Abstract
Defining how cancer-associated mutations perturb signaling networks in stem/progenitor populations that are integral to tumor formation and maintenance is a fundamental problem with biologic and clinical implications. Point mutations in RAS genes contribute to many cancers, including myeloid malignancies. We investigated the effects of an oncogenic KrasG12D allele on phosphorylated signaling molecules in primary c-kit+ lin−/low hematopoietic stem/progenitor cells. Comparison of wild-type and KrasG12D c-kit+ lin−/low cells shows that K-RasG12D expression causes hyperproliferation in vivo and results in abnormal levels of phosphorylated STAT5, ERK, and S6 under basal and stimulated conditions. Whereas KrasG12D cells demonstrate hyperactive signaling after exposure to granulocyte-macrophage colony-stimulating factor, we unexpectedly observe a paradoxical attenuation of ERK and S6 phosphorylation in response to stem cell factor. These studies provide direct biochemical evidence that cancer stem/progenitor cells remodel signaling networks in response to oncogenic stress and demonstrate that multi-parameter flow cytometry can be used to monitor the effects of targeted therapeutics in vivo. This strategy has broad implications for defining the architecture of signaling networks in primary cancer cells and for implementing stem cell–targeted interventions.
Introduction
Ras proteins are signal-switch molecules that modulate cell fates by cycling between inactive GDP-bound and active GTP-bound conformations (Ras-GDP and Ras-GTP). When GTP-bound, Ras interacts productively with downstream effectors including the Ral-GDS, Raf/MEK/ERK, and phosphoinositol 3′ (PI3) kinase/Akt pathways.1 Guanine nucleotide exchange factors promote Ras activation, and signaling is terminated when Ras-GTP is hydrolyzed to Ras-GDP by its intrinsic GTPase activity, which is markedly accelerated by GTPase-activating proteins (GAPs).2–4
Somatic KRAS2, NRAS, and HRAS mutations are found in approximately 30% of human cancers and encode mutant proteins that accumulate in the GTP-bound conformation due to reduced intrinsic GTPase activity and resistance to GAPs.5,6 Hematopoietic malignancies are tractable experimental systems for interrogating signaling networks in primary cells and for integrating biochemical, genetic, and cell biologic data. KRAS2 and NRAS are mutated frequently in myeloid malignancies, including acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), and juvenile myelomonocytic leukemia (JMML), which are classified as myeloproliferative disorders (MPDs).7–9 An important role of hyperactive Ras in myeloid leukemias is further illustrated by the fact that many patients without RAS mutation harbor other genetic lesions that deregulate Ras signaling such as loss of the NF1 tumor suppressor, point mutations that activate the SHP-2 protein tyrosine phosphatase, and the BCR-ABL fusion protein.10,11 However, despite the extensive evidence implicating hyperactive Ras in myeloid leukemogenesis, the biochemical consequences of specific mutations have heretofore not been investigated at the single-cell level. Studies of this type are particularly important to advance our understanding of MPDs and other cancers that demonstrate extensive cellular heterogeneity.
We generated Mx1-Cre, LSL-KrasG12D mice to investigate the consequences of expressing an oncogenic K-RasG12D protein in hematopoietic cells.12 In this strain, excising a “loxP-stop-loxP” (LSL) repressor cassette by Cre recombinase induces K-RasG12D production from the endogenous locus and closely models somatic KRAS mutation. Mx1-Cre, LSL-KrasG12D mice rapidly develop a fatal monocytic MPD that accurately models CMML and JMML and is characterized by myeloproliferation, splenomegaly, and profound in vitro hypersensitivity of myeloid progenitors to cytokines. These cellular phenotypes presumably result from hyperactivation of Ras-dependent signaling networks caused by elevated Ras-GTP levels. Surprisingly, however, although Ras accumulates in the GTP-bound conformation in the bone marrows of Kras mutant mice with MPDs, we did not detect elevated levels of phosphorylated ERK, MEK, or Akt (pERK, pMEK, and pAkt).12 These data, which are consistent with studies in related models of lung and pancreatic cancer, imply that the biochemical consequences of expressing K-RasG12D at endogenous levels in primary cells differ markedly from what is observed when the oncoprotein is overexpressed in immortalized cell lines.13 They also suggest that the activation of downstream effectors by oncogenic Ras may induce negative regulatory responses in primary cells. However, an inherent limitation of conventional Western blotting is that whole marrow is pooled for analysis, which might obscure abnormalities within rare cell populations that are integral to the disease phenotype. Indeed, proliferating myeloid progenitor cells represent less than 1% of normal and KrasG12D bone marrow.
Multi-parameter flow cytometric (FACS) analysis is a powerful new approach for assaying intracellular levels of phosphorylated (phospho) proteins.14,15 Although flow cytometry has been used to perform immunophenotyping for decades, harnessing this technology to measure phospho-protein levels within cells proved challenging as denaturation, dehydration, and nonspecific antibody binding are problematic following methanol permeabilization. Nolan and colleagues (Krutzik et al,16,17 Krutzik and Nolan18 ) performed extensive optimization to measure intracellular phospho-protein levels in human cells, cell lines, and murine splenocytes. These investigators recently applied multi-parameter FACS profiling to human AML samples19 and to automated signaling pathway determinations.20 In both studies, it was apparent that deregulated signaling was not readily observable at the level of basal phosphorylation but that evoked signaling revealed “hidden” pathway connectivity in cancer cells.14 Mx1-Cre, LSL-KrasG12D mice provide a tractable experimental system for extending this technology to investigate mechanisms of aberrant growth in response to hyperactive Ras signaling in primary cells.
Here we show that measuring protein phosphorylation within a defined population that is enriched for hematopoietic stem and progenitor cells reveals distinct biochemical effects of K-RasG12D expression that are difficult to detect in whole bone marrow. K-RasG12D induces elevated levels of pSTAT5, pERK, and pS6 in unstimulated stem/progenitor cells and sensitizes these cells to GM-CSF stimulation. We also find that primary stem/progenitor cells can selectively down-regulate cytokine-activated signaling networks in response to oncogenic stress. These studies also demonstrate the utility of flow cytometry for assessing the pharmacodynamic effects of a molecularly targeted agent in stem/progenitor populations, which are thought to be essential for tumor maintenance in many cancers.21,22
Materials and methods
Mice
The LSL-KrasG12D strain was generated by Jackson et al.23 C57BL/6 × 129SvJae F1 Mx1-Cre, LSL-KrasG12D mice12 received a single intraperitoneal injection of 250 μg polyinosinic-polycytidilic acid (pIpC; Sigma Aldrich USA, St Louis, MO) at 21 days of age and were analyzed at 60 to 80 days old with signs of advanced MPD, including splenomegaly, leukocytosis, and anemia. All animals were maintained in the rodent barrier facility at the University of California, San Francisco (UCSF) and the experimental procedures were approved by the UCSF Committee on Animal Research.
Bone marrow cell harvest and stimulation
Bone marrow cells were harvested from femora and tibiae into IMDM (UCSF Cell Culture Facility) containing 1% bovine serum albumin (BSA; Sigma Aldrich USA). Erythrocytes were lysed and mononuclear cells were incubated in the harvesting medium at 37°C for 2 to 2.5 hours. The cells were stimulated with murine GM-CSF 10 ng/mL or murine stem cell factor (SCF) 100 ng/mL (Peprotech, Rocky Hill, NJ). For the in vitro PI3 kinase inhibitor experiment, cells were split and those subject to inhibition were incubated for 30 minutes with LY294002 (Cell Signaling Technology, Danvers, MA) 10 μM or 50 μM prior to stimulation with GM-CSF 10 ng/mL or SCF 100 ng/mL for 10 minutes. For the MEK inhibitor experiment, CI-1040 (Pfizer, Groton, CT) or vehicle control was injected 200 mg/kg intraperitoneally 2 hours prior to killing the mice. Bone marrow cells were then harvested and stimulated with GM-CSF (10 ng/mL) for 10 minutes immediately after red blood cell lysis.
Western blotting
Following stimulation, cells were pelleted and lysed in solution containing 1% NP-40; 50 mM Tris-HCl, pH 7.4; 2 mM EDTA; 30 mM NaF; 30 mM β-glycerophosphate; 20 mM Na4P2O7; 1 mM Na3VO4; and Complete protease inhibitor cocktail (Roche, Basel, Switzerland). The protein concentration was determined using the BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL). Thirty micrograms from each lysate was separated on a 10% polyacrylamide gel (Bio-Rad, Hercules, CA) and transferred to an Immobilon-P PVDF membrane (Millipore, Billerica, MA). Membranes were blocked overnight at 4°C in TBS-T (Tris-buffered saline, 0.05% Tween-20) with 5% BSA, then incubated with primary antibodies in fresh blocking solution for 90 minutes at room temperature and washed 3 times in TBS-T having 0.15% Tween-20. Secondary detection was achieved by incubating with horseradish peroxidase–conjugated donkey anti–rabbit antibody (DAKO USA, Carpinteria, CA) for 50 minutes at room temperature followed by 3 washes and enhanced chemiluminescence (ECL) detection (GE Healthcare, Piscataway, NJ). Signal intensity was measured by densitometry using a Kodak Image Station 440 (Eastman Kodak, New Haven, CT) and analyzed by background subtraction, normalization by dividing by the associated actin signal, and scaling to a maximum of 100 units for each protein.
Flow cytometric analysis of myeloid progenitors
Common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) were analyzed and sorted as previously described.24 BrdU (5′-bromodeoxyuridine) incorporation was assessed by injecting 6- to 8-week-old mice intraperitoneally with 1 mg BrdU (Sigma Aldrich USA) 2 hours prior to harvest, followed by purification of myeloid progenitors and subsequent BrdU detection as described previously.25
Flow cytometric analysis of phospho-proteins
Following stimulation, cells were fixed with paraformaldehyde at a final concentration of 2% (16% ampules; Electron Microscopy Sciences, Hatfield, PA) at room temperature for 10 minutes. The cells were then washed twice with PBS, permeabilized with ice-cold 95% methanol (Electron Microscopy Sciences) while vortexing, placed on ice for 20 minutes, and then stored at −20°C at minimum overnight and up to 2 weeks. Upon removal from storage, the samples were washed 3 times with PBS and incubated at 4°C for 1 hour in FACS buffer (Hanks balanced salt solution containing 4% FBS; Hyclone, Logan, UT). The cells were pelleted and then incubated with anti-CD16/CD32 antibody (2.4G2; BD Biosciences, San Jose, CA) on ice for 15 minutes. Unconjugated primary antibodies were added at optimized concentrations and incubated at room temperature for 30 minutes. Samples were washed once with FACS buffer. Secondary and directly conjugated antibodies were added at optimized concentrations and incubated in the dark at room temperature for 30 minutes. Samples were washed once with PBS and analyzed on a FACScan or LSR II flow cytometer (BD Biosciences) equipped with 488-nm and 633-nm lasers. Data were collected using CellQuest or DIVA software (BD Biosciences) and analyzed using FlowJo (Tree Star, Ashland, OR).
Antibodies
Western-blotting experiments were performed using polyclonal rabbit antibodies to pMEK1/2 (Ser217/221; Cell Signaling Technology no. 9121), pSTAT5 (Tyr694; Cell Signaling Technology no. 9351), and pS6 ribosomal protein (Ser235/236; Cell Signaling Technology no. 2211). Primary antibodies used to detect phospho-proteins by flow cytometry were pERK (Thr202/Tyr204; Cell Signaling Technology no. 9101) and pS6. These were detected using donkey anti–rabbit F(ab′)2 fragment conjugated to FITC or APC (Jackson Immunoresearch, West Grove, PA). Flow-cytometry detection of phospho-STAT5 used an antibody directly conjugated to Alexa 647 (Tyr 694; BD Biosciences). Surface proteins were detected with phycoerythrin (PE)–conjugated antibodies to CD3 (17A2), CD4 (GK1.5), B220 (RA3-6B2), CD11b/Mac1 (M1/70), Gr1 (RB6-8C5), and TER-119 from BD Pharmingen and with PE-conjugated anti-CD8α (53-6.7) and PE-Cy5–conjugated anti-CD117/c-kit (2B8) from eBioscience (San Diego, CA).
Colony assays
Bone marrow cells were harvested in IMDM containing 20% FBS (Hyclone). After erythrocyte lysis and Fc receptor blocking with unlabeled anti-CD16/CD32, cells were stained for surface markers using the same directly conjugated antibodies used for phospho-specific flow cytometry. Single c-kit+ lin−/low cells were sorted into 96-well plates (1 cell per well) containing methylcellulose medium using a FACSAria (BD Biosciences) equipped with an automated cell deposition unit. GM-CSF and FLT3 ligand (Peprotech), penicillin, streptomycin, and 2-mercaptoethanol were added to M3434 methylcellulose medium, which contains IL-6, IL-3, SCF, and EPO (StemCell Technologies, Vancouver, BC, Canada). To study GM-CSF dose response, 100, 300, or 1000 c-kit+ lin−/low cells were sorted directly into 1 mL of M3231 methylcellulose medium (StemCell Technologies) supplemented with GM-CSF and 2-mercaptoethanol. For all assays, plates were incubated at 37°C in 5% CO2 and colonies were counted on day 8. Photomicrographs were taken using a TMS-F inverted microscope fitted with a 4×/0.1 objective, Ph3 annulus, and Coolpix 5000 digital camera (Nikon, Melville, NY). Cytospin cytologic preparations (10 minutes at 18 g; Thermo Fisher Scientific, Waltham, MA) were visualized through an Eclipse E400 microscope (Nikon) using a 50×/0.90 objective and photographed. Photographs were acquired and cropped using iPhoto 2.0.1 (Apple Computer, Cupertino, CA).
Results
Optimizing conditions for bone marrow Western blotting
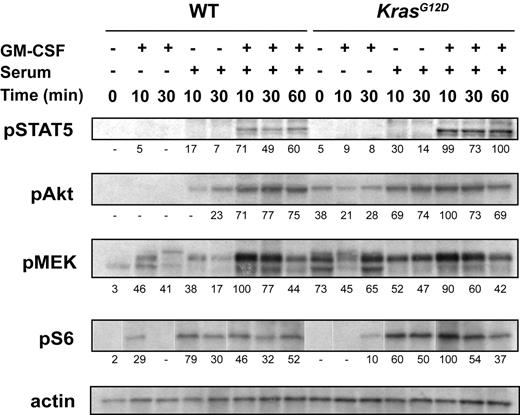
We previously assayed signal transduction in bone marrow that was harvested into IMDM containing 2% FCS, incubated for 4 hours at 37°C, and then stimulated with GM-CSF.12 We first asked if modifying these conditions might uncover subtle differences between wild-type (WT) and Mx1-Cre, KrasG12D cells. These studies showed that Mx1-Cre, KrasG12D marrows that are collected and incubated in IMDM containing 1% BSA display elevated basal levels of pMEK and pAkt as well as enhanced and prolonged phosphorylation of MEK, Akt, S6, and STAT5 in response to serum, GM-CSF, or both (Figure 1). Biochemical differences between WT and Kras mutant cells are most apparent in the presence of serum or GM-CSF alone, which supports the idea that K-RasG12D sensitizes primary cells to extracellular stimuli. Interestingly, pMEK is principally activated by GM-CSF, pAkt and pS6 levels increase in response to serum alone, and pSTAT5 is induced by both stimuli (Figure 1). However, analyses of whole bone marrow cannot discriminate among cell lineages, and the abnormally high percentage of GM-CSF–responsive myeloid cells in Mx1-Cre, KrasG12D bone marrows could underlie modest elevations in phosphorylated Ras effectors.
Phospho-protein levels in WT and Mx1-Cre, KrasG12D bone marrows. After 2 hours of serum starvation, total bone marrow cells were stimulated with GM-CSF, serum, or both for 10, 30, or 60 minutes. The cells were lysed and Western blotting was performed. Signal intensity normalized to actin is indicated below each panel, using arbitrary units. The figure shows representative data from 11 independent experiments.
Phospho-protein levels in WT and Mx1-Cre, KrasG12D bone marrows. After 2 hours of serum starvation, total bone marrow cells were stimulated with GM-CSF, serum, or both for 10, 30, or 60 minutes. The cells were lysed and Western blotting was performed. Signal intensity normalized to actin is indicated below each panel, using arbitrary units. The figure shows representative data from 11 independent experiments.
Hyperplasia and aberrant proliferation of myeloid progenitor cells in Mx1-Cre, KrasG12D mice
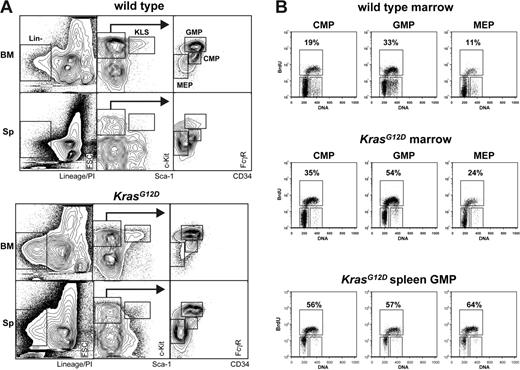
To address the inherent limitation that Western blotting “averages” the biochemical effects of K-RasG12D expression in heterogeneous tissues, we sought to use flow cytometry to interrogate signaling in pathologic stem and progenitor cells. First, we defined stem/progenitor populations that are expanded in Mx1-Cre, KrasG12D mice with MPDs, as aberrant Ras signaling in these cells is likely to contribute to the leukemic phenotype. CMPs, GMPs, and MEPs were markedly increased in the spleens of Mx1-Cre, KrasG12D mice despite normal-to-reduced numbers in bone marrows (Figure 2A), consistent with a previous report.26 Overall, there was a 1.2- to 4.5-fold expansion of the CMP, GMP, and MEP populations in mice with advanced MPDs (Table 1). We also found large numbers of splenic c-kit+ lin−/low Sca1+ (KLS) cells, which include early stem/progenitor cells. Labeling with BrdU consistently revealed elevated rates of proliferation in Kras mutant CMP, GMP, and MEP populations (Figure 2B) that were similar in marrow and spleen. Thus, K-RasG12D protein expression causes hyperproliferation of hematopoietic progenitor cells in vivo. Our data imply that K-RasG12D does not impair the ability of hematopoietic stem cells to differentiate into early or lineage-committed progenitors, and they suggest that the tumor burden in MPDs is partly due to effects of K-RasG12D on cell-cycle progression within the progenitor compartment. These studies also identify stem/progenitor cells as of particular interest for investigating the biochemical consequences of oncogenic Ras expression.
In vivo BrdU incorporation by WT and Mx1-Cre, KrasG12D bone marrow cells. (A) Gating strategy for enumerating progenitor populations in bone marrow and spleen, which are quantified in Table 1. (B) Bone marrow and spleen cells were harvested from Mx1-Cre, KrasG12D (n = 3) and littermate control (n = 3) mice 2 hours after intraperitoneal injection with BrdU. Marrow samples from animals with like genotypes were pooled. CMPs, GMPs, and MEPs were isolated from the pooled samples by FACS and stained for BrdU incorporation (vertical axis) and DNA content (PI; horizontal axis). To assess variation among animals, splenic GMPs were isolated from each of the 3 individual Mx1-Cre, KrasG12D mice and analyzed as described above (bottom panel).
In vivo BrdU incorporation by WT and Mx1-Cre, KrasG12D bone marrow cells. (A) Gating strategy for enumerating progenitor populations in bone marrow and spleen, which are quantified in Table 1. (B) Bone marrow and spleen cells were harvested from Mx1-Cre, KrasG12D (n = 3) and littermate control (n = 3) mice 2 hours after intraperitoneal injection with BrdU. Marrow samples from animals with like genotypes were pooled. CMPs, GMPs, and MEPs were isolated from the pooled samples by FACS and stained for BrdU incorporation (vertical axis) and DNA content (PI; horizontal axis). To assess variation among animals, splenic GMPs were isolated from each of the 3 individual Mx1-Cre, KrasG12D mice and analyzed as described above (bottom panel).
Myeloid progenitor populations in WT and Mx1-Cre, KrasGI2D mice
| Population and organ . | WT . | Mx1-Cre, KrasGI2D . | Ratio* . |
|---|---|---|---|
| KLS | |||
| Marrow† | 24 ± 13 | 14 ± 5.0 | |
| Spleen | 7.4 ± 5.8 | 1300 ± 36 | |
| Total‡ | 127 | 1400 | 11 |
| CMP | |||
| Marrow | 210 ± 45 | 34 ± 10 | |
| Spleen | 25 ± 9.4 | 1100 ± 250 | |
| Total | 1100 | 1300 | 1.2 |
| GMP | |||
| Marrow | 420 ± 76 | 220 ± 92 | |
| Spleen | 7.8 ± 3.9 | 5200 ± 1400 | |
| Total | 2100 | 6300 | 3.0 |
| MEP | |||
| Marrow | 210 ± 48 | 52 ± 21 | |
| Spleen | 65 ± 40 | 4900 ± 1600 | |
| Total | 1100 | 5100 | 4.6 |
| Population and organ . | WT . | Mx1-Cre, KrasGI2D . | Ratio* . |
|---|---|---|---|
| KLS | |||
| Marrow† | 24 ± 13 | 14 ± 5.0 | |
| Spleen | 7.4 ± 5.8 | 1300 ± 36 | |
| Total‡ | 127 | 1400 | 11 |
| CMP | |||
| Marrow | 210 ± 45 | 34 ± 10 | |
| Spleen | 25 ± 9.4 | 1100 ± 250 | |
| Total | 1100 | 1300 | 1.2 |
| GMP | |||
| Marrow | 420 ± 76 | 220 ± 92 | |
| Spleen | 7.8 ± 3.9 | 5200 ± 1400 | |
| Total | 2100 | 6300 | 3.0 |
| MEP | |||
| Marrow | 210 ± 48 | 52 ± 21 | |
| Spleen | 65 ± 40 | 4900 ± 1600 | |
| Total | 1100 | 5100 | 4.6 |
Stem and myeloid progenitor populations were enumerated by flow cytometry. All numbers indicate absolute numbers, in thousands, of cells recovered (n = 3; mean ± SEM).
Ratio of Mx1-Cre, KrasGI2D to WT.
Per 2 tibiae and 2 femora.
The total number of cells in each population was estimated based on the assumption that tibiae and femora comprise 20% of total marrow space.27
To assay stem and progenitor cells by phospho-FACS, we focused on cells defined by expression of c-kit and no (or low) expression of mature lineage markers (c-kit+ lin−/low). This 2-color stain identifies a population comprising 1% to 2% of nucleated marrow cells that is highly enriched for hematopoietic stem and progenitor cells24,28 while allowing for detection of phospho-proteins using other channels. Unlike whole marrow, c-kit+ lin−/low cells from WT and Mx1-Cre, KrasG12D mice have similar immunophenotypic and functional properties (Figure 3A). We found that 3% to 10% of WT and Kras mutant c-kit+ lin−/low cells formed CFU-GM colonies in methylcellulose, and the c-kit+ lin−/low population includes virtually all the CFU-GM in marrow (data not shown). Importantly, Mx1-Cre, KrasG12D myeloid progenitor colonies that are grown from single cells in a 96-well format retain the characteristic hyperproliferative phenotype and abnormal monocytic morphology of Kras mutant GFU-GM (Figure 3B). The c-kit+ lin−/low population in Mx1-Cre, KrasG12D marrow also retains the hypersensitive pattern of CFU-GM colony formation (Figure 3C) that is a hallmark of Kras-initiated MPDs. Based on these functional characteristics, the c-kit+ lin−/low population is an accessible disease-relevant compartment for analyzing the biochemical mechanism of aberrant hematopoietic growth in responses to oncogenic Ras.
Mx1-Cre, KrasG12D c-kit+ lin−/low bone marrow cells form abnormal CFU-GM colonies and are hypersensitive to GM-CSF. (A) The c-kit+ lin−/low population analyzed in these studies was isolated by first gating live, nonneutrophil cells based on scatter properties and then gating cells that expressed c-kit and had no or low expression of the mature lineage markers. (B) CFU-GM colonies grown from single c-kit+ lin−/low cells were photographed (left; original magnification × 40) and cytospin preparations were stained with Wright-Giemsa (right; original magnification × 500). (C) c-kit+ lin−/low cells (100-1000 per mL) from WT (▪) or Mx1-Cre, KrasG12D (○) mice were plated in methylcellulose medium with a range of GM-CSF concentrations.
Mx1-Cre, KrasG12D c-kit+ lin−/low bone marrow cells form abnormal CFU-GM colonies and are hypersensitive to GM-CSF. (A) The c-kit+ lin−/low population analyzed in these studies was isolated by first gating live, nonneutrophil cells based on scatter properties and then gating cells that expressed c-kit and had no or low expression of the mature lineage markers. (B) CFU-GM colonies grown from single c-kit+ lin−/low cells were photographed (left; original magnification × 40) and cytospin preparations were stained with Wright-Giemsa (right; original magnification × 500). (C) c-kit+ lin−/low cells (100-1000 per mL) from WT (▪) or Mx1-Cre, KrasG12D (○) mice were plated in methylcellulose medium with a range of GM-CSF concentrations.
Phospho-protein levels in a population enriched for hematopoietic stem/progenitor cells
Adapting the multi-parameter FACS methodology for use in mouse bone marrow required considerable optimization. Briefly, rehydrating surface proteins, blocking nonspecific antibody interactions, and performing multiple washes gave the best results. Furthermore, because neutrophils contribute significantly to nonspecific antibody binding, this population was excluded by scatter properties in all analyses. The experimental protocol that is available through the Shannon Lab website29 reliably detects the lineage-specific surface markers Mac1, Gr1, CD3, CD4, CD8, B220, and TER-119 as well as the stem and progenitor cell marker c-kit. Phospho-protein staining was validated by comparison with Western blotting in IL-3–stimulated Ba/F3 murine pro-B cells (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article) and in GM-CSF–stimulated bone marrow (Figure S2).
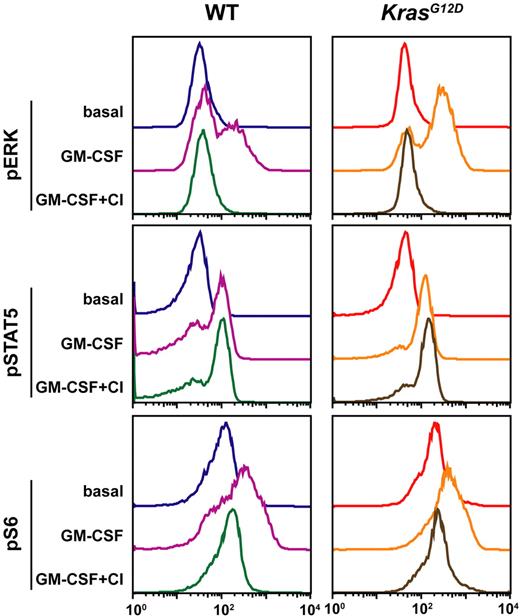
We found that c-kit+ lin−/low cells display a different phospho-protein profile than bone marrow that was gated to eliminate neutrophils (hereafter referred to as “whole” marrow; Figure 3A). FACS analysis revealed comparably low pERK levels in unstimulated WT and Mx1-Cre, KrasG12D bone marrows (Figure 4A). WT and Mx1-Cre, KrasG12D marrow cells demonstrate similar maximal pERK levels in response to GM-CSF stimulation (Figure 4A), which indicates that the modest increase in ERK phosphorylation detected in whole bone marrow by Western blotting is due to a greater proportion of cells that respond to GM-CSF rather than to a higher peak response of individual cells. By contrast, gating on the c-kit+ lin−/low population uncovered elevated basal pERK levels in Mx1-Cre, KrasG12D cells and showed that a higher proportion of cells respond to GM-CSF stimulation by phosphorylating ERK (Figure 4B). Moreover, the maximal response is greater in Kras mutant cells (10-minute time point), and pERK levels remain elevated above baseline values after 30 minutes (Figure 4B). We also found that GM-CSF stimulation increased pSTAT5 levels in WT and Mx1-Cre, KrasG12D c-kit+ lin−/low cells, which were sustained for 30 minutes. Interestingly, while the magnitude and duration of STAT5 phosphorylation was similar in cells of both genotypes, the proportion of responding c-kit+ lin−/low cells was higher in Mx1-Cre, KrasG12D mice (Figure 4B). As was true of pERK, the pattern of pS6 activation in response to GM-CSF differed markedly between WT and Mx1-Cre, KrasG12D mice. Kras mutant cells showed elevated basal pS6 levels and a large proportion of the c-kit+ lin−/low population demonstrated robust and prolonged S6 phosphorylation (Figure 4B).
c-kit+ lin−/low cells have genotype-specific signaling profiles that vary with the cytokine stimulus. Serum- and cytokine-starved whole bone marrow from WT and Mx1-Cre, KrasG12D mice was stimulated with GM-CSF (10 ng/mL) or SCF (100 ng/mL) for 10 or 30 minutes. Levels of phosphorylated ERK, STAT5, and S6 were measured using phospho-specific flow cytometry in the (A) whole marrow and (B-C) in the c-kit+ lin−/low subset. The figure shows representative data from 10 independent experiments.
c-kit+ lin−/low cells have genotype-specific signaling profiles that vary with the cytokine stimulus. Serum- and cytokine-starved whole bone marrow from WT and Mx1-Cre, KrasG12D mice was stimulated with GM-CSF (10 ng/mL) or SCF (100 ng/mL) for 10 or 30 minutes. Levels of phosphorylated ERK, STAT5, and S6 were measured using phospho-specific flow cytometry in the (A) whole marrow and (B-C) in the c-kit+ lin−/low subset. The figure shows representative data from 10 independent experiments.
To study the effects of activating a growth factor receptor that is expressed on a broad range of stem/progenitor cells, we assayed pSTAT5, pERK, and pS6 levels after exposing c-kit+ lin−/low cells to SCF. WT and Kras mutant c-kit+ lin−/low cells express similar levels of c-kit, which is the receptor for SCF (Figure 3A). Importantly and unexpectedly, Mx1-Cre, KrasG12D cells showed markedly attenuated pERK and pS6 responses to this cytokine (Figure 4C). The differential signaling responses to SCF and GM-CSF in WT and Mx1-Cre, KrasG12D c-kit+ lin−/low cells were observed within 7 days of pIpC injection (data not shown). Based on a recent report that activated PI3 kinase dominantly interferes with Egr-1 induction by oncogenic HRAS in 3T3 fibroblasts,30 we asked if exposing c-kit+ lin−/low bone marrow cells from Mx1-Cre, KrasG12D to the PI3 kinase inhibitor LY294002 might restore the pERK and pS6 responses to SCF. In these studies, WT and Kras mutant cells were incubated with 10 or 50 μM of LY294002 for 30 minutes before stimulation. WT cells that were treated with LY294002 showed a dose-dependent reduction in pERK phosphorylation in response to SCF, with little or no effect on pS6 levels (Figure 5 left). Exposing Kras mutant c-kit+ lin−/low cells to LY294002 induced a modest increase in S6 phosphorylation, but this response remained remarkably attenuated compared with WT cells (Figure 5 middle panel).
Effects of the PI3 kinase inhibitor LY294002 on pERK, pSTAT5, and pS6 activation. WT and Mx1-Cre, KrasG12D c-kit+ lin−/low cells that were incubated for 30 minutes in the presence of LY294002 (LY) were stimulated with GM-CSF (10 ng/mL) or SCF (100 ng/mL) for 10 minutes and analyzed by multi-parameter FACS. Identical results were obtained in 2 independent experiments.
Effects of the PI3 kinase inhibitor LY294002 on pERK, pSTAT5, and pS6 activation. WT and Mx1-Cre, KrasG12D c-kit+ lin−/low cells that were incubated for 30 minutes in the presence of LY294002 (LY) were stimulated with GM-CSF (10 ng/mL) or SCF (100 ng/mL) for 10 minutes and analyzed by multi-parameter FACS. Identical results were obtained in 2 independent experiments.
Pharmacodynamic monitoring of phosphorylated signaling molecules after treatment with an MEK inhibitor
CI-1040 is a potent and selective inhibitor of MEK that has been evaluated in phase 1 and 2 clinical trials.31,32 We administered CI-1040 or control vehicle to Mx1-Cre, KrasG12D mice with MPDs and to WT control animals and collected marrow for analysis 2 hours later. The cells were stimulated with GM-CSF then fixed and permeabilized for FACS analysis. WT and Mx1-Cre, KrasG12D c-kit+ lin−/low cells from mice that received the control vehicle exhibited activation of effectors as seen previously (Figure 6). By contrast, treatment with CI-1040 abrogated ERK phosphorylation in both WT and Mx1-Cre, KrasG12D cells but did not alter pSTAT5 levels (Figure 6). Interestingly, CI-1040 had dramatic effects on S6 phosphorylation. CI-1040 treatment reduced pS6 levels to baseline values in Kras mutant cells and caused a major reduction in pS6 levels in WT cells. These data show that S6 phosphorylation is predominantly MEK dependent in c-kit+ lin−/low cells and provides additional evidence that oncogenic K-Ras influences signaling network architecture. Furthermore, they demonstrate that combining FACS analysis with small-molecule inhibitors can define upstream/downstream relationships within signaling networks.
Pharmacodynamic monitoring of MEK inhibition in c-kit+ lin−/low cells by multi-parameter FACS. Bone marrow was obtained from WT and Mx1-Cre, KrasG12D mice 2 hours after they received an intraperitoneal injection of vehicle control or CI-1040. The bone marrow was quickly harvested and stimulated with GM-CSF (10 ng/mL) for 10 minutes. Levels of phosphorylated ERK, STAT5, and S6 were measured in the c-kit+ lin−/low subset. These data were confirmed in a second experimental group.
Pharmacodynamic monitoring of MEK inhibition in c-kit+ lin−/low cells by multi-parameter FACS. Bone marrow was obtained from WT and Mx1-Cre, KrasG12D mice 2 hours after they received an intraperitoneal injection of vehicle control or CI-1040. The bone marrow was quickly harvested and stimulated with GM-CSF (10 ng/mL) for 10 minutes. Levels of phosphorylated ERK, STAT5, and S6 were measured in the c-kit+ lin−/low subset. These data were confirmed in a second experimental group.
Discussion
Advancing our current understanding of cancer pathogenesis requires elucidating how oncoproteins deregulate growth and survival signals in stem and progenitor cells. These cells underlie the maintenance and growth of many cancers22 ; however, their scarcity poses significant technical barriers. Furthermore, model systems that rely on overexpressing oncogenes in transformed cells may not accurately recapitulate the flexible and dynamic nature of signaling networks in primary cells. We addressed these issues by exploiting multi-parameter FACS to characterize the biochemical responses of disease-relevant stem/progenitor cell populations to endogenous levels of K-RasG12D. This methodology overcomes the potential confounding effects of cell type–specific differences in the activation status of cellular phospho-proteins within heterogeneous tissues and enables investigators to directly compare immunophenotypically defined normal and malignant populations. A previous study that assayed 6 phospho-proteins in cryopreserved AML blasts under basal conditions and after exposure to 5 different stimuli illustrated the power of multi-parameter FACS for interrogating signaling networks in primary cancer cells.19 Remarkably, although these 30 specimens were associated with a diverse spectrum of morphologic, biologic, and clinical findings, unsupervised clustering revealed 4 distinct subgroups, which correlated with chemotherapy resistance, cytogenetic abnormalities, and the presence of internal tandem duplications in the FLT3 gene. Multi-parameter FACS also uncovered a potentiated STAT5 response of FLT3 mutant blasts to myeloid cytokines that had not been detected using conventional assays and was absent in unstimulated cells.19,33 This study of human AML, in which a heterogeneous collection of specimens was interrogated under a specific set of experimental conditions, might be viewed as a “top down” approach to mapping cancer signaling networks. Our analysis of Mx1-Cre, KrasG12D bone marrows represents a complementary “bottom up” strategy whereby multi-parameter FACS was used to determine the biochemical consequences of expressing endogenous levels of a known oncoprotein in a defined population of cells that is highly enriched for stem/progenitor activity. A recent analysis of lung cancers from LSL- KrasG12D mice identified a gene expression “signature” that was then applied to uncover a transcriptional profile that distinguished human lung cancers with KRAS mutations.34 By analogy, investigating phospho-protein “signatures” in primary cancers from genetically engineered mice will enable investigators to define how specific mutations perturb the basic architecture of signaling networks. This information will inform studies examining the effects of these mutations in the context of complex and genetically heterogeneous human cancers.
We found that a primary effect of K-RasG12D expression is to increase the proliferation of CMPs, GMPs, and MEPs in vivo. As these immature progenitors undergo extensive amplification as they differentiate within the marrow, it is likely that these increased rates of cell division contribute directly to the MPD phenotype. The c-kit+ lin−/low fraction, which is highly enriched for stem/progenitor activity, includes CMPs, GMPs, and MEPs. We selected GM-CSF as a stimulus because a hypersensitive pattern of myeloid progenitor growth is a hallmark feature of JMML, a myeloid malignancy with a high frequency of alterations in Ras pathway genes.35 Mutating the respective murine homologs is also sufficient to induce hypersensitive myeloid progenitor growth,12,26,36–38 and a study in which Nf1 and Gmcsf mutant mice were intercrossed strongly implicated aberrant GM-CSF signaling as integral to the MPD that results from homozygous Nf1 inactivation.39 Multi-parameter FACS revealed elevated levels of pSTAT5 and pERK in c-kit+ lin−/low bone marrow cells from Mx1-Cre, KrasG12D mice that were deprived of serum for 2 hours and robust activation of pSTAT5, pERK, and pS6 in response to GM-CSF. The latter finding demonstrates that Ras effector cascades are not constitutively saturated in primary Kras mutant hematopoietic cells despite elevated Ras-GTP levels.12 It is possible that GM-CSF stimulation increases the levels of phosphorylated effectors by activating wild-type Ras proteins, by increasing GTP loading on K-RasG12D, or by both mechanisms. These biochemical data imply that K-RasG12D expression does not render primary hematopoietic stem/progenitor cells independent of environmental influences but instead sensitizes these cells to stimuli within the bone marrow microenvironment. However, it is possible that the conditions used to harvest and stimulate cells in vitro may not accurately recapitulate the signaling in vivo. The observation that myeloid progenitor colony growth from Kras mutant c-kit+ lin−/low cells is enhanced by GM-CSF (Figure 3C) is also consistent with this proposed mechanism.
A major difference between WT and Mx1-Cre, KrasG12D c-kit+ lin−/low bone marrows is that a higher proportion of mutant cells increase the levels of pSTAT5, pERK, and pS6 in response to GM-CSF. ERK and S6 also display enhanced and prolonged phosphorylation compared with WT cells. These data are consistent with the idea that K-RasG12D expression lowers the threshold for activating signaling cascades downstream of the GM-CSF receptor. Alternatively, Kras mutant stem/progenitor cells may demonstrate aberrant receptor expression. Interestingly, transcription of the murine GM-CSF receptor α chain is markedly up-regulated in mice that develop MPDs due to somatic JunB inactivation.40 STAT5, which is recruited to phosphorylated tyrosine residues on the activated GM-CSF receptor, is thought to function parallel to or upstream of Ras in myeloid growth control.41 Consistent with this model, CI-1040 abrogated the ability of GM-CSF to increase pERK levels in c-kit+ lin−/low cells but had no effect on STAT5 activation. The magnitude and duration of the pSTAT5 response was similar in WT and Kras mutant c-kit+ lin−/low cells; however, a higher percentage of Mx1-Cre, KrasG12D cells phosphorylated STAT5 in response to GM-CSF. These data could indicate that K-RasG12D protein expression increases the fraction of stem/progenitor cells with sufficient numbers of GM-CSF receptors to activate JAK2/STAT5 signaling or that it decreases the threshold for STAT5 phosphorylation in cells with low receptor numbers.
The impaired pERK and pS6 responses of Kras mutant cells to SCF were completely unexpected. Although the term “oncogene addiction” is commonly used to describe the general notion that cancer cells respond to oncoproteins by remodeling signaling networks, biochemical data demonstrating that this occurs in vivo are lacking. We provide direct evidence that K-RasG12D expression markedly attenuates the ability of hematopoietic stem/progenitor cells to activate S6 through the Raf/MEK/ERK cascade in response to SCF. While the underlying mechanism is unknown, our data imply that this is not due to a dominant interfering effect of hyperactive PI3 kinase. We speculate that Kras mutant cells attenuate downstream effectors of SCF as an adaptive response that minimizes the deleterious effects of oncoprotein expression. If this is true, it is intriguing that hematopoietic stem/progenitor cells that express K-RasG12D remain hypersensitive to GM-CSF, a cytokine that is strongly implicated in the pathogenesis of JMML and some cases of CMML.11,35,42 Furthermore, because SCF is a critical regulator of survival in normal hematopoietic cells,43 specific attenuation of SCF signaling in leukemic stem/progenitor cells may make them dependent on oncogenic signaling pathways.
Using an antibody that recognizes serine residues within S6 (S235 and S236) that can be phosphorylated by ERK, p70S6K1, and p70S6K2 in murine hepatocytes,44 we found that S6 is a downstream target of the Raf/MEK/ERK cascade in hematopoietic cells. This is supported by our observations that pERK and pS6 levels change in parallel in c-kit+ lin−/low cells exposed to GM-CSF or SCF and that LY294002 does not abrogate S6 responses to these cytokines. S6 kinases 1 and 2 (p70S6K1 and p70S6K2) are believed to be the principal in vivo mediators of mammalian S6 activation in many cell types and are regulated primarily by PI3K; however, we unexpectedly discovered that S6 phosphorylation is highly MEK dependent in hematopoietic stem/progenitor cells. These data are consistent with recent studies in strains of knock-out mice showing that S6 phosphorylation can be regulated by the Raf/MEK/ERK cascade.44 The relative contributions of the Raf/MEK/ERK, PI3K/Akt, and other pathways to activating S6 in hematopoietic stem/progenitor cells can be dissected further by approaches that include using different kinase inhibitors, interrogating other network proteins, and analyzing S6 phosphorylation at other sites.
Despite the potential of multi-parameter FACS to address signaling in rare populations, there are currently some important limitations. Although the c-kit+ lin−/low population is enriched for stem/progenitor activity, Sca1 is typically used to cleanly isolate hematopoietic stem cells.24,28 We focused on the c-kit+ lin−/low fraction because we could not detect Sca1 after methanol permeabilization despite trials of multiple different clones, fluorophores, and sequential staining techniques. These data suggest that Sca1, which is a glycolipid-anchored membrane protein, may be stripped or modified by methanol treatment. If this proves true, this problem might be overcome by sorting based on the expression of other stem cell–selective markers such as SLAM family members.45 Alternatively, it may be feasible to develop a 2-step procedure in which cell populations of interest are first isolated by sorting and then stimulated, fixed, and permeabilized. We did not employ this approach in the current study based on the unpredictable effects that the first round of antibody staining and sorting might have on signaling networks.
In conclusion, we combined 2 powerful technologies in cancer biology, multi-parameter FACS and a genetically engineered strain of conditional mutant mice, to interrogate the biochemical consequences of a specific oncoprotein in a disease-relevant population of primary stem/progenitor cells. We illustrate discrete effects of K-RasG12D expression in c-kit+ lin−/low cells and show that this approach can be used to assess inhibition of biochemical targets. While further technical refinements will undoubtedly occur, these studies and the previous work of Irish et al19 in human AML establish multi-parameter FACS as a technique that has the unique capacity to define the architecture of signaling networks in primary cancer cells in the basal state and in response to perturbations such as cytokine stimulation and treatment with therapeutic agents.
Authorship
Contribution: M.E.M.V.M, E.D.-F., and B.S.B. designed and performed research and wrote the paper. J.A.A. performed research. E.P. designed and performed research. J.M.I. and N.K. analyzed data. G.P.N. contributed analytical tools, vital protocols, and reagents. K.S. designed research and wrote the paper.
Conflict-of-interest disclosure: G.P.N. and J.M.I hold patents through Stanford University related to the technology employed in the present study. G.P.N declares a financial interest in a company that holds some rights these same patents.
M.E.M.V.M. and E.D.-F contributed equally to this study.
Correspondence: Kevin Shannon and Benjamin S. Braun, Department of Pediatrics, University of California, San Francisco, 513 Parnassus Avenue, HSE 302, Box 0519, San Francisco, CA 94143; e-mail: shannonk@peds.ucsf.edu and braunb@peds.ucsf.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We are indebted to Tyler Jacks and David Tuveson for LSL-KrasG12D mice, to Judy Leopold for CI-1040, to Jacob Rozmus for technical assistance, and to Irv Weissman for insight and support.
This work was supported by National Institutes of Health (R01 CA72614, N01-HV-28183, K08 CA103868, and 8-PAI036535C), by Specialized Center of Research awards from the Leukemia and Lymphoma Society of America to Stanford University and to Cold Spring Harbor National Laboratory, by the Jeffrey and Karen Peterson Family Foundation, and by the Frank A. Campini Foundation. M.E.M.V.M. is the recipient of a research fellowship for UCSF medical Student from the Genentech Foundation. J.M.I. is a Fellow of the Leukemia and Lymphoma Society.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal