Abstract
Processing of pre-miRNA through Dicer1 generates an miRNA duplex that consists of an miRNA and miRNA* strand. Despite the general view that miRNA*s have no functional role, we further investigated miRNA* species in 10 deep-sequencing libraries from mouse and human tissue. Comparisons of miRNA/miRNA* ratios across the miRNA sequence libraries revealed that 50% of the investigated miRNA duplexes exhibited a highly dominant strand. Conversely, 10% of miRNA duplexes showed a comparable expression of both strands, whereas the remaining 40% exhibited variable ratios across the examined libraries, as exemplified by miR-223/miR-223* in murine and human cell lines. Functional analyses revealed a regulatory role for miR-223* in myeloid progenitor cells, which implies an active role for both arms of the miR-223 duplex. This was further underscored by the demonstration that miR-223 and miR-223* targeted the insulin-like growth factor 1 receptor/phosphatidylinositol 3-kinase axis and that high miR-223* levels were associated with increased overall survival in patients with acute myeloid leukemia. Thus, we found a supporting role for miR-223* in differentiating myeloid cells in normal and leukemic cell states. The fact that the miR-223 duplex acts through both arms extends the complexity of miRNA-directed gene regulation of this myeloid key miRNA.
Introduction
The canonical miRNA biogenesis pathway involves the stepwise processing of miRNA precursor transcripts that contain hairpin structures in the nucleus and in the cytoplasm.1 After processing through Drosha in the nucleus, miRNA-containing hairpins are exported into the cytoplasm and cleaved by Dicer, which results in an ∼ 21-25 nt miRNA duplex.2 Although both strands of miRNA duplexes are necessarily produced in equal amounts by enzymatic processing, their accumulation is mainly asymmetric at steady state. Depending on its frequency, the most abundant strand of a processed pre-miRNA is referred as miRNA, whereas the less abundant strand is known as “passenger strand” or miRNA* (read miRNA star),3-5 initially annotated by the first miRNA sequencing approaches.6-9 Although the mechanism of miRNA strand selection and RNA-induced silencing complex (RISC) loading are still unclear, studies on small interfering RNA (siRNA) duplexes indicated that the relative thermodynamic stability of the 2 ends of the duplex determines which strand is to be selected.3,4 The strand with relatively unstable base pairs at the 5′ end typically evades degradation.3,4 Thermodynamic stability profiling studies on miRNA precursors suggested that the same rule might apply to most, although not all, miRNAs.3
Recent profiling approaches from our group10-12 and others13,14 aimed at detecting and quantifying small RNA species, demonstrated not only the presence of miRNA* strands across species but also their high abundance for certain miRNA duplexes. The incorporation of miRNA*s into the RISC was shown recently in Drosophila melanogaster.15,16 Moreover, luciferase reporter assays have provided evidence for the functionality of both miRNA strands.17 Furthermore, single-nucleotide polymorphisms identified within the pre–miR-146 hairpin and seed region of miR-146* are purported to change its target specificity.18,19
When one considers that every miRNA duplex consists of an miRNA and miRNA*, it is interesting to note that only 8.1% of miRNA*s (80 miRNA*s of 940 miRNAs, miRBase release 15.0) in the human genome have been annotated. This is likely due in part to the low expression levels of certain miRNA*s, as well as the fact that miRNA* sequences have only recently been added to miRNA microarrays and TaqMan probe libraries. The advent of next-generation sequencing has dramatically increased the ability to sensitively, comprehensively, and quantitatively assess the pattern of miRNA and miRNA* species. To this end, we analyzed 10 deep-sequencing libraries derived from different model organisms, solid tumors, and in particular, leukemias to determine the expression levels of known and nonannotated miRNA*s compared with their cognate miRNA strands. This comprehensive approach allowed us to identify tissue- and species-independent patterns of miRNA/miRNA* expression, which suggested a novel classification for miRNA duplexes. Differential ratios of miRNA to miRNA* were also revealed, as illustrated by the dramatic differential expression of miR-223 and miR-223* in normal myeloid and pathophysiologic myeloid conditions.
Methods
Small RNA library preparation
Previously published libraries for undifferentiated and differentiated H9 human embryonic stem cells and murine ND13 (mouse leukemia 1) and ND13+Meis1 (mouse leukemia 2) cell lines were reannotated according to miRBase release 13.0.10,11 Furthermore, 3 human leukemia cell lines (leukemia 1-3), 2 libraries derived from human prostate cancer tissue (prostate 1 and prostate 2), and 1 library derived from a human colon cancer cell line (colon) were generated as published previously.10 Short RNA sequences from each library were aligned to the human genome (NCBI36) for human-derived libraries and to the mouse genome (NCBIM37) for murine-derived libraries. Small RNA sequences were filtered by considering only those sequences with alignments that overlapped with mature miRNA or miRNA* annotations as found in miRBase release 13.0. Sequence counts for each miRNA and miRNA* were considered to be the counts of the total number of sequences in the library as described previously.10,11 In some cases, we found evidence of the expression of an miRNA* sequence with no annotation in miRBase. Expression for unannotated miRNA* sequences was calculated as the count for the most abundant sequence that was approximately reverse complementary with the miRNA sequence. To qualify as a potential miRNA*, a sequence required an alignment to the region of the stem loop complementary to the mature miRNA sequence, ± 5 nucleotides at each end. Ratios between miRNA and miRNA* were calculated as (miRNA counts + 1):(miRNA* counts + 1), which provided a well-defined measurement for the possibility of miRNA* sequences with no stable expression. See supplemental Table 5 for the total number of reads and alignment efficacy (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Retroviral vectors and cDNA
All murine miR-223 constructs (miR-223, miR-223mut, and miR-223*mut) were ordered from Integrated DNA Technologies (IDT; www.idtdna.com) and cloned into a murine stem cell virus (MSCV) construct as described previously.20 In all constructs, the published internal ribosome entry-site (IRES) cassette was replaced with a phosphoglycerate kinase promoter sequence, which drove enhanced green fluorescent protein expression. All construct sequences are listed in supplemental Methods (Experimental Procedures).
Retroviral infection and clonogenic progenitor assay
Whole mouse BM from miR-223–deficient (knockout [KO]) mice and wild-type (WT) mice21 was extracted, lysed with NH4Cl, and stimulated for 48 hours in DMEM supplemented with 15% FBS, 10 ng/mL human IL-6, 6 ng/mL mouse IL-3, and 100 ng/mL mouse SCF (StemCell Technologies Inc). The cells were transduced by cocultivation with irradiated (4000 cGy) viral producers in the presence of 5 μg/mL protamine sulfate (Sigma-Aldrich) for 48 hours. Green fluorescent protein–positive cells were FACS sorted into 15-mL falcon tubes that contained 3 mL of methylcellulose (Methocult M3434; StemCell Technologies) supplemented with cytokines as described previously.22 The gene transfer ranged from 2%-5%. Colonies were counted and evaluated 7 days after plating by standard criteria.
Real-time PCR
RNA was extracted with TRIzol (Invitrogen) as described previously.11 Reverse transcription of each miRNA or sno-202 was performed with the TaqMan miRNA reverse transcription kit (Applied Biosystems, Inc [ABI]) according to the manufacturer's instructions. Each patient RNA sample was measured with a NanoDrop instrument (Thermo Scientific) and equilibrated to a concentration of 5 ng/mL. Twenty nanograms of RNA was used for miR-223*–specific reverse transcription. Five nanograms of RNA from a healthy BM donor was used as standard to calculate ΔΔCt values and fold changes. MiR-92 was used as a housekeeping gene for all human samples23 and sno-202 for murine samples. Quantitative RT-PCR was performed with miR-223 (ABI No. 4395406), miR-223* (ABI No. 4395209), miR-92 (ABI No. 4373013), and sno-202 (ABI No. 4380914) ABI TaqMan probes on an ABI 7900HT fast real-time PCR system in triplicates. Reverse transcription of total mRNA was performed with random primers and Superscript III (Invitrogen) as described previously.24 Real-time PCR on coding genes was performed with SYBR green (Invitrogen) as described previously.24 All primers are listed in supplemental Table 6.
Luciferase assays
Predicted (TargetScan custom version 5.0, hsa-miR-223* seed: GUGUAUU) miR-223*–binding regions of insulin-like growth factor 1 receptor (IGF1R; chr15:99501790-99502039) were cloned into pMIR-REPORT (Ambion) and transfected with hsa-miR-223* (Thermo Scientific Dharmacon), a scrambled control (Ambion), or a negative control miRNA (Ambion) into 293T cells. For the 3′UTR-luciferase assays, 20 ng of pMirReport-3′UTR, 10 pmol of miRNAs, and 0.17 ng of thymidine kinase–Renilla were cotransfected into 4 × 105 293T cells (48-well format) with Lipofectamine 2000 transfection reagent (Invitrogen). The assays were read in a Lumat LB 9507 tube luminometer (EG&G Berthold), and the luciferase/Renilla ratio was calculated. Student t test was used for statistical analysis, and P < .05 was considered as significant.
Retroviral infection of murine BM cells
Total BM from miR-223 KO (mir-223−/−) and WT (miR-223+/+) mice21 was extracted and prestimulated for 2 days as described previously.24 Then, the BM cells were simultaneously cotransduced with MSCV-Hoxa9-pgk-neomycin and an MSCV-Meis1-IRES-YFP by cocultivation with irradiated (4000 cGy) viral producers as described previously.24 HoxA9-Meis1–transduced cells were sorted for yellow fluorescent protein expression and continuously selected with neomycin (1.4 mg/mL).
Microarrays
RNA from the above-described BM cell lines was extracted with TRIzol (Invitrogen) as described previously.11 The extracted RNA was further cleaned with the miRNA easy kit (QIAGEN) according to the manufacturer's instructions. The Affymetrix mouse gene 1.0 ST array was used to generate gene expression data on 2 biologic replicates. Gene expression array data (CEL files) was analyzed with GeneSpring GX 11.5.1 software (Agilent Technologies). The exon RMA-16 algorithm and quantile normalization were used, and the median of all samples in each experimental setting was used for baseline transformation. Genes were then filtered according to expression of the 20th-100th percentile of the raw data and tested for significance (1-way Welch ANOVA, corrected P value cutoff = .05). MIAME (minimal information about a microarray experiment) guidelines were followed for data presentation. All microarray data are available at the Gene Expression Omnibus (GEO) under accession No. GSE29453.
Statistical analysis
Statistical analysis for overall survival and correlation of clinical parameters was performed as described previously.25 The definition of overall survival followed recommended criteria.26 In brief, acute myeloid leukemia (AML) samples were dichotomized at the median and the 25th and 75th percentiles of the miR-223* real-time expression value. The Kaplan-Meier method and log-rank test were used to estimate the distribution of overall survival and to compare differences between survival curves. Pairwise comparisons were performed by Student t test for continuous variables and by χ2 test for categorical variables. The 2-sided level of significance was set at P < .05. Statistical analyses were performed with the statistical software package SPSS 15.0 (SPSS Inc).
Further details about patient collectives and performed methods can be found in the experimental procedures detailed in supplemental Methods. For all patients, written informed consent was obtained before therapy according to the Declaration of Helsinki, and the study was approved by the institutional review board of Hannover Medical School.
Results
The abundance of miRNA*s is tissue dependent
We investigated the expression and relationship of miRNAs and their corresponding miRNA*s in 10 Illumina sequencing libraries derived from murine and human cell lines and tissues. In detail, we analyzed the miRNA expression profile of 2 murine hematopoietic libraries, a mouse preleukemic myeloid cell line (mouse leukemia 1; NUP98-HOXD13 [ND13]) and its leukemic counterpart (mouse leukemia 2; ND13+Meis1), derived from transformed mouse BM cells with an NUP98-HOXD13 fusion gene and the HOX cofactor Meis1.11 In addition, we analyzed undifferentiated and differentiated embryonic stem cell libraries,10 3 common human leukemia cell lines (human leukemia 1-3),27 2 libraries derived from human prostate cancer tissue (prostate 1 and prostate 2), and 1 library derived from a human colon cancer cell line (colon). The miRNA expression profile of each library was analyzed. The expression level of a mature miRNA or miRNA* was calculated by the total number of sequences in the library that found a best-hit alignment to that miRNA or miRNA*.
All libraries were analyzed according to the sum of all sequences10,11 and only miRNA/miRNA* pairs that originated from the same stem-loop structure were considered for analysis if either the miRNA or miRNA* showed an expression level ≥ 100. The varying sequencing depths of the libraries ranged from 666 362 to 4 661 901 miRNA reads and from 5273 to 130 652 miRNA* reads (Table 1).
Top 5 of the highest expressed annotated miRNA*s in miRBase from each library
| . | Mouse leukemia 1 . | Mouse leukemia 2 . | ES undiff . | ES diff . | Leukemia 1 . | Leukemia 2 . | Leukemia 3 . | Prostate 2 . | Prostate 1 . | Colon . |
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA/miRNA* reads† | 819 023/25 750 | 666 362/29 718 | 1 062 508/130 652 | 1 275 288/51 895 | 872 298/10 202 | 1 730 532/30 466 | 13 28 614/6483 | 1 251 565/5273 | 2 357 704/7679 | 4 661 901/42 458 |
| % miRNA* | 3.1 | 4.5 | 12.3 | 4.1 | 1.2 | 1.8 | 0.5 | 0.4 | 0.3 | 0.9 |
| Top 5 miRNA*† | mmu-miR-140* | mmu-miR-140* | hsa-miR-302a* | hsa-miR-302a* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-25* | hsa-miR-30a* | hsa-miR-221* | hsa-miR-221* |
| mmu-miR-30e* | mmu-miR-30e* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-493* | hsa-miR-223* | hsa-miR-374a* | hsa-miR-25* | hsa-miR-25* | hsa-miR-30e* | |
| mmu-miR-322* | mmu-miR-322* | hsa-miR-25* | hsa-miR-30a* | hsa-miR-181a* | hsa-miR-92a-1* | hsa-miR-30e* | hsa-miR-223* | hsa-miR-21* | hsa-miR-106b* | |
| mmu-miR-106b* | mmu-miR-106b* | hsa-miR-92b* | hsa-miR-25* | hsa-miR-221* | hsa-miR-17* | hsa-miR-221* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-374a* | |
| mmu-miR-27b* | mmu-miR-17* | hsa-miR-221* | hsa-miR-92b* | hsa-miR-25* | hsa-miR-25* | hsa-miR-223* | hsa-miR-221* | hsa-miR-155* | hsa-miR-21* |
| . | Mouse leukemia 1 . | Mouse leukemia 2 . | ES undiff . | ES diff . | Leukemia 1 . | Leukemia 2 . | Leukemia 3 . | Prostate 2 . | Prostate 1 . | Colon . |
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA/miRNA* reads† | 819 023/25 750 | 666 362/29 718 | 1 062 508/130 652 | 1 275 288/51 895 | 872 298/10 202 | 1 730 532/30 466 | 13 28 614/6483 | 1 251 565/5273 | 2 357 704/7679 | 4 661 901/42 458 |
| % miRNA* | 3.1 | 4.5 | 12.3 | 4.1 | 1.2 | 1.8 | 0.5 | 0.4 | 0.3 | 0.9 |
| Top 5 miRNA*† | mmu-miR-140* | mmu-miR-140* | hsa-miR-302a* | hsa-miR-302a* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-25* | hsa-miR-30a* | hsa-miR-221* | hsa-miR-221* |
| mmu-miR-30e* | mmu-miR-30e* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-493* | hsa-miR-223* | hsa-miR-374a* | hsa-miR-25* | hsa-miR-25* | hsa-miR-30e* | |
| mmu-miR-322* | mmu-miR-322* | hsa-miR-25* | hsa-miR-30a* | hsa-miR-181a* | hsa-miR-92a-1* | hsa-miR-30e* | hsa-miR-223* | hsa-miR-21* | hsa-miR-106b* | |
| mmu-miR-106b* | mmu-miR-106b* | hsa-miR-92b* | hsa-miR-25* | hsa-miR-221* | hsa-miR-17* | hsa-miR-221* | hsa-miR-30e* | hsa-miR-30e* | hsa-miR-374a* | |
| mmu-miR-27b* | mmu-miR-17* | hsa-miR-221* | hsa-miR-92b* | hsa-miR-25* | hsa-miR-25* | hsa-miR-223* | hsa-miR-221* | hsa-miR-155* | hsa-miR-21* |
ES undiff indicates undifferentiated embryonic stem cells; and ES diff, differentiated embryonic stem cells.
Only annotated miRNA*s.
Across all libraries, the percentage of all detected miRNA* species compared with all detected miRNAs varied from 0.3% to 12.3% (Table 1), as pictured for embryonic stem cells in supplemental Figure 1A, which suggests tissue-dependent expression levels of miRNA*s. The highest percentage of miRNA* species was detected in the undifferentiated embryonic stem cell library (12.3%), whereas the lowest percentage was detected in the prostate 2 library (0.3%; Table 1). Classification of the miRNA/miRNA* ratio into groups (Table 2) showed that ∼ 50% of all miRNA duplexes (range 40.7%-61.2%) revealed high ratios (> 100) consistent with strong preferential processing of 1 dominant miRNA strand. A significant proportion, ∼ 24% (range 18.1%-30.9%), had intermediate ratios (between 100 and 10), and strikingly, ∼ 13% (range 8.6%-17.7%) showed low ratios (between 10 and 1). In addition, ∼ 13% (range 7.1%-19.1%) showed inverted ratios (< 1; Table 2). These findings oppose the general assumption that only 1 strand is highly dominant for any given miRNA duplex. Moreover, high strand abundance also was detected for some members in the low-ratio groups, which indicates a weak correlation between strand abundance and miRNA duplex ratio (supplemental Figure 1B). In addition, the observation that > 10% of all miRNA duplexes displayed an inverse ratio is indicative that incorrect annotations are documented in miRBase (see supplemental Table 1 for all ratios across all libraries, supplemental Table 2 for all ratios with > 100 sequence counts, and supplemental Table 3 for all sequences and sequence counts for each individual library).
Distribution (%) of the various miRNA/miRNA* ratio groups in each library
| % of all ratios . | Mouse leukemia 1 . | Mouse leukemia 2 . | ES undiff . | ES diff . | Leukemia 1 . | Leukemia 2 . | Leukemia 3 . | Prostate 1 . | Prostate 2 . | Colon . | Breast . | Average for all libraries . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| > 100 | 47.9 | 50.9 | 42.0 | 43.4 | 40.7 | 47.0 | 60.6 | 61.2 | 56.4 | 54.2 | 50.5 | 50.4 |
| 100-10 | 24.4 | 25.4 | 30.9 | 27.4 | 25.2 | 28.5 | 22.2 | 18.1 | 20.8 | 19.6 | 18.2 | 23.7 |
| < 10 | 12.6 | 123 | 14.0 | 15.1 | 15.0 | 14.6 | 10.1 | 8.6 | 10.7 | 11.7 | 17.7 | 12.9 |
| < 1 | 15.1 | 11.4 | 13.1 | 13.9 | 19.1 | 9.9 | 7.1 | 12.1 | 12.1 | 14.5 | 13.6 | 12.9 |
| % of all ratios . | Mouse leukemia 1 . | Mouse leukemia 2 . | ES undiff . | ES diff . | Leukemia 1 . | Leukemia 2 . | Leukemia 3 . | Prostate 1 . | Prostate 2 . | Colon . | Breast . | Average for all libraries . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| > 100 | 47.9 | 50.9 | 42.0 | 43.4 | 40.7 | 47.0 | 60.6 | 61.2 | 56.4 | 54.2 | 50.5 | 50.4 |
| 100-10 | 24.4 | 25.4 | 30.9 | 27.4 | 25.2 | 28.5 | 22.2 | 18.1 | 20.8 | 19.6 | 18.2 | 23.7 |
| < 10 | 12.6 | 123 | 14.0 | 15.1 | 15.0 | 14.6 | 10.1 | 8.6 | 10.7 | 11.7 | 17.7 | 12.9 |
| < 1 | 15.1 | 11.4 | 13.1 | 13.9 | 19.1 | 9.9 | 7.1 | 12.1 | 12.1 | 14.5 | 13.6 | 12.9 |
ES undiff indicates undifferentiated embryonic stem cells; and ES diff, differentiated embryonic stem cells.
miRNA*s can be classified according to their abundance in relation to the corresponding miRNA
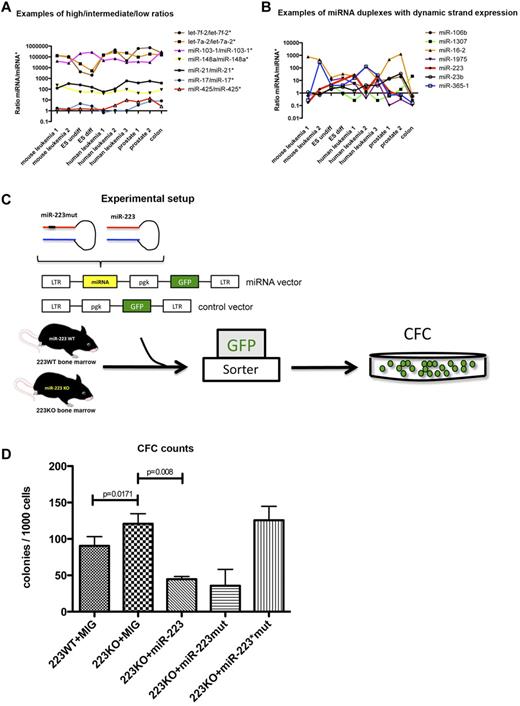
On the basis of the finding that the distribution of miRNA/miRNA* ratios was grouped similarly in all libraries, we questioned whether the ratios for individual miRNA duplexes remained constant or changed across all investigated tissues. Indeed, most miRNA duplexes preserved their miRNA/miRNA* distribution across the different libraries (Figure 1A; supplemental Tables 1-3). On this basis, it is possible to classify miRNA duplexes into α-duplexes having a dominant strand with a ratio > 10, such as the let-7 family, and β-duplexes exhibiting relatively balanced ratios ≥ 0.1 and ≤ 10, spanning ∼ 1 order of magnitude, such as miR-17 and miR-425 (Figure 1A; Table 3). However, 7 miRNAs were characterized by a dynamic arm expression, with ratios ranging between > 10 and < 1 and therefore interchanging dominant miRNA arms between the different tissues (Figure 1B; Table 4). This phenomenon was independent of conservation, as demonstrated by oscillating miRNA arm expression between the investigated libraries for both poorly conserved miRNA duplexes such as miR-1307 and broadly conserved duplexes such as miR-223 (supplemental Figure 1D).
Deep-sequencing profile of 10 miRNA libraries. (A) Examples of miRNA/miRNA* duplexes with high, intermediate, and low ratios across all tissues. The x-axis indicates all sequencing libraries, and the y-axis indicates all calculated miRNA:miRNA* ratios. (B) Examples of miRNA duplexes with dynamic arm expression. The x-axis indicates all sequencing libraries, and the y-axis indicates all calculated miRNA:miRNA* ratios. (C) Experimental setup to test the activity of miR-223 and miR-223* in colony-forming cell assays. (D) Colony counts for each experimental arm (n = 3). ES undiff indicates undifferentiated embryonic stem cells; ES diff, differentiated embryonic stem cells; LTR, long terminal repeat; pgk, phosphoglycerate kinase; GFP, green fluorescent protein; CFC, colony-forming cell; and MIG, MSCV (murine stem-cell virus)–IRES-GFP.
Deep-sequencing profile of 10 miRNA libraries. (A) Examples of miRNA/miRNA* duplexes with high, intermediate, and low ratios across all tissues. The x-axis indicates all sequencing libraries, and the y-axis indicates all calculated miRNA:miRNA* ratios. (B) Examples of miRNA duplexes with dynamic arm expression. The x-axis indicates all sequencing libraries, and the y-axis indicates all calculated miRNA:miRNA* ratios. (C) Experimental setup to test the activity of miR-223 and miR-223* in colony-forming cell assays. (D) Colony counts for each experimental arm (n = 3). ES undiff indicates undifferentiated embryonic stem cells; ES diff, differentiated embryonic stem cells; LTR, long terminal repeat; pgk, phosphoglycerate kinase; GFP, green fluorescent protein; CFC, colony-forming cell; and MIG, MSCV (murine stem-cell virus)–IRES-GFP.
Examples of α- and β-duplexes
| . | α-duplexes . | β-duplexes . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio (miRNA/miRNA*) | let-7a | miR-103 | miR-320a | miR-107 | miR-101 | miR-30e | miR-17 | miR-30a | miR-32 | miR-625 |
| Mouse leukemia 1 | 15773.5 | 40465 | 9354 | 21195 | 1881 | 0.754216867 | 1.703703704 | N/E | N/E | N/E |
| Mouse leukemia 2 | 31822.66667 | 26642 | 5505 | 16623 | 1237 | 0.828804348 | 1.508274232 | N/E | N/E | N/E |
| ES undiff | 11179 | 207069 | 25214 | 20119 | 8012 | 0.141397289 | 4.563953488 | 2.751046025 | 0.888888889 | 0.581699346 |
| ES diff | 4986 | 269177 | 2228 | 19099 | 3880 | 0.365884431 | 2.691432904 | 3.246661102 | 1.636363636 | 0.520958084 |
| Leukemia 1 | 6349 | 46489 | 37114 | 32102 | 19329 | 0.013737836 | 0.655882353 | 0.5 | 3 | 1.474358974 |
| Leukemia 2 | 5364.58 | 68834 | 8378.5 | 17958 | 6974 | 1.716995682 | 1.081261596 | 2.735955056 | 5.538461538 | 0.785714286 |
| Leukemia 3 | 2701.321839 | 31017 | 3095 | 11571 | 5629 | 0.849372385 | 1.05952381 | 0.230769231 | 3.75 | 0.388888889 |
| Prostate 1 | 47738 | 35289 | 2782 | 7347 | 2555 | 0.235474006 | 3.344827586 | 2.492537313 | 2.166666667 | 1.4 |
| Prostate 2 | 14021.92308 | 31233 | 2090 | 9151 | 1676 | 0.153729072 | 7.416149068 | 1.145530146 | 1.8 | 6.428571429 |
| Colon | 16389.34043 | 309168 | 109730 | 6041 | 25246 | 0.974157734 | 8.263414634 | 3.492307692 | 1.6 | 1.125 |
| . | α-duplexes . | β-duplexes . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio (miRNA/miRNA*) | let-7a | miR-103 | miR-320a | miR-107 | miR-101 | miR-30e | miR-17 | miR-30a | miR-32 | miR-625 |
| Mouse leukemia 1 | 15773.5 | 40465 | 9354 | 21195 | 1881 | 0.754216867 | 1.703703704 | N/E | N/E | N/E |
| Mouse leukemia 2 | 31822.66667 | 26642 | 5505 | 16623 | 1237 | 0.828804348 | 1.508274232 | N/E | N/E | N/E |
| ES undiff | 11179 | 207069 | 25214 | 20119 | 8012 | 0.141397289 | 4.563953488 | 2.751046025 | 0.888888889 | 0.581699346 |
| ES diff | 4986 | 269177 | 2228 | 19099 | 3880 | 0.365884431 | 2.691432904 | 3.246661102 | 1.636363636 | 0.520958084 |
| Leukemia 1 | 6349 | 46489 | 37114 | 32102 | 19329 | 0.013737836 | 0.655882353 | 0.5 | 3 | 1.474358974 |
| Leukemia 2 | 5364.58 | 68834 | 8378.5 | 17958 | 6974 | 1.716995682 | 1.081261596 | 2.735955056 | 5.538461538 | 0.785714286 |
| Leukemia 3 | 2701.321839 | 31017 | 3095 | 11571 | 5629 | 0.849372385 | 1.05952381 | 0.230769231 | 3.75 | 0.388888889 |
| Prostate 1 | 47738 | 35289 | 2782 | 7347 | 2555 | 0.235474006 | 3.344827586 | 2.492537313 | 2.166666667 | 1.4 |
| Prostate 2 | 14021.92308 | 31233 | 2090 | 9151 | 1676 | 0.153729072 | 7.416149068 | 1.145530146 | 1.8 | 6.428571429 |
| Colon | 16389.34043 | 309168 | 109730 | 6041 | 25246 | 0.974157734 | 8.263414634 | 3.492307692 | 1.6 | 1.125 |
N/E indicates not expressed; ES undiff, undifferentiated embryonic stem cells; and ES diff, differentiated embryonic stem cells..
miRNA duplexes with dynamic arm expression
| Ratio (miRNA/miRNA*) . | miR-106b . | miR-1307 . | miR-16 . | miR-1975 . | miR-223 . | miR-23b . | miR-365 . |
|---|---|---|---|---|---|---|---|
| Mouse leukemia 1 | 0.62 | n/e | 767.75 | n/e | 0.17 | 1.16 | 0.28 |
| Mouse leukemia 2 | 1.00 | n/e | 449.00 | n/e | 2.02 | 0.68 | 287.00 |
| ES undiff | 1.89 | 4.41 | 17.01 | 10.06 | n/e | 2.40 | 4.00 |
| ES diff | 3.61 | 0.97 | 32.88 | 15.21 | n/e | 2.98 | 4.06 |
| Leukemia 1 | 5.67 | 0.26 | 20.59 | 5.80 | 28.28 | 1.51 | 15.50 |
| Leukemia 2 | 2.68 | 1.49 | 121.18 | 0.39 | 1.41 | 4.24 | 126.00 |
| Leukemia 3 | 8.94 | 0.26 | 11.62 | 9.36 | 21.82 | 3.92 | 26.00 |
| Prostate 1 | 12.65 | 0.58 | 464.13 | 0.10 | 1.41 | 14.00 | 0.67 |
| Prostate 2 | 19.67 | 1.97 | 1262.44 | 0.31 | 0.52 | 34.37 | 0.74 |
| Colon | 0.28 | 21.69 | 0.58 | 0.10 | n/e | 0.55 | 0.16 |
| Ratio (miRNA/miRNA*) . | miR-106b . | miR-1307 . | miR-16 . | miR-1975 . | miR-223 . | miR-23b . | miR-365 . |
|---|---|---|---|---|---|---|---|
| Mouse leukemia 1 | 0.62 | n/e | 767.75 | n/e | 0.17 | 1.16 | 0.28 |
| Mouse leukemia 2 | 1.00 | n/e | 449.00 | n/e | 2.02 | 0.68 | 287.00 |
| ES undiff | 1.89 | 4.41 | 17.01 | 10.06 | n/e | 2.40 | 4.00 |
| ES diff | 3.61 | 0.97 | 32.88 | 15.21 | n/e | 2.98 | 4.06 |
| Leukemia 1 | 5.67 | 0.26 | 20.59 | 5.80 | 28.28 | 1.51 | 15.50 |
| Leukemia 2 | 2.68 | 1.49 | 121.18 | 0.39 | 1.41 | 4.24 | 126.00 |
| Leukemia 3 | 8.94 | 0.26 | 11.62 | 9.36 | 21.82 | 3.92 | 26.00 |
| Prostate 1 | 12.65 | 0.58 | 464.13 | 0.10 | 1.41 | 14.00 | 0.67 |
| Prostate 2 | 19.67 | 1.97 | 1262.44 | 0.31 | 0.52 | 34.37 | 0.74 |
| Colon | 0.28 | 21.69 | 0.58 | 0.10 | n/e | 0.55 | 0.16 |
MiR-223 and miR-223* accumulation is a dynamic process
The finding that certain miRNAs displayed tissue-dependent miRNA arm selection prompted interest in the possible biologic function of selective accumulation of miRNA* strands. Of the reported miRNAs in Figure 1B, miR-223, a known regulator of myeloid differentiation,21,28,29 is the best functionally characterized miRNA in hematopoietic tissue. Lentiviral overexpression of hsa-miR-223 in NB4 cells, a promyelocytic leukemia cell line, led to further differentiation of these cells,29,30 which underscores its role in myeloid differentiation. Interestingly, miR-223 and miR-223* were differentially expressed in the investigated tissues, especially in the ND13 (mouse leukemia 1) and ND13+Meis1 (mouse leukemia 2) leukemia progression model (Figure 1B). Specifically, miR-223* was enriched by ∼ 30-fold in the ND13 myeloid progenitor line compared with its leukemic counterpart ND13+Meis1 (3366 vs 115 counts), and miR-223* sequence counts also exceeded miR-22311 (Figure 1B; supplemental Figure 1C). This dynamic pattern could be observed across all BM-derived libraries, regardless of human or mouse origin (supplemental Figure 1C), which implies that miR-223*s contribute to the function of miR-223. In contrast, other BM-specific miRNAs showed a more constant expression pattern (supplemental Figure 1C). Variable miR-223* levels and the fact that miR-223 as well as miR-223* remained broadly conserved in vertebrates (supplemental Figure 1D) pointed toward an miRNA duplex with 2 functional arms. Because ectopic expression of miR-223 involves overexpression of pre-miR-223,28,29 which includes expression of both miRNA duplex arm, it remains unknown whether miR-223* contributes to the observed phenotype.
miR-223* decreases the colony-forming capacity of miR-223–deficient BM cells
To test possible functions of both miR-223 and miR-223* in hematopoietic cells, we exploited a murine miR-223KO model.21 Previous analysis of miR-223–deficient BM revealed an enhanced number of myeloid progenitors and impaired differentiation of granulocytes.21 To assess the influence of miR-223* in myeloid progenitor cells, we used a retroviral construct engineered to render the seed region of the miR-223 arm inactive (miR-223mut; Figure 1C). WT and miR-223 KO BM transduced with miR-223 or miR-223mut were plated, and their colony-forming capacity was assessed (Figure 1D). Colony-forming cell progenitor numbers were elevated in the miR-223 KO background (P = .0171). Overexpression of miR-223* with the miR-223mut construct significantly reduced the colony-forming cell output (P = .008), which highlights the regulatory potential of miR-223* (Figure 1D). These findings suggest that miR-223 and miR-223* might have separate functions that complement each other. Specific knockdown of the miR-223 strand has been shown to impair differentiation in granulocytes,21 whereas the present results imply a regulatory role of miR-223* in proliferation or self-renewal of myeloid progenitor cells. These results demonstrate a role for miR-223* in primary hematopoietic cells.
miR-223* and miR-223 target the IGF1R pathway
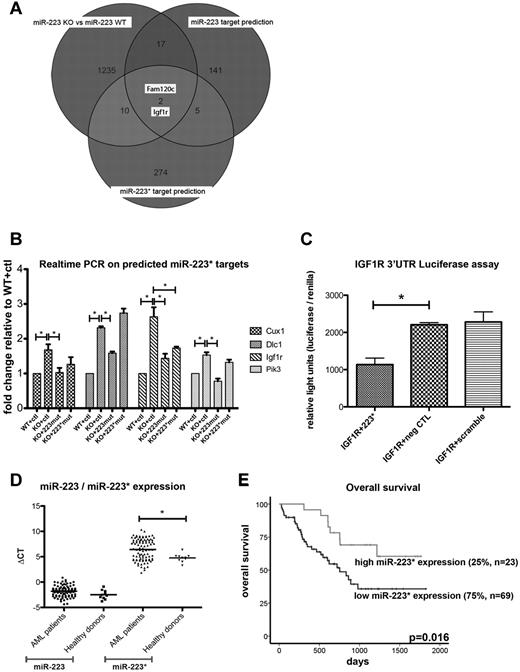
To investigate further possible functions of miR-223*, we used bioinformatics to predict conserved targets with TargetScan custom (Version 5.0, www.targetscan.org) using the 7-nt seed sequence (GUGUAUU) of hsa-miR-223* and the predicted targets of hsa-miR-223 (supplemental Figure 2A and supplemental Table 4). Interestingly, both arms are predicted to regulate IGF1R signaling, a pathway implicated in normal and malignant hematopoiesis. Therefore, to further identify differentially expressed targets between miR-223 WT and KO cells, we performed an mRNA microarray expression analysis on immortalized miR-223 BM cells. In our analysis, we found a set of 1264 differentially expressed genes (743 genes up-regulated, 521 down-regulated) between miR-223KO and miR-223WT cells (n = 2, P < .05; supplemental Table 4). When we compared the differentially expressed genes with predicted targets for mmu-miR-223 and mmu-miR-223* (www.targetscan.org), 2 genes (Figure 2A), Fam120c (1.078-fold, P = .006), a gene linked to autism, and Igf1r (1.37-fold, P = .04), a gene linked to the IGF1R/phosphatidylinositol 3-kinase (PIK3) axis and involved in hematopoietic malignancies,31-34 overlapped between the 2 prediction lists. These findings suggest a shared regulation of the IGF1R/PI3K axis by miR-223 and miR-223*. In further support of this, the effect of miR-223 depletion was significantly reversed for Igf1r by retroviral overexpression of miR-223* (miR-223mut, 1.13-fold, P = .043) and selectively with miR-223 (miR-223*mut, 1.11-fold, P = .043; supplemental Table 4). To confirm and extend these findings, we used quantitative real-time PCR analysis35 to quantify 12 predicted miR-223* targets (Cux1, Igf1r, Neo1, Pi3kcd, Dlc1, Lyn, Ube2, Fgf7, Sema3a, Slc2a, Pten, and Lpp) implicated in both normal and leukemic development. After filtering, using a threshold of a 1.5-fold increase in miR-223KO cells, Cux1, Dlc1, Pik3cd, and Igf1r remained as possible targets of miR-223* (Figure 2B). In further support of this, the effect of miR-223 depletion was significantly reversed for all 4 targets by retroviral overexpression of miR-223* (using the miR-223mut construct; Figure 2B; supplemental Figure 2B). As predicted, with a similar approach with an inactivated miR-223* arm (miR-223*mut) to thus selectively overexpress miR-223, Igf1r levels exhibited a significant decrease, which indicates a regulatory role for both miR-223 and miR-223* (P = .029 and P = .04, respectively) in the expression of Igf1r. However, the present in vitro data also indicate that miR-223 might have a higher binding affinity than miR-223*, because lower miR-223 levels showed a similar Igf1r knockdown (Figure 2B; supplemental Figure 2B). The regulatory potential of miR-223* on IGF1R was also confirmed through an in vitro luciferase assay (P = .018; Figure 2C). Combined, these results raise the intriguing possibility that miR-223* plays a cooperative role with miR-223 by targeting the same transcript within the IGF1R/PI3K axis, a key pathway for developmental and malignant processes.36,37
Quantification of miR-223* and miR-223* targets. (A) Venn diagram comparing differentially expressed genes (shown as transcript cluster IDs), assessed by mRNA microarrays with predicted mRNA targets (shown as transcript cluster IDs). (B) Quantification of Cux1, Igf1r, Dlc1, and Pik3cd by real-time PCR in miR-223KO cells expressing strand-inactivating constructs (n = 3). ctl indicates control. (C) IGF1R luciferase assay. neg CTL indicates negative control. (D) Comparison of miR-223 and miR-223* levels in healthy donors (n = 10) and AML patients with normal cytogenetics (n = 92). The y-axis depicts ΔCT (miRNA−miR-92) values. (E) miR-223* levels measured in 92 AML patients with normal cytogenetics by real-time PCR. Overall survival was plotted on miR-223* levels dichotomized to the 75th percentile. *P < .05
Quantification of miR-223* and miR-223* targets. (A) Venn diagram comparing differentially expressed genes (shown as transcript cluster IDs), assessed by mRNA microarrays with predicted mRNA targets (shown as transcript cluster IDs). (B) Quantification of Cux1, Igf1r, Dlc1, and Pik3cd by real-time PCR in miR-223KO cells expressing strand-inactivating constructs (n = 3). ctl indicates control. (C) IGF1R luciferase assay. neg CTL indicates negative control. (D) Comparison of miR-223 and miR-223* levels in healthy donors (n = 10) and AML patients with normal cytogenetics (n = 92). The y-axis depicts ΔCT (miRNA−miR-92) values. (E) miR-223* levels measured in 92 AML patients with normal cytogenetics by real-time PCR. Overall survival was plotted on miR-223* levels dichotomized to the 75th percentile. *P < .05
miR-223* levels correlate with prognostic markers in AML patients
Considering that in vitro inhibition of IGF1R signaling also affects leukemic blasts33 and leads to the inhibition of colony formation of AML blasts,38 we examined miR-223 and miR-223* expression levels in normal and malignant cells and whether they correlated with clinical parameters of AML patients. We quantified miR-223* and miR-223 expression by real-time PCR in 92 AML patients < 60 years of age with normal cytogenetics (Table 5) and 10 healthy donors. In line with our sequencing libraries, miR-223* levels in AML samples and in normal hematopoietic samples only showed a moderate positive correlation with the expression of miR-223 (R2 = 0.2351, R2 = 0.0729, respectively; Figure S2C), which underscores its variable expression. We found miR-223* to be significantly more highly expressed in healthy donors (P = .016) than in AML patient samples (Figure 2D). Interestingly, with a dichotomization to the 75th percentile, high miR-223* expression levels were significantly associated with superior overall survival (P = .016; Figure 2E). In contrast, miR-223 was not associated with any survival parameters, regardless of dichotomization. Neither miRNA correlated with any other clinical parameter (Table 5), which indicates that the correlation between high miR-223* levels and increased survival was not confounded by other clinical variables.
Clinical characteristics and correlation of prognostic markers in 92 profiled AML patients dichotomized to the 75% percentile
| Clinical parameter . | Low miR223* . | High miR223* . | P . |
|---|---|---|---|
| Cases | (n = 69) | (n = 23) | |
| Age, y | 0.28 | ||
| median | 48 | 48 | |
| range | 17-60 | 25-60 | |
| Sex | 0.55 | ||
| male, no. (%) | 38 (55) | 11 (48) | |
| female, no. (%) | 31 (45) | 12 (52) | |
| FAB-subtype | 0.73 | ||
| M0, no. (%) | 1 (2) | 0 (0) | |
| M1, no. (%) | 7 (10) | 2 (9) | |
| M2, no. (%) | 16 (23) | 4 (17) | |
| M4, no. (%) | 30 (43) | 11 (48) | |
| M5, no. (%) | 11 (16) | 3 (13) | |
| M6, no. (%) | 1 (2) | 2 (9) | |
| M7, no. (%) | 0 | 0 (0) | |
| missing data, no. (%) | 3 (4) | 1 (4) | |
| Percentage blasts in sample | 0.25 | ||
| median (%) | 80 | 75 | |
| missing data, no. (%) | 5 | 0 | |
| Type of AML | 0.85 | ||
| de novo. no. (%) | 61 (88) | 20 (870 | |
| post MDS/secondary, no. (%) | 8 (12) | 3 (13) | |
| WBC count | 0.78 | ||
| median (×109/l) | 27 | 29 | |
| range (×109/l) | 1-328 | 1-170 | |
| ECOG performance status | 0.14 | ||
| 0 or 1, no. (%) | 66 (96) | 20 (87) | |
| 2, no. (%) | 3 (4) | 3 (13) | |
| FLT3-ITD | 0.42 | ||
| mutated, no. (%) | 21 (30) | 5 (22) | |
| NPM1 | 0.33 | ||
| mutated, no. (%) | 36 (52) | 15 (65) | |
| missing, no. (%) | 5 (7) | 1 (4) | |
| NPM1 mutated/FLT3-ITD negative | 0.25 | ||
| low risk, no. (%) | 23 (33) | 11 (48) | |
| missing, no. (%) | 5 (7) | 1 (4) | |
| CEBPA | 0.51 | ||
| mutated, no. (%) | 10 (14) | 5 (22) | |
| missing, no. (%) | 5 (7) | 0 (0) |
| Clinical parameter . | Low miR223* . | High miR223* . | P . |
|---|---|---|---|
| Cases | (n = 69) | (n = 23) | |
| Age, y | 0.28 | ||
| median | 48 | 48 | |
| range | 17-60 | 25-60 | |
| Sex | 0.55 | ||
| male, no. (%) | 38 (55) | 11 (48) | |
| female, no. (%) | 31 (45) | 12 (52) | |
| FAB-subtype | 0.73 | ||
| M0, no. (%) | 1 (2) | 0 (0) | |
| M1, no. (%) | 7 (10) | 2 (9) | |
| M2, no. (%) | 16 (23) | 4 (17) | |
| M4, no. (%) | 30 (43) | 11 (48) | |
| M5, no. (%) | 11 (16) | 3 (13) | |
| M6, no. (%) | 1 (2) | 2 (9) | |
| M7, no. (%) | 0 | 0 (0) | |
| missing data, no. (%) | 3 (4) | 1 (4) | |
| Percentage blasts in sample | 0.25 | ||
| median (%) | 80 | 75 | |
| missing data, no. (%) | 5 | 0 | |
| Type of AML | 0.85 | ||
| de novo. no. (%) | 61 (88) | 20 (870 | |
| post MDS/secondary, no. (%) | 8 (12) | 3 (13) | |
| WBC count | 0.78 | ||
| median (×109/l) | 27 | 29 | |
| range (×109/l) | 1-328 | 1-170 | |
| ECOG performance status | 0.14 | ||
| 0 or 1, no. (%) | 66 (96) | 20 (87) | |
| 2, no. (%) | 3 (4) | 3 (13) | |
| FLT3-ITD | 0.42 | ||
| mutated, no. (%) | 21 (30) | 5 (22) | |
| NPM1 | 0.33 | ||
| mutated, no. (%) | 36 (52) | 15 (65) | |
| missing, no. (%) | 5 (7) | 1 (4) | |
| NPM1 mutated/FLT3-ITD negative | 0.25 | ||
| low risk, no. (%) | 23 (33) | 11 (48) | |
| missing, no. (%) | 5 (7) | 1 (4) | |
| CEBPA | 0.51 | ||
| mutated, no. (%) | 10 (14) | 5 (22) | |
| missing, no. (%) | 5 (7) | 0 (0) |
Discussion
Here, we have exploited the recent availability of next-generation deep-sequencing data from 10 different libraries to enable a comprehensive, quantitative analysis of miRNAs and the corresponding miRNA*s. Our results provide evidence that relative miRNA and miRNA* expression is conserved between various tissues and cell lines, which allows for a novel classification and prediction of possible functional miRNA and miRNA* strands derived from 1 duplex. Intriguingly, for select miRNAs such as miR-223/miR-223*, the miRNA/miRNA* ratios exhibited considerable variability across libraries. Further investigation of miR-223* in vitro suggests a functional role in concert with miR-223 in myeloid cells; in addition, high miR-223* levels were associated with a higher overall survival in AML patients with normal cytogenetics.
Across all examined libraries, the proportion of miRNA*s to miRNAs ranged between 0.3% and 12.3%, which indicates that the overall miRNA* expression is tissue and species dependent. Comparison of all the sequencing libraries derived from different tissues revealed conserved patterns between miRNA and the corresponding miRNA* arms. In addition to instances with highly dominant miRNA arms, such as the highly abundant let-7 family, we also found instances with balanced expression of the miRNA and miRNA* species, such as miR-30e, which confirms previous findings by Ro et al.17 We termed miRNA duplexes that gave rise to a dominant strand “α-duplexes” and duplexes that gave rise to a more balanced strand expression “β-duplexes.” This novel classification carries with it the important prediction of whether 1 or, in case of a β-duplex, both miRNA strands are functionally active. In addition, we found several miRNAs in which the annotated miRNA* arm was strongly dominant over the miRNA arm, which indicates improper annotation in miRBase, such as miR-129* and miR-517*. Furthermore, we were able to unambiguously annotate the miRNA* strand of miRNAs such as miR-423 and miR-371. Other recent miRNA profiling efforts have found differentially expressed miRNA*s, such as miR-9*,39 miR-199*,40,41 miR-126*,42,43 miR-363*,44 miR-18*,45 miR-29c*,46 and miR-155*,47 in the pathogenesis of Waldenström macroglobulinemia, lung cancer, and a metastasis model, as well as in developmental processes such as organ adhesion, respectively. In our libraries, hsa-miR-199 showed features of an α-duplex, whereas hsa-miR-9 and hsa-miR-363 displayed a more balanced expression (supplemental Table 2). These results are in contrast to published data suggesting that miR-199* is expressed at detectable levels in fibroblasts,41 which might be attributable to differences in the profiling methodologies and cell types studied.
Thus far, the mechanisms by which strand selection occurs have not been completely resolved. Recent studies have indicated that the relative thermodynamic stability of the 2 ends of the duplex determines strand selection.3,4 In general, the strand with relatively unstable base pairs at the 5′ end evades degradation; however, this concept has been challenged by studies describing tissue-dependent paired expression of miRNAs.15-17 This could be because of tissue-dependent stability of miRNAs and miRNA*s, as well as unknown extrinsic factors, as suggested by the developmentally controlled arm switch observed for miR-2015 in the embryonic and adult stages of sponges.48 We could not verify such a phenomenon in the present comparison of undifferentiated and differentiated embryonic stem cells. Another possibility involves the presence of an miRNA or miRNA* target in a cell leading to a target-dependent strand selection or cell-specific modification of RISC cofactors such as TRBP, which might influence selection of the active miRNA arm. Recently, Chatterjee et al49 demonstrated that mRNAs can stabilize their cognate miRNAs, which suggests coordinated RISC assembly depending on an miRNA and its target levels. Given that miR-223 and miR-223* have Igf1r as a common target, varying Igf1r mRNA levels in myeloid cells could theoretically contribute to the described inconsistent miR-223/miR-223* ratios. However, only a few miRNA duplexes exhibited inconstant expression patterns, which demonstrates that in general, miRNA strand selection is a highly preserved mechanism.
Analysis of the dynamics of miRNA strand accumulation and the specific expression of miR-223* (supplemental Figure 2D) in mature myeloid cells suggested that miR-223* is relevant in myeloid differentiation. Current approaches to overexpress an miRNA use miRNA mimics to produce a short-term effect in cell lines. In general, BM cells are very difficult to transfect, and changes are usually detected after a delay of several days. We therefore took advantage of a retroviral approach by creating retroviral vectors that contained mutations in either the miR-223 or the miR-223* seed region. A vector in which the miR-223* seed region was expressed but miR-223 was mutated (inactive) was sufficient to partially reverse (rescue) the enhanced colony-forming cell–plating efficiency of miR223KO BM cells and thus implicates a functional role for miR-223*.
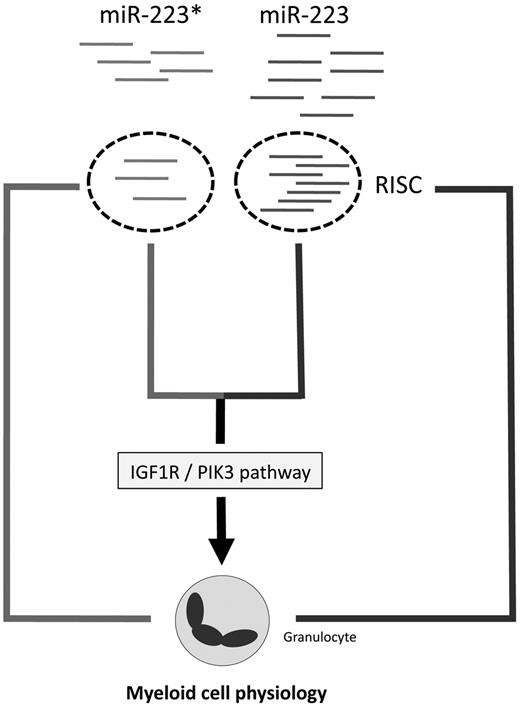
The relatively high abundance of miR-223* evident from our sequencing libraries strongly argues that miR-223* is able to enter the RISC and therefore is functionally active. This possibility is corroborated by the frequent enrichment of even less abundant miRNA* strands in RISC15 and our miR-223* target screen. Here, we demonstrated that Pik3cd, Cux1, Dlc1, and Igf1r were targeted by miR-223*, which led to their mRNA degradation. This is especially interesting when one considers that a tightly regulated IGF1R/PI3K axis, which affects PTEN and SHIP, among other inositol phosphatases, is necessary for normal BM development50,51 and myeloid malignancies.38 Therefore, deregulated miR-223* and miR-223 expression might be associated with clinical parameters in AML patients. Indeed, high miR-223* expression levels correlate with a better overall survival in AML patients with normal cytogenetics, which strongly highlights a role of miR-223* in AML. We hypothesize that increased miR-223 and miR-223* levels might activate different programs, with miR-223 affecting myeloid differentiation29 and miR-223* complementing miR-223 function by possibly activating apoptosis and/or inhibiting self-renewal or proliferation of progenitor cells (Figure 3), and we also considered the possibility of novel SNPs in miR-223/miR-223*, as shown for miR-146. However, we could not detect any miR-223* polymorphisms in 95 profiled AML patients, which might be because of a nonsignificant number of profiled patients.
The present data revealed that miRNA arm accumulation underlies conserved patterns, but not exclusively the dominance of one miRNA duplex strand. Depending on the tissue, miRNA*s can be more abundant than previously assumed, which implies a functional role for highly expressed miRNA*s. An important functional role for miRNA*s, as illustrated by the results obtained for miR-223* in normal and malignant myeloid cells, points toward a broader and thus more complicated interaction of miRNAs and disease-related pathways than previously assumed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Patty Rosten, Anisa Salmi, and Lisa Yue for invaluable advice and assistance. They also thank all members of the Humphries' laboratory for stimulating discussions.
This work was supported by grants from the Terry Fox Foundation, Genome Canada/BC, the Canadian Institutes of Health, the Cancer Research Society, and the Canadian NCE Stem Cell Network.
Authorship
Contribution: F.K., S.M.M., M.H., J.R., A.R., T.B., D.L., D.T.S., and K.D. performed experiments; F.K., A.M., L.B., R.D.M., D.T.S., A.K., A.W., K.D., and L.P. analyzed results and created the figures; and F.K., C.J.E., Y.W., S.A.A., H.D., M.A.M., F.D.C., C.B., and R.K.H. designed the research and wrote the paper.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: R. Keith Humphries, Terry Fox Laboratory, BC Cancer Agency, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: khumphri@bccrc.ca.