribund stage versus 1451 × 103/μL in controls). Similarly as Nras G12D/G12D mice at 2 days after pI-pC treatment, moribund mice showed a significantly expanded myeloid compartment in both bone marrow and spleen (Figure 4E).
All the Nras G12D/G12D mice die with a severe myeloproliferative disease. After pI-pC injections, control and Nras G12D/G12D mice were kept for an extended period of time until Nras G12D/G12D mice reached a moribund stage. (A) Kaplan-Meier comparative survival analysis of control and Nras G12D/G12D mice. Cumulative survival was plotted against days after the first pI-pC injection. (B) Splenomegaly in Nras G12D/G12D mice. Results are presented as averages of spleen weights + SDs. (C) Representative histologic H&E sections from spleen show an extensive infiltration of myelomonocytic cells and extramedullary hematopoiesis in Nras G12D/G12D mice. (D) Complete blood count was performed on peripheral blood samples drawn from control and Nras G12D/G12D mice. *P < .05. (E-F) Quantitative analysis of myeloid progenitor (MP) compartment in bone marrow (E) or spleen (F) of control and moribund Nras G12D/G12D mice as described in Figure 3. Results are presented as averages + SDs. *P < .05.
All the Nras G12D/G12D mice die with a severe myeloproliferative disease. After pI-pC injections, control and Nras G12D/G12D mice were kept for an extended period of time until Nras G12D/G12D mice reached a moribund stage. (A) Kaplan-Meier comparative survival analysis of control and Nras G12D/G12D mice. Cumulative survival was plotted against days after the first pI-pC injection. (B) Splenomegaly in Nras G12D/G12D mice. Results are presented as averages of spleen weights + SDs. (C) Representative histologic H&E sections from spleen show an extensive infiltration of myelomonocytic cells and extramedullary hematopoiesis in Nras G12D/G12D mice. (D) Complete blood count was performed on peripheral blood samples drawn from control and Nras G12D/G12D mice. *P < .05. (E-F) Quantitative analysis of myeloid progenitor (MP) compartment in bone marrow (E) or spleen (F) of control and moribund Nras G12D/G12D mice as described in Figure 3. Results are presented as averages + SDs. *P < .05.
Recipient mice transplanted with Nras G12D/G12D bone marrow cells develop TALL with a 100% penetrance
To study the hematopoietic cell-autonomous role of oncogenic Nras signaling in leukemogenesis, we transplanted 2.5 × 105 bone marrow cells (CD45.2+) isolated from control, Nras G12D/+, or Nras G12D/G12D mice along with same number of WT competitor cells (CD45.1+) into lethally irradiated recipient mice (Figure 5A). Approximately 95% of recipient mice transplanted with Nras G12D/+ cells developed CMML 6-24 months after transplantation and ∼ 8% of recipient mice developed acute TALL (TALL-het) 6-7 months after transplantation (some of the animals developed both of the diseases; Figure 5A-B).19 In contrast, all of the recipient mice transplanted with Nras G12D/G12D cells died of TALL (TALL-homo) 5-9 months after transplantation (Figure 5A-B) and none of them developed sustained CMML-like phenotypes nor did they develop AML.
Oncogenic Nras initiates TALL in a dose-dependent manner. Lethally irradiated mice (CD45.1+) were transplanted with 2.5 × 105 total bone marrow cells of control, Nras G12D/+, or Nras G12D/G12D mice along with same number of competitor cells. (A) Kaplan-Meier survival curves of reconstituted mice. Cumulative survival was plotted against days after transplantation. (B) Disease distribution patterns in recipient mice transplanted with Nras G12D/+ or Nras G12D/G12D cells. (C) Hepatosplenomegaly and enlarged thymi in TALL-het and -homo mice. (Top) Enlarged thymus in a representative recipient mouse that developed a TALL disease. (Bottom) Results are presented as averages of spleen, liver, or thymus weights + SDs. Student t test was performed: *P < .001. (D) Representative histologic H&E sections of thymus from control, TALL-het, and TALL-homo mice. (E) Flow cytometric analysis of total thymocytes of representative control, TALL-het, and TALL-homo mice.
Oncogenic Nras initiates TALL in a dose-dependent manner. Lethally irradiated mice (CD45.1+) were transplanted with 2.5 × 105 total bone marrow cells of control, Nras G12D/+, or Nras G12D/G12D mice along with same number of competitor cells. (A) Kaplan-Meier survival curves of reconstituted mice. Cumulative survival was plotted against days after transplantation. (B) Disease distribution patterns in recipient mice transplanted with Nras G12D/+ or Nras G12D/G12D cells. (C) Hepatosplenomegaly and enlarged thymi in TALL-het and -homo mice. (Top) Enlarged thymus in a representative recipient mouse that developed a TALL disease. (Bottom) Results are presented as averages of spleen, liver, or thymus weights + SDs. Student t test was performed: *P < .001. (D) Representative histologic H&E sections of thymus from control, TALL-het, and TALL-homo mice. (E) Flow cytometric analysis of total thymocytes of representative control, TALL-het, and TALL-homo mice.
We characterized both TALL-het and TALL-homo mice (Figure 5C-E). Both groups of animals showed variable hepatosplenomegly with T-cell infiltration at the moribund stage, whereas their thymi were consistently enlarged 8- to 10-fold over controls (Figure 5C). Compared with control thymus, which showed a distinct pattern of cortical and medullar architecture and was filled with maturing T cells with dense nuclear staining, the TALL-het and -homo thymus demonstrated a completely effaced thymic architecture filled with actively proliferating T-cell blasts and scattered tangible-body macrophages (Figure 5D). We further analyzed both TALL-het and -homo thymocytes with various T-cell markers (Figure 5E). Both groups of tumor samples were double positive for CD4 and CD8 and positive for CD44, similar to the T-cell disease initiated by endogenous oncogenic Kras mutation.24,37
To examine the development of T-cell malignancies, we analyzed recipient mice transplanted with Nras G12D/G12D cells 4 months after transplantation (supplemental Figure 4). At this pre-TALL stage, the thymus size was normal compared with controls. However, there was significant expansion of CD4−CD8− cells. These double-negative cells started to show up-regulation of CD44.
Oncogenic Nras-initiated T-cell diseases are transplantable to secondary recipients
To determine whether oncogenic Nras-initiated T-cell malignancies are transplantable, we isolated bone marrow cells from individual diseased mice that had been transplanted with Nras G12D/+ or Nras G12D/G12D cells (supplemental Figure 5). Bone marrow cells (1 × 104) were transplanted into sublethally irradiated secondary recipient mice. All secondary recipient mice developed TALL and died within 2 months after transplantation (supplemental Figure 5A). Further fractionation of tumor cells demonstrated that the activity of tumor initiating cells is restricted in CD8+ T cells (data not shown), and particularly enriched in CD8+ Sca1− cKit− cells (supplemental Figure 5B).
Both TALL-het and TALL-homo tumors are predominantly monoclonal and acquire clonal secondary Notch1 mutations
To determine whether the oncogenic Nras-initiated TALL is clonal in origin, we performed genomic Southern analysis of the TCRβ locus in multiple TALL-het and TALL-homo tumor samples (Figure 6A).24 The probe hybridizes to the Vβ region of the T-cell receptor β locus.28 Our results indicate that the tumors are predominantly monoclonal.
Oncogenic Nras-initiated TALL tumors contain clonal Notch1 mutations. (A) Southern blot analysis of genomic DNA obtained from TALL-het and TALL-homo tumors. The blot was hybridized with a probe to the Vβ region of T-cell receptor β. (B) Sequence analysis of the exon 34 of Notch1 in control, TALL-het, and TALL-homo thymocytes.
Oncogenic Nras-initiated TALL tumors contain clonal Notch1 mutations. (A) Southern blot analysis of genomic DNA obtained from TALL-het and TALL-homo tumors. The blot was hybridized with a probe to the Vβ region of T-cell receptor β. (B) Sequence analysis of the exon 34 of Notch1 in control, TALL-het, and TALL-homo thymocytes.
We next sequenced the Notch1 gene for mutations in the heterodimerization (exons 26 and 27) or PEST (exon 34) domains that have been previously observed in both human and murine models of TALL. No mutations were detected at the heterodimerization cleavage site, but 2 of 4 TALL-het tumors and 3 of 4 TALL-homo tumors harbored clonal PEST domain mutations (Figure 6B). These mutations are all insertional mutations, which lead to frame shifts and premature stops in the PEST domain. Compared with TALL-het and TALL-homo, oncogenic Kras-initiated TALL has a significantly shorter latency; the diseased animals die 1.5-4 months after transplantation.24,37,38 Similarly as shown before,37 these tumors are predominantly oligoclonal (supplemental Figure 6A) and 5 of 10 tumors contain Notch1 mutations in the PEST domain (supplemental Figure 6B). Consistent with a recent report,39 4 of 5 Notch1 mutation-positive tumors contain heterogeneous Notch1 mutations (supplemental Figure 6C).
Oncogenic Nras-initiated TALL cells are sensitive to γ-secretase inhibitor and Ras downstream effector inhibitors
To determine whether the growth of TALL-het and -homo cells depends on abnormal Notch1 signaling, we treated various TALL cells with GSI XXI, a small molecule inhibitor of γ-secretase. The tested cells included TALL-het primary tumor cells, and cell lines derived from TALL-het and TALL-homo tumors, respectively. All of them carried a Notch1 mutation in the PEST domain. All of the tested cells were sensitive to inhibition of Notch signaling at concentrations as low as 0.05μM (supplemental Figure 7A). The decrease in cell growth resulted from increased apoptotic cell death as demonstrated by a dose-dependent increase in annexin V-positive cells (data not shown).
To investigate the effects of inhibition of Ras downstream pathways, we also treated the TALL-homo cell line with the MEK inhibitor, U0126 (supplemental Figure 7B). Treatment for 48 hours caused a dose-dependent inhibition of cell growth. Moreover, because both Notch1 and Ras signaling pathways affect PI3K signaling,40,41 we treated TALL-homo cells with rapamycin (supplemental Figure 7C) and LY294002 (supplemental Figure 7D). Again, both inhibition treatments resulted in a dose-dependent suppression of cell growth. The specificity of XXI to inhibit Notch signaling and U0126 to block ERK1/2 activation is shown in supplemental Figure 7E.
Endogenous Nras G12D/+ signaling promotes TALL in the presence of an intact WT Nras allele
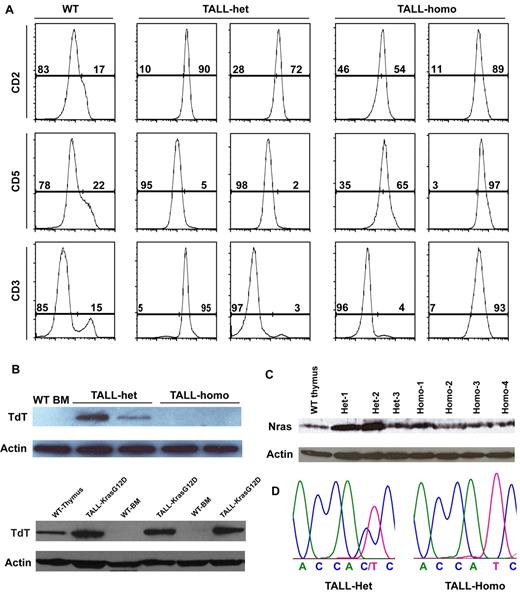
We characterized both TALL-het and -homo tumors with T-cell markers expressed at different developmental stages (Figure 7A). TALL-het tumors are surface CD2+ CD5− CD3+/−, whereas TALL-homo tumors are CD2+ CD5+ CD3+/−, suggesting that TALL-het and -homo cells may be transformed at different developmental stages. We further analyzed the expression of terminal deoxynucleotidyl transferase (TdT) in both types of tumor cells (Figure 7B). TdT is usually expressed in primitive T-lymphocytes in the normal thymus as well as in most of cases of TALL in both human and mouse.42,43 We found that despite being morphologically similar and consistent with human TALL, both oncogenic Kras-initiated TALL and oncogenic Nras-initiated TALL-het are TdT-positive, whereas oncogenic Nras-initiated TALL-homo tumors are TdT-negative. This suggests a potential role for a dose-dependent survival/differentiation signal of oncogenic Nras in neoplastic T-cell precursors.
TALL-het tumors are generated through a distinct genetic mechanism from TALL-homo tumors. (A) Flow cytomytric analysis of TALL-het and -homo tumor cells with T-cell markers expressed at different developmental stages. Two representative tumors of 5 for each tumor type are shown. (B) Western blot analysis of TdT expression levels in control cells, oncogenic Nras mutation-initiated TALL-het and -homo tumors (top panels), and oncogenic Kras mutation-initiated TALL-KrasG12D tumors (bottom panels). (C) Western blot analysis of total Nras expression levels in control, TALL-het, and -homo thymocytes. (D) Direct sequencing of RT-PCR amplified products in TALL-het and -homo tumors at the Nras G12 codon. Results are representative of 5 animals for each tumor type.
TALL-het tumors are generated through a distinct genetic mechanism from TALL-homo tumors. (A) Flow cytomytric analysis of TALL-het and -homo tumor cells with T-cell markers expressed at different developmental stages. Two representative tumors of 5 for each tumor type are shown. (B) Western blot analysis of TdT expression levels in control cells, oncogenic Nras mutation-initiated TALL-het and -homo tumors (top panels), and oncogenic Kras mutation-initiated TALL-KrasG12D tumors (bottom panels). (C) Western blot analysis of total Nras expression levels in control, TALL-het, and -homo thymocytes. (D) Direct sequencing of RT-PCR amplified products in TALL-het and -homo tumors at the Nras G12 codon. Results are representative of 5 animals for each tumor type.
Because up-regulation of oncogenic Nras protein through genetic (eg UPD of the oncogenic Nras allele) and/or epigenetic mechanisms is often observed during tumor development in both human and mouse,14,19,44 we tested whether this mechanism is also involved in the formation of TALL-het and/or TALL-homo tumors. Western blot analysis showed that TALL-homo tumors express comparable levels of Nras as control thymocytes, whereas 2 of 3 TALL-het tumors express significantly higher levels of Nras than controls (Figure 7C). Further sequencing analysis of mRNA transcripts and genomic DNA demonstrated that both WT and oncogenic Nras alleles are intact and transcribed in TALL-het tumor cells (Figure 7D, n = 5). Therefore, the up-regulation of oncogenic Nras protein in TALL-het tumors is associated with concomitant up-regulation of WT Nras protein.
Discussion
In this study, we created a gradient of oncogenic Nras signaling ranging from 25%-200% of endogenous monoallelic expression of Nras G12D (supplemental Figure 8). First, we show that expressing up to 80% of Nras G12D/+ neither affects normal mouse embryonic development nor results in tumors. In contrast, early embryonic expression of Nras G12D/+ leads to embryonic lethality. Second, at its normal endogenous level, the tumor transforming capability of oncogenic Nras is not only dose-dependent but also cell type–dependent. Third, although both Nras G12D/+ and Nras G12D/G12D initiate TALL, tumor cells appear to be transformed at different developmental stages and through distinct genetic mechanisms of tumor progression.
Somatic expression of Nras G12D/+ versus Nras G12D/G12D
Although the Ras genes have long been established as proto-oncogenes, substantial evidence indicates that WT Ras genes act as tumor suppressor genes in different tumor models.45,46 Compared with Nras G12D/+, Nras G12D/G12D not only doubles the dose of oncogenic Nras but also loses the potential restraint from the WT Nras gene. The latter might also contribute to the acute MPD phenotypes in Nras G12D/G12D mice.
Somatic versus bone marrow-specific expression of Nras G12D/G12D
In our new model, we found that widespread expression of Nras G12D/G12D in mice leads to acute MPD with a complete penetrance (Figures 2,Figure 3–4), whereas bone marrow-specific expression of Nras G12D/G12D in recipient mice results in 100% penetrant TALL (Figure 5). A similar phenotypic switch is also seen in the case of endogenous oncogenic Kras.24,34,35,37,38 Several possibilities might account for this observation. First, the MPD phenotypes in primary Nras G12D/G12D mice are likely complicated by systemic interferon-mediated responses, induced Nras G12D/G12D expression in nonhematopoietic cells, and the simultaneous expression of Nras G12D/G12D in 80%-90% of myeloid cells. Thus, the initiation of MPD phenotypes with 100% penetrance in the primary mice might be a transient phenomenon attributable to microenvironmental factors and does not necessarily imply the long-term maintenance in a hematopoietic-cell autonomous manner. Thus, we believe that it is critical to adopt the bone marrow transplantation system to study the cell-autonomous role of genes in leukemogenesis. Second, it is possible that either the MPD state or the pI-pC treatment hinders the engraftment of MPD-initiating cells and thus contributes to our results. Third, it is also likely that MPD phenotypes are maintained by genetically altered hematopoietic stem cells (HSCs). Nras G12D/G12D signaling might substantially alter HSC behaviors so that these HSCs no longer sustain MPD phenotypes in recipient mice until they reach a lethal stage.
It is nevertheless surprising that thus far none of the recipient mice with Nras G12D/G12D cells has succumbed to a myeloid disease. At the dose of 2.5 × 105 cells and under the same experimental procedure, 10 of 72 recipient mice transplanted with Kras G12D cells died with JMML-like phenotypes (B.M. and J.D., unpublished data, April 2011), whereas 0 of 64 recipient mice of Nras G12D/G12D cells developed a sustained MPD (P = .002). It is likely that Nras G12D/G12D signaling is insufficient to initiate a strong myeloid disease in a timely manner after transplantation.
Tumor transforming activity of oncogenic Nras is both dose-dependent and cell type–dependent
Overexpression of oncogenic Nras (at least 6-12 fold over endogenous Nras G12D/+) in mouse bone marrow cells efficiently induces CMML/AML within 3-6 months after transplantation.17 Moreover, increasing overexpression levels of oncogenic Nras leads to stronger phenotypes and significantly shorter disease latency, suggesting that stronger oncogenic Nras signaling results in greater tumor transforming potential. Contrary to results from oncogenic Nras overexpression, our bone marrow transplant models show that the tumor transforming potential of oncogenic Nras signaling is not only dose-dependent but also cell type-dependent. In myeloid cells, compared with endogenous Nras G12D/+, Nras G12D/G12D does not accelerate CMML formation nor lead to CMML transformation to AML. The failure to accelerate CMML formation by Nras G12D/G12D might be attributed to the confounding emergence of fatal TALL. In contrast, in the T-cell lineage, Nras G12D/G12D signaling dramatically increases the TALL penetrance but does not significantly shorten the disease latency (Figure 5). Apparently, T cells require stronger signaling than myeloid cells during leukemia development. Our observation is consistent with the case of different BCR-ABL forms; p190 BCR-ABL shows stronger tyrosine kinase activity and is often associated with ALL, whereas p210 BCR/ABL displays weaker tyrosine kinase activity and are predominantly identified in CML patients.47 Our data emphasize the importance of constructing and studying physiologic mouse models for human cancers.
T-cell leukemia/lymphoma initiating cells
It remains controversial whether TALL is maintained by a rare population of tumor cells called tumor-initiating cells (TICs) or by the majority of tumor cells. Pten deficiency-initiated TALL is driven by TICs, whose activity is enriched in a CD3+ cKitmid subpopulation of tumor cells.48 In contrast, TALL isolated from Eμ-Nras transgenic mice is sustained by virtually all the tumor cells.49 Interestingly, our TALL models show intermediate frequencies of TICs (supplemental Figure 3). Collectively, these results suggest that the frequency of TICs in TALL is genetic alteration-dependent. Considerably more work is required to address this issue and the results might vary from case to case.
Differential mechanisms underlying the generation of TALL-het and -homo tumors
Although both Nras G12D/+ and Nras G12D/G12D initiate TALL, the tumor cells appear to be transformed through distinct mechanisms. Two of 3 TALL-het tumors acquired enhanced oncogenic Nras signaling through up-regulation of both WT and oncogenic Nras proteins (Figure 7C). However, such mechanism is not detected in TALL-homo tumors. This mechanism is also distinct from the findings in Nras G12D/+ initiated CMML, in which 2 of 5 mice up-regulated the expression of oncogenic Nras protein through UPD of the oncogenic Nras allele.19 It is not clear what leads to the differential mechanism(s) of up-regulating oncogenic Nras signaling in myeloid cells versus T cells. Nonetheless, we anticipate that oncogenic NRAS mutations are rare in TALL because of the low incidence of TALL-het and rare biallelic NRAS mutations. Indeed, we and others found that oncogenic NRAS mutations occur in < 5% of TALL patients (Bos,4 Rodenhuis,11 Neri,12 Lubbert13 ; Z.L., unpublished results, April 2011).
Nras G12D/+ initiated versus Kras G12D/+ initiated TALL
Morphologically and phenotypically, Nras G12D/+ initiated T-cell diseases (TALL-het) closely resemble those initiated by Kras G12D/+.24,37 Both of them are CD4, CD8 double positive but prone to be CD8 single positive during lymphoma development, CD44 positive, TdT positive, and transplantable to secondary recipient mice. Notch1 mutations in the PEST domain are identified in ∼ 50% of each tumor type and tumor cells containing Notch1 mutations are sensitive to γ-secretase inhibitor XXI. On the other hand, TALL-het occurs in only ∼ 8% of recipient mice 6-7 months after transplantation, whereas oncogenic Kras initiates T-cell diseases 1.5-3.5 months after transplantation with an almost complete penetrance. Although both types of tumors contain Notch1 mutations, such mutations in TALL-het tumors are clonal but heterogeneous in oncogenic Kras-initiated T-cell diseases. The clonality of Notch1 mutations correlates to the clonality of the TALL tumors. The differences observed in these 2 TALL models regarding tumor latency, penetrance, and clonality likely result from the dramatic signaling strengths elicited by endogenous Nras G12D/+ signaling and Kras G12D/+ signaling.19,36
Taken together, our study shows a dose- and cell type–dependent role of endogenous oncogenic Nras signaling in hematopoietic malignancies (supplemental Figure 8).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Harvey F. Lodish and Dr Rudolf Jaenisch for their generous help and support of this project at the early stages. We thank Dr Zefeng Wang for computational modeling and Dr Laurie Jackson-Grusby for providing us with the STOP cassette construct. We are grateful to Drs Kevin Haigis and Tyler Jacks for providing us the conditional oncogenic Nras mice. We thank Drs Norman Drinkwater, Lily Huang, and Shannon Kenney for helpful discussion and critical comments on the manuscript. We are grateful to Dr Norman Drinkwater for his help in statistical analysis.
This work was supported by a Howard Temin Award from the National Cancer Institute, a Shaw Scientist Award from the Greater Milwaukee Foundation, a research grant from the Elsa Pardee Foundation and from the Wendy Will Case Cancer Fund, an ASH Scholar Award from the American Society of Hematology, and a pilot project grant from the American Cancer Society Institutional Research Grant to J.Z. This project was also supported in part by the UW Institute for Clinical and Translational Research, funded through a National Institutes of Health/NCRR Clinical and Translational Science Award, 1UL1RR025011.
National Institutes of Health
Authorship
Contribution: J.W. was responsible for experimental design, execution, and writing the manuscript; Z.W. and Q.C. were responsible for ES cell culture and tetraploid injection to generate LSL Nras G12Dhypo allele; Z.L., Y.L., L.X.T., J.D., M.-J.R., and B.M. were responsible for experimental execution; K.H.Y. and E.A.R. were responsible for histopathologic analysis and editing of the manuscript; and J.Z. was responsible for experimental design and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K.H.Y. is Department of Hematopathology, University of Texas M. D. Anderson Cancer Center, Houston, TX.
Correspondence: Jing Zhang, McArdle Laboratory for Cancer Research, 1400 University Ave, Rm 417A, University of Wisconsin-Madison, Madison, WI; e-mail: zhang@oncology.wisc.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal