Abstract
While miRNAs are increasingly linked to various immune responses, whether they can be targeted for regulating in vivo inflammatory processes such as endotoxin-induced Gram-negative sepsis is not known. Production of cytokines by the dendritic cells (DCs) plays a critical role in response to endotoxin, lipopolysaccharide (LPS). We profiled the miRNA and mRNA of CD11c+ DCs in an unbiased manner and found that at baseline, miR-142-3p was among the most highly expressed endogenous miRs while IL-6 was among the most highly expressed mRNA after LPS stimulation. Multiple computational algorithms predicted the IL-6 3′ untranslated region (UTR) to be a target of miR-142-3p. Studies using luciferase reporters carrying wild-type (WT) and mutant IL-6 3′UTR confirmed IL-6 as a target for miR-142-3p. In vitro knockdown and overexpression studies demonstrated a critical and specific role for miR142-3p in regulating IL-6 production by the DCs after LPS stimulation. Importantly, treatment of only WT but not the IL-6–deficient (IL-6−/−) mice with locked nucleic acid (LNA)–modified phosphorothioate oligonucleotide complementary to miR 142-3p reduced endotoxin-induced mortality. These results demonstrate a critical role for miR-142-3p in regulating DC responses to LPS and provide proof of concept for targeting miRs as a novel strategy for treatment of endotoxin-induced mortality.
Introduction
Sepsis from Gram-negative infections is a major health problem. Despite the current advances, it is associated with significant mortality.1 The development of sepsis results from an exuberant systemic inflammation.
The microRNAs (miRs) are the highly conserved small, single-stranded noncoding RNAs.2 They are key regulators of gene expression that control various aspects of cellular responses.2 They suppress gene expression by binding to partially complementary sequences in the 3′ untranslated region (UTR) of mRNAs, and inhibit their translation into protein or accelerate their degradation.2 Emerging research suggests that miRs can modulate key pathways involved in the innate and adaptive immune responses.3,4 Their contribution is, however, largely unknown and just beginning to be understood.
The innate immune system is an important first line of defense against infectious agents and it includes many types of cells. Among these, dendritic cells (DCs) are pivotal for both recognition of Ags and control of an array of immune responses.5 DCs recognize microbes through distinct pattern recognition receptors (PRRs).6 The first microbial component to be studied in detail and known to cause septic shock is endotoxin (lipopolysaccharide [LPS])7 which is recognized via TLR-4.7 LPS causes many changes in the DCs, but the elicitation of cytokine production is perhaps the one with clear biologic relevance.6
The inflammatory cytokine response by the DCs is regulated at both the transcriptional and translational levels.8-13 Recent studies have suggested that the stability and translation of cytokine-encoding mRNA may be related to certain miR-mediated mRNA destability.3,13-16 But the complete miR repertoire and their role in modulating DC responses and the subsequent impact on endotoxin-induced shock are not well known.
Here, we report a novel mechanism of miR-mediated modulation of IL-6 expression by the DCs in response to stimulation by LPS. miRNA and mRNA profiling of DCs demonstrated that at baseline miR-142-3p was among the most highly expressed endogenous miRs, while IL-6 was among the most highly expressed mRNA after LPS stimulation. Computational algorithms predicted IL-6 3′UTR to be targeted by miR-142-3p, which is highly conserved across species. Experiments with luciferase reporters carrying wild-type (WT) and various truncated forms of IL-6 3′UTR demonstrated that miR-142-3p specifically targets IL-6. In vitro knockdown of endogenous and overexpression of miR-142-3p in primary DCs confirmed the functional relevance of miR-142-3p in regulating IL-6 expression in DCs. Furthermore, experiments with LNA-modified oligonucleotides specific for miR-142-3p in the WT and IL-6−/− mice validated the in vivo specificity for IL-6 and the functional role of miR-142-3p in regulating mortality from endotoxemia.
Methods
Mice and DCs
WT or IL-6–deficient (IL-6−/−) C57BL/6 (B6) female mice, aged at 8 to 12 weeks, were purchased from The Jackson Laboratory and cared for under the regulations of the guidelines by the University of Michigan's University Laboratory Animal Medicine (ULAM). To obtain mouse DCs, BM cells were cultured with murine recombinant GM-CSF (10 ng/mL; BD PharMingen) and IL-4 (10 ng/mL; PeproTech) for 7 days and harvested as described previously.17 DCs were harvested and positively selected by the autoMACS Pro Separator (Miltenyi Biotec) for CD11c+ cells. To separate DC subpopulations, the CD11c+ DCs were processed for FACS sorting to obtain the CD11c+/CD8+ and CD11c+/CD8− subpopulations.
Human cells
Studies with human cells were performed after obtaining informed consent from the participants, in accordance with the Declaration of Helsinki, and were approved by the University of Michigan Medical School Institutional Review Board. Peripheral blood from healthy volunteers was used to isolate DCs and T cells17 by sedimentation using the Ficoll-Hypaque technique (Pharmacia Biotech). The dermal microvascular endothelial cells (HMVEC-D), the human skin fibroblasts (ATCC), breast cancer cell line MCF7, and prostate cancer cell line PC-3 were all obtained (ATCC) and cultured as in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All the cells were processed as in supplemental Methods.
Exiqon microRNA arrays
Microarray experiments were conducted as double-channel Hy3/Hy5 in duplicate on Exiqon's miRCURY LNA microRNA Array Version 10.0 (Exiqon). The miRNeasy Mini Kit (QIAGEN) was used to purify total RNA, including miRNA and other small RNA molecules from mouse DCs. The samples were analyzed and the images were captured and analyzed by GAL-file (Exiqon) which covers miRNA ID annotated by Version 10.0 of miRBase (http://microrna.sanger.ac.uk) as described previously18,19 and in supplemental Methods.
Affymetrix microarrays
Mouse DCs were left untreated or treated with LPS (500 ng/mL) for 12 hours, tcRNA were extracted with TRIzol reagent and cleaned over Qiagen RNeasy columns (QIAGEN). Synthesis and labeling of cRNA and hybridization of arrays was conducted as described previously.20 Stained arrays (430 2.0) were scanned on an Agilent Gene Array Scanner (Affymetrix).
MicroRNA prediction tools
To identify miRNAs targeting IL-6, we integrated the output results of multiple prediction programs; Miranda (http://www.microrna.org/microrna/), MicroCosm Targets (http://microrna.sanger.ac.uk), and PITA TOP (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html).18,21,22 We imported the various output formats from each of the target prediction programs, and identified the miRNAs that are expressed in DCs, and subsequently integrated the results to find common overlaps. The candidate miRNAs were sorted based on the predefined rankings from each respective program.
Plasmid reporters, oligonucleotides, and transfection
psiCHECK-2 vector, a dual-luciferase plasmid containing both firefly luciferase gene and Renilla luciferase gene, was obtained from Promega. miRNA target sequences (see Figure 2A) were inserted in the 3′UTR of the hRluc gene. Cleavage target sites are reverse complements of their respective predicted mature miRNAs. The fragments of IL-6 3′UTR were cloned from the mouse small intestine DNA library by PCR using specific primer pairs (supplemental Table 6). The reporter constructs were transfected into DCs using N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate [DOTAP]) as described previously and in supplemental Methods.17 The LNA-modified in vivo knockdown anti-miRs for miR-142-3p and scrambled control were purchased from Exiqon, and delivered by IP administration (supplemental Table 6 and supplemental Methods).
miRNA quantitative real-time PCR assays
For miRNA quantitative PCR, total RNA including small RNA was isolated using the miRNeasy Mini Kit (QIAGEN) from mouse DCs (including CD11c+ DCs that were treated with diluent or with LPS 500 ng/mL, for 12 hours), CD11c+/CD8+ DCs, and CD11c+/CD8− DCs, mouse splenocytes treated with in vivo scramble or knockdown probes, CD11c+ DCs (from spleen or lymphoid nodes), T cells, B cells, kidney, and brain tissues), and human samples (including DCs, T cells, endothelial cells, fibroblast cells, and MCF7 and PC3 cells). Specific RT primers for miR142-3p, miR-142-5p, miR-155, miR-223, miR-7a, miR-27a, miR-744, miR-668, miR-720, miR-146a, and snoRNA135 (Applied Biosystems) were used and QPCR was performed as described in supplemental Methods.
Cytokine ELISAs
TNF-α, IL-12, IL-6, and IL-10 were measured in culture supernatants or mouse sera by ELISA with specific anti-mouse mAbs for capture and detection. The appropriate standards were purchased from BD Systems (TNF-α and IL-6) or BD OptEIA (IL-12p70 and IL-10). Assays were performed according to the manufacturer's protocol and read at 450 nm using a microplate reader (Bio-Rad).17
Mixed lymphocyte cultures
Splenic T cells from naive B6 or BALB/c mice or CD11c+ DCs derived from B6 mice BM were isolated by autoMACS. DCs were transfected with 50-nm miR-142-3p-LNA KD or scramble control probes using DOTAP as described previously. After 24 hours of transfection, a total of 2 × 105 BALB/c or B6 T cells were cultured with 5 × 103 transfected B6 DCs for 96 hours. Incorporation of 3H-thymidine (1 μCi/well) by proliferating T cells during the final 18 hours of culture was measured by a Betaplate reader.
Immunoblot analyses
After 48 hours of transfection, cells lysates were prepared and immunoblot analyses were performed as in supplemental Methods.
In vivo experiments
B6 mice were administered once with saline-formulated miR-142-3p-antimiR or scrambled control using an injection volume of 10 mL/kg with IP doses ranging from 5 to 25 mg/kg. After 2 days, mice spleens were harvested and analyzed for expression of miRNA-142-3p, miR-142-5p, and miR-155 by Q-PCR. For cytokine analyses after endotoxemia, 10 mg/kg miR-142-3p-antimiR or scrambled control was given (IP) at 46 hours followed by an IP dose of LPS (35 mg/kg in 700 μL). Two to 3 hours after LPS injection, that is, ∼ 48 hours after treatment with anti-miRs the spleens/serum from B6 WT or IL-6−/− were harvested for analyses of mRNA levels of cytokine TNF-α and IL-6. ELISA was used for detection of serum cytokine levels. For survival experiments, WT or IL-6−/− B6 mice were administered once on day −2 and day 0 with saline-formulated LNA-anti-miR-142-3p, or LNA mismatch control by the injection volume of 200 μL with an IP dose of 10 mg/kg. On day 0, 4 hours after IP injection with in vivo KD probes or scramble control, the mice were administrated IP with LPS at a dose of 35 mg/kg in volume of 700 μL, and survival was recorded every 3-4 hours for the first 72 hours and then daily for the next 4-7 days.
Statistical analyses
For miR array data, the median signal was used as the expression value. The expression levels were calculated with signals above the background (F median signal/B median signal). Affymetrix microarray data were published and analyzed using R statistical environment (Version 2.7; http://cran.r-project.org/) provided by Bioconductor (http://www.bioconductor.org/). The probe sets were selected based on a 2-fold difference with the added constraint that at least one sample had to have an expression value of 26 or greater. This protects us from choosing probe sets with a large fold change that is based on 2 small numbers. The expression values after log2 transformed for each gene were calculated using a robust multiarray average (RMA),23 which is a modeling strategy that converts the PM probe values into an expression value for each gene. All comparisons between expression values of miRNA, mRNA, or protein across sample classes were made using the paired 2-sample Student t test. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE28340.
Results
Profiling and identification of miR-142-3p as a temporally regulated miR in DCs
We first analyzed the expression profile of miRs in BM-derived murine DCs from C57BL/6 (B6) female mice, aged at 8 to 12 weeks. We used the Exiqon miRCURY LNA microarray platform, which contains probes specific for 511 mouse miRs annotated in miR Base 11.0 V. We averaged the relative Cy3 or Cy5 intensity levels from 4 separate experiments. Eighty-eight miRs were detected in mouse BMDCs (supplemental Table 1). Of these 88 detected, 33 miRs were highly expressed (over 10- to 85-fold over background), 19 were moderately expressed (5- to 10-fold), and 36 were expressed 3- to 5-fold over the background (supplemental Table 1).
Because miRs regulate protein translation and/or mRNA destabilization,19,24 we next correlated changes in gene expression with the miR profile of the DCs after LPS stimulation. Using Affymetrix chip array (contains more than 45 100 probes), we compared the gene expression profiles of DCs after LPS or diluent stimulation (supplemental Table 2). From the 100 significantly up-regulated transcripts after LPS stimulation (supplemental Table 3), the top 10 genes that were the most highly expressed were the prostaglandin-endoperoxide synthase 2, chemokine (C-X-C motif) ligand 10, NO synthase 2, IFN-inducible GTPase 1, cDNA sequence U90926, chemokine (C-X-C motif) ligand 11 and 1, CD69, IL-6, and SOCS3 (Table 1).
Ten significantly up-regulated transcripts by LPS
| Symbol . | Gene full name . | Fold change . | Primary function . |
|---|---|---|---|
| Ptgs2 | Prostaglandin | 7.677 | VEGF signaling pathway |
| Cxcl 10 | Chemokine (C-X-C motif) ligand 10 | 7.62 | Immune response |
| Nos 2 | NO synthase 2, inducible, macrophage | 6.331 | Response to hypoxia |
| Ligp 1 | IFN-inducible GTPase 1 | 5.945 | Cytokine and chemokine-mediated signaling pathway |
| U90926 | cDNA sequence U90926 | 5.878 | Extracellular space |
| Cxcl 11 | Chemokine (C-X-C motif) ligand 11 | 5.639 | Immune response |
| Cxcl 1 | Chemokine (C-X-C motif) ligand 1 | 5.432 | Immune response |
| CD69 | CD69 Ag | 5.351 | Immune response |
| Il6 | Interleukin 6 | 5.33 | Immune response |
| Socs 3 | Suppressor of cytokine signaling 3 | 5.149 | Jak-STAT signaling pathway |
| Symbol . | Gene full name . | Fold change . | Primary function . |
|---|---|---|---|
| Ptgs2 | Prostaglandin | 7.677 | VEGF signaling pathway |
| Cxcl 10 | Chemokine (C-X-C motif) ligand 10 | 7.62 | Immune response |
| Nos 2 | NO synthase 2, inducible, macrophage | 6.331 | Response to hypoxia |
| Ligp 1 | IFN-inducible GTPase 1 | 5.945 | Cytokine and chemokine-mediated signaling pathway |
| U90926 | cDNA sequence U90926 | 5.878 | Extracellular space |
| Cxcl 11 | Chemokine (C-X-C motif) ligand 11 | 5.639 | Immune response |
| Cxcl 1 | Chemokine (C-X-C motif) ligand 1 | 5.432 | Immune response |
| CD69 | CD69 Ag | 5.351 | Immune response |
| Il6 | Interleukin 6 | 5.33 | Immune response |
| Socs 3 | Suppressor of cytokine signaling 3 | 5.149 | Jak-STAT signaling pathway |
Most significantly up-regulated mRNA transcripts by LPS. The top 10 most significantly up-regulated transcripts in response to LPS treatment as determined by Affymetrix microarrays. Data shown are the average values from 2 separate experiments.
LPS indicates lipopolysaccharide.
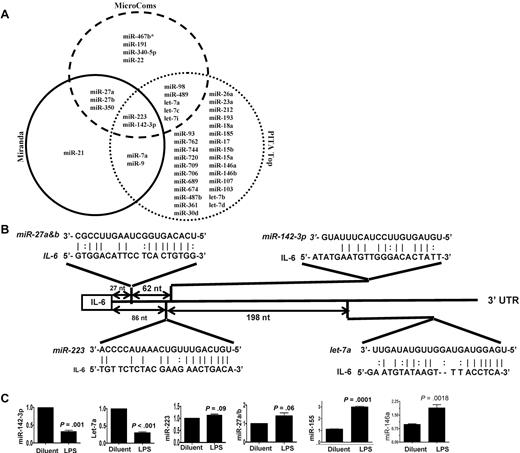
Because IL-6 was among the most up-regulated genes after LPS stimulation,25,26 we next focused on determining the key miRs that may be involved in regulating an IL-6 in DCs. To this end, we computationally nominated miRs that might contribute to IL-6 regulation. Because intersecting the results of multiple prediction algorithms can increase specificity, we integrated the results of 3 prediction software programs to detect potential miRs that may target IL-6 3′UTR.18 From the miRs that were detected in the DCs, 43 miRs were predicted to target IL-6 3′UTR by at least 1 of the 3 algorithms (Figure 1A and supplemental Table 4). However, only 2 miRs, miRNA-223 and miR-142-3p, were predicted by all 3 algorithms (Figure 1A and supplemental Table 4) suggesting a high likelihood for targeting IL-6. Analyses of the IL-6 3′UTR indicated that the targeting sites for these miRs exist upstream of the region of AREs (Figure 1B and supplemental Figure 1). The miR array profiling showed high but different endogenous expression levels of these miRs. Specifically miR-142-3p, miR-223, miR-27a, miR-27b, and let-7a were expressed at 79-, 27-, 9-, 15-, and 50-fold over the background signal, respectively (Table 2 and supplemental Table 1).
Validation of DC miR profile by RT-PCR. (A) Computational prediction of miRs targeting IL-6 3′UTR by the miRanda, MicroCosm Target, and PITA Top programs The miR-142-3p and miR-223 were predicted by all 3 programs. (B) The potential miR target sequences in the 3′UTR of IL-6 mRNA that are computationally predicted in DCs are shown and the seed sequence pairings are indicated by the lines. (C) Validation of miR expression pattern at baseline and the response to LPS. DCs were treated with either LPS or diluent for 12 hours and analyzed. The levels of miRNA-142-3p, miR-223, Let-7a, miR-27a/b, miR-155, and miR-146a were analyzed by TaqMan quantitative RT-PCR. Expression of miRNAs is presented relative to snoRNA135. Data are representative of 2 to 6 separate experiments with similar results (mean ± SEM).
Validation of DC miR profile by RT-PCR. (A) Computational prediction of miRs targeting IL-6 3′UTR by the miRanda, MicroCosm Target, and PITA Top programs The miR-142-3p and miR-223 were predicted by all 3 programs. (B) The potential miR target sequences in the 3′UTR of IL-6 mRNA that are computationally predicted in DCs are shown and the seed sequence pairings are indicated by the lines. (C) Validation of miR expression pattern at baseline and the response to LPS. DCs were treated with either LPS or diluent for 12 hours and analyzed. The levels of miRNA-142-3p, miR-223, Let-7a, miR-27a/b, miR-155, and miR-146a were analyzed by TaqMan quantitative RT-PCR. Expression of miRNAs is presented relative to snoRNA135. Data are representative of 2 to 6 separate experiments with similar results (mean ± SEM).
Thirty-four highly expressed miRNAs in mouse BMDCs
| miRNA name . | Mean endogenous level . | miRNA name . | Mean endogenous level . |
|---|---|---|---|
| mmu-miR-23a | 84.84 | mmu-miR-223 | 27.14 |
| mmu-miR-21 | 83.65 | mmu-miR-19b | 21.92 |
| mmu-miR-142-3p | 79.52 | mmu-let-7i | 19.64 |
| mmu-miR-690 | 54.52 | mmu-miR-30b | 18.52 |
| mmu-let-7a | 50.33 | mmu-miR-147 | 16.96 |
| mmu-miR-494 | 50.14 | mmu-miR-27b | 15.36 |
| mmu-miR-16 | 49.85 | mmu-miR-15b | 13.93 |
| mmu-miR-709 | 48.64 | mmu-miR-222 | 13.58 |
| mmu-miR-29a | 48.36 | mmu-miR-15a | 13.31 |
| mmu-miR-22 | 41.43 | mmu-miR-378 | 12.31 |
| mmu-miR-142-5p | 38.36 | mmu-let-7c | 11.85 |
| mmu-miR-146b | 32.92 | mmu-miR-26a | 11.72 |
| mmu-miR-711 | 31.8 | mmu-miR-762 | 11.27 |
| mmu-miR-23b | 29.37 | mmu-miR-17 | 10.8 |
| mmu-miR-24 | 28.9 | mmu-miR-720 | 10.74 |
| mmu-miR-155 | 28.38 | mmu-miR-19a | 10.26 |
| mmu-let-7d | 27.47 | mmu-miR-27a | 9.82 |
| miRNA name . | Mean endogenous level . | miRNA name . | Mean endogenous level . |
|---|---|---|---|
| mmu-miR-23a | 84.84 | mmu-miR-223 | 27.14 |
| mmu-miR-21 | 83.65 | mmu-miR-19b | 21.92 |
| mmu-miR-142-3p | 79.52 | mmu-let-7i | 19.64 |
| mmu-miR-690 | 54.52 | mmu-miR-30b | 18.52 |
| mmu-let-7a | 50.33 | mmu-miR-147 | 16.96 |
| mmu-miR-494 | 50.14 | mmu-miR-27b | 15.36 |
| mmu-miR-16 | 49.85 | mmu-miR-15b | 13.93 |
| mmu-miR-709 | 48.64 | mmu-miR-222 | 13.58 |
| mmu-miR-29a | 48.36 | mmu-miR-15a | 13.31 |
| mmu-miR-22 | 41.43 | mmu-miR-378 | 12.31 |
| mmu-miR-142-5p | 38.36 | mmu-let-7c | 11.85 |
| mmu-miR-146b | 32.92 | mmu-miR-26a | 11.72 |
| mmu-miR-711 | 31.8 | mmu-miR-762 | 11.27 |
| mmu-miR-23b | 29.37 | mmu-miR-17 | 10.8 |
| mmu-miR-24 | 28.9 | mmu-miR-720 | 10.74 |
| mmu-miR-155 | 28.38 | mmu-miR-19a | 10.26 |
| mmu-let-7d | 27.47 | mmu-miR-27a | 9.82 |
Expression levels of miRs in DCs. The endogenous expression value was obtained by median signals over relative background signals averaged from 4 biologically separate arrays and was extracted from supplemental Table 1.
We next validated the array expression patterns of a select few miRs that were computationally predicted to target IL-6 3′ UTR with the TaqMan quantitative RT-PCR (Q-RT-PCR). The expression pattern of miR-142-3p, miR-let-7a, miR-223, miR-27a/b, the miR-146a and miR-155 at baseline was consistent with the miR array data (Figure 1C). However, their expression levels were differentially regulated on LPS treatment. The expression of miR-155, and miR-146a known LPS-inducible miRs14 were expectedly up-regulated while the expression of miR142-3p and miR-let-7a was significantly down-regulated and no change was observed in the expression of miR-223 and miR-27a/b after treatment with LPS (Figure 1C).
Endogenous miR-142-3p targets IL-6 3′ UTRs in DCs
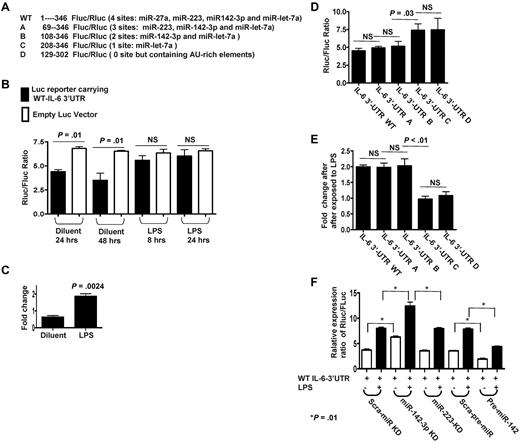
To directly analyze the miRs that specifically target IL-6 in the DCs we used the vector psiCHECK reporter system that contains modified versions of both Renillar reniformis (sea pansy) luciferase (hRluc) and Photinus pyalis (firefly) luciferase (hluc+) on a single plasmid (supplemental Figure 2).27 Based on the computational prediction, we cloned the cDNA fragment from 1 to 346 nt in mouse WT IL-6 3′UTR containing the potential miR binding sites for miR-27a, miR-223, miR-142-3p, and let-7a into the MCS of psiCHECK reporter right after hRluc stop codon, named as WT reporter (Figure 2A). The WT reporter or empty psiCHECK were transfected into DCs and the ratios of Rluc/Fluc were calculated after treatment with LPS or the control diluents. The expression ratios of Rluc vs Fluc in WT reporter were significantly reduced at 24 or 48 hours transfection compared with vector controls, P < .01 (Figure 2B), suggesting that the IL-6 3′UTR was likely being targeted by the above endogenous miRs. In contrast, on treatment with LPS, the ratio of Rluc vs Fluc in WT reporter was significantly increased (P = .0024; Figure 2C). Collectively, these data indicate that Rluc expression driven by the IL-6 3′UTR reporter was repressed by the endogenous miRs in diluent control DCs and was de-repressed on LPS treatment.
Endogenous miR-142-3p directly targets IL-6 3′ UTRs in DCs. (A) Schematic of the psiCHECK-2 constructs inserted with WT or truncated fragments of IL-6 3′UTR with the distinct miR target sites. (B) Differential regulatory repressions of psiCHECK-2 reporter carrying WT IL-6 3′UTR by endogenous miRNAs in DCs treated with or without LPS. Data shown are representative of results from 3 independent experiments (mean ± SEM). (C) Significantly de-repressed Luc expressions following LPS treatment. Fold changes of expression levels of WT reporters were calculated after first normalizing to empty vector control and were then compared between DCs treated with or without LPS. Data shown are representative of results from 3 separate experiments (mean ± SEM). (D) Differentially suppressed expression ratios of Luc reporters carrying WT or the various truncated IL-6 3′UTR that are mutated at various target sites for the endogenous miRs. Data are representative of 5 separate experiments (mean ± SEM). (E) Differentially regulated de-repression of Luc reporters by LPS treatment. Fold changes were calculated after normalized to empty vector first then compared between DCs treated with or without LPS. Data were obtained over 4 independent experiments (mean ± SEM). (F) Endogenous miR-142-3p predominantly targets IL-6 3′ UTR. Expression levels of Luc reporters carrying WT IL-6 3′UTR were measured after knockdown of either miR-142-3p or miR-223 and after overexpression of miR-142 in DCs treated with or without LPS. The results are from 4 separate experiments (mean ± SEM).
Endogenous miR-142-3p directly targets IL-6 3′ UTRs in DCs. (A) Schematic of the psiCHECK-2 constructs inserted with WT or truncated fragments of IL-6 3′UTR with the distinct miR target sites. (B) Differential regulatory repressions of psiCHECK-2 reporter carrying WT IL-6 3′UTR by endogenous miRNAs in DCs treated with or without LPS. Data shown are representative of results from 3 independent experiments (mean ± SEM). (C) Significantly de-repressed Luc expressions following LPS treatment. Fold changes of expression levels of WT reporters were calculated after first normalizing to empty vector control and were then compared between DCs treated with or without LPS. Data shown are representative of results from 3 separate experiments (mean ± SEM). (D) Differentially suppressed expression ratios of Luc reporters carrying WT or the various truncated IL-6 3′UTR that are mutated at various target sites for the endogenous miRs. Data are representative of 5 separate experiments (mean ± SEM). (E) Differentially regulated de-repression of Luc reporters by LPS treatment. Fold changes were calculated after normalized to empty vector first then compared between DCs treated with or without LPS. Data were obtained over 4 independent experiments (mean ± SEM). (F) Endogenous miR-142-3p predominantly targets IL-6 3′ UTR. Expression levels of Luc reporters carrying WT IL-6 3′UTR were measured after knockdown of either miR-142-3p or miR-223 and after overexpression of miR-142 in DCs treated with or without LPS. The results are from 4 separate experiments (mean ± SEM).
To determine whether only the miR-142-3p and/or other miRs also target IL-6 3′UTR, we systematically mutated and cloned the various miR target sites in the IL-6 3′UTR fragments using the reporter system as shown (Figure 2A). In addition to the WT construct, the following constructs were included: construct A containing target sites for miR-223, 142-3p, and let-7a; construct B containing target sites for only miR-142-3p and miR-let-7a; construct C containing target site for only let-7a; and construct D, contained no recognized miR target sites (Figure 2A). At baseline, constructs A and B displayed similar Rluc/Fluc ratios as the WT reporter (Figure 2D). Furthermore, treatment with LPS increased the ratio of Rluc/Fluc by 2-fold compared with diluent controls in the WT, A and B, constructs (Figure 2E). In contrast, construct C (contains target site for only let-7a) and construct D (that has no recognized miR target sites) displayed significantly high expression ratios of Rluc/Fluc compared with WT reporter but showed no change in expression ratios when treated with LPS (Figure 2D-E). Because only constructs A and B were as functional as the WT reporter, these data collectively indicate that only the site specific for miR-142-3p is critical for the regulation of IL-6 3′UTR by the endogenous miRs.
Next, to further confirm the specificity of the regulation of IL-6 3′UTR by miR-142-3p, we first knocked-down endogenous miR-142-3p or miR-223 as a control with LNA-knockdown probes. The miR-142-3p and miR-223 were efficiently knocked down (supplemental Figure 3A-B). The suppression of miR-142-3p but not miR-223 increased the ratio by ∼ 2-fold, P < .01 (Figure 2F).
We next transfected DCs with pre-miR-142 to evaluate the effect of an increase in the baseline expression of miR-142-3p (overexpression) on the ratio of Luc reporters carrying WT IL-6 3′UTR. The basal expression ratio of Luc reporters carrying WT IL-6 3′UTR was significantly reduced on overexpression of exogenous miR-142-3p in the DCs (Figure 2F). Furthermore, LPS treatment increased the expression ratio of the IL-6 3′UTR reporter in all groups but was significantly greater in the miR-142-3p knockdown group (P < .01) compared with the scrambled control or the miR-223 knockdown groups (Figure 2F). These data demonstrate that miR-142-3p directly targets and regulates IL-6 3′UTR in DCs.
Knockdown and overexpression of miR-142-3p regulate endogenous IL-6 expression
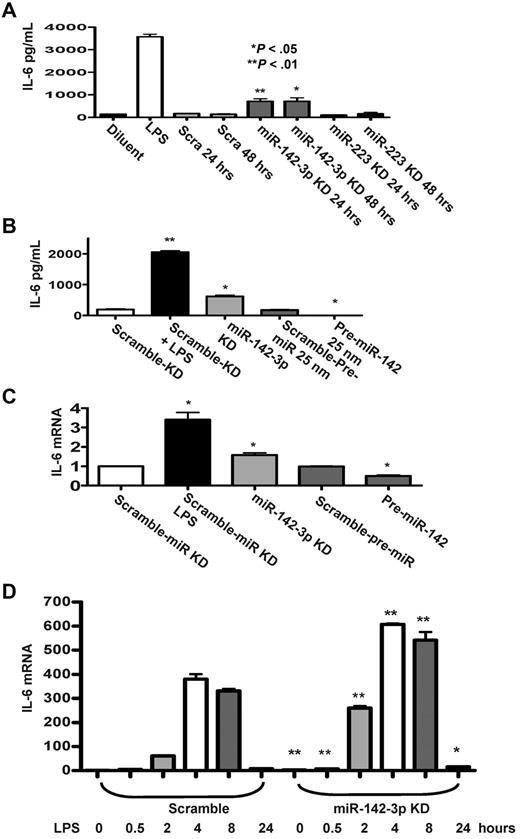
In light of the prediction by the computational algorithms, we next examined the functional relevance of miR-142-3p on the expression of endogenous IL-6 by DCs. First, we examined the effect of the repression of miR-142-3p. The miR-142-3p LNA-knockdown probe or miR-223-LNA knockdown probe was transfected into BMDCs and the level of IL-6 was measured by ELISA. A significantly higher level of IL-6 was detected in DCs after miR-142-3p knockdown for 24 or 48 hours (P < .05 and P < .01) compared with the scrambled or miR-223-LNA knockdown transfected DCs (Figure 3A).
Effect of miR-142-3p on expression of endogenous IL-6 protein and mRNA levels in DCs. (A) Knockdown of miR-142-3p de-represses IL-6 expression: IL-6 levels were measured by ELISA in the supernatants from DCs treated with or without LPS and those transfected with either miR-142-3p or miR-223 knockdown or scramble probes for 24 or 48 hours. Data shown are the pooled results from 4 separate experiments (mean ± SEM). (B) IL-6 protein levels are reduced by the overexpression of miR-142-3p: IL-6 was measured by the ELISA in the supernatants of DCs treated with LPS or diluents and ELISA in DCs transfected with either miR-142-3p knockdown or scramble probes or pre-miR-142 duplex or scramble control for 48 hours. Data shown are the pooled results from 3 separate experiments. (C) IL-6 mRNA expression is altered with either knockdown or overexpression of miR-142-3p: IL-6 mRNA levels were analyzed by quantitative RT-PCR in DCs that were transfected with either miR-142-3p knockdown or pre-miR-142 overexpression or scramble controls for 48 hours. Data are combined from 5 experiments with similar results (mean ± SEM). (D) Kinetics of IL-6 mRNA expression following miR-142-3p knockdown and LPS treatment: DCs were transfected with miR-142-3p knockdown or scramble probes for 24 hours, and then treated with LPS for the indicated time. IL-6 mRNA levels were analyzed as in panel C. Data shown are combined results from 3 similar experiments (mean ± SEM).
Effect of miR-142-3p on expression of endogenous IL-6 protein and mRNA levels in DCs. (A) Knockdown of miR-142-3p de-represses IL-6 expression: IL-6 levels were measured by ELISA in the supernatants from DCs treated with or without LPS and those transfected with either miR-142-3p or miR-223 knockdown or scramble probes for 24 or 48 hours. Data shown are the pooled results from 4 separate experiments (mean ± SEM). (B) IL-6 protein levels are reduced by the overexpression of miR-142-3p: IL-6 was measured by the ELISA in the supernatants of DCs treated with LPS or diluents and ELISA in DCs transfected with either miR-142-3p knockdown or scramble probes or pre-miR-142 duplex or scramble control for 48 hours. Data shown are the pooled results from 3 separate experiments. (C) IL-6 mRNA expression is altered with either knockdown or overexpression of miR-142-3p: IL-6 mRNA levels were analyzed by quantitative RT-PCR in DCs that were transfected with either miR-142-3p knockdown or pre-miR-142 overexpression or scramble controls for 48 hours. Data are combined from 5 experiments with similar results (mean ± SEM). (D) Kinetics of IL-6 mRNA expression following miR-142-3p knockdown and LPS treatment: DCs were transfected with miR-142-3p knockdown or scramble probes for 24 hours, and then treated with LPS for the indicated time. IL-6 mRNA levels were analyzed as in panel C. Data shown are combined results from 3 similar experiments (mean ± SEM).
Next, we transfected DCs with pre-miR-142 to evaluate the effect of miR-142-3p over expression on the endogenous levels of IL-6. The basal expression levels of IL-6 disappeared in DCs that were transfected with pre-miR-142 compared with scrambled controls (Figure 3B). To further assess whether miR-142-3p also impacts the stability of IL-6 mRNA, the DCs were treated as in Figure 3B, and the samples were processed for Q-RT-PCR. Consistent with the above data, significantly increased levels of IL-6 mRNA expression were observed in DCs transfected with miR-142-3p-LNA knockdown probe compared with the DCs transfected with the scrambled control probe (Figure 3C), while overexpression with pre-miR-142 transfection reduced the levels of IL-6 mRNA compared with scrambled controls (Figure 3C).
To understand the functional role of miR-142-3p on IL-6 induction after LPS stimulation, DCs were transfected with either miR-142-3p-LNA knockdown probe or the scrambled control for 24 hours. The cells were then treated with LPS for varying duration (ranging from 30 minutes to 24 hours). As shown in Figure 3D, knockdown of miR-142-3p significantly increased IL-6 mRNA expression both at baseline and after treatment with LPS (P < .01 or P < .04, respectively, Figure 3D). This enhanced induction of IL-6 mRNA by the DCs after LPS stimulation after miR-142-3p knockdown was observed at 30 minutes and lasted even at 24 hours (Figure 3D).
miR-142-3p specifically targets only IL-6 expression in DCs
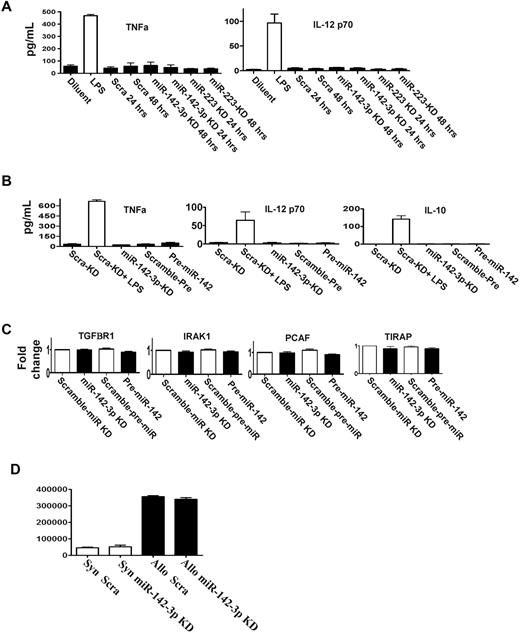
DCs stimulated by LPS also secrete other cytokines such as TNF-α, IL-12, and IL-10.17,22 We therefore analyzed whether miR-142-3p or miR-223 regulated their expression. DCs were transfected with the LNA knockdown probes for either miR-142-3p or miR-223 or pre-miR-142 or the scrambled control. The expression of TNFα, IL-12, and IL-10 was increased after LPS stimulation compared with the baseline regardless of transfection of the above probes (Figure 4A-B).
miR-142-3p does not affect the endogenous expression of other cytokine responses by the DCs. (A) Knockdown of either miR-142-3p and miR-223 does not affect other proinflammatory cytokines: levels of TNFα and IL-12 were analyzed by ELISA in the supernatants of DCs treated with LPS or diluents and transfected with either miR-142-3p knockdown or scramble probes or miR-223 knockdown probes for 48 hours. Data shown are combined results from 3 similar experiments (mean ± SEM). (B) The expression of cytokines TNFα, IL-12, and IL-10 are not affected by either the overexpression or knockdown of miR-142-3p: TNFα, IL-12, and IL-10 analyzed by ELISA in the supernatants of DCs treated with LPS or diluent and transfected with either miR-142-3p knockdown or scramble probes or pre-miR-142 duplex for 48 hours. Data shown are combined results from 3 similar experiments (mean ± SEM). (C) miR-142-3p has no direct impact on the expression of other immune molecules: the mRNA levels of TGFBR1, IRAK1, PCAF, and TIRAP were analyzed by quantitative RT-PCR in the DCs that were transfected with either miR-142-3p knockdown or pre-miR-142 duplex or scramble control for 48 hours. Data are combined from 3 experiments with similar results (mean ± SEM). (D) Knockdown of miR-142-3p in DCs does not affect their ability to stimulate allogeneic T cells: allogeneic (BALB/c) T cells were cultured with B6 BM DCs that were transfected with either miR-142-3p knockdown or scramble probes. T-cell proliferation was evaluated at 96 hours following pulsing with 3H for the last 18 hours. Data shown are results from 1 of 2 similar experiments (mean ± SEM).
miR-142-3p does not affect the endogenous expression of other cytokine responses by the DCs. (A) Knockdown of either miR-142-3p and miR-223 does not affect other proinflammatory cytokines: levels of TNFα and IL-12 were analyzed by ELISA in the supernatants of DCs treated with LPS or diluents and transfected with either miR-142-3p knockdown or scramble probes or miR-223 knockdown probes for 48 hours. Data shown are combined results from 3 similar experiments (mean ± SEM). (B) The expression of cytokines TNFα, IL-12, and IL-10 are not affected by either the overexpression or knockdown of miR-142-3p: TNFα, IL-12, and IL-10 analyzed by ELISA in the supernatants of DCs treated with LPS or diluent and transfected with either miR-142-3p knockdown or scramble probes or pre-miR-142 duplex for 48 hours. Data shown are combined results from 3 similar experiments (mean ± SEM). (C) miR-142-3p has no direct impact on the expression of other immune molecules: the mRNA levels of TGFBR1, IRAK1, PCAF, and TIRAP were analyzed by quantitative RT-PCR in the DCs that were transfected with either miR-142-3p knockdown or pre-miR-142 duplex or scramble control for 48 hours. Data are combined from 3 experiments with similar results (mean ± SEM). (D) Knockdown of miR-142-3p in DCs does not affect their ability to stimulate allogeneic T cells: allogeneic (BALB/c) T cells were cultured with B6 BM DCs that were transfected with either miR-142-3p knockdown or scramble probes. T-cell proliferation was evaluated at 96 hours following pulsing with 3H for the last 18 hours. Data shown are results from 1 of 2 similar experiments (mean ± SEM).
Each miR has the potential to repress the expression of many genes, which may be predicted by computational algorithms.2 In addition to IL-6, the computational algorithms predicted that the following known immunologically relevant genes may be potential targets for miR-142-3p: namely, TIRAP, IRAK1, TGFBR1, and PCAF.28 To analyze the relevance of miR-142-3p in the regulation of these molecules, DCs were transfected with miR-142-3p LNA knockdown probes, the pre-miR-142 duplexes or the relevant scrambled controls for 48 hours. In contrast to IL-6 regulation, no significant changes were observed in the mRNA and protein expression of TIRAP, IRAK, TGFBR1, or PCAF in any of the groups (Figure 4C, supplemental Figure 4).
In addition to secretion of cytokines, DCs also play a key role in stimulating T cells. Therefore, we next examined whether repression of endogenous miR-142-3p will modulate their allogeneic T-cell stimulatory functions. In contrast to the impact on the secretion of IL-6, transfection of DCs with miR-142-3p knockdown probes did not alter their allostimuatory responses in an MLR reaction compared with DCs transfected with control probes (Figure 4D).
Targeting miR-142-3p reduces endotoxin induced mortality
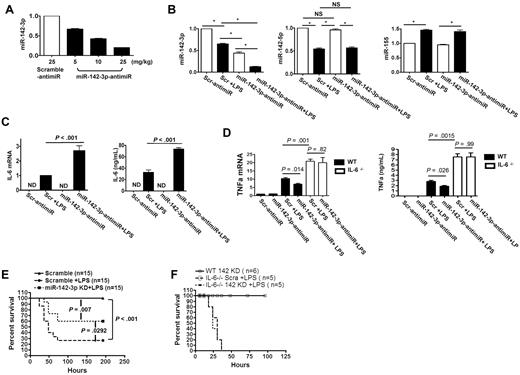
We next examined the role of miR-142-3p in regulating in vivo IL-6 responses. We used high-affinity LNA-anti-miR with phosphorothioate modifications, which has been shown to facilitate silencing of miRNAs in vivo.29 We first confirmed the efficiency and specificity of the miR-142-3p–anti-miR probes in vitro (supplemental Figure 5). The baseline expression of endogenous miR-142-3p was significantly repressed by miR-142-3p–anti-miR in a dose-dependent manner (5 mg/kg, 10 mg/kg, or 25 mg/kg IP injection) compared with the scrambled control (25 mg/kg; Figure 5A).
miR-142-3p regulates IL-6 expression in vivo. To analyze the effect of miR-142-3p, B6 mice were injected with either high-affinity LNA-anti–miR-142-3p with phosphorothioate modifications or scrambled control at various doses. The splenocytes were then harvested and used for the analyses of the expression of miR-142-3p as described in “Methods.” (A) Dose-dependent in vivo knockdown of miR-142-3p by anti–miR-142-3p in splenocytes measured by TaqMan quantitative RT-PCR. Data shown are from 1 of 2 experiments with 3-4 mice/group (mean ± SEM). (B) anti–miR-142-3p specifically knocked down miR-142-3p but did not affect off-target miRs. Expression levels of miR-142-3p and other miRs such as miR-142-5p or miR-155 were measured by TaqMan quantitative RT-PCR. Data shown are from 1 of 2 experiments with 3 mice/group (mean ± SEM). (C) In vivo knockdown of miR-142-3p by anti–miR-142-3p de-repressed LPS-induced IL-6 expression at both mRNA and protein levels: IL-6 mRNA levels in splenocytes were determined by quantitative RT-PCR and the protein levels was measured in the sera by ELISA. Data shown are from 1 of 2 experiments with 4-5 mice/group. (D) In vivo knockdown of miR-142-3p by anti–miR-142-3p does not affect LPS-induced TNFα expression at both mRNA and protein levels: TNFα mRNA levels in splenocytes were determined by quantitative RT-PCR and the protein levels was measured in the sera by ELISA of IL-6−/− and WT B6 mice following administration of LPS. Data shown are from 2 experiments with 6 mice/group. (E) Silencing of mir-142-3p mitigated endotoxin-induced mortality. Treatment of mice with LNA-modified oligonucleotide complementary to miR-142-3p (anti–miR-142-3p) followed by LPS injection (■) significantly reduced mortality compared with animals that were treated with scrambled anti-miR and LPS (●). P = .029. Data shown are combined from 2 different experiments with similar results. (F) Silencing of mir-142-3p does not alter endotoxin-induced mortality in IL-6−/− mice. Treatment of IL-6−/− mice with LNA-modified oligonucleotide complementary to miR-142-3p followed by LPS injection (dotted line, n = 6) did not affect the rate and overall mortality compared with IL-6−/− animals that were treated with scrambled anti-miR and LPS (solid line, n = 6). P = NS.
miR-142-3p regulates IL-6 expression in vivo. To analyze the effect of miR-142-3p, B6 mice were injected with either high-affinity LNA-anti–miR-142-3p with phosphorothioate modifications or scrambled control at various doses. The splenocytes were then harvested and used for the analyses of the expression of miR-142-3p as described in “Methods.” (A) Dose-dependent in vivo knockdown of miR-142-3p by anti–miR-142-3p in splenocytes measured by TaqMan quantitative RT-PCR. Data shown are from 1 of 2 experiments with 3-4 mice/group (mean ± SEM). (B) anti–miR-142-3p specifically knocked down miR-142-3p but did not affect off-target miRs. Expression levels of miR-142-3p and other miRs such as miR-142-5p or miR-155 were measured by TaqMan quantitative RT-PCR. Data shown are from 1 of 2 experiments with 3 mice/group (mean ± SEM). (C) In vivo knockdown of miR-142-3p by anti–miR-142-3p de-repressed LPS-induced IL-6 expression at both mRNA and protein levels: IL-6 mRNA levels in splenocytes were determined by quantitative RT-PCR and the protein levels was measured in the sera by ELISA. Data shown are from 1 of 2 experiments with 4-5 mice/group. (D) In vivo knockdown of miR-142-3p by anti–miR-142-3p does not affect LPS-induced TNFα expression at both mRNA and protein levels: TNFα mRNA levels in splenocytes were determined by quantitative RT-PCR and the protein levels was measured in the sera by ELISA of IL-6−/− and WT B6 mice following administration of LPS. Data shown are from 2 experiments with 6 mice/group. (E) Silencing of mir-142-3p mitigated endotoxin-induced mortality. Treatment of mice with LNA-modified oligonucleotide complementary to miR-142-3p (anti–miR-142-3p) followed by LPS injection (■) significantly reduced mortality compared with animals that were treated with scrambled anti-miR and LPS (●). P = .029. Data shown are combined from 2 different experiments with similar results. (F) Silencing of mir-142-3p does not alter endotoxin-induced mortality in IL-6−/− mice. Treatment of IL-6−/− mice with LNA-modified oligonucleotide complementary to miR-142-3p followed by LPS injection (dotted line, n = 6) did not affect the rate and overall mortality compared with IL-6−/− animals that were treated with scrambled anti-miR and LPS (solid line, n = 6). P = NS.
We next used the 10 mg/kg dose of the control and miR-142-3p–anti-miR to determine the effect of in vivo repression of miR–142-3p on IL-6 after LPS stimulation. The expression of miR–142-3p expression in spleens was reduced by 40% in the control anti-miR-treated-mice after LPS administration compared with diluent-injected mice. The expression of miR-142-3p was decreased by 50% in mice administered with miR-142-3p–anti-miR alone and decreased even further (80%) in mice that received both miR-142–3p-anti-miR and LPS (Figure 5B). Furthermore, the expression of miR-142-5p, miR-155 (Figure 5B) or miR-146a (supplemental Figure 5) was not altered demonstrating in vivo specificity.
We next examined whether the in vivo changes in miR-142-3p expression by miR-142-3p–anti-miR with or without LPS administration altered IL-6 mRNA expression in the spleens (Q-RT-PCR) or the IL-6 protein levels in sera (ELISA). Consistent with in vitro observations, LPS increased the expression of IL-6 in both control-anti-miR or miR-142-3p–anti-miR injected mice compared with non-LPS controls. Importantly, the highest expression of IL-6 mRNA (2.5-fold) and protein (2-fold) was observed in the mice that were injected with both miR-142-3p–anti-miR and LPS (Figure 5C). In contrast, expression of TNFα was increased after LPS treatment but was not significantly different demonstrating the specificity for induction of IL-6 (Figure 5D).
Next, as shown in Figure 5E, silencing of miR-142-3p in the WT B6 animals significantly reduced endotoxin-mediated mortality compared with animals that were treated with scrambled controls (60% vs 25%, P < .03). However, targeting miR-142-3p did not completely abrogate mortality from LPS suggesting that other additional factors might be contributing to overall mortality from endotoxin.
To confirm the specificity of IL-6 in mediating the protection against endotoxin-induced mortality by targeting miR-142-3p, we next used IL-6−/− animals. The IL-6−/− mice were injected with LPS and treated with either the control, scrambled or the miR-142-3p-anti-miR. All of the IL-6−/− mice that were injected with LPS and received the control anti-miR died (Figure 5F). However, the IL-6−/− animals that were administered with miR-142-3p–anti-miR, in contrast to similarly treated WT animals (Figure 5E), also died from endotoxin injection demonstrating the specificity of IL-6 for the protection (Figure 5F). To further confirm the specificity, we also analyzed the expression of TNFα mRNA (another cytokine that is typically elevated in endotoxemia) in the spleens of both WT and IL-6−/− animals early after LPS administration. The expression of TNFα was significant increased in both the WT and IL-6−/− animals regardless of whether they were treated with the control or miR-142-3p-anti-miR (Figure 5D). The levels of TNFα were also elevated at the protein level as measured by ELISA in the sera of the both WT and IL-6−/− animals that were injected with LPS, regardless of anti-miR administration (Figure 5D).
Human and murine hematopoietic cells express miRNA-142-3p
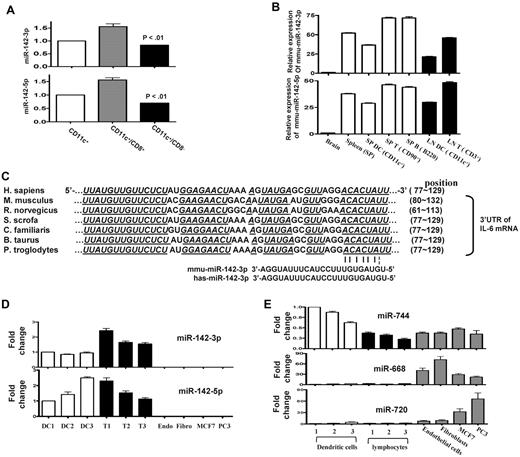
Both miR-142-3p and miR-142-5p are highly expressed in mouse BMDCs (supplemental Table 1). Given that DCs are a heterogeneous population, we next determined whether the DC subsets demonstrated differential expression of miR-142. To this end, we analyzed CD8+ and CD8− subsets of DCs and found that miR-142 was expressed (Figure 6A). Consistent with previous reports,30,31 we confirmed that both miR-142-3p and miR-142-5p were also highly expressed in the T and B lymphocytes of naive mice (Figure 6B) and also in the peritoneal macrophages (data not shown).
Human and murine hematopoietic cells express miR-142. (A) Endogenous expression of miR-142 in CD11c DC subsets: basal expression levels of miRNA-142-3p and 142-5p was analyzed by in purified FACS sorted CD11c+/CD8+ DC and CD11c+/CD8− DC subsets with quantitative RT-PCR. Data shown are from 1 of 3 similar experiments (mean ± SEM). (B) miR-142s are specific for hematopoietic cells in mice. miR-142 expression was analyzed in the brain, whole splenocytes, DCs harvested from spleen, and the T and B lymphocytes harvested from the spleen or lymphoid nodes of naive mice. Data were obtained from 1 of 2 experiments with n = 3/group. (C) The miR-142-3p target sequence in the 3′UTR of IL-6 mRNA is conserved across several mammalian species. Seed sequence pairing is indicated by lines. (D) miR-142-3p and miR-142-5p are highly expressed in human hematopoietic cells: DCs and T lymphocytes were harvested from the PBMCs that were freshly isolated from normal healthy human volunteers (n = 3). The human cell lines—such as the endothelial cells, fibroblasts, and some cancers cells such as MCF7 and PC3—were used for controls as the nonhematopoietic cells. miR-142-3p and miR-142-5p expression was analyzed by quantitative RT-PCR. Results shown are from 1 of 2 experiments. (E) Control miR expression analyses in human cells: The above cells were also concomitantly analyzed for expression of miR-744, 668, and 720 as additional controls. miR-744 was extensively expressed in hematopoietic, nonhematopoietic, and cancer cells. Both miR-668 and miR-720 were expressed in nonhematopoietic and cancer cells but not in hematopoietic cells.
Human and murine hematopoietic cells express miR-142. (A) Endogenous expression of miR-142 in CD11c DC subsets: basal expression levels of miRNA-142-3p and 142-5p was analyzed by in purified FACS sorted CD11c+/CD8+ DC and CD11c+/CD8− DC subsets with quantitative RT-PCR. Data shown are from 1 of 3 similar experiments (mean ± SEM). (B) miR-142s are specific for hematopoietic cells in mice. miR-142 expression was analyzed in the brain, whole splenocytes, DCs harvested from spleen, and the T and B lymphocytes harvested from the spleen or lymphoid nodes of naive mice. Data were obtained from 1 of 2 experiments with n = 3/group. (C) The miR-142-3p target sequence in the 3′UTR of IL-6 mRNA is conserved across several mammalian species. Seed sequence pairing is indicated by lines. (D) miR-142-3p and miR-142-5p are highly expressed in human hematopoietic cells: DCs and T lymphocytes were harvested from the PBMCs that were freshly isolated from normal healthy human volunteers (n = 3). The human cell lines—such as the endothelial cells, fibroblasts, and some cancers cells such as MCF7 and PC3—were used for controls as the nonhematopoietic cells. miR-142-3p and miR-142-5p expression was analyzed by quantitative RT-PCR. Results shown are from 1 of 2 experiments. (E) Control miR expression analyses in human cells: The above cells were also concomitantly analyzed for expression of miR-744, 668, and 720 as additional controls. miR-744 was extensively expressed in hematopoietic, nonhematopoietic, and cancer cells. Both miR-668 and miR-720 were expressed in nonhematopoietic and cancer cells but not in hematopoietic cells.
The region targeted by miRNA-142-3p in the 3′UTR of IL-6 mRNA is conserved in both humans and mice. Therefore, we next examined whether miR-142-3p is also expressed in human DCs (Figure 6C). Consistent with murine data, both miR-142-3p and miR-142-5p were highly expressed in human PBMC-derived DCs and T cells (Figure 6D) but were not expressed in the nonhematopoietic human cells such as the endothelial cells, fibroblasts, and some cancer cells (Figure 6D) in contrast to the control miR-744 expressed in all cells and the nonhematopoietic miR-668 and miR-720s (Figure 6E).
Discussion
Gram-negative sepsis induced by endotoxin is a major cause of death in critically ill patients.1,32 Endotoxemia is characterized, initially, by a massive inflammatory response. DCs play an important role in this response.7 Endotoxin (LPS) is detected by TLR-4 after which distinct signaling pathways are activated that lead to the secretion of several inflammatory cytokines such as IL-6 and TNF-α.6,7 The role of miRs in modulating the DC functions and their impact on in vivo endotoxin-induced mortality are unknown.3,8 The objective of our study was the identification of the miRs controlling a prototypical TLR-mediated inflammatory response, namely the endotoxin-induced gene expression program in DCs and to determine whether targeting a specific miR can regulate endotoxin-induced mortality.
We used microarray technology to identify miRNAs that are expressed at baseline in primary murine DCs. To analyze the genes that might be regulated by the miRNAs, we also simultaneously profiled the mRNAs at baseline and after LPS stimulation. Because IL-6 is one of the most up-regulated genes and is known to be involved in many DC-mediated responses,33 we computationally nominated the miRNAs that might contribute to IL-6 regulation. The intersecting results of multiple prediction algorithms suggested a strong association between only a few miRs and IL-6 expression. Among these overlapping miRs that were predicted to target IL-6 3′UTR, our studies with the reporters carrying IL-6 3′UTR demonstrated that miR-142-3p might be the critical suppressor for IL-6. Furthermore, studies with knockdown and exogenous expression of miR-142-3p demonstrated that it is specific and critical for suppressing IL-6 expression, having no identifiable impact on the other LPS-responsive cytokines such as TNF-α, IL-12, and IL-10, or on the molecules we examined that are related to immune-responsive pathways, such as TGFβR1, IRAK1, and TIRAP.
An important observation is the specificity of miR-142-3p for the IL-6 3′ UTR. This might be because of the integration of multiple regulatory processes known to be relevant in miRNA-mediated gene regulation such as (1) limitation of target-site accessibility to other miRNAs,34,35 (2) binding within the 3′UTR away from the stop codon,36 or (3) the composition of the AU-rich element (ARE) clusters near the site.2 However, the processes that lead to miR-142-3p down-regulation after LPS stimulation remain to be investigated.
The miR-142 has previously been shown to be expressed in murine BM, spleen, and thymus and was especially high in B cells and T-cell subsets.30,31,37 Ectopic expression of miR-142 has been found to modulate T-lymphoid lineage differentiation.31 It has also been associated with malignancies of hematopoietic lineage, especially those involving t(8;17) translocations.38,39 However, the functions of endogenous miR-142 are largely unknown. Our data demonstrate that miR-142-3p and miR-142-5p are highly expressed in murine and human DCs but not in human endothelial or epithelial cells, suggesting hematopoietic cell-specific expression. Thus, the reduction of IL-6 and the in vivo survival benefit is likely because of the effect of miR142-3p on not only DCs but also the other hematopoietic-derived cells such as macrophages and lymphocytes. Nonetheless, the specific effects of miR-142 in addition to regulation of IL-6 in the regulation of other genes and function of macrophages, the T and B cells will be explored in future studies.
Several recent studies have demonstrated a role for miRs in regulating inflammatory responses to LPS.3,8,14,15,40,41 These studies were done primarily in cell lines and macrophages. The macrophage inflammatory response to pathogens has been shown to involve up-regulation of several miRs, such as miR-155, miR-146, miR-147, miR-21, and miR-9.3,8,14,15 We now extend these studies to primary DCs and to our knowledge, for the first time, demonstrate in vivo relevance to LPS-induced inflammatory response. The microarray profiling data from our study suggest that the inflammatory responses of macrophages and DCs may share many similarities and also certain differences. We show that LPS-induced inflammatory response regulated the expression of miR-142-3p in DCs. In contrast, we found that IFNγ, dsRNA (TLR-3 agonist) induced significant increases in miR-142-3p expression reflecting that the pathways and factors are distinct and may involve products of the secondary responsive genes. More importantly, we found that targeting miR-142-3p mitigated LPS-induced mortality in an IL-6–specific manner. Nonetheless, it is important to point out a key caveat to these observations. The observed in vivo effects are likely due the consequence of targeting miR-142-3p in all hematopoietic cells and do not represent effects that are specific to DCs or to any particular hematopoietic cellular subsets. Future studies using mice that are specifically deficient in IL-6–producing DCs and/or those with deficiency of miR-142-3p that is limited to DCs will help dissect the in vivo DC specificity of our observations.
In addition to DCs, IL-6 is also expressed by several other types of cells. Previous reports have suggested that IL-6 expression is regulated by miR-26 in a Zcchc 11 dependent manner in lung epithelial and hepatic cells while its expression was likely regulated by Let-7 in mammary epithelial cells.42,43 In contrast, we demonstrate for the first time that IL-6 expression in immune cells, specifically in DCs, is directly modulated by miR-142-3p. We further show that miR-142-3p is expressed by DCs from both mice and humans and that its target site in IL-6 3′UTR is highly conserved between mice and humans. Our data thus confirm and extend the previous observations on the role of miRs in regulating IL-6 and suggest that the expression pattern of miRs and the regulation of specific genes are cell/tissue-specific and might depend on environmental cues.
IL-6 is known to be a key component of LPS-induced endotoxemia and septic shock.33,44-46 Depending on the context, IL-6 can act as either as a pro- or an anti-inflammatory cytokine.45,47 Despite its proinflammatory nature, expression of endogenous IL-6 can paradoxically reduce endotoxin-induced mortality.46 The failure of anti-inflammatory therapies for sepsis in clinical trials clearly raises the question of whether mortality in sepsis is only a consequence of uncontrolled proinflammatory response.48 Although the initial, proinflammatory phase of sepsis can cause mortality, the overall mortality from sepsis is a consequence of both the initial hyperinflammatory state and the later immunosuppressive state.1,50 Importantly, our study also demonstrates a role for miR-142-3p in the in vivo regulation of LPS-induced endogenous expression of IL-6. Using high-affinity LNA-modified oligonucleotides with a complete phosphorothioate backbone to achieve higher knockdown efficiency,29 we successfully knocked down miR-142-3p and modulated IL-6 expression after endotoxemia. More importantly, this significantly reduced endotoxin-induced mortality. Our data suggest that targeting miR-142-3p in vivo with LNA-modified oligonucleotides may have therapeutic potential in endotoxemia. In addition, because IL-6 can also be induced by other noninfectious inflammatory stimuli26 and its deregulation has also implicated in the pathology of several other diseases such as multiple myeloma, rheumatoid arthritis, systemic-onset juvenile chronic arthritis, and GVHD,26,50 our data suggest that targeting miR-142-3p may also have potential ramifications for several inflammatory diseases.
In summary, we provide evidence for a novel miR-142-3p in specifically modulating LPS-induced DC responses and mortality, and demonstrate that targeting miRs may represent a novel strategy for mitigating endotoxin-mediated sepsis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rebecca Evers for the technical help with animal care issues.
This work was supported by National Institutes of Health grants AI-075284 (P.R.) and HL-090775 (P.R.).
P.R. is a recipient of the Scholar in Clinical Research from the Leukemia & Lymphoma Society and the Basic Science Investigator Award from American Society of Transplantation.
National Institutes of Health
Authorship
Contribution: Y.S. designed and performed experiments, analyzed data, and wrote the paper; S.V. designed and performed experiments and analyzed data; C.A.M. performed experiments and analyzed data; Q.C. performed experiments and analyzed data; P.C., T.T., C.M., E.N., I.T., and Y.W. performed experiments; P.A.W. analyzed data and wrote the paper; and A.C. and P.R. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pavan Reddy, Department of Internal Medicine, University of Michigan Comprehensive Cancer Center, 3312 CCC, 1500 East Medical Center Dr, Ann Arbor, MI 48109-0942; e-mail: reddypr@umich.edu.