Abstract
Two types of mutations of a transcription factor CCAAT-enhancer binding protein α (C/EBPα) are found in leukemic cells of 5%-14% of acute myeloid leukemia (AML) patients: N-terminal mutations expressing dominant negative p30 and C-terminal mutations in the basic leucine zipper domain. Our results showed that a mutation of C/EBPα in one allele was observed in AML after myelodysplastic syndrome, while the 2 alleles are mutated in de novo AML. Unlike an N-terminal frame-shift mutant (C/EBPα-Nm)–transduced cells, a C-terminal mutant (C/EBPα-Cm)–transduced cells alone induced AML with leukopenia in mice 4-12 months after bone marrow transplantation. Coexpression of both mutants induced AML with marked leukocytosis with shorter latencies. Interestingly, C/EBPα-Cm collaborated with an Flt3-activating mutant Flt3-ITD in inducing AML. Moreover, C/EBPα-Cm strongly blocked myeloid differentiation of 32Dcl3 cells, suggesting its class II mutation-like role in leukemogenesis. Although C/EBPα-Cm failed to inhibit transcriptional activity of wild-type C/EBPα, it suppressed the synergistic effect between C/EBPα and PU.1. On the other hand, C/EBPα-Nm inhibited C/EBPα activation in the absence of PU.1, despite low expression levels of p30 protein generated by C/EBPα-Nm. Thus, 2 types of C/EBPα mutations are implicated in leukemo-genesis, involving different and cooperating molecular mechanisms.
Introduction
The CCATT/enhancer binding protein α (C/EBPα) transcription factor is a critical regulator of proliferation and differentiation in myeloid cells.1,2 C/EBPα consists of an N-terminal transcriptional activation domain and a C-terminal basic leucine zipper (bZIP) domain.3-5 Two isoforms of C/EBPα proteins are generated from different translation start sites: a full-length 42-kDa protein (p42) and a truncated 30-kDa protein (p30) that lacks an N-terminal transcriptional activation domain. C/EBPα-p30 isoform inhibits C/EBPα-p42–mediated transcription.6-8 Importantly, C/EBPα promotes differentiation both by up-regulation of lineage-specific gene products9-11 and by proliferation arrest.12 Recent studies have indicated that C/EBPα-induced growth arrest is regulated by its interaction with other molecules involved in growth control: E2F,13-15 Max,16 and SWI/SNF chromatin remodeling complexes.17 For example, repression of E2F activity by E2F-C/EBPα interaction results in the down-regulation of c-Myc, leading to granulocytic differentiation.18,19
According to several studies, CEBPA mutations are found in 5%-14% of acute myeloid leukemia (AML) patients belonging to the French-American-British subtypes M1, M2, or in some cases M4.8,20-22 The mutations of the CEBPA gene can be largely categorized into 2 types: one is an N-terminal frame-shift mutation disrupting p42 and producing p30 as a major product, and the other is a C-terminal in-frame mutation disrupting the bZIP region. Interestingly, most AML patients with CEBPA mutations have both mutations simultaneously,23-25 and such patients displayed a favorable outcome.22,26 On the other hand, AML patients with single CEBPA mutations did not express a distinctive signature, presumably due to a variety of associating gene alterations, including Flt3 activating mutations. Related to this, involvement of single CEBPA mutations with myelodysplastic syndrome (MDS) remains to be clarified.27,28
Analysis of mice with genetic alterations in the CEBPA locus has contributed to delineation of molecular mechanisms by which CEBPA mutations induce leukemia. Conditional deficiency of C/EBPα led to a differentiation block at the transition between common myeloid progenitors and granulocyte/monocyte progenitors but not to development of leukemia.1 However, CebpaL/L mice, expressing only p30 as C/EBPα protein, developed myeloid leukemia with complete penetration.29 On the other hand, CebpaBRM2/BRM2 mice, carrying a point mutation in the bZIP domain that dampened E2F interaction, showed only preleukemic features.14 Interestingly, the same group has recently reported combinational effects of the C-terminal and the N-terminal mutations on leukemogenesis by using CebpaL/L mice, CebpaK/K mice carrying the K313 duplication in the C-terminal domain, and CebpaK/L mice.29,30 They proposed that efficient leukemogenesis is caused by the combination of both premalignant HSC expansion induced by C-terminal CEBPA mutation and residual myeloid lineage commitment maintained by the N-terminal CEBPA mutation.30
Causative gene alterations in hematologic malignancies have been extensively studied, and it is now recognized that multiple mutations contribute to development of leukemia. These gene alterations are categorized into 2 groups, class I and class II mutations.31,32 Class I mutations include activating mutations of FLT3, C-KIT, JAK2, SHP2, and RAS, or inactivating mutations of TP53 and NF-1, and induce proliferation or block apoptosis of hemopoietic cells. On the other hand, class II mutations disrupt normal functions of transcription factors and chromosome-modifying enzymes including MLL, RUNX1, RARA, and PU.1 and hamper differentiation of hemopoietic cells. Combinations of class I and class II mutations are frequently observed in patients' leukemic cells.33,34 In addition, we and others presented evidence that class I and class II mutations collaborate in the development of leukemia in mouse models.35,36 Among a variety of gene alterations found in leukemia, CEBPA mutations are unique because different CEBPA mutations are frequently found on different alleles in leukemic cells of de novo AML.22-26
In the present study, we searched for mutations of the CEBPA gene in patients with myeloid malignancies, and found N- and C-terminal double mutations in patients with de novo AML. In patients with MDS/AML or therapy-related AML or MDS, only N- or C-terminal single mutation was identified. We chose a C-terminal mutation 304_323dup (hereafter called C/EBPα-Cm) and an N-terminal mutation (T60fsX159) (hereafter C/EBPα-Nm) for further analysis that had been isolated as double CEBPA mutations in a de novo AML patient. To evaluate the effects of these mutations on leukemogenesis, we used a mouse bone marrow transplantation (BMT) model. Interestingly, unlike the phenotype in CebpaK/K, CebpaK/+, CebpaBRM2/BRM2, or CebpaBRM2/+ mice,14,29,30 C/EBPα-Cm alone induced AML with leukopenia in transplanted mice after BMT. We also confirmed the efficient induction of AML by coexpression of C/EBPα-Cm and C/EBPα-Nm. We will discuss the possible molecular mechanisms by which C/EBPα-Nm worked in concert with C/EBPα-Cm in accelerating leukemogenesis.
Methods
Patients and samples
We chose patients with hematologic diseases (224 MDS/AML patients, 71 therapy-related AML or MDS patients, and 89 de novo AML patients, who had been diagnosed at Hiroshima University Hospital between 1985 and 2007, are not a consecutive series of patients). All studies were approved by the Institutional Review Board at Hiroshima University and the ethics committee of the University of Tokyo (approval no. 20-10-0620). Patients' informed consents were obtained in accordance with the Declaration of Helsinki. CEBPA mutation screening by polymerase chain reaction (PCR)–single strand conformation polymorphism analysis and identification of CEBPA, AML1, N-RAS, FLT3, PTPN11, C-KIT, and TP53 mutations was performed as described previously.37
Retroviral vectors
We used 2 C/EBPα mutants, Nm or Cm, as well as C/EBPα wild-type (WT) and C/EBPα N-terminal truncated p30 (p30). C/EBPα-WT, p30, Nm, or Cm, which was tagged with a FLAG or Myc epitope at the C terminus, was inserted upstream of the internal ribosome entry site-enhanced green fluorescent protein (IRES-EGFP) cassette of pMYs-IG to generate pMYs-FLAG or Myc-tagged CEBPα-WT, p30, Nm, or Cm-IG, respectively. Similarly, these fragments were subcloned into pMXs-IRES-puro (pMXs-IP), pMXs-IRES-blasticidin (pMXs-IB), or pMYs-IRES-dsRED (pMYs-IR). Flt3-ITD cDNA, which was derived from patient's leukemic cells harboring a 20-amino acid tandem duplication called M338,39 was subcloned into pMYs-IG to generate pMYs-Flt3-ITD-IG. Human granulocyte colony-stimulating factor receptor (G-CSF-R) cDNA, a kind gift from Dr Shigekazu Nagata (Kyoto University, Kyoto, Japan), was subconed into pMXs-IB to generate pMXs-G-CSF-R-IB.
Retroviral infection was done as described previously.40 Briefly, retroviruses were generated by transient transfection of Plat-E packaging cells with FuGENE 6 (Roche Diagnostics).41,42 Growth of transduced 32Dcl3 cells, which were subject to the drug selection with 1 μg/mL puromycin or 10 μg/mL blasticidin, was estimated by quantitating luminescence as described previously.43
Flow cytometric analysis
Briefly, cells were stained with phycoerythrin-conjugated antibodies (Abs) or biotinylated Abs and phycoerythrin/Cy5-streptavidin (eBioscience). Flow cytometric analysis of the stained cells was performed with FACSCalibur flow (BD Biosciences) equipped with FlowJo Version 7.2.4 software (TreeStar).
Real-time reverse-transcription PCR
Real-time reverse-transcription (RT) PCR was performed as described previously.40 Reaction was subject to one cycle of 95°C for 30 seconds, 45 cycles of PCR at 95°C for 5 seconds, 55°C for 10 seconds, and 72°C for 10 seconds. The following primer pairs were used: 5′-AAGGCCCAGTGTGTCTCTGT-3′ (forward), and 5′-TACCAGCCCCAACTCAAAAC-3′ (reverse) for G-CSF-R, 5′-AGAGGGAAATCGTGCGTGAC-3′ (forward), and 5′-CAATAGTGATGACCTGGCCGT-3′ (reverse) for β-actin, 5′-GCCCCTAGTGCTGCATGAG-3′ (forward) and 5′-CCACAGACACCACATCAATTTCTT-3′ (reverse) for c-Myc.
Western blot analysis
Equal numbers of cells were lysed and Western blotting was performed as described previously.35,40 Anti-Flag (M2) Ab (Sigma-Aldrich), anti-c-Myc (9E10) Ab (Roche Diagnostics), anti-C/EBPα (14AA) or (N-19) Ab (Santa Cruz Biotechnology), ERK1/2, signal transducer and activator of transcription (STAT)3, STAT5, AKT1, or Flt3 Abs, and phospho-STAT3 Ab (Santa Cruz Biotechnology), anti-phospho mitogen-activated protein kinase and phospho-AKT Abs (Cell Signaling Technology), and phospho-STAT5 Ab (BD Biosciences) were used.
Luciferase assay
The 293T cells were transiently transfected with the luciferase reporter plasmid p(C/EBP)2TK (kindly provided by Atsushi Iwama, Chiba University, Japan), pMXs-C/EBPα-WT or mutants-IP, and pEF-BOS/PU.1 for C/EBPα transcriptional activity, or E2Fx6-TATA-LUC, pCMV-E2F1, and pCMV-DP1 (kindly provided by Claus Nerlov, EMBL Mouse Biology Unit, Italy) for E2F transcriptional activity.15 Luciferase assays were performed by Dual luciferase assay systems (Promega).
Immunostaining
Immunostaining of 293T cells transiently transfected with retrovirus constructs was performed as described previously.35 After fixation with 1.5% paraformaldehyde, cells were immunostained with rabbit anti-Flag Ab or fluorescein isothiocyanate–conjugated mouse anti–c-Myc Ab (Sigma-Aldrich). The cells were then stained with Alexa Fluor 546–conjugated goat anti–rabbit immunoglobulin G secondary Ab (Molecular Probes). Nuclei were counterstained with Hoechst (H33342). Fluorescent images were analyzed on a confocal microscope (FLUOVIEW FV300 scanning laser biological microscope JX70 system; Olympus) equipped with SenSys/0L cold charge-coupled device (CCD) camera (Olympus). The objective lens (an LCPlanFI 60×/1.40 NA oil) was used.
Gel shift assay
Nuclear extracts from transfected 293T cells were incubated with 2 μg of polydeoxyinosinic-deoxycytidylic acid and then with double-stranded CSF3R promoter oligonucleotide labeled with γ32P-adenosine triphosphate. Cold competition and a super-shift reaction were carried out by adding a 40-fold excess of cold CSF3R oligo or 1.5 μg of anti-C/EBP (14AA)X Ab (Santa Cruz Biotechnology), respectively. The resulting complexes were resolved on 4.5% polyacrylamide gel.44
Colony assay
Infected mouse bone marrow (BM) mononuclear cells (1 × 104) were plated in methylcellulose medium (StemCell Technologies) supplemented with 50 ng/mL each of interleukin (IL)–3, IL-6, stem cell factor, and granulocyte/macrophage-colony-stimulating factor (GM-CSF; R&D Systems) in the presence of 1 μg/mL puromycin. Colonies were counted after 1-week culture, and single-cell suspensions (104 cells) of drug-resistant colonies were subsequently replated.
Mouse BMT
Mouse BMT was performed as described previously.40 Briefly, BM mononuclear cells were isolated from the femurs and tibias of C57BL/6 (Ly-5.1) donor mice 4 days after intraperitoneal administration of 150 mg/kg 5-fluorouracil. The cells were stimulated with 50 ng/mL of mouse stem cell factor, mouse FLT3 ligand, mouse IL-6, and human thrombopoietin (all cytokines were from R&D Systems). The prestimulated cells were infected for 60 hours with the retroviruses harboring pMYs-C/EBPα-Cm-IG, pMYs-C/EBPα-Nm-IG, pMYs-Myc-tagged C/EBPα-Cm-IG, pMYs-Flag-tagged C/EBPα-Nm-IR, pMYs-Flt3-ITD-IG, pMYs-IG, or pMYs-IR, using 6-well dishes coated with RetroNectin (Takara Bio). Then, 3-5 × 105 of the infected BM cells (that had not been sorted after either single or double infection) were injected into sublethally γ-irradiated C57BL/6 (Ly-5.2) recipient mice. Overall survival of transplanted mice were estimated using the Kaplan-Meier method. All animal studies were approved by the Animal Care Committee of the Institute of Medical Science, The University of Tokyo.
Statistical analysis
Statistical significance was calculated using the Student t test for independent variables. P values < .05 were considered statistically significant.
Results
CEBPA mutations in patients with myeloid malignancies
After we performed single-strand conformation polymorphism analysis to screen for CEBPA mutations in patients with hematologic disorders, CEBPA mutations were identified in 7 of 224 MDS/AML patients, 8 of 71 therapy-related AML or MDS patients, and 5 of 89 de novo AML patients (Table 1). Although the number of the de novo AML patients with CEBPA mutations were small, they all carried both an N-terminal mutation and a C-terminal bZIP in-frame mutation on the different alleles as reported previously.20-26 On the other hand, most MDS/AML or therapy-related AML or MDS patients had single CEBPA mutations. As exceptions, we found both an N-terminal mutation and a C-terminal frame-shift mutation in 1 case of AML after MDS (patient [Pt] #806) and homozygous N-terminal frame-shift mutations in one case of therapy-related MDS/RAEB (Pt #811). Examination of other genes (RUNX1, N-RAS, FLT3, PTPN11, C-KIT, and TP53 gene) in these patients demonstrated that one case of MDS/AML (Pt #769) had both RUNX1 and PTPN11 mutations and that patients with therapy-related AML or MDS (Pt #59 or #346) had a mutation of RUNX1 or of N-RAS, respectively. Consistent with recent reports,22,26 our clinical data showed that overall survival was better in de novo AML patients with double CEBPA mutations compared with others with single CEBPA mutations (Table 1). These results suggested that double CEBPA mutations were able to induce AML, whereas single CEBPA mutation would lead to more aggressive AML, in concert with other gene alterations that have not been fully characterized.
Clinical features and genetic findings of the patients with CEBPA mutations
| Patient no. . | Age, y/sex . | Diagnosis . | N terminal C terminal . | Karyotype . | Other gene mutation* . | Survival (years) . | ||
|---|---|---|---|---|---|---|---|---|
| p30 type . | bZIP inframe . | Frameshift . | ||||||
| MDS/AML | ||||||||
| 142 | 79/F | AML following MDS | I62fsX160 | - | - | 46,XY[20/20] | - | 1.0 |
| 829 | 70/M | AML following MDS | P51fsX160 | - | - | 45,XY,der(17;18) (q10;q10)[2/20]/46,XY[18/20] | - | 0.4 |
| 769 | 69/M | AML following MDS | - | R297P | - | 46,XY[20/20] | AML1, PTPN11 | 1.6 |
| 896 | 77/M | AML following MDS | - | K313del | - | 46,XY[20/20] | - | 0.6 |
| 22 | 71/M | AML following MDS | - | - | G176fsX317 | 47,XY,+1,der(1;15) (q10;q10)[20/20] | - | 0.8 |
| 679 | 89/M | MDS(RAEB) | - | - | P235fsX318 | 46,XY[20/20] | - | 1.9 |
| 806 | 77/M | AML following MDS | G38fsX107 | - | R291fsX313 | 46,XY[20/20] | - | 1.9 |
| Therapy-related AML or MDS | ||||||||
| 59 | 80/F | AML(M4) | L19fsX159 | - | - | 47,XX,+8,t(9;11) (p22;q23)[20/20] | AML1 | 1.7 |
| 158 | 72/F | AML following MDS | F106fsX154 | - | - | 46,XX[20/20] | - | 1.0 |
| 811 | 76/M | MDS(RAEB) | E59X† | - | - | 46,XY[20/20] | - | 1.7 |
| 1068 | 66/M | MDS(RAEB) | - | Q305P | - | 46,XY[20/20] | - | > 1.5 |
| 346 | 59/M | AML following MDS | - | - | S190fsX320 | 43,XX,del(5)(q31), -7,-15,-18,-21, +mar [20/20] | N-RAS | 0.8 |
| 577 | 56/F | AML following MDS | - | - | C213X | 46,XX[20/20] | - | 2.4 |
| 629 | 89/F | AML following MDS | - | - | L350fsX360 | 46,XX[20/20] | - | 1.2 |
| 920 | 69/F | AML(M5) | - | - | S348fsX422 | 46,XX,t(11;17) (p15;q21)[20/20] | - | 0.2 |
| De novo AML(M2) | ||||||||
| 40 | 68/F | AML(M2) | A111fsX166 | S299_L304dup | - | 46,XX[20/20] | - | > 10.5 |
| 292 | 75/F | AML(M2) | F33fsX107 | R297P | - | 46,XX[20/20] | - | > 8.4 |
| 662 | 58/F | AML(M2) | S65fsX167 | K313dup | - | 46,XX[20/20] | - | > 5.0 |
| 888 | 70/F | AML(M2) | A111fsX166 | N321D | - | 47,XX,+8[20/20] | - | > 3.8 |
| 941 | 31/F | AML(M2Eo) | T60fsX159 | 304_323dup | - | 46,XX,del(7)(q32)[20/20] | - | > 3.5 |
| Patient no. . | Age, y/sex . | Diagnosis . | N terminal C terminal . | Karyotype . | Other gene mutation* . | Survival (years) . | ||
|---|---|---|---|---|---|---|---|---|
| p30 type . | bZIP inframe . | Frameshift . | ||||||
| MDS/AML | ||||||||
| 142 | 79/F | AML following MDS | I62fsX160 | - | - | 46,XY[20/20] | - | 1.0 |
| 829 | 70/M | AML following MDS | P51fsX160 | - | - | 45,XY,der(17;18) (q10;q10)[2/20]/46,XY[18/20] | - | 0.4 |
| 769 | 69/M | AML following MDS | - | R297P | - | 46,XY[20/20] | AML1, PTPN11 | 1.6 |
| 896 | 77/M | AML following MDS | - | K313del | - | 46,XY[20/20] | - | 0.6 |
| 22 | 71/M | AML following MDS | - | - | G176fsX317 | 47,XY,+1,der(1;15) (q10;q10)[20/20] | - | 0.8 |
| 679 | 89/M | MDS(RAEB) | - | - | P235fsX318 | 46,XY[20/20] | - | 1.9 |
| 806 | 77/M | AML following MDS | G38fsX107 | - | R291fsX313 | 46,XY[20/20] | - | 1.9 |
| Therapy-related AML or MDS | ||||||||
| 59 | 80/F | AML(M4) | L19fsX159 | - | - | 47,XX,+8,t(9;11) (p22;q23)[20/20] | AML1 | 1.7 |
| 158 | 72/F | AML following MDS | F106fsX154 | - | - | 46,XX[20/20] | - | 1.0 |
| 811 | 76/M | MDS(RAEB) | E59X† | - | - | 46,XY[20/20] | - | 1.7 |
| 1068 | 66/M | MDS(RAEB) | - | Q305P | - | 46,XY[20/20] | - | > 1.5 |
| 346 | 59/M | AML following MDS | - | - | S190fsX320 | 43,XX,del(5)(q31), -7,-15,-18,-21, +mar [20/20] | N-RAS | 0.8 |
| 577 | 56/F | AML following MDS | - | - | C213X | 46,XX[20/20] | - | 2.4 |
| 629 | 89/F | AML following MDS | - | - | L350fsX360 | 46,XX[20/20] | - | 1.2 |
| 920 | 69/F | AML(M5) | - | - | S348fsX422 | 46,XX,t(11;17) (p15;q21)[20/20] | - | 0.2 |
| De novo AML(M2) | ||||||||
| 40 | 68/F | AML(M2) | A111fsX166 | S299_L304dup | - | 46,XX[20/20] | - | > 10.5 |
| 292 | 75/F | AML(M2) | F33fsX107 | R297P | - | 46,XX[20/20] | - | > 8.4 |
| 662 | 58/F | AML(M2) | S65fsX167 | K313dup | - | 46,XX[20/20] | - | > 5.0 |
| 888 | 70/F | AML(M2) | A111fsX166 | N321D | - | 47,XX,+8[20/20] | - | > 3.8 |
| 941 | 31/F | AML(M2Eo) | T60fsX159 | 304_323dup | - | 46,XX,del(7)(q32)[20/20] | - | > 3.5 |
RAEB indicates refractory anemia with excess blasts.
Indicates that no mutation was detected in AML1, N-RAS, FLT3, PTPN11, C-KIT, and TP53 genes.
Homozygous mutation.
The C-terminal but not N-terminal mutations of C/EBPα inhibited G-CSF–induced differentiation of 32Dcl3 cells into mature neutrophils
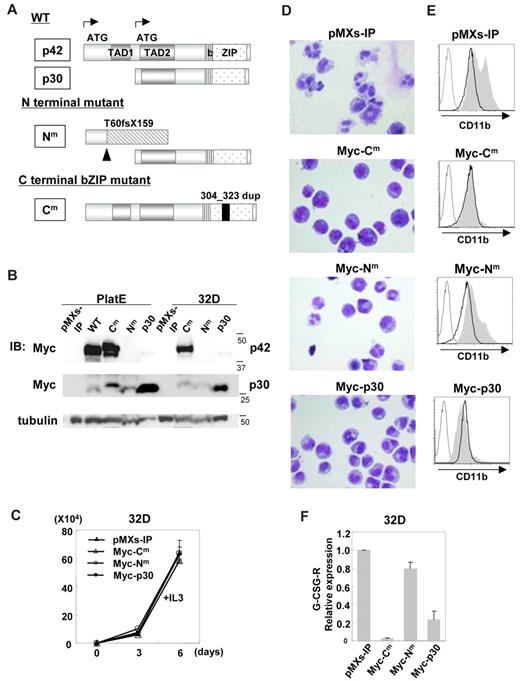
For further analysis, we chose an N-terminal mutant producing p30 designated as C/EBPα-Nm and a C-terminal b-ZIP in-frame mutant designated as C/EBPα-Cm (Figure 1A). C/EBPα-WT, C/EBPα-Nm, C/EBPα-Cm, C/EBPα-p30, or mock (pMXs-IP) was expressed in Plat-E cells. Expression of p42 protein or p30 protein generated by Myc-tagged C/EBPα-WT and mutants was verified by using anti-Myc Ab as bands corresponding to expected molecular weights (Figure 1B). In addition, we confirmed that N-terminal polypeptide produced by C/EBPα-Nm was detected by anti-C/EBPα Ab recognizing the N-terminal portion of C/EBPα (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Notably, the expression levels of p30 generated by C/EBPα-Nm were lower than those generated by C/EBPα-p30 (Figure 1B), indicating that deletion of the N-terminal part including the first start codon might increase the expression levels of p30 protein. We then infected 32Dcl3 cells with retroviruses harboring C/EBPα-WT or mutants, and the infected cells were subjected to drug selection. The 32Dcl3 cells expressing detectable levels of C/EBPα-WT were not obtained, presumably due to its strong inhibitory effect on proliferation. Western blot analysis showed that 32Dcl3 cells transduced with C/EBPα-Cm expressed the full length of C/EBPα-Cm at high levels (Figure 1B). C/EBPα-Nm or C/EBPα p30 transduced into 32Dcl3 cells was detected as a band (30 kDa), but the expression level of the former was much lower than that of the latter (Figure 1B). Growth speed was comparable among these transfectants in the presence of IL-3 (Figure 1C). However, the potential of these transfectants to differentiate in response to G-CSF varied; G-CSF treatment induced terminal differentiation of 32Dcl3 cells transduced with mock, as indicated by the appearance of polymorphonucleated neutrophils and up-regulation of CD11b on the surface (Figure 1D-E). G-CSF–induced granulocytic differentiation of 32Dcl3 cells was profoundly inhibited by C/EBPα-Cm, while it was only weakly inhibited by C/EBPα-Nm (Figure 1D-E). The differentiation was also attenuated by C/EBPα-p30, as reported previously.45 We reasoned that the difference of 32Dcl3 cells expressing C/EBPα-Nm or C/EBPα-p30 in the granulocytic differentiation levels was due to the dissimilar expression levels of a short form of C/EBPα (30 kDa). As for the expression levels of G-CSF-R transcripts, a target of C/EBPα, they were extremely or moderately decreased in 32Dcl3 cells expressing C/EBPα-Cm or C/EBPα-p30, respectively, compared with other transfectants (Figure 1F), implicating G-CSF-R in induction of granulocytic differentiation. However, when human G-CSF-R was transduced into C/EBPα-Cm-expressing 32Dcl3 cells, G-CSF–induced granulocytic differentiation was not completely recovered (supplemental Figure 2). These results indicated that the differentiation block in 32Dcl3 cells expressing C/EBPα-Cm was presumably due to the suppression of C/EBPα activation, but not simply due to the decreased expression of G-CSF-R downstream of C/EBPα.46
C/EBPα-Cm had the strong ability to block myeloid differentiation. (A) Schematic diagram of C/EBPα-WT (p42 and p30) and mutants, T60fsX159 (C/EBPα-Nm) and 304_323 dup (C/EBPα-Cm). TAD indicates the transcriptional activation domain; bZIP, basic region leucine zipper domain. (B) Expression of C/EBPα-WT and its mutants in Plat-E cells transiently transfected with a Myc-tagged C/EBPα-WT, C/EBPα-Cm, C/EBPα-Nm, or C/EBPα-p30 or an empty vector (pMXs-IP) and expression of C/EBPα mutants in 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Cell lysates were subject to immunoblotting with anti-Myc Ab or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (C) The growth of 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) in the presence of 1 ng/mL IL-3. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. (D-E) 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) were cultured in the presence of 50 ng/mL G-CSF for 6 days. (D) Morphology of these cells was assessed by Giemsa staining. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×40. (E) Surface expression of CD11b in these transfectants after incubation with 1 ng/mL IL-3 (bold histograms) or 50 ng/mL G-CSF (filled histograms) for 6 days was analyzed by flow cytometry. The result of control staining is shown as a thin-lined histogram. Data are representative of 3 independent experiments. (F) Relative expression levels of G-CSF-R in 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) were estimated by using real-time PCR. Data are representative of 3 independent experiments.
C/EBPα-Cm had the strong ability to block myeloid differentiation. (A) Schematic diagram of C/EBPα-WT (p42 and p30) and mutants, T60fsX159 (C/EBPα-Nm) and 304_323 dup (C/EBPα-Cm). TAD indicates the transcriptional activation domain; bZIP, basic region leucine zipper domain. (B) Expression of C/EBPα-WT and its mutants in Plat-E cells transiently transfected with a Myc-tagged C/EBPα-WT, C/EBPα-Cm, C/EBPα-Nm, or C/EBPα-p30 or an empty vector (pMXs-IP) and expression of C/EBPα mutants in 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Cell lysates were subject to immunoblotting with anti-Myc Ab or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (C) The growth of 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) in the presence of 1 ng/mL IL-3. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. (D-E) 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) were cultured in the presence of 50 ng/mL G-CSF for 6 days. (D) Morphology of these cells was assessed by Giemsa staining. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×40. (E) Surface expression of CD11b in these transfectants after incubation with 1 ng/mL IL-3 (bold histograms) or 50 ng/mL G-CSF (filled histograms) for 6 days was analyzed by flow cytometry. The result of control staining is shown as a thin-lined histogram. Data are representative of 3 independent experiments. (F) Relative expression levels of G-CSF-R in 32Dcl3 cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP) were estimated by using real-time PCR. Data are representative of 3 independent experiments.
C/EBPα-Cm or C/EBPα-Nm suppressed the transcriptional activity of C/EBPα-WT by different mechanisms
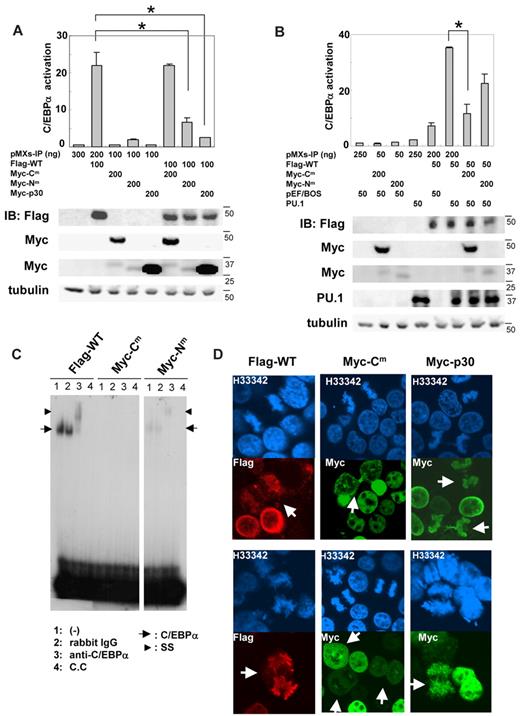
We next analyzed the transcriptional activation of C/EBPα-WT and mutants in 293T cells using a luciferase construct harboring 2 C/EBPα binding sites. As expected, C/EBPα-WT strongly activated this promoter, while neither C/EBPα-Cm, C/EBPα-Nm, nor C/EBPα-p30 showed any transcriptional activation (Figure 2A). We next examined whether these C/EBPα mutants affected the transcriptional activation of C/EBPα-WT. Although C/EBPα-Cm reduced G-CSF-R expression and inhibited G-CSF–induced myeloid differentiation of 32Dcl3 cells even more efficiently than C/EBPα-Nm (Figure 1D-F), C/EBPα-Nm as well as C/EBPα-p30 but not C/EBPα-Cm decreased promoter activity in the luciferase assay in 293T cells (top panel in Figure 2A), which was in accordance with the data shown by Gombart et al.20 Expression of C/EBPα WT and mutants in the transfected 293T cells was verified by Western blot analysis (bottom panel in Figure 2A). These results indicated that C/EBPα-Cm and C/EBPα-p30 suppressed expression of G-CSF-R by different mechanisms. Interestingly, transcriptional activation of C/EBPα-WT drastically increased in 293T cells when coexpressed with PU.1, although PU.1 itself did not stimulate the same promoter (top panel in Figure 2B). Notably, this synergistic effect was suppressed by C/EBPα-Cm and weakly by C/EBPα-Nm with low expression levels of p30 protein (top panel in Figure 2B). Expression of C/EBPα and PU.1 was also confirmed by Western blot analysis (bottom panel in Figure 2B). Therefore, we assumed that C/EBPα-Cm was interacting with other transcription factors such as PU.1, thereby suppressing the activation of C/EBPα-WT in hematopoietic cells. We also tested whether the C/EBPα mutants inhibit E2F activity. However, neither C/EBPα-Cm nor C/EBPα-Nm repressed E2F1/DP1-mediated transcription (supplemental Figure 3). In this regard, there was no difference between C/EBPα-Cm and C/EBPα-Nm. We then compared the DNA-binding ability of C/EBPα-WT and mutants by electrophoresis mobility shift assay. As shown in Figure 2C, C/EBPα-WT protein bound to the CSF3R (G-CSF-R) probe, and C/EBPα-p30 protein generated by C/EBPα-Nm less efficiently bound to the same probe. Binding was verified by super-shift of the DNA-protein complex by the anti-C/EBPα Ab. Remarkably, C/EBPα-Cm failed to bind the CSF3R probe (Figure 2C). Next, we examined subcellular localization of C/EBPα-WT and mutants. In line with previous reports, C/EBPα-WT and mutants localized in the nucleus of the interphase cells (Figure 2D). However, it was noteworthy that unlike C/EBPα-WT and C/EBPα-p30, C/EBPα-Cm was not localized on chromosome during the mitotic phase (Figure 2D). It is possible that C/EBPα-Cm without a DNA binding ability is stealing some interacting protein from chromosome. Taken together, these results indicated that C/EBPα-Nm suppressed the transcriptional activation of C/EBPα-WT by its direct binding to the promoter or by its heterodimerization with C/EBPα-WT, while C/EBPα-Cm showed the suppressive effect indirectly, probably through interaction with other transcription factors such as PU.1.
C/EBPα-Cm or C/EBPα-Nm inhibited the transcriptional activation of C/EBPα-WT by different mechanisms. (A-B top) 293T cells were transiently transfected with indicated amounts of expression plasmids (pMXs-Flag-tagged C/EBPα WT-IP, pMXs-Myc-tagged C/EBPα mutants-IP, pMXs-IP, pEF-BOS/PU.1, pEF-BOS) together with 100 ng of the luciferase reporter plasmid p(C/EBP)2TK. The total amount of plasmid for each transfection was adjusted by adding empty plasmids (pMXs-IP or pEF-BOS). Results represented the average values for relative luciferase activity that were normalized using the activity of EF1 vector as an internal control. All transfection groups were normalized with a Renilla luciferase vector as an internal control. All data points correspond to the mean and the standard deviation (SD). Data are representative of 3 independent experiments. Statistically significant differences are shown. *P < .05. (Bottom) Expression of C/EBPα-WT, C/EBPα-mutants, or PU.1 in 293T cells transiently transfected as above described. Cell lysates were subject to immunoblotting with anti-Flag Ab, anti-Myc Ab, anti-PU.1 Ab, or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (C) DNA binding of C/EBPα-WT and mutants. Electrophoresis mobility shift assay was performed with 32P-labeled oligonucleotides containing the C/EBPα binding site derived from CSF3R promoter and nuclear extracts from 293T cells transiently transfected with pMXs-Flag–tagged C/EBPα WT-IP, pMXs-Myc–tagged C/EBPα-Cm-IP, or pMXs-Myc–tagged C/EBPα-Nm-IP. Data are representative of 3 independent experiments. Lane 1: none (−); lane 2: control rabbit immunoglobulin G was added; lane 3: anti-C/EBPα Ab was added; lane 4: cold competitor (C.C) was added. Ss indicates supershifted bands. (D) 293T cells transiently transfected with pMXs-Flag–tagged C/EBPα-WT-IP, pMXs-Myc-tagged C/EBPα-Cm-IP, or pMXs-Myc–tagged C/EBPα-p30-IP were immunostained with anti-Flag Ab (red) or anti–c-Myc Ab (green) and stained with Hoechst (H33342; blue). Data are representative of 4 independent experiments (total of 15 mitotic cells were examined for each transfectant). Fluorescence images by confocal microscopy were obtained with IX70 (Olympus). Original magnification ×60.
C/EBPα-Cm or C/EBPα-Nm inhibited the transcriptional activation of C/EBPα-WT by different mechanisms. (A-B top) 293T cells were transiently transfected with indicated amounts of expression plasmids (pMXs-Flag-tagged C/EBPα WT-IP, pMXs-Myc-tagged C/EBPα mutants-IP, pMXs-IP, pEF-BOS/PU.1, pEF-BOS) together with 100 ng of the luciferase reporter plasmid p(C/EBP)2TK. The total amount of plasmid for each transfection was adjusted by adding empty plasmids (pMXs-IP or pEF-BOS). Results represented the average values for relative luciferase activity that were normalized using the activity of EF1 vector as an internal control. All transfection groups were normalized with a Renilla luciferase vector as an internal control. All data points correspond to the mean and the standard deviation (SD). Data are representative of 3 independent experiments. Statistically significant differences are shown. *P < .05. (Bottom) Expression of C/EBPα-WT, C/EBPα-mutants, or PU.1 in 293T cells transiently transfected as above described. Cell lysates were subject to immunoblotting with anti-Flag Ab, anti-Myc Ab, anti-PU.1 Ab, or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (C) DNA binding of C/EBPα-WT and mutants. Electrophoresis mobility shift assay was performed with 32P-labeled oligonucleotides containing the C/EBPα binding site derived from CSF3R promoter and nuclear extracts from 293T cells transiently transfected with pMXs-Flag–tagged C/EBPα WT-IP, pMXs-Myc–tagged C/EBPα-Cm-IP, or pMXs-Myc–tagged C/EBPα-Nm-IP. Data are representative of 3 independent experiments. Lane 1: none (−); lane 2: control rabbit immunoglobulin G was added; lane 3: anti-C/EBPα Ab was added; lane 4: cold competitor (C.C) was added. Ss indicates supershifted bands. (D) 293T cells transiently transfected with pMXs-Flag–tagged C/EBPα-WT-IP, pMXs-Myc-tagged C/EBPα-Cm-IP, or pMXs-Myc–tagged C/EBPα-p30-IP were immunostained with anti-Flag Ab (red) or anti–c-Myc Ab (green) and stained with Hoechst (H33342; blue). Data are representative of 4 independent experiments (total of 15 mitotic cells were examined for each transfectant). Fluorescence images by confocal microscopy were obtained with IX70 (Olympus). Original magnification ×60.
Retroviral transduction of C/EBPα-Cm, but not C/EBPα-Nm, immortalized BM hematopoietic cells
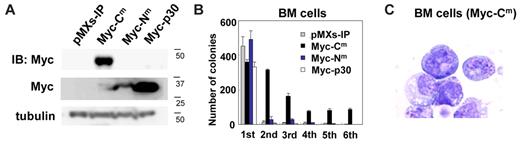
To determine the effect of C/EBPα-Cm and C/EBPα-Nm on differentiation and proliferation of hematopoietic cells, we performed serial colony-forming assays as described previously.45 Mouse BM mononuclear cells were transduced with C/EBPα-Cm, C/EBPα-Nm, or C/EBPα-p30. Expression of C/EBPα mutants in the transduced BM cells was verified by Western blot analysis (Figure 3A). We also confirmed that the expression levels of p30 protein generated by C/EBPα-p30 are higher than those by C/EBPα-Nm (Figure 3A). Irrespective of different expression levels of p30, most C/EBPα-Nm- and C/EBPα-p30–transduced BM cells did not make secondary colonies after replating (Figure 3B). On the other hand, BM cells expressing C/EBPα-Cm formed colonies after 6 rounds of replating in the presence of cytokine cocktail (Figure 3B). Cytospin preparations of these cells showed blastlike morphologies (Figure 3C). In addition, C/EBPα-Cm–transduced BM cells remained immature and were immortalized in a liquid culture containing IL-3 after several rounds of the replating in semisolid cultures.
C/EBPα-Cm, but not C/EBPα-Nm, immortalized BM cells. (A) Expression of C/EBPα-Cm, C/EBPα-Nm, or C/EBPα-p30 in BM cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Cell lysates were subject to immunoblotting with anti-Myc Ab or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (B) Colony-forming assay from BM cells transduced with C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Bars represent the number of colonies obtained per 104 cells after each round of plating in methylcellulose supplemented with stem cell factor, thrombopoietin, IL-3, and IL-6. Data are representative of 3 independent experiments. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. (C) Cytospin preparations of immortalized BM cells transduced with C/EBPα-Cm were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100.
C/EBPα-Cm, but not C/EBPα-Nm, immortalized BM cells. (A) Expression of C/EBPα-Cm, C/EBPα-Nm, or C/EBPα-p30 in BM cells transduced with Myc-tagged C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Cell lysates were subject to immunoblotting with anti-Myc Ab or anti-tubulin Ab as control. The results shown are representative of 3 independent experiments. (B) Colony-forming assay from BM cells transduced with C/EBPα-Cm, C/EBPα-Nm, C/EBPα-p30, or mock (pMXs-IP). Bars represent the number of colonies obtained per 104 cells after each round of plating in methylcellulose supplemented with stem cell factor, thrombopoietin, IL-3, and IL-6. Data are representative of 3 independent experiments. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. (C) Cytospin preparations of immortalized BM cells transduced with C/EBPα-Cm were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100.
Transduction with C/EBPα-Cm into BM cells caused AML in a mouse BMT model
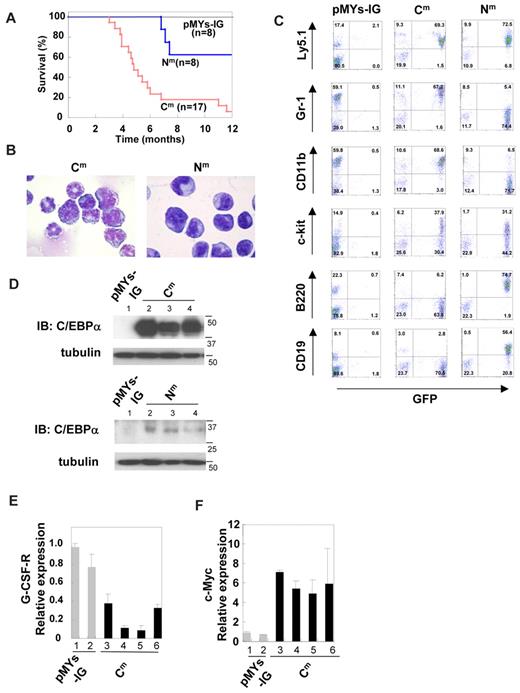
To test whether a single C/EBPα mutant induces hematopoietic abnormality, Ly-5.1 murine BM mononuclear cells, infected with retroviruses harboring C/EBPα-Cm, C/EBPα-Nm, or mock (pMYs-IG), were transplanted into irradiated syngenic Ly-5.2 mice. We confirmed efficient retrovirus infection: 50%-65% of BM cells transduced with C/EBPα-Nm or mock (pMYs-IG) and 35%-50% of BM cells transduced with C/EBPα-Cm were positive for GFP expression before transplantation. Mice receiving transplants of mock (pMYs-IG)–transduced cells (hereafter referred to as mice/pMYs-IG) remained healthy over the observation period (n =8/8) (Figure 4A). Notably, most of the mice that received transplants of C/EBPα-Cm–transduced cells (hereafter referred to as mice/Cm) developed AML within 4-12 months after transplantation (n = 16/17) (Figure 4A). These morbid mice presented similar phenotypes, characterized by hepatosplenomegaly and pancytopenia (Table 2). BM and spleen were occupied with myeloblasts and myelocytes (Figure 4B). In some cases, leukemic cells displayed morphologic aberrations such as abnormal lobular and ring-shaped nucleus. GFP-positive leukemic cells expressed CD11b and Gr-1 at high levels and c-kit at intermediate to high levels (middle panel in Figure 4C). One of the mice/Cm developed T-cell lymphoma with thymoma (data not shown). We next asked if the integration of retroviruses influenced the outcomes in the BMT model. Southern blot analysis of BM cells of mice/Cm showed a single or several integrations (supplemental Figure 4), and either 1 or 2 integration sites were identified in these samples, based on the inverse PCR method (supplemental Table 1).47 We found several common integration sites and integrations of the retroviruses in the intron of MN1 in 2 of 15 cases examined (supplemental Table 1). Considering the recent works published by Hasemann et al48 and by ourselves,40 retrovirus integration might in part influence the phenotypes of the recipient mice in our BMT models. For example, integration of the retrovirus vector into MN1 may enhance cell growth.40 However, integration sites do not seem to play major roles in the experiments of this study; C/EBPα-Cm transduction induced AML with similar phenotypes in most cases after a relatively long latency in the BMT model. On the other hand, 5 of 8 mice that received transplants of C/EBPα-Nm–transduced cells (hereafter referred to as mice/Nm) remained healthy during the observation period. Three of 8 mice/Nm developed B-cell acute lymphoblastic leukemia (B-ALL) with hepatosplenomegaly with latencies of 7 to 12 months after transplantation (Figure 4A). BM was occupied with blastlike cells, and the morbid mice exhibited leukocytosis, anemia, and thrombocytopenia (Figure 4B and data not shown). GFP-positive leukemic cells expressed B220 and CD19 at high levels and c-kit at intermediate to high levels (right panel in Figure 4C). One of the mice/Nm developed AML with splenomegaly 13 months after transplantation (data not shown). The reason why C/EBPα-Nm tend to induce B-ALL is not clear. However, we must notice a point that mouse BMT models may not always mimic human diseases.35,40 Expression of C/EBPα-Cm in spleen cells of mice/Cm with AML or p30 protein generated by C/EBPα-Nm in spleen cells of mice/Nm with B-ALL was confirmed by Western blot analysis (Figure 4D). Collectively, C/EBPα-Cm has a potential to strongly induce AML in a BMT model. Because the latency is relatively long and the leukemic cells seem to be clonal, additional events should have worked with C/EBPα-Cm in inducing leukemia. In addition, the in vivo suppressive effect of C/EBPα-Cm on the activation of endogenous C/EBPα was confirmed by the finding that G-CSF-R expression was down-regulated and c-Myc expression was up-regulated in BM samples of mice/Cm compared with mice/pMYs-IG (Figure 4E-F).
Transduction with C/EBPα-Cm alone induced AML in a mouse BMT model. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with C/EBPα-Cm-IG (Cm, n = 17), C/EBPα-Nm-IG (Nm, n = 8), or mock (pMYs-IG, n = 8). (B) Cytospin preparations of BM cells derived from mice/C/EBPα-Cm (left) or mice/Nm (right) were stained with Giemsa. A representative photograph is shown. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100. (C) Flow cytometric analysis of BM cells derived from mice/C/EBPα-Cm (middle), mice/C/EBPα-Nm (right), or mice/pMYs-IG (left). The dot plots show Ly5.1, Gr-1, CD11b, c-kit, B220, or CD19 labeled with phycoerythrin-conjugated monoclonal Ab versus expression of GFP. (D) Expression of C/EBPα-Cm protein and p30 protein generated by C/EBPα-Nm in spleen cells of mice/pMYs-IG (lane 1) and mice/Cm (lanes 2-4) (top) or in spleen cells of mice/pMYs-IG (lane 1) and mice/Nm (lanes 2-4) (bottom). Cell lysates were subject to immunoblotting with anti-C/EBPα (14AA) Ab or anti-tubulin Ab as control. Data are representative of 3 independent experiments. (E-F) Real-time PCR for G-CSF-R (E) or c-Myc (F) in BM cells derived from mice/Cm or mice/pMYs-IG. Expression levels were normalized by β-actin mRNA. The relative expression level of BM derived from mice/mock (lane 1) was defined as 1. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. Lanes 1-2: mice/pMYs-IG; lanes 3-6: mice/Cm.
Transduction with C/EBPα-Cm alone induced AML in a mouse BMT model. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with C/EBPα-Cm-IG (Cm, n = 17), C/EBPα-Nm-IG (Nm, n = 8), or mock (pMYs-IG, n = 8). (B) Cytospin preparations of BM cells derived from mice/C/EBPα-Cm (left) or mice/Nm (right) were stained with Giemsa. A representative photograph is shown. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100. (C) Flow cytometric analysis of BM cells derived from mice/C/EBPα-Cm (middle), mice/C/EBPα-Nm (right), or mice/pMYs-IG (left). The dot plots show Ly5.1, Gr-1, CD11b, c-kit, B220, or CD19 labeled with phycoerythrin-conjugated monoclonal Ab versus expression of GFP. (D) Expression of C/EBPα-Cm protein and p30 protein generated by C/EBPα-Nm in spleen cells of mice/pMYs-IG (lane 1) and mice/Cm (lanes 2-4) (top) or in spleen cells of mice/pMYs-IG (lane 1) and mice/Nm (lanes 2-4) (bottom). Cell lysates were subject to immunoblotting with anti-C/EBPα (14AA) Ab or anti-tubulin Ab as control. Data are representative of 3 independent experiments. (E-F) Real-time PCR for G-CSF-R (E) or c-Myc (F) in BM cells derived from mice/Cm or mice/pMYs-IG. Expression levels were normalized by β-actin mRNA. The relative expression level of BM derived from mice/mock (lane 1) was defined as 1. All data points correspond to the mean and the standard deviation (SD) of 3 independent experiments. Lanes 1-2: mice/pMYs-IG; lanes 3-6: mice/Cm.
Characteristics of AML caused by C/EBPα mutants
| . | pMYs-IG/pMYs-IR (n = 8) . | Myc-Cm/pMYs-IR (n = 6) . | Myc-Cm/Flag-Nm (n = 8) . |
|---|---|---|---|
| WBC (/μL) | 9060 ± 1648 | 5816 ± 3128 | 36 675 ± 22 956 |
| Hb (g/dL) | 16.4 ± 3.2 | 12.4 ± 2.2 | 10.7 ± 2.3 |
| Plt (× 104/μL) | 79.4 ± 43.1 | 7.2 ± 4.5 | 19.8 ± 13.1 |
| BM count (× 107) | 3.34 ± 0.73 | 1.69 ± 0.30 | 2.83 ± 0.88 |
| Leukemic cells (%) | - | 60-92 | 72-94 |
| Liver weight (mg) | 1433 ± 153 | 2071 ± 1281 | 2441 ± 1315 |
| Spleen weight (mg) | 113 ± 24 | 476 ± 220 | 549 ± 239 |
| . | pMYs-IG/pMYs-IR (n = 8) . | Myc-Cm/pMYs-IR (n = 6) . | Myc-Cm/Flag-Nm (n = 8) . |
|---|---|---|---|
| WBC (/μL) | 9060 ± 1648 | 5816 ± 3128 | 36 675 ± 22 956 |
| Hb (g/dL) | 16.4 ± 3.2 | 12.4 ± 2.2 | 10.7 ± 2.3 |
| Plt (× 104/μL) | 79.4 ± 43.1 | 7.2 ± 4.5 | 19.8 ± 13.1 |
| BM count (× 107) | 3.34 ± 0.73 | 1.69 ± 0.30 | 2.83 ± 0.88 |
| Leukemic cells (%) | - | 60-92 | 72-94 |
| Liver weight (mg) | 1433 ± 153 | 2071 ± 1281 | 2441 ± 1315 |
| Spleen weight (mg) | 113 ± 24 | 476 ± 220 | 549 ± 239 |
Averages and standard deviations are shown. BM cells were isolated from both tibias and femurs.
WBC indicates white blood cell; Hb, hemoglobin; and Plt, platelets.
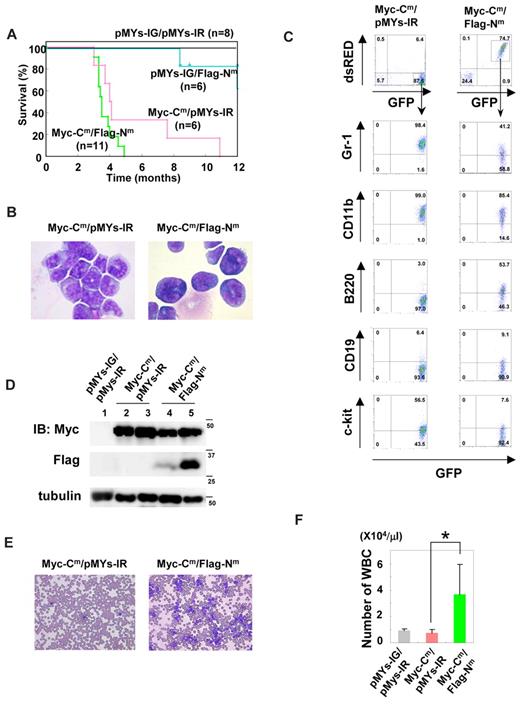
Transduction with both C/EBPα-Cm and C/EBPα-Nm induced more aggressive AML with leucocytosis
To next ask whether the combination of both C/EBPα-Cm and C/EBPα-Nm would induce AML more efficiently, we performed BMT, using murine BM mononuclear cells infected with retroviruses harboring Myc-tagged C/EBPα-Cm-IRES-GFP and Flag-tagged C/EBPα-Nm-IRES-dsRED. BM mononuclear cells expressing both mutants were recognized as GFP- and dsRED-double positive cells, 10%-22% of BM cells before the transplantation. Notably, mice that had received transplants of BM cells expressing both mutants (hereafter referred to as mice/Myc-Cm/Flag-Nm) developed AML with hepatosplenomegaly with shorter latencies (3-5 months) compared with mice that had received transplants of BM cells expressing both Myc-Cm-IRES-GFP and mock (pMYs-IR, hereafter referred to as mice/Myc-Cm/pMYs-IR; Figure 5A). Of note, there was no significant difference of the phenotypes between mice/Cm and mice/Myc-Cm/pMYs-IR or between mice/Nm and mice that had received transplants of BM cells expressing both mock (pMYs-IG) and Flag-Nm-IRES-dsRED (hereafter referred to as mice/pMYs-IG/Flag-Nm; Figures 4–5 and data not shown). The percentages of the immature blast ranged from 72%-94% in mice/Myc-Cm/Flag-Nm (Table 2) compared with 62%-92% in C/EBPα-Cm–induced leukemia (Table 2 and Figure 5B). Morphologies of the leukemic blasts are more immature in mice/Myc-Cm/Flag-Nm than mice/Myc-Cm/pMYs-IR (Figure 5B), consistent with the lower expression of Gr-1 in the former (Figure 5C and data not shown). Flow cytometric analysis delineated that most leukemic cells of mice/Myc-Cm/Flag-Nm expressed both GFP and dsRED (Figure 5C) and invariable markers: CD11b-inntermediate and Gr-1, B-220, c-kit-low (Figure 5C). Expression of both C/EBPα-Cm protein and p30 protein generated by C/EBPα-Nm in leukemic cells of mice/Myc-Cm/Flag-Nm was confirmed by Western blot analysis (Figure 5D). Expression levels of p30 protein generated by C/EBPα-Nm were not correlated with the disease latency in mice/Myc-Cm/Flag-Nm (Figure 5A,D). The morbid mice/Myc-Cm/Flag-Nm suffered from anemia and thrombocytopenia-like mice/Myc-Cm/pMYs-IR; however, it was of note that unlike mice/Myc-Cm/pMYs-IR, most mice/Myc-Cm/Flag-Nm exhibited marked leukocytosis (Figure 5E-F and Table 2). These results suggested that C/EBPα-Nm either confers a proliferative advantage on immature myeloid cells or collaborates with C/EBPα-Cm in blocking differentiation of myeloid cells in vivo. It is of note that this collaborative effect was induced by relatively low levels of p30 protein generated by C/EBPα-Nm.
Coexpression of both C/EBPα-Cm and C/EBPα-Nm led to AML with leukocytosis with shorter latencies. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with both Myc-tagged C/EBPα-Cm-IG and pMYs-IR (Myc-Cm/pMYs-IR, n = 6), both pMYs-IG and Flag-tagged C/EBPα-Nm-IR (pMYs-IG/Flag-Nm, n = 6), both Myc-tagged C/EBPα-Cm-IG and Flag-tagged C/EBPα-Nm-IR (Myc-Cm/Flag-Nm, n = 11), or mock (pMYs-IG/pMYs-IR, n = 8). (B) Cytospin preparations of BM cells derived from mice/My-Cm/pMYs-IR or mice/Myc-Cm/Flag-Nm were stained with Giemsa. A representative photograph is shown. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100. (C) Flow cytometric analysis of BM cells derived from mice/Myc-Cm/pMYs-IR (left) or mice/Myc-Cm/Flag-Nm (right). The dot plots show expression of dsRED versus expression of GFP (1st panel). In the indicated gating, the dot plots show expression of Gr-1, CD11b, B220, CD19, or c-kit labeled with phycoerythrin-Cy5–conjugated streptavidin versus expression of GFP. (D) Expression of Myc-tagged C/EBPα-Cm protein and p30 protein generated by Flag-tagged C/EBPα-Nm in BM cells derived from mice/pMYs-IG/pMYs-IR (lane 1), mice/Myc-Cm/pMYs-IR (lanes 2-3), or mice/Myc-Cm/Flag-Nm (lanes 4-5) was detected by using anti-Myc monoclonal Ab (top) and ant-Flag mAb (middle), respectively, in Western blot analysis. Equal loading was evaluated by probing the immunoblots with anti-tubulin Ab (bottom). Data are representative of 3 independent experiments. (E) Peripheral blood smears obtained from mice/Myc-Cm/pMYs-IR (left) or mice/Myc-Cm/Flag-Nm (right) were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×20. (F) Counts of white blood cells (WBC) obtained from mice/Myc-Cm/pMYs-IR (n = 6), mice/Myc-Cm/Flag-Nm (n = 8), or mice/pMYs-IG/pMYs-IR (n = 8). All data points correspond to the mean and the standard deviation (SD). Statistically significant differences are shown. *P < .05.
Coexpression of both C/EBPα-Cm and C/EBPα-Nm led to AML with leukocytosis with shorter latencies. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with both Myc-tagged C/EBPα-Cm-IG and pMYs-IR (Myc-Cm/pMYs-IR, n = 6), both pMYs-IG and Flag-tagged C/EBPα-Nm-IR (pMYs-IG/Flag-Nm, n = 6), both Myc-tagged C/EBPα-Cm-IG and Flag-tagged C/EBPα-Nm-IR (Myc-Cm/Flag-Nm, n = 11), or mock (pMYs-IG/pMYs-IR, n = 8). (B) Cytospin preparations of BM cells derived from mice/My-Cm/pMYs-IR or mice/Myc-Cm/Flag-Nm were stained with Giemsa. A representative photograph is shown. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×100. (C) Flow cytometric analysis of BM cells derived from mice/Myc-Cm/pMYs-IR (left) or mice/Myc-Cm/Flag-Nm (right). The dot plots show expression of dsRED versus expression of GFP (1st panel). In the indicated gating, the dot plots show expression of Gr-1, CD11b, B220, CD19, or c-kit labeled with phycoerythrin-Cy5–conjugated streptavidin versus expression of GFP. (D) Expression of Myc-tagged C/EBPα-Cm protein and p30 protein generated by Flag-tagged C/EBPα-Nm in BM cells derived from mice/pMYs-IG/pMYs-IR (lane 1), mice/Myc-Cm/pMYs-IR (lanes 2-3), or mice/Myc-Cm/Flag-Nm (lanes 4-5) was detected by using anti-Myc monoclonal Ab (top) and ant-Flag mAb (middle), respectively, in Western blot analysis. Equal loading was evaluated by probing the immunoblots with anti-tubulin Ab (bottom). Data are representative of 3 independent experiments. (E) Peripheral blood smears obtained from mice/Myc-Cm/pMYs-IR (left) or mice/Myc-Cm/Flag-Nm (right) were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×20. (F) Counts of white blood cells (WBC) obtained from mice/Myc-Cm/pMYs-IR (n = 6), mice/Myc-Cm/Flag-Nm (n = 8), or mice/pMYs-IG/pMYs-IR (n = 8). All data points correspond to the mean and the standard deviation (SD). Statistically significant differences are shown. *P < .05.
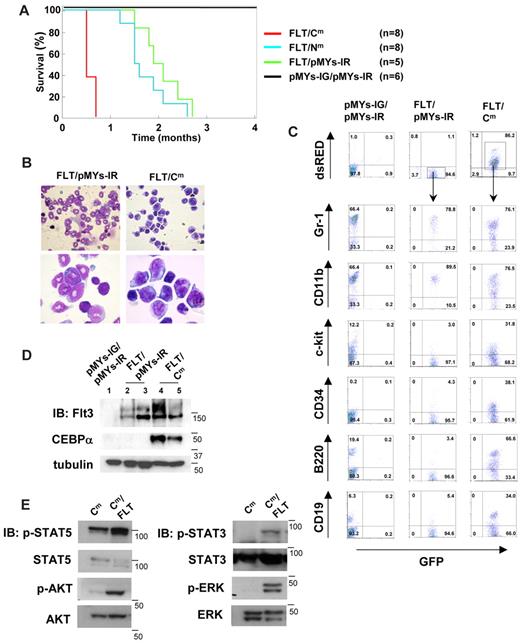
C/EBPα-Cm, but not C/EBPα-Nm, collaborated with Flt3-ITD in inducing AML in a BMT model
Because C/EBPα-Cm possessed the potential to strongly suppress myeloid differentiation, this mutation could be categorized into class II mutations. We speculated that AML would be efficiently induced by combining C/EBPα-Cm with class I gene alterations. To test this, murine BM mononuclear cells, transduced with both Flt3-ITD and either C/EBPα-Cm or C/EBPα-Nm, were transplanted into the recipient mice. BM mononuclear cells expressing both mutants were recognized as GFP- and dsRED-double positive cells, 10%-20% of BM cells before the transplantation. As reported previously,49 mice receiving transplants of BM cells expressing both Flt3-ITD-IRES-GFP and mock (pMYs-IR) (mice/FLT/pMYs-IR) developed myeloproliferative neoplasm (MPN) within 1.5-3 months after transplantation (Figure 6A). BM and spleen were occupied with increased numbers of mature myeloid cells expressing CD11b at high levels and Gr-1 at intermediate to high levels (Figure 6B-C). Intriguingly, mice transplanted with BM cells expressing both Flt3-ITD-IRES-GFP and C/EBPα-Cm-IRES-dsRED (mice/FLT/Cm) developed aggressive leukemia within 2-3 weeks after transplantation (Figure 6A). Histologic examination of mice/FLT/Cm showed that BM was occupied with the 2 populations: large and small blastlike cells (Figure 6B). However, flow cytometric analysis demonstrated that both populations, double positive for GFP and dsRED, similarly expressed B220, CD19, Gr-1, and CD11b and could not be differentiated (Figure 6C and data not shown). Thus, mice/FLT/Cm invariably developed biphenotypic leukemia. Western blot analysis demonstrated that both Flt3-ITD and C/EBPα-Cm proteins were expressed in spleen cells of mice/FLT/Cm (Figure 6D). On the other hand, mice that received transplants of BM cells expressing both Flt3-ITD-IRES-GFP and C/EBPα-Nm-IRES-dsRED (mice/FLT/Nm) developed MPN with latencies comparable with those of MPN developed by mice/FLT/pMYs-IR, although some lymphoid blast cells were observed in 2 mice/FLT/Nm (Figure 6A and data not shown). Thus, C/EBPα-Nm did not significantly collaborate with Flt3-ITD in leukemogenesis in the present BMT model. Finally, leukemic cells derived from mice/FLT/Cm proliferated independently of IL-3 in the culture, while those from mice/Cm still required IL-3 for their growth. Leukemic cells derived from mice/FLT or mice/FLT/Nm did not survive even in the presence of IL-3. Moreover, we found stronger activation of STAT5, STAT3, AKT, and ERK in leukemic cell lines derived from mice/Cm/FLT compared with those from mice/Cm (Figure 6E). These results indicated that Flt3-ITD conferred additional proliferative potentials as a class I mutation on the cells expressing C/EBPα-Cm alone, thereby inducing aggressive leukemia.
C/EBPα-Cm, but not C/EBPα-Nm, collaborated with Flt3-ITD in inducing aggressive AML. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with both Flt3-ITD-IG and pMYs-IR (FLT/pMYs-IR, n = 5), both Flt3-ITD-IG and C/EBPα-Cm-IR (FLT/Cm, n = 8), both Flt3-ITD-IG and C/EBPα-Nm-IR (FLT/Nm, n = 8), or mock (pMYs-IG/pMYs-IR, n = 8). (B) Cytospin preparations of BM cells derived from mice/FLT/pMYs-IR or mice/FLT/Cm were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×40 (top), ×100 (bottom). (C) Flow cytometric analysis of BM cells derived from mice/pMYs-IG/pMYs-IR, mice/FLT/pMYs-IR, or mice/FLT/Cm. The dot plots show expression of dsRED versus expression of GFP (first panel). In the indicated gating, the dot plots show expression of Gr-1, CD11b, c-kit, CD34, B220, or CD19 labeled with phycoerythrin-Cy5–conjugated streptavidin versus expression of GFP. (D) Expression of C/EBPα-Cm or Flt3-ITD in spleen cells of mice/pMYs-IG/pMYs-IR (lane 1), mice/FLT/pMYs-IG (lanes 2-3), or mice/FLT/Cm (lanes 4-5) was detected by using anti-C/EBPα (14AA) Ab (middle) or anti-Flt3 Ab (top), respectively, in Western blot analysis. Equal loading was evaluated by probing the immunoblots with anti-tubulin Ab. Data are representative of 3 independent experiments. (E) Immortalized leukemic cells derived from mice/FLT/Cm had increased phosphorylation of STAT5, AKT, STAT3, and ERK compared with those from mice/Cm. Whole-cell extracts of the former cells (immortalized in the absence of IL-3) or the latter (immortalized in the presence of IL-3) were immunoblotted with phospho-specific Abs as described in the Methods. Equal loading was evaluated by reprobing the immunoblots with anti-STAT5, anti-AKT, anti-STAT3, or anti-ERK Abs. Data are representative of 3 independent experiments.
C/EBPα-Cm, but not C/EBPα-Nm, collaborated with Flt3-ITD in inducing aggressive AML. (A) Kaplan-Meier analysis for the survival of mice that received transplants of BM cells transduced with both Flt3-ITD-IG and pMYs-IR (FLT/pMYs-IR, n = 5), both Flt3-ITD-IG and C/EBPα-Cm-IR (FLT/Cm, n = 8), both Flt3-ITD-IG and C/EBPα-Nm-IR (FLT/Nm, n = 8), or mock (pMYs-IG/pMYs-IR, n = 8). (B) Cytospin preparations of BM cells derived from mice/FLT/pMYs-IR or mice/FLT/Cm were stained with Giemsa. Images were obtained with a BX51 microscope and a DP12 camera (Olympus); objective lens, UplanFl (Olympus); original magnification ×40 (top), ×100 (bottom). (C) Flow cytometric analysis of BM cells derived from mice/pMYs-IG/pMYs-IR, mice/FLT/pMYs-IR, or mice/FLT/Cm. The dot plots show expression of dsRED versus expression of GFP (first panel). In the indicated gating, the dot plots show expression of Gr-1, CD11b, c-kit, CD34, B220, or CD19 labeled with phycoerythrin-Cy5–conjugated streptavidin versus expression of GFP. (D) Expression of C/EBPα-Cm or Flt3-ITD in spleen cells of mice/pMYs-IG/pMYs-IR (lane 1), mice/FLT/pMYs-IG (lanes 2-3), or mice/FLT/Cm (lanes 4-5) was detected by using anti-C/EBPα (14AA) Ab (middle) or anti-Flt3 Ab (top), respectively, in Western blot analysis. Equal loading was evaluated by probing the immunoblots with anti-tubulin Ab. Data are representative of 3 independent experiments. (E) Immortalized leukemic cells derived from mice/FLT/Cm had increased phosphorylation of STAT5, AKT, STAT3, and ERK compared with those from mice/Cm. Whole-cell extracts of the former cells (immortalized in the absence of IL-3) or the latter (immortalized in the presence of IL-3) were immunoblotted with phospho-specific Abs as described in the Methods. Equal loading was evaluated by reprobing the immunoblots with anti-STAT5, anti-AKT, anti-STAT3, or anti-ERK Abs. Data are representative of 3 independent experiments.
Discussion
The present results on CEBPA mutations of AML patients confirmed previous reports20-28 ; CEBPA mutations are found in 5%-14% of de novo AML, and most of them harbor 2 distinct mutations on different alleles and have good prognosis. In addition, our results suggested that mutations of CEBPA are found only in one allele in most cases of therapy-related AML or MDS, and AML progressed from MDS harboring CEBPA mutations (8/71 and 7/224). While we did not find additional mutations in other genes in de novo AML patients with double CEBPA mutations, we detected 3 additional mutations in 15 patients with therapy-related AML or MDS and MDS/AML. These results indicate that a CEBPA mutation collaborates with either a different type of CEBPA mutations or mutations in different genes in inducing leukemia.
Analysis of CEBPA mutations in in vitro assays provided novel insights concerning the role of CEBPα in blood cells. C/EBPα-Nm and p30, but not C/EBPα-Cm, suppressed transcriptional activation of C/EBPα-WT in a luciferase assay using 293T cells (Figure 2A) as reported previously.20 Curiously, expression of G-CSF-R, a major target of C/EBPα, was profoundly suppressed by C/EBPα-Cm but not by C/EBPα-Nm in 32Dcl3 cells (Figure 1F). C/EBPα-Cm suppressed G-CSF–induced granulocytic differentiation of 32Dcl3 cells more efficiently than C/EBPα-Nm (Figure 1D-E). It is possible that insufficient suppression of G-CSF–induced differentiation of 32Dcl3 cells by C/EBPα-Nm despite its inhibitory activity on transcriptional activation of C/EBPα-WT in 293T cells may be because of the low expression of C/EBPα-Nm in 32Dcl3 cells (Figure 1B). In fact, C/EBPα-p30 moderately suppressed the expression of G-CSF-R and inhibited G-CSF–induced differentiation of 32Dcl3 cells (Figure 1D-F). However, this does not explain why C/EBPα-Cm efficiently blocks the differentiation of 32Dcl3 cells despite its inability to suppress C/EBPα activation in the luciferase assay. Therefore, we speculated that C/EBPα mutants behave differently in epithelial 293T cells and hematopoietic 32Dcl3 cells and tested whether hemotopoietic cell–specific transcription factors play some role in 32Dcl3 cells. Because it was reported that PU.1 plays important roles in macrophage differentiation, which is hampered by its interaction with C/EBPα through its C-terminal bZIP domain,50 we investigated whether PU.1 plays some role in C/EBPα-Cm–mediated transcription. The present result indicated that C/EBPα synergized with exogenously expressed PU.1 in stimulating transcription of the target genes in 293T cells, which was profoundly inhibited by C/EBPα-Cm but not by C/EBPα-Nm (Figure 2B), and suggested that C/EBPα-Cm hampers PU.1 from interacting with other molecules including C/EBPα in hemopoietic cells, leading to inhibition of granulocytic differentiation.
As for cooperation between C/EBPα-Nm and C/EBPα-Cm in leukemogenesis, we demonstrated using BMT models that C/EBPα-Nm and C/EBPα-Cm in combination induced AML with shorter latencies compared with transplantation of C/EBPα-Cm–transduced BM cells alone. In addition, combining both mutations resulted in increased number of leukemic cells, implicating C/EBPα-Nm in expansion of the cells whose differentiation was blocked by C/EBPα-Cm. Thus, these results suggested that C/EBPα-Cm works as a class II mutation while C/EBPα-Nm works as a class I mutation in inducing leukemia.31-36 To test this hypothesis, we built another BMT model where BM cells transduced with Flt3-ITD and either C/EBPα-Cm or C/EBPα-Nm were transplanted to lethally irradiated mice. Flt3-ITD dramatically shortened the latency of leukemia induced by C/EBPα-Cm but not by C/EBPα-Nm, indicating that C/EBPα-Cm worked as a class II mutation in inducing leukemia. Transplantation of BM cells transduced with both C/EBPα-Cm and Flt3-ITD quickly induced leukemia in just 2 weeks after transplantation. Most of the transplanted mice seemed to develop biphenotypic leukemia as assessed on the morphology and surface marker expressions (Figure 6B-C). In our hands, BM cells transduced with Flt3-ITD sometimes induce lymphoid malignancies in addition to myeloproliferative disease,49 bringing some complexity to the experiment. Nonetheless, dramatically shortened latencies with the combination of C/EBPα-Cm and Flt3-ITD strongly indicated that C/EBPα-Cm works as a class II mutation in inducing leukemia. On the other hand, because the expression levels of C/EBPα-Nm was low in our experiments, further experiments will be required to firmly demonstrate that C/EBPα-Nm plays a class I-like role. One possible experiment is to test a combination between C/EBPα-Nm and a known class II mutation. Nonetheless, a class I-like role of C/EBPα-Nm was suggested by the marked increase in the number of leukemic cells in mice/Myc-Cm/Flag-Nm compared with mice/Myc-Cm/pMYs-IR. In relation to this, although the “2-hit theory” well explain many clinical observations, additional classes of mutations may be required for the comprehensive understanding of leukemogenesis as proposed by Renneville et al32 In fact, we detected more than 3 mutations including mutations, chromosomal translocations, or deletions in 5 of 20 patients with leukemia and MDS (Table 1).
Concerning the in vivo effects of CEBPA mutations, several different results were reported.29,30,51-54 Bereshchenko et al30 have recently published a report using knock-in mice that C/EBPα-p30 and a C-terminal mutation collaborated in inducing leukemia. Our results basically agreed with those by Bereshchenko et al. However, while our results implicated C-terminal mutations of C/EBPα in differentiation, leading to leukemia with relatively long latencies, Bereshchenko et al30 suggested premalignant HSC expansion by C-terminal mutations. The reason for the disparity is not clear, but was partly caused by the difference in the strength or functions of different C-terminal mutants or in the expression levels of mutants in knock-in mice and BMT models. Concerning the experimental systems, knock-in mice are superior to mouse BMT models in several aspects as indicated previously.29,30 Most importantly, expression of the mutant C/EBPα is driven by the authentic promoter in knock-in mice while it is over-driven by an external promoter in BMT models. Moreover, replacement of both alleles with different C/EBPα mutations closely mimics human leukemia, as it lacks the WT C/EBPα unlike the BMT model. In addition, in BMT models, retrovirus integration sites sometimes modify the phenotype of the disease. However, BMT models do have some advantages. First, in contrast to knock-in mice where all hemopoietic cells express the mutant allele, only some cells can be of a leukemia origin, which would more faithfully mimic human pathologic situations. In addition, various mutants can be readily tested in vivo. Bereshchenko et al30 used K313dup as a C-terminal CEBPA mutation, demonstrating that miceK313dup/+ did not develop leukemia. K313dup was a weak inducer of leukemia in our BMT model, where only 1 of the 4 transplanted mice developed myeloid leukemia in 10 months (data not shown). On the other hand, C/EBPα-Cm, a C-terminal mutation with 304-323dup that we used in the present study, induced leukemia in most transplanted mice (Figure 4A). Thus, knock-in mice models and BMT models can complement with each other in investigating in vivo leukemogenesis.
To summarize, we have presented a series of evidence, including clinical data, in vitro experiments, and mouse BMT models, showing that 2 different mutations of CEBPA, C/EBPα-Nm and C/EBPα-Cm, play distinct roles in leukemogenesis. Moreover, our results strongly indicated that C/EBPα-Cm is able to play as a class II mutation in concert with Flt3-ITD in inducing leukemia. Further elucidation of the molecular mechanism of CEBPA mutations–induced leukemia may pave a novel way to treating patients with leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Atsushi Iwama, Dr Claus Nerlov, and Dr Shigekazu Nagata for kindly providing plasmids. We are grateful to Dr Dovie Wylie for her excellent editing of the English.
This work was supported by the Ministry of Education, Science, Technology, Sports and Culture, Japan and in part supported by Global COE Program “Center of Education and Research for the Advanced Genome-Based Medicine, for personalized medicine and the control of worldwide infectious diseases,” MEXT, Japan.
Authorship
Contribution: N.K. did all the experiments and participated in writing the manuscript; J.K. oversaw all the experiments and actively participated in manuscript writing; N.D., Y.K., N.W.O., K.T., F.N., T.O., and Y.E. technically supported BMT; Y.F. and H.N. provided plasmids and reagents; Y.H. and H.H. provided and analyzed human samples; and T.K. conceived the project, secured funding, and actively participated in manuscript writing.
Conflict-of-interest disclosure: T.K. serves as a consultant for R&D Systems. The remaining authors declare no competing financial interests.
Correspondence: Toshio Kitamura, Division of Cellular Therapy and Division of Stem Cell Signaling, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: kitamura@ims.u-tokyo.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal