Mutations that activate the N-ras oncogene are among the most frequently detected genetic alterations in human acute myeloid leukemias (AMLs), Philadelphia chromosome-negative myeloproliferative disorders (MPDs), and myelodysplastic syndromes (MDSs). However, because N-ras has not been shown to induce these disorders in an in vivo model, the role of N-ras in the evolution of myeloid leukemia is unclear. To investigate the potential of N-ras to induce myeloid leukemia, lethally irradiated mice were reconstituted with bone marrow (BM) cells infected with a retroviral vector carrying activated N-ras. Approximately 60% of these mice developed hematopoietic disorders, including severe MPDs resembling human chronic myelogenous leukemia (CML) or AML with differentiation (French-American-British [FAB] classification M2). Other reconstituted mice succumbed to hematopoietic defects that were pathologically similar to human MDSs. The latter disorders appeared to be due to a myeloid impairment that was demonstrated by enumeration of day-12 colony-forming units-spleen (CFU-S) and by in vitro colony assays. A high level of apoptosis associated with thymic atrophy and peripheral blood (PB) lymphopenia was also evident in N-rasreconstituted mice. Our results are consistent with a model in which antiproliferative effects are a primary consequence of N-rasmutations and secondary transforming events are necessary for the development of myeloid leukemia. This is the first report of an in vivo model for N-ras induced MPD and leukemia.
THE N-ras ONCOGENE is one member of the ras super gene family that also includes the closely related oncogenes c-Ki-ras and c-Ha-ras.1These oncogenes all encode 21-kD guanine nucleotide binding proteins that operate as molecular switches to regulate the transduction of physiological signals from the cell membrane to the nucleus.2 Receptors for a number of cytokines, including platelet-derived growth factor (PDGF), epidermal growth factor, macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-2 (IL-2), and IL-3 transmit mitogenic signals via ras. ras is also an effector molecule for a number of oncoproteins, includingsrc, fes, neu, and the bcr-abl fusion protein.3 Activation of ras can stimulate a number of divergent downstream pathways via various (putative) target molecules such as raf,4 phosphotidylinositol-3-OH kinase,5 RalGDS,6 and Jun N-terminal kinase.7 The phenotypic consequences of rasactivation are varied and appear to be cell-type specific and dependent on the initiating signal. Downstream divergence probably accounts for the potential of ras signal transduction pathways to elicit multiple cellular outcomes.
ras signal transduction pathways become constitutively activated when a cell acquires a specific point mutation in one of theras genes.8 Mutations that activate the N-ras oncogene are among the most frequently detected genetic alterations in human myeloid disorders. N-ras mutations have been detected in acute myeloid leukemia (AML), Philadelphia chromosome-negative myeloproliferative disorders (MPDs), and myelodysplastic syndromes (MDSs) at frequencies that vary from approximately 25% to 40%.9-11ras signal transduction pathways are also activated in some myeloid leukemias that retain wild-type ras alleles. For example, the bcr-ablfusion protein expressed in Philadelphia chromosome-positive MPDs activates a ras signal transduction pathway that appears necessary for transformation.12,13ras is also activated in juvenile chronic myelogenous leukemia (JCML) as a consequence of loss of the neurofibromatosis (NF1) gene that normally functions to negatively regulate ras.14,15In chronic myelomonocytic leukemia (CMML), ras may be activated by the t(5,12) translocation that fuses PDGF receptor andtel.16
Although recent investigations have provided much insight into the molecular mechanisms involved in ras signal transduction, the role of N-ras in the development of myeloid leukemia remains unclear. The high incidence of N-ras mutations in preleukemic conditions such as MDSs and MPDs suggests that activation of N-ras may play an initiating or predisposing role in hematopoietic neoplasia. In contrast, N-ras mutations in AML samples are sometimes only detected in a subset of the leukemic population,17 suggesting that N-ras mutations may be involved in the progression rather than initiation of leukemia. To date, most of the studies that have sought to define the role ofras disregulation in hematopoietic neoplasia have used models in which the Ha-ras oncogene is constitutively activated.18-22 However, the significance of these studies is unclear, because Ha-ras does not appear to be involved in human myeloid disorders,8 and it was recently shown that Ha-ras and N-ras have different transformation capabilities in hematopoietic cells.23 This may explain the induction of lymphoid, rather than myeloid disorders in mice that were repopulated with bone marrow (BM) cells expressing activated Ha-ras.20,22 Transgenic mice carrying activated N-ras developed mammary tumors, reticulum cell sarcomas, and lymphomas after a long latency.24,25 However, in humans, these tumor types rarely harbor N-ras mutations. The incidence of N-ras mutations in lymphomas is apparently significantly lower than the frequency of ras mutations in myeloid disorders.26 The malignancies that developed in these transgenic animals are more likely to reflect the tissue specificity of the promoters used to drive the transgenes rather than the transformation specificity of mutant N-ras.
To investigate the role of N-ras mutations in myeloid leukemia, we have reconstituted mice with BM progenitor cells that express mutationally activated N-ras. Myeloid disorders resembling human pathologies associated with N-ras mutations, including MDS, chronic myelogenous leukemia (CML), and AML, arose in the reconstituted mice. These investigations demonstrate that mutational activation of N-ras may be a predisposing event, and they provide the first report of a relevant in vivo model for studying the evolution of human myeloid leukemia.
MATERIALS AND METHODS
Retroviral vectors and packaging cell lines.
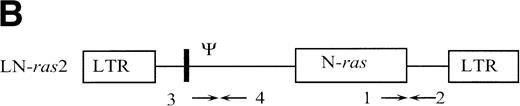
The retroviral vector termed LK (shown in Fig 1) was constructed by replacing neo within the retroviral vector pLNL627with a polylinker. The polylinker contains recognition sites forEcoRI, Bgl II, Sac I, Sma I,BamHI, Xba I, and HindIII. To construct LN-ras2, a 650-bp HindIII/BamHI fragment of human N-ras cDNA containing an activating mutation at codon 12 was excised from pZipN-ras (Der and Tainsky, unpublished data) and subcloned by blunt-end ligation into theBamHI site of pLK. The integrity and orientation of the resultant pLN-ras2 plasmid was checked by polymerase chain reaction (PCR) analysis and restriction enzyme digestion. The codon 12 mutation within the N-ras cDNA was confirmed by sequence analysis. The LN-ras2 vector is shown in Fig 1B.
Retroviral vectors and packaging cell lines. (A) LK; (B) LN-ras2. LTR, M-MLV long terminal repeat; Ψ, packaging region; thick vertical line, viral splice donor site; parallel vertical lines, polylinker; arrows, primers used for PCR. (C and D) Immunohistochemical staining for human N-ras protein. (C) ΨLN-ras215 cells show cytoplasmic staining of N-ras protein. (D) ΨLN-ras2 nonproducers (LN-ras2 transfectant that did not make detectable virus). Mophology of (E), ΨLN-ras215 producers; (F) ΨLK09 producers (original magnification ×250).
Retroviral vectors and packaging cell lines. (A) LK; (B) LN-ras2. LTR, M-MLV long terminal repeat; Ψ, packaging region; thick vertical line, viral splice donor site; parallel vertical lines, polylinker; arrows, primers used for PCR. (C and D) Immunohistochemical staining for human N-ras protein. (C) ΨLN-ras215 cells show cytoplasmic staining of N-ras protein. (D) ΨLN-ras2 nonproducers (LN-ras2 transfectant that did not make detectable virus). Mophology of (E), ΨLN-ras215 producers; (F) ΨLK09 producers (original magnification ×250).
To establish producer cell lines, the plasmids pLK and pLN-ras2 were each cotransfected, along with pMolneo,28 into Ψ-cre packaging cells.29 Cotransfections were performed using 10 μg of retroviral vector plasmid and 1 μg of pMolneo in a standard calcium phosphate precipitation procedure.30 The pMolneo vector does not include any packaging sequence or any sequence encoding retroviral proteins that would potentiate the generation of replication competent retroviruses (RCRs). Transfectants were selected in 1 mg/mL G418, cloned using cloning cylinders, and assayed for viral titer by viral RNA dot blot analysis. For isolation of viral RNA, viral particles were precipitated from 900 μL of clarified viral supernatant by the addition of 225 μL of 40% polyethylene glycol and 128 μL NaCl, followed by incubation on ice for 1 hour. After centrifugation at 13,000g for 10 minutes, the pelleted viral particles were resuspended in 250 μL RNase-free TE and extracted with phenol/chloroform/IAA (24:24:1). Generally, viral RNA samples were extracted in triplicates. Viral RNA samples were applied to Zeta probe nylon membranes (Bio-Rad, Hercules, CA) using a dot blot apparatus (Bio-Rad) according to the manufacturer’s instructions. Membranes were hybridized to an α-32P-dCTP-labeled packaging region (Ψ) probe using standard procedures. The Ψ probe is a 505-bp PCR product (primers 3 and 4, Fig 1) that extends from the viral splice donor site of pLK into gag. After hybridization and autoradiography, viral dot blots were quantitated by densitometry and standardized against dilutions of RNA prepared from the supernatant of the Ψ2AV viral producer cell line.31 Using this method, the titers of the producer clones used in this study, ΨLK09 and ΨLN-ras215, were determined to be of the order 105colony-forming units (CFU)/mL and 106 CFU/mL, respectively. Southern and Northern blot analysis using both the Ψ probe and an N-ras cDNA confirmed that both of these clones harbored full-length proviruses and expressed transcripts of the expected size. The N-ras probe hybridized to the same ΨLN-ras215 transcripts and provirus as the Ψ probe. The ΨLN-ras215 and ΨLK09 cell lines were assayed for RCR using the marker rescue assay. The titer of virus in the supernatant from a positive control (NIH3T3 cells infected with replication competent Moloney murine leukemia virus [M-MLV]) was determined to be approximately 105 CFU/mL, and no RCR was detected in undiluted supernatant from either of the viral producer clones.
BM infections and reconstitution of irradiated mice.
BM was flushed from the femurs of 8- to 10-week-old female Balb/c mice that were injected intraperitoneally 4 days before with 5-flurouracil (5-FU; 150 mg/kg body weight). For retroviral infection, BM cells were resuspended in supernatant collected from retroviral producers and supplemented with 10% fetal bovine serum (FBS), 8 μg/mL polybrene, 30 μg/mL transferrin, 1 mg/mL bovine serum albumin (BSA), and 20% WEHI conditioned media (WCM; a source of IL-3), 1 ng/mL IL-6, and, in the second series, 20 ng/mL kit ligand (KL). Two rounds of infection were performed over a 40-hour period.
Recipient mice (11- to 14-week-old female Balb/c) were irradiated with 8.5 Gy from a linear accelerator. Retrovirally infected BM cells suspended in serum-free Iscove’s modified Dulbecco’s medium (IMDM) were injected into irradiated mice by intravenous injection within 5 hours of irradiation. BM from one donor mouse was used per irradiated recipient mouse.
Colony assays.
BM obtained from mice pretreated with 5-FU was infected with either LN-ras2 or LK as described above. The infected cells were maintained in IMDM supplemented with IL-3 and 10% FBS for 48 hours before seeding 105 cells/mL in 0.8% methylcellulose with 25% horse serum, 20% WCM, 100 ng/mL IL-6, 2 U/mL erythropoietin (Epo), 1 mg/mL BSA, and 10−4 mol/L β-mercaptoethanol. Resulting colonies were scored on day 10 to 14 after benzidine staining for visualization of erythroid colonies.
Colony-forming units-spleen (CFU-S) assay.
BM progenitor cells (2 × 105) infected with either LN-ras2 or LK virus as described above were transplanted into lethally irradiated recipient mice. The recipient mice were killed 12 days after transplantation and CFU-S were microscopically determined and enumerated.
Cytospun cell preparations, blood smears, and histology.
Cytospun cell preparations and blood smears were stained with 0.25% May-Grunwald stain and counterstained with 10% Giemsa stain. Tissue samples for histology were preserved in cold 4% formaldehyde in phosphate-buffered saline (PBS). Embedding, sectioning, and staining with hematoxylin and eosin was performed by the Veterinary Histopathology Department, University of Sydney (Sydney, Australia).
Fluorescence-activated cell sorting (FACS) analysis.
Approximately 105 spleen, BM, or thymus cells were washed once in PBS plus 2% FBS and then resuspended in 50 μL of diluted conjugated antibody (Becton Dickinson, Mountain View, CA) and incubated on ice for 30 minutes. The cells were then washed twice in PBS plus 2% FBS, resuspended in 1 mL PBS, and analyzed on a Becton Dickinson FACSort. Gates were set to exclude mature erythrocytes and dead cells from the analysis.
TUNEL analysis.
Paraffin-embedded sections mounted on polylysine slides (prepared by the Veterinary Histopathology Department, University of Sydney) were analyzed using the TUNEL reaction essentially as described.32 Briefly, after deparaffinization and rehydration, the sections were incubated in 20 μg/mL proteinase K for 15 minutes at room temperature, washed in dH2O, incubated in 2% H2O2 for 5 minutes at room temperature, and rinsed again in dH2O. The terminal deoxynucleotydyl transferase (TdT) reaction was performed using a terminal transferase kit and biotinylated dUTP (Sigma, St Louis, MO). The reaction was terminated by rinsing the sections in 2× SSC for 15 minutes, then rinsing in dH2O, and finally washing in PBS for 5 minutes. The color reaction and counterstaining were performed using a universal antimouse staining kit (Sigma) according to the manufacturer’s instructions.
Immunohistochemistry.
Paraffin-embedded sections mounted on polylysine slides were deparaffinized and rehydrated for staining with a human N-ras–specific antibody, F155 (Santa Cruz, Santa Cruz, CA). Exogenous peroxidase activity was quenched in a 5-minute incubation with 2% H2O2, followed by blocking with 2% BSA. The primary antibody was incubated overnight at 4°C and visualized using the Univeral Antimouse Staining Kit (Sigma). Counterstaining was performed with hematoxylin. Producer cells were stained for N-ras protein on a cytocentrifuge preparation after fixation with methanol:acetone (1:1) at −20°C for 5 minutes.
PCR analysis.
Genomic DNA was isolated by phenol/chloroform extraction as described previously.30 Cell lysates were prepared by resuspending 105 to 106 cells in 500 μL H2O, boiling for 10 minutes, and collecting the supernatant after centrifugation at 13,000g for 5 minutes. PCR amplification was performed using 1 U Taq polymerase (Cetus Perkin Elmer, Norwalk, CT) with primers at a concentration of 200 nmol/L and 150 to 250 ng of genomic DNA or 10 μL of cell lysate in the manufacturer supplied buffer. The thermal cycling program used was: 3 minutes at 95°C for 1 cycle; then 30 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 2 minutes; and a final elongation step of 72°C for 10 minutes.
Primers used for PCR (shown in Fig 1) were as follows: (1) 5′ CGAAGGCTTCCTCTGTGTAT 3′ (human N-ras); (2) 5′ GGCTTCAGCTGGTGATATTG 3′ (MLV env); (3) 5′ TGGCCAGCAACTTATCTGTGT 3′ (Ψ region M-MLV/MSV); and (4) 5′ TCTTGACATCTACCGACTGG 3′ (M-MLV gag).
Northern analysis.
RNA extraction and Northern analysis was performed as previously described.30 Nylon membranes (Hybond N; Amersham, Arlington Heights, IL) were hybridized to an α-32P[dCTP]-labeled Ψ probe (described above).
RESULTS
Retroviral vectors and packaging cell lines.
The retroviral vectors and packaging cell lines used in these studies are shown in Fig 1 and are further described in Materials and Methods. The N-ras cDNA within LN-ras2 has a single activating point mutation at codon 12 that replaces the wild-type glycine residue with aspartate. This mutation is one of the most common mutations detected in human myeloid leukemias.8 Because internal promoters and selectable markers were previously associated with vector rearrangements and appeared to suppress retroviral expression in vivo,33-35 no additional sequence was included in these constructs. The titers of the retroviral producer clones, ΨLN-ras215 and ΨLK09, used in these studies were determined to be of the order of 106 and 105 CFU/mL, respectively. Immunohistochemistry using a human N-ras–specific antibody showed high levels of human N-ras protein in the ΨLN-ras215 producer clone (Fig 1C and D). Because of expression of mutant N-ras, ΨLN-ras215 cells are morphologically transformed. As shown in Fig 1, ΨLN-ras215 cells are rounded and highly refractile and grow loosely attached to the culture flask without cell-cell contact inhibition. ΨLN-ras215 cells can be grown to very high density, enabling collection of highly concentrated virus. Indeed, the titer of some ΨLN-ras215 viral supernatants was estimated to be around 1 × 107 CFU/mL. In contrast to ΨLN-ras215, ΨLK09 cells (as well as Ψ-cre parental cells and ΨN-ras2 nonproducers) are flat, spindle-shaped, achorage-dependent, and contact-inhibited. Despite the difference in growth kinetics and viral titer, the transduction efficiency of ΨLN-ras215 and ΨLK09 as assayed on 5-FU–treated BM was comparable. PCR analysis of individual colonies plucked from methylcellulose cultures indicated that up to 80% of colony-forming units-cells (CFU-C) could be transduced with either ΨLN-ras215 or ΨLK09 (data not shown). Similar BM transduction efficiencies were also determined for additional retroviruses generated in our laboratory. The viral titer of the latter vectors were also variable, within the range of 105 to 107.36 37
Proliferative disorders and impaired reconstitution in LN-ras2 mice.
To investigate the consequences of N-ras activation in an in vivo model, lethally irradiated mice were transplanted with retrovirally transduced BM progenitor cells. For the first reconstitution series, retroviral transduction was performed in the presence of IL-3 and IL-6. In this experiment, mice that received LN-ras2–infected BM were termed Nr1 to Nr15 and mice that received LK-infected marrow were termed LK1 to LK7. BM for infection with either LNras2 or LK was harvested from one donor mouse per recipient such that mice receiving LN-ras2 or LK-infected marrow would receive the same number of stem cells. Ten of the 15 Nr mice became moribund and were killed or had died by week 14. In contrast, mice that were infused with LK-infected BM remained healthy throughout the course of the experiment. The mice termed Nr11 to Nr15 were healthy when killed at the end of the experiment (31 weeks after transplantation). The disease latencies and a description of pathologies of the Nr mice are listed in Table 1.
LN-ras2 Reconstitution Summary
| . | Latency (d) . | Pathology . | Diagnosis . |
|---|---|---|---|
| Nr1 | 10 | Unrelated illness | |
| Nr2 | 11 | Reduced splenic hematopoiesis, hypoplastic BM. | Impaired reconstitution |
| Nr3 | 12 | Reduced splenic hematopoiesis, hypoplastic BM. | Impaired reconstitution |
| Nr4 | 14 | Reduced splenic hematopoiesis, hypoplastic BM, anemia, small thymus. | Impaired reconstitution |
| Nr5 | 14 | Reduced splenic hematopoiesis, hypoplastic BM, anemia, small thymus. | Impaired reconstitution |
| Nr6 | 26 | Reduced splenic hematopoiesis, hypoplastic BM, anemia. | Impaired reconstitution |
| Nr7 | 37 | Splenic nodes, erythroid hyperplasia in spleen, apoptosis in spleen, hypoplastic BM, anemia. | Impaired reconstitution, erythroid hyperplasia |
| Nr8 | 49 | Splenic nodes, erythroid hyperplasia in spleen, erythropoiesis in BM, aberrant erythrocytes, small thymus, apoptosis in the spleen. | Impaired reconstitution, erythroid hyperplasia |
| Nr9 | 50 | Splenic nodes, erythroid hyperplasia in spleen, mast cells in the spleen, hypoplastic BM, anemia, abberant erythrocytes, apoptosis in the spleen and thymus. | Impaired reconstitution, erythroid hyperplasia |
| Nr10 | 98 | ND | ND |
| Nr16 | 107 | Splenomegaly, small thymus, myelomonocytic hyperplasia, infiltrates within spleen, thymus, lung, liver, kidney, BM and PB, intestinal nodules, PB granulocytosis, apoptosis. | Myeloproliferative disorder |
| Nr17 | 141 | Splenomegaly, small thymus, mast cell hyperplasia, erythroid hyperplasia, mast cell infiltrates in spleen, liver, lung, BM and PB, BM hypoplasia, PB granulocytosis, anemia, aberrant erythrocytes, hematoma. | Mast cell disorder, erythroid hyperplasia |
| Nr18 | 144 | Splenomegaly, small thymus, myelomonocytic hyperplasia, infiltrates within spleen, thymus, lung, liver, kidney, BM and PB, lung tumor, PB granulocytosis, apoptosis. | Myeloproliferative disorder |
| Nr19 | 163 | ND | ND |
| Nr20 | 192 | Splenic nodes, small thymus, enlarged liver, mast cell hyperplasia, infiltrates within BM, liver and lung, PB blasts and monocytosis. | Monoblastic leukemia, mast cell disorder |
| Nr21 | 308 | Erythroid hyperplasia within spleen and BM. | Erythroid hyperplasia |
| Nr22 | 385 | Hind leg paralysis, splenomegaly, myelomonocytic hyperplasia, infiltrates within spleen, BM and lung, aberrant erythrocytes, apoptosis. | Myeloproliferative disorder |
| . | Latency (d) . | Pathology . | Diagnosis . |
|---|---|---|---|
| Nr1 | 10 | Unrelated illness | |
| Nr2 | 11 | Reduced splenic hematopoiesis, hypoplastic BM. | Impaired reconstitution |
| Nr3 | 12 | Reduced splenic hematopoiesis, hypoplastic BM. | Impaired reconstitution |
| Nr4 | 14 | Reduced splenic hematopoiesis, hypoplastic BM, anemia, small thymus. | Impaired reconstitution |
| Nr5 | 14 | Reduced splenic hematopoiesis, hypoplastic BM, anemia, small thymus. | Impaired reconstitution |
| Nr6 | 26 | Reduced splenic hematopoiesis, hypoplastic BM, anemia. | Impaired reconstitution |
| Nr7 | 37 | Splenic nodes, erythroid hyperplasia in spleen, apoptosis in spleen, hypoplastic BM, anemia. | Impaired reconstitution, erythroid hyperplasia |
| Nr8 | 49 | Splenic nodes, erythroid hyperplasia in spleen, erythropoiesis in BM, aberrant erythrocytes, small thymus, apoptosis in the spleen. | Impaired reconstitution, erythroid hyperplasia |
| Nr9 | 50 | Splenic nodes, erythroid hyperplasia in spleen, mast cells in the spleen, hypoplastic BM, anemia, abberant erythrocytes, apoptosis in the spleen and thymus. | Impaired reconstitution, erythroid hyperplasia |
| Nr10 | 98 | ND | ND |
| Nr16 | 107 | Splenomegaly, small thymus, myelomonocytic hyperplasia, infiltrates within spleen, thymus, lung, liver, kidney, BM and PB, intestinal nodules, PB granulocytosis, apoptosis. | Myeloproliferative disorder |
| Nr17 | 141 | Splenomegaly, small thymus, mast cell hyperplasia, erythroid hyperplasia, mast cell infiltrates in spleen, liver, lung, BM and PB, BM hypoplasia, PB granulocytosis, anemia, aberrant erythrocytes, hematoma. | Mast cell disorder, erythroid hyperplasia |
| Nr18 | 144 | Splenomegaly, small thymus, myelomonocytic hyperplasia, infiltrates within spleen, thymus, lung, liver, kidney, BM and PB, lung tumor, PB granulocytosis, apoptosis. | Myeloproliferative disorder |
| Nr19 | 163 | ND | ND |
| Nr20 | 192 | Splenic nodes, small thymus, enlarged liver, mast cell hyperplasia, infiltrates within BM, liver and lung, PB blasts and monocytosis. | Monoblastic leukemia, mast cell disorder |
| Nr21 | 308 | Erythroid hyperplasia within spleen and BM. | Erythroid hyperplasia |
| Nr22 | 385 | Hind leg paralysis, splenomegaly, myelomonocytic hyperplasia, infiltrates within spleen, BM and lung, aberrant erythrocytes, apoptosis. | Myeloproliferative disorder |
Abbreviation: ND, not determined.
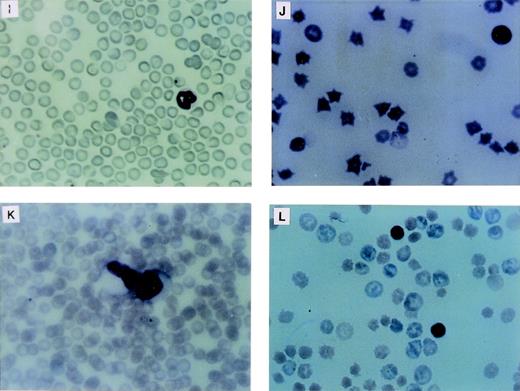
The LN-ras2–reconstituted mice that became moribund within the first 4 weeks (Nr2-6) succumbed to hematopoietic deficiencies characterized by hypoplastic BM, reduced splenic hematopoiesis, small thymuses, and anemia. The Nr mice that were killed at or near day 12 had fewer macroscopic spleen colonies than a time-matched LK control mouse. The spleens of Nr mice that became moribund between days 37 and 50 were enlarged and had macroscopic nodal outgrowths. The histology and a cytocentrifuged preparation of spleen cells from one of the latter mice is shown in Fig2B and F, respectively. The normal follicular architecture of the spleen from this mouse was disrupted by an expansion of erythroid progenitor cells. Aberrant erythrocytes were evident within the peripheral blood (PB) of these mice (Fig 2J), indicating that erythroid maturation was defective. Blast cells were also present in the PB of these mice. Cytocentrifuged preparations also showed an increase in mast cells and a depletion of mature granulocytes and lymphocytes within their spleens. FACS analysis of spleen cell preparations confirmed these observations (data not shown). We conclude that these mice developed erythroid hyperplasia associated with a hematopoietic impairment.
Proliferative disorders in LN-ras2 reconstituted mice. (A) LK8 spleen section. Normal splenic architecture, follicular organization of the lymphoid cells, marginal zones, and red pulp composed of erythroid cells are shown. (B) Nr9 spleen section. The splenic architecture is disrupted by darkly stained hematopoietic cells (arrow). (C) Nr18 spleen section. Large foci of myelomonocytic cells that obliterate the normal splenic architecture (light staining) are shown. (D) Nr17 spleen section. Sheets of abnormal mast cells (light staining) that have disrupted the normal splenic architecture are shown. (E) LK8 spleen cells. Normal splenocytes, large and small lymphocytes, myeloid cells, and erythroid progenitor cells are shown. (F) Nr9 spleen cells. Abundant erythroid progenitors (arrows) are shown. (G) Nr16 spleen cells. Increased numbers of large monocytes (X) and myeloid cells (arrow) are shown. (H) Nr17 spleen cells. Darkly granulated mast cells (X) and increased numbers of erythroid progenitors (arrows) are shown. (I) LK8 blood. Normal erythrocytes and a neutrophilic granulocyte are shown. (J) Nr9 blood. Polychromatic and spiculated erythrocytes are demonstrated. (K) Nr18 blood. Two abnormal myelomonocytic cells are shown. (L) Blood from Nr17. Aberrant erythrocytes are shown. Histologic sections (A through D) were stained with hematoxylin and eosin (original magnification ×50). Cytospun preparations (E through H) and blood smears (I through L) were stained with May-Grunwald and Giemsa (original magnification ×500).
Proliferative disorders in LN-ras2 reconstituted mice. (A) LK8 spleen section. Normal splenic architecture, follicular organization of the lymphoid cells, marginal zones, and red pulp composed of erythroid cells are shown. (B) Nr9 spleen section. The splenic architecture is disrupted by darkly stained hematopoietic cells (arrow). (C) Nr18 spleen section. Large foci of myelomonocytic cells that obliterate the normal splenic architecture (light staining) are shown. (D) Nr17 spleen section. Sheets of abnormal mast cells (light staining) that have disrupted the normal splenic architecture are shown. (E) LK8 spleen cells. Normal splenocytes, large and small lymphocytes, myeloid cells, and erythroid progenitor cells are shown. (F) Nr9 spleen cells. Abundant erythroid progenitors (arrows) are shown. (G) Nr16 spleen cells. Increased numbers of large monocytes (X) and myeloid cells (arrow) are shown. (H) Nr17 spleen cells. Darkly granulated mast cells (X) and increased numbers of erythroid progenitors (arrows) are shown. (I) LK8 blood. Normal erythrocytes and a neutrophilic granulocyte are shown. (J) Nr9 blood. Polychromatic and spiculated erythrocytes are demonstrated. (K) Nr18 blood. Two abnormal myelomonocytic cells are shown. (L) Blood from Nr17. Aberrant erythrocytes are shown. Histologic sections (A through D) were stained with hematoxylin and eosin (original magnification ×50). Cytospun preparations (E through H) and blood smears (I through L) were stained with May-Grunwald and Giemsa (original magnification ×500).
Further reconstitution experiments were performed using KL in addition to IL-3 and IL-6 in the viral infection medium. When added to BM cultures in combination with IL-3 and IL-6, KL increases CFU-S numbers and the long-term repopulating ability of BM cells.38 Apart from the addition of KL, all other constituents in the infection culture, the period of infection, and the ratio of donor mice to recipients remained the same in the second reconstitution series as in the first. The second set of reconstituted mice are referred to as Nr16 to Nr 27 and LK8 to LK13 (Table 1). In contrast to the mice in the first series, all Nr mice in the second series survived the initial (4 weeks) reconstitution period and 58% (7/12) of the Nr mice later succumbed to severe proliferative disorders. None of the LK control mice developed hematopoietic disorders.
The hematopoietic disorders that were characterized in Nr mice in this reconstitution series were more severe, in terms of cell expansion and invasiveness, than the disorders in the first reconstitution series. The spleens of moribund Nr mice were enlarged up to fourfold, mottled pink, and, in some cases (eg, Nr20), had large white nodules. Additional macroscopic abnormalities included a lung tumor in Nr18 and white nodal growths along the intestine of Nr16. Examination of histologic sections showed large foci of abnormal hematopoietic cells throughout the spleen, liver, lung, kidney, thymus, and BM of these Nr mice. The intestinal nodes in the Nr16 mouse and the lung tumor in Nr18 were confirmed as hematopoietic outgrowths. Representative histology and cytospun preparations of spleen cells from these mice are shown in Fig 2C, D, G, and H. Abundant, abnormally large myelomonocytic cells that were at various stages of differentiation were evident in cytospun preparations of spleen and BM cells from several of these mice (Fig2G). The ratio of myelocytes versus monocytic cells within the spleen and BM varied among these mice. Severe erythroid and mast cell disorders also afflicted Nr mice in this experiment (Fig 2H). Mast cell disorders were confirmed by staining tissue sections with toluidine blue (data not shown).
Differential counts of PB leukocytes for representative mice are shown in Table 2. These data show a relative elevation of myeloid cells, particularly granulocytes, at the expense of lymphoid cells in the Nr mice that succumbed to hematopoietic disorders. Generally, the granulocytes in these mice were morphologically mature, although some abnormal myeloblasts were identified in the blood of Nr16, Nr17, and Nr18 (Fig 2K). As in the first reconstitution series, PB erythrocytes in Nr mice with erythroid hyperplasias were abnormal, exhibiting polychromasia and irregular shapes (Fig 2L). Abundant blast cells and increased numbers of monocytes were evident in the PB of Nr20. The disorders in Nr16, Nr18, and Nr21 are referred to as MPDs because the abnormal myelomonocytic cells appeared to be differentiating, whereas the large numbers of blast cells in Nr20 were indicative of overt leukemia. The variable pathologies and long latencies required for the development of MPDs and leukemia in Nr mice suggests that a secondary molecular event was necessary for malignant evolution. It is unlikely that mutagenesis due to retroviral insertion accounts for the second oncogenic hit, because (1) vector stocks were free of RCR and (2) thymomas, which are typically induced by replication competent Moloney murine leukemia virus, were not evident in Nr mice.
PB Leukocyte Differential Counts
| . | Lymphocytes . | Monocytes . | Granulocytes . | Other . |
|---|---|---|---|---|
| LK8 | 62 | 10 | 28 | |
| LK9 | 67 | 3 | 30 | |
| Nr16 | 16 | 3 | 76 | 5* |
| Nr17 | 27 | 1 | 72 | |
| Nr18 | 47 | 3 | 48 | 2* |
| Nr20 | 32 | 29 | 5 | 33† |
| . | Lymphocytes . | Monocytes . | Granulocytes . | Other . |
|---|---|---|---|---|
| LK8 | 62 | 10 | 28 | |
| LK9 | 67 | 3 | 30 | |
| Nr16 | 16 | 3 | 76 | 5* |
| Nr17 | 27 | 1 | 72 | |
| Nr18 | 47 | 3 | 48 | 2* |
| Nr20 | 32 | 29 | 5 | 33† |
Myeloblasts.
Undifferentiated blast cells.
FACS analysis of spleen and BM cells confirmed the phenotype of neoplastic cells in mice with hematopoietic disorders. Representative results from these investigations are summarized in Table 3. Erythroid hyperplasia (eg, Nr17) was characterized by an increase in the proportion of nucleated erythroid cells (TER119 reactive) that was offset by a decrease in T-lymphoid and B-lymphoid cells (Thy1.2+ and B220+, respectively) as well as a decrease in monocytic cells (Mac-1+). FACS analysis of cell suspensions from mice with MPDs (eg, Nr16 and Nr18) did not show drastic changes, although in the Nr18 mouse, increases in the percentage of monocytic and granulocytic (α-Gr+) cells were detected in spleen and a modest elevation of monocytes was detected in BM.
FACS Analysis of Primary Spleen (S) and BM Cells From Reconstituted Mice
| . | TER1193-150 . | Mac-13-151 . | Thy 1.23-152 . | B2203-153 . | a-Gr# . |
|---|---|---|---|---|---|
| LK9S | 8.9 | 14.4 | 25.32 | 55.0 | 0.2 |
| Nr16S | 13.1 | 8.21 | 13.6 | 54.0 | ND |
| Nr17S | 68.4 | 6.2 | 2.9 | 6.7 | 0.1 |
| Nr18S | 19.5 | 24.0 | 28.5 | 48.4 | 2.5 |
| LK9BM | 20.1 | 59.1 | 5.5 | 12.9 | 51.9 |
| Nr16BM | 18.0 | 49.0 | 7.1 | 8.7 | ND |
| Nr17BM | 60.6 | 39.5 | 1.6 | 38.2 | 35.1 |
| Nr18BM | 15.4 | 64.4 | 3.6 | 17.1 | 62.2 |
| . | TER1193-150 . | Mac-13-151 . | Thy 1.23-152 . | B2203-153 . | a-Gr# . |
|---|---|---|---|---|---|
| LK9S | 8.9 | 14.4 | 25.32 | 55.0 | 0.2 |
| Nr16S | 13.1 | 8.21 | 13.6 | 54.0 | ND |
| Nr17S | 68.4 | 6.2 | 2.9 | 6.7 | 0.1 |
| Nr18S | 19.5 | 24.0 | 28.5 | 48.4 | 2.5 |
| LK9BM | 20.1 | 59.1 | 5.5 | 12.9 | 51.9 |
| Nr16BM | 18.0 | 49.0 | 7.1 | 8.7 | ND |
| Nr17BM | 60.6 | 39.5 | 1.6 | 38.2 | 35.1 |
| Nr18BM | 15.4 | 64.4 | 3.6 | 17.1 | 62.2 |
Values represent positive cells as a percentage of the total cell population.
Abbreviation: ND, not determined.
Reacts with erythroid cells.
Reacts with monocytic cells, granulocytes, and natural killer cells.
Reacts with T-lymphoid cells.
Reacts with B-lymphoid cells.
#Reacts with granulocytes.
Apoptosis in LN-ras2 mice.
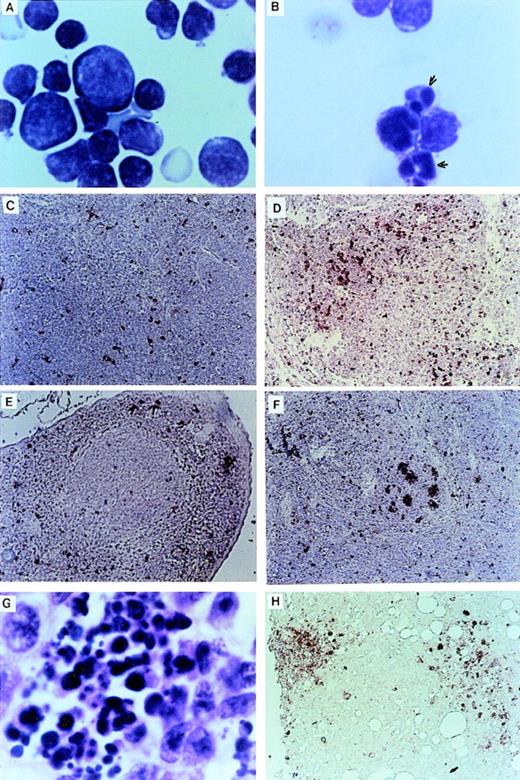
The thymuses of Nr mice were small in comparison to the thymuses of LK animals. Examination of cytocentrifuged preparations showed many thymocytes with apoptotic features such as condensed and fragmented nuclei (Fig 3A and B). TUNEL analysis32 confirmed that there was massive apoptotic cell death within the thymic cortex of some Nr mice (Fig 3C and D). There was also a higher incidence of apoptotic cells in the spleens of Nr mice in the first reconstitution series (Fig 3E through G). Apoptotic cells were predominantly confined to the lymphoid follicles in Nr9 and were scattered throughout the spleen of Nr8. Macrophages that have engulfed cell debris that may stain with the TUNEL reaction are sometimes found within germinal centers of animals harboring certain malignancies. However, high-power magnification of the spleen of Nr7 shows no phagocytic macrophages (Fig 3G). The localization of apoptotic bodies within the thymic cortex and lymphoid follicle of Nr mice strongly suggests that the cells undergoing programmed cell death were lymphoid; however, we cannot rule out the possibility that some other cell types were TUNEL-positive. Apoptosis of lymphocytes in Nr mice could account for the reduced numbers of lymphoid cells seen in the PB (Table 2) and the absence of lymphoid malignancies in Nr mice. Extensive apoptosis was also evident among transformed foci of myelomonocytic cells (Fig 3H). However, because we speculate that additional oncogenes were activated in these cells, we do not necessarily assume that apoptosis of transformed cells was a direct consequence of the LN-ras2 vector.
Apoptosis in LN-ras2 reconstituted mice. (A) LK3 thymocytes demonstrate normal morphology. (B) Nr9 thymocytes have condensed and fragmented nuclei (arrows). (C) LK2 TUNEL-stained thymus section. Small numbers of apoptotic cells (stained red) dispersed throughout the cortex are shown. (D) Nr9 TUNEL-stained thymus section. An elevation of apoptotic cells is shown. (E) LK3 TUNEL-stained spleen section. Small numbers of apoptotic cells are dispersed throughout the spleen. (F) Nr9 TUNEL-stained spleen section. Apoptotic cells clustered within lymphoid follicles are shown. (G) Spleen section from Nr9 showing morphology of apoptotic cells. (H) Nr16 TUNEL-stained section of intestinal node. Cytospun preparations (A and B) were stained with May-Grunwald and Giemsa (original magnification ×1,250). Tissue section (G) was stained with hematoxylin and eosin (original magnification ×1,250). TUNEL-stained tissue sections (C through F and H) apoptotic cells are stained red (original magnification ×125).
Apoptosis in LN-ras2 reconstituted mice. (A) LK3 thymocytes demonstrate normal morphology. (B) Nr9 thymocytes have condensed and fragmented nuclei (arrows). (C) LK2 TUNEL-stained thymus section. Small numbers of apoptotic cells (stained red) dispersed throughout the cortex are shown. (D) Nr9 TUNEL-stained thymus section. An elevation of apoptotic cells is shown. (E) LK3 TUNEL-stained spleen section. Small numbers of apoptotic cells are dispersed throughout the spleen. (F) Nr9 TUNEL-stained spleen section. Apoptotic cells clustered within lymphoid follicles are shown. (G) Spleen section from Nr9 showing morphology of apoptotic cells. (H) Nr16 TUNEL-stained section of intestinal node. Cytospun preparations (A and B) were stained with May-Grunwald and Giemsa (original magnification ×1,250). Tissue section (G) was stained with hematoxylin and eosin (original magnification ×1,250). TUNEL-stained tissue sections (C through F and H) apoptotic cells are stained red (original magnification ×125).
Impaired myelopoiesis and altered differentiation of LN-ras2–transduced BM progenitor cells.
To further investigate the apparent defect in myelopoiesis caused by expression of mutant N-ras, day-12 CFU-S generated from retrovirally transduced BM cells were evaluated. Mice transplanted with LN-ras2–transduced cells developed significantly fewer day-12 CFU-S (P = .001, n = 4, Student’s t-test) than mice transplanted with LK-infected cells. The average number of CFU-S in LN-ras2 and LK mice was 5 ± 0.8 and 10 ± 1.4, respectively. Thus, it appears that LN-ras2 impairs the multipotent myeloid progenitor cells that give rise to day-12 CFU-S. Because day-12 CFU-S confer short-term (6 weeks) radioprotection on irradiated hosts,39 it is likely that LN-ras2–mediated inhibition of myeloid progenitor cells acted to impair repopulation of Nr mice in the first reconstitution. Colony assays were also performed to assess the effects of mutant N-ras expression on myelopoiesis. The results in Table 4 demonstrate that BM cells infected with LN-ras2 generated fewer colonies than LK-infected BM (P = .01, Student’s t-test). The reduction in colony formation was highly significant for colony-forming unit granulocyte, erythroid, monocyte, megakaryocyte (CFU-GEMM) and colony-forming unit granulocyte, monocyte, megakaryocyte (CFU-GMM; P< .005). Colony-forming unit–granulocyte-macrophage (CFU-GM) generated by LN-ras2–infected BM was also significantly reduced (P = .05). No erythroid colonies were detected in LN-ras2 cultures. This may be due to defects in erythroid differentiation that were evident in vivo (Fig 2J and L). A similar observation was made by Darley et al,40 who also demonstrated that human erythroid progenitor cells transduced by a retroviral vector carrying activated N-ras have a drastically reduced ability to form colonies and fail to differentiate in vitro. The results from clonogenic assasy shown in Table 4 are consistent with our results from CFU-S and the first series of repopulation studies and further demonstrate that transduction by LN-ras2 inhibits proliferation of hematopoietic progenitor cells.
Proliferative Inhibition of Myeloid Progenitors Transduced With LN-ras2
| . | No. of Colonies4-150 . | ||||
|---|---|---|---|---|---|
| CFU-GM4-151 . | CFU-GMM‡ . | CFU-GEMM‡ . | Erythroid . | Total4-153 . | |
| LK | 171.3 ± 38.7 | 12.0 ± 3.3 | 8.0 ± 1.7 | 5.7 ± 2.2 | 192.5 ± 36.5 |
| LN-ras2 | 131.0 ± 8.7 | 5.7 ± 1.0 | 1.3 ± 1.8 | 0 | 137.0 ± 9.3 |
| . | No. of Colonies4-150 . | ||||
|---|---|---|---|---|---|
| CFU-GM4-151 . | CFU-GMM‡ . | CFU-GEMM‡ . | Erythroid . | Total4-153 . | |
| LK | 171.3 ± 38.7 | 12.0 ± 3.3 | 8.0 ± 1.7 | 5.7 ± 2.2 | 192.5 ± 36.5 |
| LN-ras2 | 131.0 ± 8.7 | 5.7 ± 1.0 | 1.3 ± 1.8 | 0 | 137.0 ± 9.3 |
Average calculated from triplicate plates in two distinct experiments.
P = .05.
P < .005.
P = .01.
Retrovirally transduced BM cells were also cultured in liquid media supplemented with WCM (as a source of IL-3). FACS analysis of nonadherent cells at day 27 showed reduced expression of mature myeloid markers on nonadherent LN-ras2 cells compared with LK cells (Fig 4A through D). Altered maturation of LN-ras2 cells was also evident by morphologic examination of colonies plucked from methlycellulose cultures (Fig 4E and F). After 4 weeks in liquid culture, mature macrophages predominated in colonies derived from LK cultures. In contrast, LN-ras2 colonies contained abundant immature monocytic cells. Thus, it appears that transduction with LN-ras2 impairs both differentiation and proliferation.
Altered differentiation of LN-ras2–transduced BM cells. (A through D) FACS analysis of transduced BM cells after 27 days in liquid culture. (A) LK cells stained with Mac-1 antibody (macrophage specific). (B) LK cells stained with Gr-1 (granulocyte specific). Underlayed (open curves represent isotype controls). (C) LN-ras2 cells stained with Mac-1. (D) LN-ras2 cells stained with Gr-1. (E and F) Cytocentrifuge preparations stained with May-Grunwald and Giemsa showing morphologic appearance of methylcellulose colonies after 7 days in liquid culture. View shows representative cells from pooled colonies from LK (E) and LN-ras2 (F) cultures (original magnification ×500).
Altered differentiation of LN-ras2–transduced BM cells. (A through D) FACS analysis of transduced BM cells after 27 days in liquid culture. (A) LK cells stained with Mac-1 antibody (macrophage specific). (B) LK cells stained with Gr-1 (granulocyte specific). Underlayed (open curves represent isotype controls). (C) LN-ras2 cells stained with Mac-1. (D) LN-ras2 cells stained with Gr-1. (E and F) Cytocentrifuge preparations stained with May-Grunwald and Giemsa showing morphologic appearance of methylcellulose colonies after 7 days in liquid culture. View shows representative cells from pooled colonies from LK (E) and LN-ras2 (F) cultures (original magnification ×500).
Molecular analysis of hematopoietic cells from reconstituted mice.
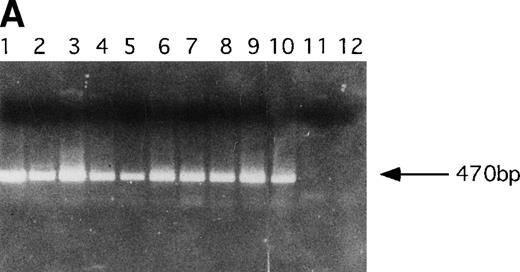
PCR analysis was performed to detect LN-ras2 and LK proviral integrants in retrovirally infected BM and reconstituted mice. Figure 5A shows PCR analysis performed using primers 1 and 2 (Fig 1A) to detect LN-ras2. LNras2 PCR products of the predicted size (470 bp) were amplified from ΨLN-ras215 DNA (lane 1) and from hematopoietic cells of all Nr mice that succumbed to hematopoietic disorders (examples shown in lanes 2 through 9). LN-ras2 sequence was also detected in BM cells that were used for in vitro experiments (lane 10). An LK fragment of the expected size was amplified from ΨLK09 DNA from cells from the LK reconstituted mice (data not shown).
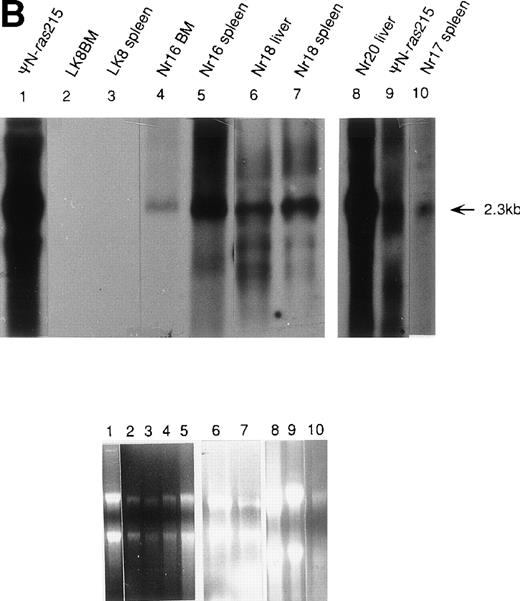
Molecular analysis of reconstituted mice. (A) Proviral integrants. Ethidium bromide-stained agarose gel showing PCR products amplified from lysates of cells from reconstituted mice or BM cultures. Primers 1 and 2 (Fig 1) were used for amplification of LN-ras2 sequence. Lane 1, ΨLN-ras215 (positive control); lane 2, Nr1 spleen; lane 3, Nr4 thymus; lane 4, Nr5 spleen; lane 5, Nr7 spleen; lane 6, Nr7 lung; lane 7, Nr8 lung; lane 8, Nr8 spleen; lane 9, Nr9 lung; lane 10, LN-ras2–infected BM (in vitro analysis); lane 11, negative control DNA; lane 12, no sample. (B) Proviral expression. Autoradiograph of a Northern blot hybridized to a Ψ region probe (described in Materials and Methods). RNA samples are indicated above each well. Transcript sizes are indicated to the right. The RNA samples on ethidium bromide-stained agarose gels are shown below the autoradiograph. Lane numbers above the gel correspond to the autoradiograph.
Molecular analysis of reconstituted mice. (A) Proviral integrants. Ethidium bromide-stained agarose gel showing PCR products amplified from lysates of cells from reconstituted mice or BM cultures. Primers 1 and 2 (Fig 1) were used for amplification of LN-ras2 sequence. Lane 1, ΨLN-ras215 (positive control); lane 2, Nr1 spleen; lane 3, Nr4 thymus; lane 4, Nr5 spleen; lane 5, Nr7 spleen; lane 6, Nr7 lung; lane 7, Nr8 lung; lane 8, Nr8 spleen; lane 9, Nr9 lung; lane 10, LN-ras2–infected BM (in vitro analysis); lane 11, negative control DNA; lane 12, no sample. (B) Proviral expression. Autoradiograph of a Northern blot hybridized to a Ψ region probe (described in Materials and Methods). RNA samples are indicated above each well. Transcript sizes are indicated to the right. The RNA samples on ethidium bromide-stained agarose gels are shown below the autoradiograph. Lane numbers above the gel correspond to the autoradiograph.
Northern analysis and reverse transcriptase-PCR (RT-PCR) were performed to investigate proviral expression within the reconstituted animals. Representative results from Northern analyses are shown in Fig 5B. A 2.3-kb transcript was detected in samples from the Nr reconstituted mice (lanes 4 through 8 and 10), indicating expression of full-length LN-ras2 in hematopoietic organs of these mice. In the analysis shown in Fig 5B, no LK-derived transcript was detected in the spleen or BM of the LK8 mouse (lanes 2 and 3). This may be a consequence of the lower titer of LK compared with LN-ras2. The lower titer of LK decreases the ability of this virus to transduce long-term repopulating cells. However, LK-derived transcripts were detected by RT-PCR in spleen and thymus of the majority of the LK mice that were tested. Also, LK-derived PCR products were detected in some LK mice 385 days after transplantation (data not shown).
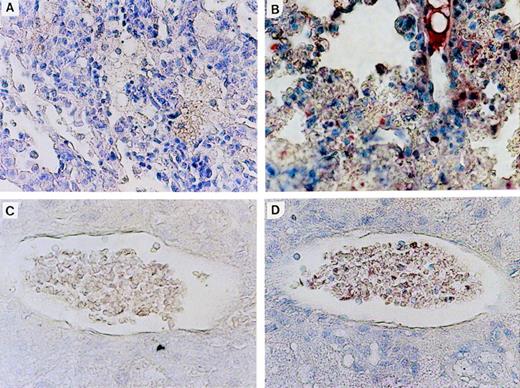
Expression of N-ras protein in Nr mice was confirmed by immunohistochemical staining of tissue sections with an N-ras–specific antibody. Figure 6shows N-ras protein expression within mononuclear cells in the PB and lung of mice with myeloproliferative disorders. Expression of human N-ras protein was also detected in LN-ras2–infected BM cells used for in vitro analyses (data not shown).
N-ras protein expression in reconstituted mice. Immunohistochemical staining with human N-ras antibody. (A) LK8 lung. (B) Nr16 lung. Stained hematopoietic cells are shown. (C) LK 8 blood vessel in liver. (D) Nr18 blood vessel in liver. Stained mononuclear cells within a blood vessel are shown.
N-ras protein expression in reconstituted mice. Immunohistochemical staining with human N-ras antibody. (A) LK8 lung. (B) Nr16 lung. Stained hematopoietic cells are shown. (C) LK 8 blood vessel in liver. (D) Nr18 blood vessel in liver. Stained mononuclear cells within a blood vessel are shown.
DISCUSSION
In this report, expression of mutant N-ras was shown to impair myelopoiesis as well as promote leukemogenesis and induce apoptosis. Maturation of LN-ras2–transduced myeloid cells was shown to be perturbed in in vitro assays and aberrant erythroid differentiation was evident from blood smears of transplanted mice. LN-ras2–transduced BM cells also exhibited an impaired ability to reconstitute lethally irradiated mice. This defect was only partially overcome by the inclusion of KL in addition to IL-6 and IL-3 in the retroviral infection cocktail. Cells transduced with LN-ras2 in the presence of KL rescued mice from irradiation but generated fewer day-12 CFU-S in vivo and fewer myeloid and erythroid colonies in vitro than BM transduced by the control vector. The inclusion of KL in the infection media may have influenced pathologic outcome by either promoting survival of transduced cells or by altering the transduction efficiency and/or target cell populations of LN-ras2. Further investigations could explore these possibilities.
The myelopoietic defect associated with LNras2 transduction is contrary to previous demonstrations of ras-induced growth enhancement of hematopoietic cell lines.19,21,41 This apparent discrepancy may reflect differental effects of ras expression in primary and immortalized cells. It was previously shown that, althoughras transforms rodent fibroblast cell lines, coexpression of an additional oncogene is necessary for ras-induced transformation of primary rat fibroblasts.42,43 Indeed, in the absence of a cooperating oncogene, ras induces cell cycle arrest of primary fibroblasts.44 Similarly, N-ras expression in primary human erythroblasts appears to slow cell cycling and thereby inhibit proliferation and alter differentiation.40 A recent report also indicates that ras signal transduction pathways can induce G1 cell cycle arrest by upregulating p21cip/waf1.45 Our results are consistent with these studies and suggest that N-ras expression may have impaired myelopoiesis by altering cell cycle regulation. Additional genetic alterations, such as those that confer an immortal phenotype, appear necessary for ras-mediated transformation of hematopoietic cells.
Several Nr mice developed hematopoietic disorders that resembled human MPD, such as those seen in CML, which are characterized by excessive proliferation of myeloid cells that continue to differentiate.46 However, in comparison to the MPD seen in human CML, the pathology of the Nr mice was more aggressive; nonhematopoietic organs were infiltrated and extramedullary tumors composed of myelomonocytic cells arose. Hence, with regard to aggressiveness, the myeloid disorders in Nr mice were also similar to AML with differentiation (French-American-British [FAB] classified M2).47 It is significant that N-ras codon 12 mutations are detected at a relatively high frequency in both human AMLs9,17 and in bcr-abl–negative CML.11 N-ras mutations are also frequent in human MDS and correlate with a high risk of leukemic transformation.48,49 MDSs are preleukemic conditions that arise as a consequence of BM dysfunction and are characterized by anemia, erythroid hyperplasia, and, in some cases, BM hypoplasia and apoptosis.50 51 Because similar disorders were characterized in Nr7, Nr8, and Nr9, the myeloid impairment induced by expression of mutant N-ras in these mice may describe an underlying defect in human MDS.
The neoplastic disorders characterized in the Nr mice are phenotypically similar to MPDs that developed in bothbcr-abl–reconstituted mice52-54 and mice that were transplanted with hematopoietic cells from NF1-null mice.55bcr-abl–reconstituted mice also succumbed to erythroid disorders that were sometimes coincident with mast cell accumulations and granulocytosis,56 an apparently similar disorder to the pathology of Nr17 in this study. Becauseras-dependent pathways are necessary forbcr-abl–induced oncogenesis12,13 and are also activated in JCML as a consequence of loss of the NF1gene,14,15 these studies suggest that N-rasactivation, bcr-abl translocation, and NF1 loss have some redundant functions in myeloid cell transformation. The absence ofras mutations in myeloid leukemic cells with either thebcr-abl translocation11 or NF1 gene loss57 is consistent with this hypothesis.
This is the first report of a relevant in vivo model for N-ras–induced MPD or myelomonocytic leukemia. The long latencies indicate a predisposing, yet nonetheless causal, role for N-ras in these pathologies. Previously, in two independent investigations, BM repopulated mice that carried mutationally activated Ha-ras developed thymic lymphomas20,22 and B-cell leukemias.22 The development of lymphoid rather than myeloid disorders in the Ha-ras mice may be due to the different transformation abilities of Ha-ras versus N-ras in hematopoietic cells23 or a consequence of inefficient expression of the Ha-ras within the myeloid compartment. Although the retroviral vector used in the investigation of Dunbar et al20 was expressed within the thymus, no vector expression was detected in the spleen or BM of the reconstituted mice. Regardless of the reason for the lymphoid phenotype, the significance of these Ha-ras investigations is unclear, because Ha-ras mutations are not detected in human hematopoietic disorders.8 The development of lymphomas in transgenic mice expressing activated N-ras24,25 is probably a reflection of the specificity of the different transgene promoters rather than an indication of the transformation specificity of N-ras. In the study by Harris et al,24 transgenic expression was directed by an Ig heavy chain (Eμ) enhancer that operates most efficiently in lymphoid cells rather than in myeloid cells.58 The mouse mammary tumor virus (MMTV) promoter used by Mangues et al25 is primarily expressed in epithelial cells and is not as efficient in hematopoietic cells.59
Apoptosis of lymphocytes may also account for the absence of lymphoid leukemias in LN-ras2–reconstituted mice. A high level of apoptosis was observed within the thymus and spleen of mice that succumbed to hematopoietic deficiencies. PB lymphocytes were scarce in these animals. Because LN-ras2 vector sequences were detected in both thymus and spleen of Nr mice and elevated apoptosis was not seen in any of the control mice, apoptotic cell death within these organs appears to be a consequence of N-ras expression. Other investigators have provided evidence of ras-induced apoptosis of embryonic fibroblasts60 and neuronal cells61 in vitro. Neuronal apoptosis was also demonstrated in mice with a null mutation in the gene encoding p120-rasGAP, a negative regulator ofras signal transduction pathways.62 Recently, erythroblasts expressing mutant N-ras were also shown to be susceptible to apoptosis.40 The present investigation extends these findings by providing in vivo evidence for N-rasinduced apoptosis of hematopoietic cells.
In summary, these studies demonstrate that mutant N-rasexpression impairs myelopoiesis and erythopoiesis and confers susceptibility to hematopoietic disorders that resemble human MDS, MPD, and AML. A long latency was required for the development of invasive MPD and overt leukemia, suggesting that activation of additional oncogenes was necessary for the evolution of these disorders. The N-ras–associated myeloid impairment may represent a preleukemic phenotype such as MDS. Extensive apoptosis was also observed in mice reconstituted with BM cells expressing mutant N-ras. Our results suggest that the primary consequences of N-ras mutations are antiproliferative and that secondary transforming events are necessay for the development of myeloid leukemia. These studies are consistent with the multistep hypothesis for the development of leukemia and provide the first relevant in vivo model of N-ras–induced myeloid disorders.
ACKNOWLEDGMENT
The authors thank C. Der for the pZip-N-ras plasmid, W. Gerlach (Johnson and Johnson Research Laboratories) for access to facilities, and A. Todd (Johnson and Johnson Research Laboratories) and P.B. Rowe (Children’s Medical Research Institute) for helpful discussion. We also thank Terry Rothwell (Sydney University) for expert advice on histopathology and Julie Ferguson (Garvan Institute) for animal husbandry.
Supported by a project grant from the National Health and Medical Research Council of Australia. G.S. is an NHMRC Senior Research Fellow.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to G. Symonds, PhD, Johnson and Johnson Research Laboratories, GPO BOX 3331, Sydney, NSW 2001, Australia.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal