Abstract
Gene expression profiling of 207 uniformly treated children with high-risk B-progenitor acute lymphoblastic leukemia revealed 29 of 207 cases (14%) with markedly elevated expression of CRLF2 (cytokine receptor-like factor 2). Each of the 29 cases harbored a genomic rearrangement of CRLF2: 18 of 29 (62%) had a translocation of the immunoglobulin heavy chain gene IGH@ on 14q32 to CRLF2 in the pseudoautosomal region 1 of Xp22.3/Yp11.3, whereas 10 (34%) cases had a 320-kb interstitial deletion centromeric of CRLF2, resulting in a P2RY8-CRLF2 fusion. One case had both IGH@-CRLF2 and P2RY8-CRLF2, and another had a novel CRLF2 rearrangement. Only 2 of 29 cases were Down syndrome. CRLF2 rearrangements were significantly associated with activating mutations of JAK1 or JAK2, deletion or mutation of IKZF1, and Hispanic/Latino ethnicity (Fisher exact test, P < .001 for each). Within this cohort, patients with CRLF2 rearrangements had extremely poor treatment outcomes compared with those without CRLF2 rearrangements (35.3% vs 71.3% relapse-free survival at 4 years; P < .001). Together, these observations suggest that activation of CRLF2 expression, mutation of JAK kinases, and alterations of IKZF1 cooperate to promote B-cell leukemogenesis and identify these pathways as important therapeutic targets in this disease. This trial was registered at www.clinicaltrials.gov as #NCT00005603.
Introduction
Pediatric acute lymphoblastic leukemia (ALL) is a heterogeneous group of diseases with a complex spectrum of genetic abnormalities, including recurring chromosomal translocations and genomic deletions and amplifications.1-8 Over the past 30 years, dramatic improvements in survival have been achieved in this disease through the progressive intensification of standard chemotherapy and the development of detailed risk classification schemes that tailor the intensity of therapy to the predicted risk of relapse.4-6 In Children's Oncology Group (COG) ALL clinical trials, children with B-precursor ALL are first assigned to “standard” or “high” risk groups based on National Cancer Institute criteria of presenting age and white blood cell count.9,10 The presence or absence of specific recurring cytogenetic abnormalities1-3,8 and measures of minimal residual disease at the end of induction therapy11,12 are then used to refine risk classification, placing children into “low” (high hyperdiploidy with favorable chromosome trisomies or t(12;21)(ETV6-RUNX1)), “standard/intermediate,” “high,” or “very high” (hypodiploidy or t(9;22)(BCR-ABL1)) risk groups. More recently, assessment of DNA copy number abnormalities has revealed that deletion or mutation of genes, such as IKZF1 (IKAROS), are also highly predictive of a poorer outcome in pediatric ALL.13
Using current risk classification schema incorporating clinical and genetic variables, more than 25% of children initially diagnosed with ALL are classified as being at high risk of treatment failure. Moreover, many children with high-risk features lack sentinel chromosomal alterations known to be associated with poor outcome. As outcome of high-risk ALL remains poor despite intensive therapy, new therapeutic approaches are needed. To develop novel therapies directed against rational targets, a thorough understanding of the molecular alterations responsible for treatment resistance in ALL is required. To characterize genetic alterations in high-risk ALL and identify novel prognostic markers and therapeutic targets in ALL, we have performed genomic profiling (including genome-wide profiling of DNA copy number alterations, gene expression profiling, and gene resequencing) as part of the National Cancer Institute's Therapeutically Applicable Research to Generate Effective Targets Initiative (http:/target.cancer.gov). These analyses were performed on diagnostic samples obtained from more than 200 children with high-risk ALL (as defined by age, sex, and presentation peripheral blood leukocyte count) enrolled on the COG P9906 trial. All cases lacked known sentinel chromosomal alterations associated with very-high-risk disease (BCR-ABL1, hypodiploidy) and were uniformly treated with a regimen incorporating augmented postinduction therapy.14
To identify aberrantly expressed genes that might be targets of novel genetic alterations in this cohort, we performed cancer outlier profile analysis15-17 and an alternative procedure, Recognition of Outliers by Sampling Ends (R.C.H., C.G.M., X. Wang, K.K.D., G. S. Davidson, E. J. Bedrick, I.-M.C., S. R. Atlas, H.K., K. Ar, C. S. Wilson, W.W., M. Murphy, M.D., A.J.C., M. J. Borowitz, W.P.B., J.R.D., M. V. Relling, J. Yang, D. Bhojwan, W.L.C., B.M.C., G.H.R., M.A.S., S.P.H., C.L.W., manuscript submitted, August 2009). One gene overexpressed in a subset of high-risk ALL cases lacking known common chromosomal rearrangements was CRLF2 (cytokine receptor-like factor 2), also termed TSLPR (thymic stromal lymphopoietin receptor).18-22 We recently reported focal DNA copy number alterations adjacent to the CRLF2 locus in the pseudoautosomal region 1 (PAR1) at Xp22.3/Yp11.3 in high-risk ALL cases harboring Janus kinase (JAK1, JAK2, and JAK3) mutations.23 These cases also exhibited CRLF2 overexpression, suggesting that genetic alterations at the CRLF2 locus result in overexpression, CRLF2 forms a heterodimeric receptor for thymic stromal lymphopoietin (TLSP), and CRLF2-mediated signaling may promote B lymphoid survival and proliferation.24-29 Together, these observations suggest that genetic alterations dysregulating CRLF2 expression may contribute to the pathogenesis of ALL.
Consistent with this, 2 recent reports have described CRLF2 rearrangements in ALL.30,31 Russell et al identified translocation of the immunoglobulin heavy chain locus (IGH@) to CRLF2 at Xp22.3 or Yp11.3, or deletion upstream of CRLF2 in approximately 5% of patients with B-progenitor ALL.31 We have shown that the PAR1 deletion upstream (centromeric) of CRLF2 juxtaposes the first noncoding exon of P2RY8 to the entire coding region of CRLF2, resulting in a P2RY8-CRLF2 fusion, and that this fusion occurs in more than 50% of cases of childhood ALL associated with Down syndrome (DS-ALL).30 CRLF2 rearrangements were also shown to be associated with concomitant mutation of JAK1 and JAK2, and these lesions together were shown to be transforming in vitro.30 In the present study, we now demonstrate that CRLF2 genetic abnormalities are particularly frequent in children with high-risk B-progenitor ALL, where they are very significantly associated with Hispanic/Latino ethnicity, JAK1 or JAK2 mutations, IKAROS/IKZF1 deletions, and a very poor outcome.
Methods
Patient cohort
Pretreatment leukemia specimens were obtained from a cohort of 207 children with high-risk B-precursor ALL enrolled in the COG P9906 study.12 This study enrolled 272 patients from 2000 to 2003 who were treated with an augmented Berlin-Frankfurt-Münster regimen.14 Patient eligibility was based on clinical criteria (age, sex, and presentation leukocyte count) predictive of very poor outcome on prior trials (44% 4-year relapse-free survival).9 Patients with central nervous system disease (CNS3) or testicular leukemia were eligible for the trial regardless of age or white blood cell count at diagnosis. Patients with “very high risk” ALL subtypes (BCR-ABL1 or hypodiploidy) were excluded, as were those with trisomies of chromosomes 4 and 10 or t(12;21)(ETV6-RUNX1) unless they had central nervous system or testicular leukemia. The clinical characteristics of this subgroup were not significantly different from the overall cohort (data not shown). Informed consent was obtained from patients and/or their guardians according the Declaration of Helsinki, and the treatment protocol was approved by the National Cancer Institute and the institutional review boards of the participating institutions.
Gene expression profiling and genetic analyses
Gene expression profiling using Affymetrix U133 Plus 2.0 microarrays and genome-wide analysis of genetic alterations using Affymetrix 250K Sty and Nsp microarrays were performed in this cohort as previously described.13,23 This gene expression dataset may be accessed via the National Cancer Institute caArray portal (https://array.nci.nih.gov/caarray) or at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession no. GSE11877).32 Affymetrix single nucleotide polymorphism (SNP) array data are available at http://target.cancer.gov/data.13 Resequencing of all JAK1, JAK2, JAK3, and TYK2 coding exons was performed for 187 cases in the cohort with available DNA, gene expression data, and SNP microarray data. These data have been previously described in detail.13,23
FISH
Fluorescence in situ hybridization (FISH) analyses to detect rearrangement and structural alterations of the IGH@ and CRLF2 were performed on cells stored in Carnoy fixative. Disruption of the IGH@ locus was determined using the LSI IGH Dual Color Break-Apart Rearrangement Probe (Abbott Molecular). Three additional FISH probe mixtures were used. An IGH@-CRLF2 single-fusion extra-signal probe mixture was used, consisting of 2 bacterial artificial chromosome (BAC) clones (RP11-309M23 and RP11-261P4) flanking CRLF2 at Xp22.33/Yp11.32 labeled with a red fluor (AlexaFluor 568), and a third clone (RP11-815P21) centromeric to IGH@ at 14q32.33 labeled with a green fluor (AlexaFluor 488). Two CRLF2 break-apart mixtures (mix A and B) were also used, both of which contained clone RP11-309M23 telomeric to CRLF2 labeled with a green fluor (AlexaFluor 488) and 1 of 2 different clones centromeric to CRLF2 labeled with a red fluor (AlexaFluor 568; RP11-261P4 in mix A or the more centromeric clone RP11-74L17 in mix B).
BAC clones, fluorescent labels, and nick-translation materials were obtained through Invitrogen. Total BAC DNA was isolated using a plasmid midi-prep kit (Invitrogen). Slide hybridization and washes were performed using standard FISH protocols. Slides were counterstained with 4,6-diamidino-2-phenylindole and analyzed with an Olympus BX61 microscope (Olympus America) equipped with the appropriate filter combination and a CCD camera, and coupled to the CytoVision image analysis system. A total of 25 to 200 interphase cells were scored for each probe.
Flow cytometry
Flow cytometric measurement of CRLF2 expression of leukemic cells was performed by washing thawed, previously cryopreserved leukemic cells and staining them with conjugated anti–human TSLPR (clone 1A6; eBioscience), phycoerythrin-Cy5.5 conjugated anti–human CD45, and fluorescein isothiocyanate-conjugated anti–human CD34 (Invitrogen). Data were acquired using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with CellQuest Pro Software. Leukemia blast cells were identified by scatter gating (forward scatter vs side scatter) plus CD45 and CD34 expression.
PCR of P2RY8-CRLF2
Reverse-transcribed polymerase chain reaction (RT-PCR) for the P2RY8-CRLF2 fusion transcript and genomic PCR for the PAR1 deletion that results in P2RY8-CRLF2 rearrangement were performed as previously described.30 Briefly, primers for amplification of products from genomic DNA are: C1423 (5′-CGGTTTGGGGACTTTCAGAGCACAA; X/Y 1294242-1294266) and C1445 (5′-TCACCTGCTACTTCTGCCGCTGCTT; X/Y 1615822-1615846). Direct sequencing of genomic PCR products was performed with C1450 (5′-GGCATGAGCCACC GCGCCCCGCCCAATGC; X/Y 1294984-1295012). Amplification of full-length P2RY8-CRLF2 was performed with primers C1459 (5′-GCGGCCGCCTTTGCAAGGTTGC; P2RY8 exon 1, GenBank accession NM_178129, nucleotides 1-22) and C1349 (5′-GTGTCCATCACAACGCCACGTAGGA; CRLF2 exon 7, accession NM_022148, nucleotides, 1115-1139). Quantitative PCR of CRLF2 transcripts was performed by TaqMan with Hs00845692_m1 (Applied Biosystems) using a previously described method.30
Rapid amplification of cDNA ends
Rapid amplification of cDNA ends was performed using the GeneRacer kit (Invitrogen). First-strand cDNA was made using a CRLF2-specific primer (5′-GTCTCCTAGTCCTACCATCATTGGC). cDNA was amplified using the GeneRacer primer and a second nested CRLF2 gene-specific primer (5′-CTCTTTCTCCTCGGTCTGTGGG). Products were cloned and sequenced.
Statistical analyses
Associations between categorical variables were examined using Fisher exact test. Associations between clinical and genetic variables and outcome (relapse) was performed using log-rank or Gray test33 as previously described.13 The proportional hazards model of Fine and Gray was used to adjust for age, presentation leukocyte count, cytogenetic subtype, and genetic variables.34 Analyses were performed using Prism Version 5.0 (GraphPad), R (Version 2.9.1; www.r-project.org),35 SAS (Version 9.1.2; SAS Institute), Stata (Version 10; StataCorp), and SPLUS (Version 7.0; Insightful Corp).
Results
CRLF2 overexpression linked to genomic rearrangements activating CRLF2 expression
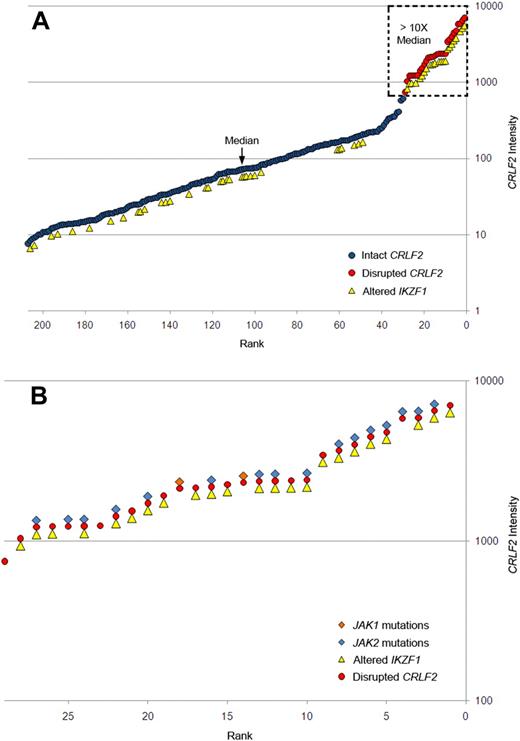
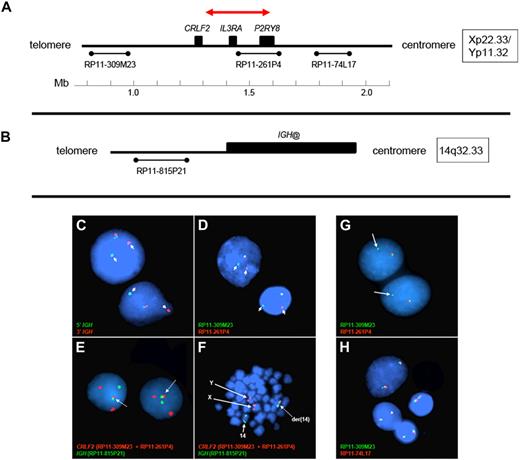
Gene expression profiling identified 29 cases within the POG 9906 high-risk ALL study population32 with markedly elevated expression of CRLF2 (Table 1), all of whom lacked known recurring sentinel chromosomal rearrangements (Figure 1A-B). FISH, using labeled BAC probes flanking the CRLF2 locus (RP11-309M23 and RP11-261P4; Figure 2A), demonstrated disruption of CRLF2 (either as a “break-apart” or deleted signal) in all 29 cases with high CRLF2 expression (Table 2; Figure 2). Twenty of the 29 cases had break-apart signals, whereas 10 of 29 cases had deletions upstream (centromeric) of CRLF2 (Table 2; Figure 2G-H). Focal PAR1 deletion upstream of CRLF2 in these 10 cases was demonstrated by deletion of probe RP11-261P4 in one chromosome and retention of the normal colocalization of probes RP11-309M23 and RP11-261P4 on the remaining sex chromosome (Figure 2G). Using a second more centromeric BAC clone (RP11-74L17; Figure 2A), probes RP11-309M23 and RP11-74L17 were colocalized in these 10 cases (Figure 2H). These findings are compatible with the previously identified focal PAR1 deletion upstream of CRLF2 that is commonly observed in ALL cases with CRLF2 rearrangement30,31 and P2RY8-CRLF2 fusion.30
Association of clinical and genetic features in CRLF2-rearranged cases in high-risk B-progenitor ALL
| Characteristic . | Disrupted CRLF2 . | Intact CRLF2 . | Total* . | P† . |
|---|---|---|---|---|
| No. of patients | 29 | 178 | 207 | |
| Age | ||||
| 10 y or older | 21 | 111 | 132 | .405 |
| Younger than 10 y | 8 | 67 | 75 | |
| Median age, y | 14.2 | 12.8 | 13.1 | > .030¶ |
| DS cases‡ | ||||
| DS | 2 | 6 | 8 | > .314 |
| Not DS | 21 | 134 | 155 | |
| Sex | ||||
| Male | 21 | 116 | 137 | .529 |
| Female | 8 | 62 | 70 | |
| WBC, ×1000/μL | ||||
| 50 or more | 16 | 92 | 108 | < .842 |
| Less than 50 | 13 | 86 | 99 | |
| Median WBC | 92.7 | 59.6 | 62.3 | < .198¶ |
| Ethnicity§ | ||||
| Hispanic/Latino | 18 | 33 | 51 | < .001 |
| Other | 11 | 143 | 154 | |
| BCR-ABL1–like GEP | ||||
| Yes | 18 | 20 | 38 | < .001 |
| No | 11 | 158 | 169 | |
| IKZF1 alteration‖ | ||||
| Deletion/mutation | 21 | 35 | 56 | < .001 |
| Normal | 5 | 126 | 131 | |
| JAK mutations‖ | ||||
| Mutation | 18 | 2 | 20 | < .001 |
| Normal | 8 | 159 | 167 | |
| CNS blasts | ||||
| No blasts | 19 | 141 | 160 | < .196 |
| Less than 5 | 6 | 20 | 26 | |
| 5 or more | 4 | 17 | 21 | |
| Probability of 4-year RFS | 35.3 (9.5) | 71.3 (3.6) | 66.3 (3.5) | .001# |
| Characteristic . | Disrupted CRLF2 . | Intact CRLF2 . | Total* . | P† . |
|---|---|---|---|---|
| No. of patients | 29 | 178 | 207 | |
| Age | ||||
| 10 y or older | 21 | 111 | 132 | .405 |
| Younger than 10 y | 8 | 67 | 75 | |
| Median age, y | 14.2 | 12.8 | 13.1 | > .030¶ |
| DS cases‡ | ||||
| DS | 2 | 6 | 8 | > .314 |
| Not DS | 21 | 134 | 155 | |
| Sex | ||||
| Male | 21 | 116 | 137 | .529 |
| Female | 8 | 62 | 70 | |
| WBC, ×1000/μL | ||||
| 50 or more | 16 | 92 | 108 | < .842 |
| Less than 50 | 13 | 86 | 99 | |
| Median WBC | 92.7 | 59.6 | 62.3 | < .198¶ |
| Ethnicity§ | ||||
| Hispanic/Latino | 18 | 33 | 51 | < .001 |
| Other | 11 | 143 | 154 | |
| BCR-ABL1–like GEP | ||||
| Yes | 18 | 20 | 38 | < .001 |
| No | 11 | 158 | 169 | |
| IKZF1 alteration‖ | ||||
| Deletion/mutation | 21 | 35 | 56 | < .001 |
| Normal | 5 | 126 | 131 | |
| JAK mutations‖ | ||||
| Mutation | 18 | 2 | 20 | < .001 |
| Normal | 8 | 159 | 167 | |
| CNS blasts | ||||
| No blasts | 19 | 141 | 160 | < .196 |
| Less than 5 | 6 | 20 | 26 | |
| 5 or more | 4 | 17 | 21 | |
| Probability of 4-year RFS | 35.3 (9.5) | 71.3 (3.6) | 66.3 (3.5) | .001# |
ALL indicates acute lymphoblastic leukemia; DS, Down syndrome; WBC, white blood cell count; GEP, gene expression profiling; CNS, central nervous system; and RFS, relapse-free survival.
All 207 patients were used unless otherwise noted.
P values were calculated using Fisher exact test unless otherwise noted.
DS status was available for 163 patients.
Ethnicity information was available for 205 patients.
Sequence information was available for 187 patients.
Mann-Whitney test was used to calculate WBC and age continuous variables.
Log-rank test used to calculate survival P value.
Association of elevated gene expression of CRLF2 with JAK mutations and IKZF1 alteration in high-risk B-progenitor ALL. (A) CRLF2 expression levels generated by Affymetrix gene expression profiling (circles) in 207 high-risk B-precursor ALL patients. y-axis represents CRLF2 intensity; x-axis, 207 ALL patients ordered from lowest to highest CRLF2 expression levels. Red circles represent samples with CRLF2 rearrangement; blue circles, patients with no detectable CRLF2 rearrangements; and yellow triangles, patients with IKZF1 alterations. Dashed box highlights samples with CRLF2 expression levels more than 10-fold higher than the median level. (B) Expanded view of the 29 ALL cases with CRLF2 rearrangement showing the association of CRLF2 rearrangement with IKZF1 alterations (yellow triangles), and mutations in JAK1 (orange diamonds) or JAK2 (blue diamonds). Detailed information on each of the 29 cases may be found in Table 2.
Association of elevated gene expression of CRLF2 with JAK mutations and IKZF1 alteration in high-risk B-progenitor ALL. (A) CRLF2 expression levels generated by Affymetrix gene expression profiling (circles) in 207 high-risk B-precursor ALL patients. y-axis represents CRLF2 intensity; x-axis, 207 ALL patients ordered from lowest to highest CRLF2 expression levels. Red circles represent samples with CRLF2 rearrangement; blue circles, patients with no detectable CRLF2 rearrangements; and yellow triangles, patients with IKZF1 alterations. Dashed box highlights samples with CRLF2 expression levels more than 10-fold higher than the median level. (B) Expanded view of the 29 ALL cases with CRLF2 rearrangement showing the association of CRLF2 rearrangement with IKZF1 alterations (yellow triangles), and mutations in JAK1 (orange diamonds) or JAK2 (blue diamonds). Detailed information on each of the 29 cases may be found in Table 2.
Design of FISH assays to detect genomic CRLF2 rearrangements. (A-B) The 3 BAC clones flanking the CRLF2 locus and 1 centromeric to IGH@ on chromosome 14, respectively. The red arrow in panel A highlights the PAR1 region involved in the deletions that join P2RY8 to CRLF2. The colors and names of the BAC probes used to perform FISH in panels C to H are shown in the lower left of each panel. (C-D) Results from the IGH and the CRLF2 break-apart assays, respectively. Arrows indicate the split signals. (E-F) The IGH-CRLF2 fusion probes on interphase and metaphase cells, respectively. Arrows in panel E highlight the fusion signal, whereas arrows in panel F indicate normal signals from X, Y, and 14 as well as the fusion signal on der14. (G) The loss of 261P4 in 1 sample resulting from PAR1 deletion. (H) The same sample shows that the signal is regained when a more centromeric BAC (74L17) is used, and 2 normal fusion signals are seen. Areas of cellular debris and nuclei lacking red/green signals in the same focal plane as the other cells were masked during image capture.
Design of FISH assays to detect genomic CRLF2 rearrangements. (A-B) The 3 BAC clones flanking the CRLF2 locus and 1 centromeric to IGH@ on chromosome 14, respectively. The red arrow in panel A highlights the PAR1 region involved in the deletions that join P2RY8 to CRLF2. The colors and names of the BAC probes used to perform FISH in panels C to H are shown in the lower left of each panel. (C-D) Results from the IGH and the CRLF2 break-apart assays, respectively. Arrows indicate the split signals. (E-F) The IGH-CRLF2 fusion probes on interphase and metaphase cells, respectively. Arrows in panel E highlight the fusion signal, whereas arrows in panel F indicate normal signals from X, Y, and 14 as well as the fusion signal on der14. (G) The loss of 261P4 in 1 sample resulting from PAR1 deletion. (H) The same sample shows that the signal is regained when a more centromeric BAC (74L17) is used, and 2 normal fusion signals are seen. Areas of cellular debris and nuclei lacking red/green signals in the same focal plane as the other cells were masked during image capture.
CRLF2 expression, rearrangement, and association with genomic alterations
| Case no. . | CRLF2 Expression: U133 Plus 2.0 . | FISH/PCR detection of CRLF2 genomic rearrangements . | Deletions/mutations/expression patterns . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CRLF2 break-apart probe . | IGH@-CRLF2 . | IGH@-CRLF2 cells, percentage . | P2RY8-CRLF2 . | CRLF2 rearrangement . | BCR-ABL1–like . | JAK mutation . | IKZF1 alteration . | ||
| 9906_258 | 7020 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_234 | 6504 | Positive | Positive | 98 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_161 | 5876 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_020 | 5841 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | JAK2 R867Q | No |
| 9906_038 | 4793 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | JAK2 I682F | Yes |
| 9906_047* | 4471 | Positive | Positive | 80 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_222 | 4005 | Positive: deletion | Negative | 0 | ND2 | P2RY8-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_257 | 3664 | Positive | Positive | 99 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_205 | 3445 | Positive | Positive | 87 | Negative | IGH-CRLF2 | Yes | ND | Yes |
| 9906_151* | 2406 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK2 QGinsR683 | Yes |
| 9906_064 | 2379 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_113 | 2373 | Positive | Negative | 0 | Negative | Unknown partner | Yes | JAK2 R683G | Yes |
| 9906_174 | 2366 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | JAK2 P933R | Yes |
| 9906_052 | 2320 | Positive | Positive | 63 | Negative | IGH-CRLF2 | No | JAK1 V658F | No |
| 9906_215 | 2258 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | Negative | Yes |
| 9906_090 | 2177 | Positive | Positive | 97 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_196 | 2145 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | Negative | Yes |
| 9906_144 | 2127 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK1 L624_R629> W | No |
| 9906_217 | 1919 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | Negative | Yes |
| 9906_168 | 1721 | Positive | Positive | 90 | Negative | IGH-CRLF2 | Yes | JAK2 R683S | Yes |
| 9906_030 | 1540 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_192 | 1427 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | JAK2 D873N | Yes |
| 9906_200 | 1244 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | ND | ND |
| 9906_225 | 1237 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_110 | 1235 | Positive: deletion | Negative | 0 | ND | P2RY8-CRLF2 | No | JAK2 R683G | No |
| 9906_048 | 1229 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | Negative | Yes |
| 9906_170 | 1217 | Positive: deletion | Positive | > 90 | ND | P2RY8 & IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_109 | 1035 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | ND | Yes |
| 9906_094 | 744 | Positive | Positive | 67 | Negative | IGH-CRLF2 | No | Negative | No |
| Case no. . | CRLF2 Expression: U133 Plus 2.0 . | FISH/PCR detection of CRLF2 genomic rearrangements . | Deletions/mutations/expression patterns . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CRLF2 break-apart probe . | IGH@-CRLF2 . | IGH@-CRLF2 cells, percentage . | P2RY8-CRLF2 . | CRLF2 rearrangement . | BCR-ABL1–like . | JAK mutation . | IKZF1 alteration . | ||
| 9906_258 | 7020 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_234 | 6504 | Positive | Positive | 98 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_161 | 5876 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_020 | 5841 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | JAK2 R867Q | No |
| 9906_038 | 4793 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | JAK2 I682F | Yes |
| 9906_047* | 4471 | Positive | Positive | 80 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_222 | 4005 | Positive: deletion | Negative | 0 | ND2 | P2RY8-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_257 | 3664 | Positive | Positive | 99 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_205 | 3445 | Positive | Positive | 87 | Negative | IGH-CRLF2 | Yes | ND | Yes |
| 9906_151* | 2406 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK2 QGinsR683 | Yes |
| 9906_064 | 2379 | Positive | Positive | 96 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_113 | 2373 | Positive | Negative | 0 | Negative | Unknown partner | Yes | JAK2 R683G | Yes |
| 9906_174 | 2366 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | JAK2 P933R | Yes |
| 9906_052 | 2320 | Positive | Positive | 63 | Negative | IGH-CRLF2 | No | JAK1 V658F | No |
| 9906_215 | 2258 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | Negative | Yes |
| 9906_090 | 2177 | Positive | Positive | 97 | Negative | IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_196 | 2145 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | Negative | Yes |
| 9906_144 | 2127 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK1 L624_R629> W | No |
| 9906_217 | 1919 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | Negative | Yes |
| 9906_168 | 1721 | Positive | Positive | 90 | Negative | IGH-CRLF2 | Yes | JAK2 R683S | Yes |
| 9906_030 | 1540 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | Yes | Negative | Yes |
| 9906_192 | 1427 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | JAK2 D873N | Yes |
| 9906_200 | 1244 | Positive | Positive | > 90 | Negative | IGH-CRLF2 | No | ND | ND |
| 9906_225 | 1237 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_110 | 1235 | Positive: deletion | Negative | 0 | ND | P2RY8-CRLF2 | No | JAK2 R683G | No |
| 9906_048 | 1229 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | Negative | Yes |
| 9906_170 | 1217 | Positive: deletion | Positive | > 90 | ND | P2RY8 & IGH-CRLF2 | Yes | JAK2 R683G | Yes |
| 9906_109 | 1035 | Positive: deletion | Negative | 0 | Positive | P2RY8-CRLF2 | No | ND | Yes |
| 9906_094 | 744 | Positive | Positive | 67 | Negative | IGH-CRLF2 | No | Negative | No |
FISH/PCR indicates fluorescence in situ hybridization/polymerase chain reaction; and ND, not determined.
Down syndrome.
Using a FISH assay for IGH@-CRLF2 rearrangement, 18 of 19 cases with CRLF2 rearrangement but no PAR1 deletion had IGH@-CRLF2 translocation (Table 2; Figure 2B,E-F). The remaining case (9906_113, Table 2) had a novel structural rearrangement of CRLF2 involving an unknown partner. One case (9906_170, Table 2) had both a PAR1 deletion (resulting in expression of P2RY8-CRLF2) and IGH@-CRLF2 translocation. In 2 cases with high CRLF2 expression and IGH-CRLF2 translocation, rapid amplification of cDNA ends revealed full-length CRLF2 transcripts, consistent with overexpression of wild-type CRLF2.
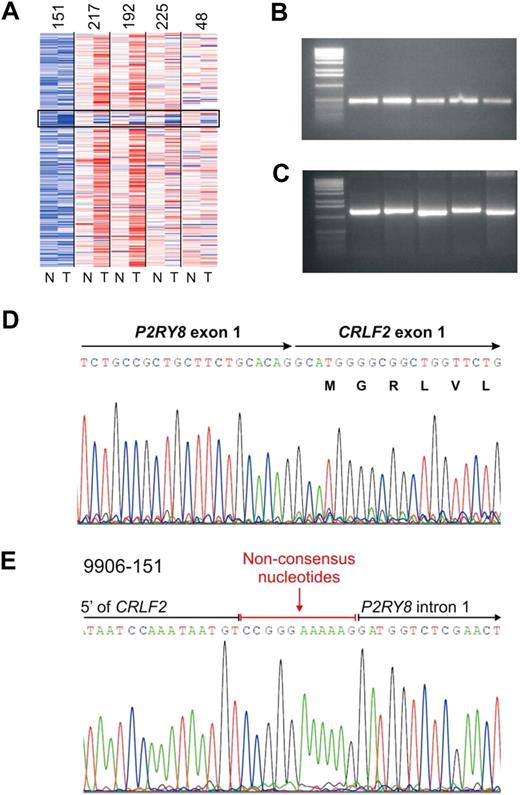
We previously identified a focal interstitial PAR1 deletion centromeric to CRLF2 in B-progenitor ALL36 and observed that this alteration was common in high-risk ALL cases in the current cohort that harbored activating JAK mutations.23 Recently, in a study focused primarily on cases of DS-ALL, we demonstrated that this interstitial PAR1 deletion juxtaposes the first noncoding exon of P2RY8 to the coding region of CRLF2, resulting in a P2RY8-CRLF2 fusion.30 Comparable results were observed in the cases in the current cohort with SNP array and FISH evidence of PAR1 deletion. Although coverage of this region on the Affymetrix 250K SNP arrays is sparse, the region of deletion appeared identical in each case (Figure 3A) and was accompanied by evidence of P2RY8-CRLF2 rearrangement on RT-PCR (Figure 3B) and/or genomic PCR (Figure 3C). Sequencing of PCR-amplified fusion transcripts demonstrated splicing of the first, noncoding exon of P2RY8 to the entire coding region of CRLF2 (Figure 3D). The fusion junction was identical in each case (Figure 3D) and arises from fusion of canonical splice donor and acceptor regions in P2RY8 and CRLF2, respectively.30 Genomic sequencing demonstrated that the breakpoints occurred within the first intron of P2R8Y and in the 5′ flanking region of CRLF2 (Figure 3E). Additional nonconsensus nucleotides were introduced at the junction, compatible with aberrant recombinase activating gene-mediated V(D)J recombination and the action of terminal deoxynucleotidyl transferase.
Characterization of PAR1 deletion centromeric to CRLF2 and PCR detection of the P2RY8-CRLF2 fusion. (A) Representative log2 ratio SNP 6.0 microarray DNA copy number data for 5 samples with PAR1 deletions. SNP array data derived from matched normal DNA is indicated by “N” and the leukemic sample with “T.” The black box highlights the region of PAR1 deletions. (B) RT-PCR demonstrating the fusion transcript of P2RY8 and CRLF2. Full-length cDNA transcripts were subjected to PCR with primers from P2RY8 and CRLF2 to generate the fusion products. (C) Genomic PCR of the chromosomal breakpoints joining CRLF2 and P2RY8. (D) Sequence of the RT-PCR product showing the junction of exon1 of P2RY8 with exon 1 of CRLF2. P2RY8 exon 1 is noncoding. (E) Germline sequence of the breakpoint. The exact junction of 5′ flanking sequence of CRLF2 to intron 1 of P2RY8 is shown. Several nonconsensus nucleotides are present at the junction.
Characterization of PAR1 deletion centromeric to CRLF2 and PCR detection of the P2RY8-CRLF2 fusion. (A) Representative log2 ratio SNP 6.0 microarray DNA copy number data for 5 samples with PAR1 deletions. SNP array data derived from matched normal DNA is indicated by “N” and the leukemic sample with “T.” The black box highlights the region of PAR1 deletions. (B) RT-PCR demonstrating the fusion transcript of P2RY8 and CRLF2. Full-length cDNA transcripts were subjected to PCR with primers from P2RY8 and CRLF2 to generate the fusion products. (C) Genomic PCR of the chromosomal breakpoints joining CRLF2 and P2RY8. (D) Sequence of the RT-PCR product showing the junction of exon1 of P2RY8 with exon 1 of CRLF2. P2RY8 exon 1 is noncoding. (E) Germline sequence of the breakpoint. The exact junction of 5′ flanking sequence of CRLF2 to intron 1 of P2RY8 is shown. Several nonconsensus nucleotides are present at the junction.
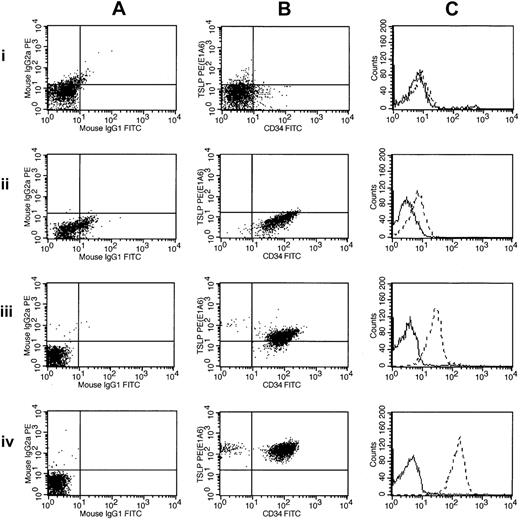
Thus, of the 29 high-risk B-precursor ALL cases with high CRLF2 expression, 18 (62%) cases had only an IGH@-CRLF2 translocation, 9 (31%) had only an interstitial deletion resulting in a P2RY8-CRLF2 fusion, 1 had both IGH@-CRLF2 and P2RY8-CRLF2, and 1 had a novel, uncharacterized CRLF2 rearrangement. Importantly, CRLF2 FISH performed on the next 37 consecutive cases in the cohort ranked by CRLF2 expression was normal. Thus, CRLF2 rearrangement is associated with markedly elevated CRLF2 expression. In contrast to the high frequency of P2RY8-CRLF2 fusions reported in DS-ALL,30 the majority of children with high-risk B-precursor ALL who have CRLF2 rearrangements have IGH-CRLF2 translocations. Moreover, there was a trend toward higher CRLF2 expression in cases with IGH@-CRLF2 translocations (Table 2). Fifteen of 20 of the highest CRLF2-expressors (as assessed by gene expression profiling or CRLF2 quantitative RT-PCR) had IGH@-CRLF2 translocations. Flow cytometric analysis of CRLF2 expression also demonstrated elevated CRLF2 expression in cases harboring IGH@-CRLF2 or P2RY8-CRLF2 (Figure 4).
Flow cytometric detection of CRLF2 protein expression. Flow cytometric measurements of CRLF2 protein expression are shown for 3 ALL samples and a normal control: 1 indicates normal buffy coat; 2, 9906_019; 3, 9906_055; and 4, 9906_258. These are ordered by their increasing levels of CRLF2 expression (determined by microarray). The leftmost panels (A) show scatter plots of the control antibodies (IgG1 and IgG2a). The middle panels (B) represent scatter plots for CRLF2 (y-axis) and CD34 (x-axis). The right panels (C) chart the relative intensity shift by histogram overlay, with the dashed lines corresponding to CRLF2. Sample 9906_258 (A-C) has the highest expression of CRLF2 in the cohort and contains the CRLF2-IGH@ translocation. The other 2 samples (9906_019 and 9906_055) have CRLF2 levels less than median.
Flow cytometric detection of CRLF2 protein expression. Flow cytometric measurements of CRLF2 protein expression are shown for 3 ALL samples and a normal control: 1 indicates normal buffy coat; 2, 9906_019; 3, 9906_055; and 4, 9906_258. These are ordered by their increasing levels of CRLF2 expression (determined by microarray). The leftmost panels (A) show scatter plots of the control antibodies (IgG1 and IgG2a). The middle panels (B) represent scatter plots for CRLF2 (y-axis) and CD34 (x-axis). The right panels (C) chart the relative intensity shift by histogram overlay, with the dashed lines corresponding to CRLF2. Sample 9906_258 (A-C) has the highest expression of CRLF2 in the cohort and contains the CRLF2-IGH@ translocation. The other 2 samples (9906_019 and 9906_055) have CRLF2 levels less than median.
Association of CRLF2 rearrangements with clinical and genetic variables
The median age of the 29 patients with CRLF2 rearrangement was not significantly different from the nonrearranged cases (12.8 vs 14.2 years; Table 1). Only 2 of the 8 DS patients in the COG 9906 high-risk ALL cohort were among the CRLF2-rearranged cases (Tables 1–2). The presence of CRLF2 rearrangements was significantly associated with Hispanic/Latino ethnicity (P < .001), the presence of JAK mutations (P < .001), and deletion or sequence mutation of IKZF1 (P < .001; Tables 1–2; Figure 1B). Eighteen (35.3%) of 51 Hispanic/Latino patients had CRLF2 rearrangements, compared with 11 (7.1%) of 154 cases of other declared ethnicity (ethnic data available for 205 patients). Twenty-one (80.8%) CRLF2-rearranged cases had concomitant deletions or sequence mutations of IKZF1, compared with 35 (21.7%) without deletions (sequence data available for 187 patients). Eighteen (69.2%) CRLF2-rearranged cases harbored JAK mutations, compared with 2 (1.2%) cases lacking CRLF2 alterations. This finding is consistent with recent observations of a highly significant association between CRLF2 alterations and JAK mutations in both DS-ALL and non–DS-ALL.30,31 The 2 JAK mutations occurring in cases without CRLF2 disruption were a JAK1 S646F mutation and a nonrecurring JAK3 mutation that lies outside the key JAK3 functional domains and, as such, may represent a nonpathogenic passenger mutation.
We previously identified a subset of high-risk Philadelphia chromosome–negative ALL cases in this cohort characterized by a lack of known recurring chromosomal rearrangements, genetic alteration of IKZF1, poor outcome, and a gene expression profile highly similar to that of BCR-ABL1–positive ALL,31,32 in which deletion of IKZF1 is also common.37 This suggested that the gene expression profile of these “BCR-ABL1–like” ALL cases may be driven, at least in part, by IKZF1 alteration and/or the presence of unknown mutation resulting in kinase activation. More than one-third of these cases harbor activating JAK mutations.23 As expected, given the strong association between CRLF2 rearrangement and JAK mutations, we observed a significant association between the presence of CRLF2 alteration and BCR-ABL1–like gene expression signature (Table 1). Using the top 100 differentially expressed probe sets in BCR-ABL1 ALL,38 18 of CRLF2-rearranged cases had a BCR-ABL1–like gene expression profile. Notably, several CRLF2-rearranged cases lacking JAK mutations had a BCR-ABL1–like signature (Table 2; cases 9906_258, 9906_205, 9906_64, and 9906_30), suggesting the presence of additional unidentified mutations resulting in activated kinase signaling. In contrast, 3 CRLF2-rearranged cases had JAK mutations but lacked a BCR-ABL1–like gene expression profile (Table 2; cases 9906_174, 9906_05, 9906_192, and 9906_110). Notably, 3 of these cases had less common JAK2 mutations (P933R and D873N) or the JAK1 V658F mutation (the homolog of the JAK2 V617F mutation observed in myeloproliferative disease39 ), in contrast to mutations at or near JAK2 R683 that are more frequently observed in B-progenitor ALL.23,40
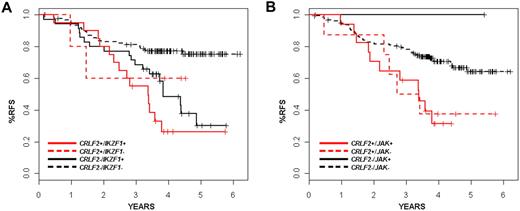
The presence of CRLF2 rearrangement was associated with a very poor treatment outcome. The predicted relapse-free survival (RFS) at 4 years for patients with disrupted CRLF2 was 35.3% plus or minus 9.5% in contrast to 71.3% plus or minus 3.6% for patients lacking the CRLF2 alteration (P = .001; Table 1). We did not observe a significant difference in outcome according to the type of CRLF2 genomic rearrangement (IGH translocation vs PAR1 deletion). On univariate analysis, Hispanic/Latino ethnicity, IKZF1 alteration, and BCR-ABL1–like gene expression signature were also associated with increased risk of relapse. In one multivariable analysis, considering age, presentation leukocyte count, Hispanic/Latino ethnicity, CRLF2 rearrangement, BCR-ABL1–like gene expression profile, IKZF1 alteration, JAK mutation, and stepwise variable selection (P < .5 for model entry and P < .25 to remain in the mode), only IKZF1 status remained significantly associated with risk of relapse at a P value threshold of .05 (hazard ratio [HR] = 2.5; 95% confidence interval [CI], 1.47-4.24, P < .001; data not shown). Age older than 10 years (HR = 0.67; 95% CI, 0.41-1.09, P = .11), Hispanic/Latino ethnicity (HR = 1.37; 95% CI, 0.83-2.22, P = .22), and the BCR-ABL1–like gene expression signature (HR = 1.5; 95% CI, 0.9-2.51, P = .11) were nonsignificantly associated with risk of relapse in this model. In a second stratified analysis (Table 3), the independent prognostic significance of CRLF2 was retained relative to Hispanic/Latino ethnicity (log-rank, P < .012), JAK mutations (P = .018), but not relative to BCR-ABL1–like expression signatures (P < .063) or IKAROS/IKZF1 deletions (P = .09). The lack of independent association of CRLF2 genomic rearrangements with relapse is explained in part by the co-occurrence of several of these genetic alterations. Of the 29 cases with CRLF2 alteration (26 of which had had comprehensive genomic data), 21 also harbored alterations of IKZF1 and 18 had JAK mutations. These observations suggest that these lesions cooperate in leukemogenesis. Notably, many IKZF1-altered cases lack CRLF2 rearrangement (35 of 187 cases, 18.7%), and these cases also frequently fail therapy, suggesting that alteration of IKZF1 alone promotes resistance to therapy. Nevertheless, the survival curves for IKZF1 and CRLF2 do suggest earlier failure for those patients with both lesions, although this trend did not reach significance (Figure 5A; Table 3). Likewise, the rate of relapse in CRLF2-altered cases was nearly identical, independent of JAK mutational status (Figure 5B; Table 3). This suggests that additional kinase-activating events are present in the CRLF2 rearranged cases lacking JAK mutations.
RFS of children with CRLF2 rearrangements stratified on the presence of IKZF1 alterations, JAK mutations, BCR-ABL1–like gene expression signatures, or ethnicity
| Genetic features or race (stratifying variable) . | Estimates for RFS at 4 years . | P value of stratified log-rank test . | |
|---|---|---|---|
| CRLF2 rearrangement, percentage . | No CRLF2 rearrangement, percentage . | ||
| IKZF1 alteration | .096 | ||
| Yes | 26.4 (10.5) (N = 21) | 48.4 (9.6) (N = 35) | |
| No | 60.0 (21.9) (N = 5) | 77.2 (3.8) (N = 126) | |
| JAK mutation | < .019 | ||
| Yes | 31.4 (12.0) (N = 18) | 100% (N = 2) | |
| No | 37.5 (17.1) (N = 8) | 70.5 (3.8) (N = 159) | |
| BCR-ABL1 signature | < .063 | ||
| Yes | 27.8 (11.7) (N = 17) | 60.0 (11.0) (N = 20) | |
| No | 44.4 (16.6) (N = 9) | 72.5 (4.0) (N = 141) | |
| Ethnicity | < .012 | ||
| Hispanic/Latino | 28.6 (11.8) (N = 17) | 61.4 (9.4) (N = 30) | |
| Other | 41.7 (17.3) (N = 9) | 72.8 (4.1) (N = 129) | |
| Genetic features or race (stratifying variable) . | Estimates for RFS at 4 years . | P value of stratified log-rank test . | |
|---|---|---|---|
| CRLF2 rearrangement, percentage . | No CRLF2 rearrangement, percentage . | ||
| IKZF1 alteration | .096 | ||
| Yes | 26.4 (10.5) (N = 21) | 48.4 (9.6) (N = 35) | |
| No | 60.0 (21.9) (N = 5) | 77.2 (3.8) (N = 126) | |
| JAK mutation | < .019 | ||
| Yes | 31.4 (12.0) (N = 18) | 100% (N = 2) | |
| No | 37.5 (17.1) (N = 8) | 70.5 (3.8) (N = 159) | |
| BCR-ABL1 signature | < .063 | ||
| Yes | 27.8 (11.7) (N = 17) | 60.0 (11.0) (N = 20) | |
| No | 44.4 (16.6) (N = 9) | 72.5 (4.0) (N = 141) | |
| Ethnicity | < .012 | ||
| Hispanic/Latino | 28.6 (11.8) (N = 17) | 61.4 (9.4) (N = 30) | |
| Other | 41.7 (17.3) (N = 9) | 72.8 (4.1) (N = 129) | |
Values are mean (SE).
RFS indicates relapse-free survival; and N, no. of patients.
Kaplan-Meier survival curves showing interaction of CRLF2 with IKZF1 alterations and JAK mutations. (A) Survival for patients with or without CRLF2 rearrangements with or without JAK mutations is shown. x-axis is time in years; y-axis, probability of RFS. The CRLF2-rearranged samples are shown in red, whereas the nonrearranged samples are in black. Solid lines indicate JAK mutations; and dashed lines, no detected JAK mutations. (B) Survival for patients with or without CRLF2 rearrangements with or without IKZF1 mutations/deletions is shown. x-axis represents time in years; y-axis, probability of RFS. The CRLF2-rearranged samples are shown in red, whereas the nonrearranged samples are in black. Solid lines represent IZKF1 alterations; dashed lines, none.
Kaplan-Meier survival curves showing interaction of CRLF2 with IKZF1 alterations and JAK mutations. (A) Survival for patients with or without CRLF2 rearrangements with or without JAK mutations is shown. x-axis is time in years; y-axis, probability of RFS. The CRLF2-rearranged samples are shown in red, whereas the nonrearranged samples are in black. Solid lines indicate JAK mutations; and dashed lines, no detected JAK mutations. (B) Survival for patients with or without CRLF2 rearrangements with or without IKZF1 mutations/deletions is shown. x-axis represents time in years; y-axis, probability of RFS. The CRLF2-rearranged samples are shown in red, whereas the nonrearranged samples are in black. Solid lines represent IZKF1 alterations; dashed lines, none.
Discussion
Genome-wide profiling of genomic alterations has proven remarkably powerful in identifying novel genetic alterations in ALL. Using alternative, complementary approaches, we and others have identified rearrangement of CRLF2 as a new recurring chromosomal alteration in B-progenitor ALL. Using FISH-based screening of large numbers of ALL cases for novel rearrangements of IGH@, Russell et al first identified IGH@-CRLF2 rearrangement, and deletions upstream of CRLF2, in approximately 5% of B-progenitor ALL cases.31 By gene expression profiling13,32 and mapping of PAR1 deletions in high-risk and DS-associated ALL,30 we have also identified IGH@-CRLF2 rearrangements, and we have precisely characterized the extent and consequences of the PAR1 deletion, which results in a novel fusion, P2RY8-CRLF2. This fusion probably places expression of CRLF2 under control of the P2RY8 promoter. P2RY8 is expressed at high levels in hematopoietic and leukemic cells, and a similar mechanism of P2RY8-mediated gene dysregulation has been reported for SOX5 in follicular lymphoma.41 Both IGH@-CRLF2 and P2RY8-CRLF2 result in overexpression of full-length CRLF2 and elevated expression of CRLF2 protein by leukemic cells.
There are several important differences between this report and other recent studies.30,31 Overall, CRLF2 rearrangement occurs at relatively low frequency in unselected B-progenitor ALL cases (5%-7%). Our analyses of the present cohort show that CRLF2 rearrangement is more frequent in high-risk ALL patients enrolled in the COG P9906 study (14.0%, 29 of 207 cases). CRLF2 is most frequently rearranged in DS-ALL patients, in which more than 50% of cases have this abnormality.30,31 The frequency of each type of CRLF2 rearrangement (IGH@ or alternative translocation and interstitial PAR1 deletion resulting in P2RY8-CRLF2 fusion) is also highly dependent on the nature of the patient cohort. In unselected B-progenitor ALL cases, PAR1 deletion is more common than CRLF2 translocation (∼ 2:1 in unselected childhood B-progenitor ALL31 and > 9:1 in DS-ALL cases with CRLF2 rearrangement).30 In contrast, IGH@-CRLF2 alteration is much more frequent than PAR1 deletions in the present cohort. Of the 29 samples with CRLF2 rearrangement, 20 cases had translocations (18 with IGH@ and 1 to an unknown partner) and 10 PAR1 deletion resulting in P2RY8-CRLF2 (1 sample had both a deletion and a translocation). This 2:1 ratio of translocation to deletion is the reverse of the pattern seen in the report from Russell et al (1:5 in unselected childhood ALL).31 The reasons for this variability are unknown, although an interesting observation is that many cases with PAR1 deletion have duplication of the X chromosome and FISH analysis demonstrates that this is accompanied by duplication of the PAR1 deletion30 (data not shown).
The clinical characteristics of patients with CRLF2 alteration also vary substantially between studies. In contrast to Russell et al,31 who found a significant difference in median ages between the patients with IGH@-CRLF2 translocations and PAR1 deletions (16 vs 4 years), we did not observe a significant difference median age in this cohort (median ages, 14.2 vs 11.1 years, respectively, P = .18). However, it should be noted that our cohort is highly selected for B-progenitor ALL cases with high-risk features, including older age, and the median age of the cohort is higher than unselected childhood ALL cohorts. It remains possible that the type of CRLF2 alteration is age dependent and that this may also contribute to the high relative frequency of IGH@-CRLF2 alteration in the present cohort. An additional clinical characteristic not found in the previous studies is the association of CRLF2 rearrangements with ethnicity. The high proportion of Hispanic/Latino children in the disrupted CRLF2 patients is remarkable (P < .001). Whether this elevated frequency of CRLF2 rearrangements in pediatric ALL is unique to the higher risk samples remains to be determined.
A unique feature of our study is the availability of comprehensive data characterizing concomitant genetic alterations, gene expression profiling, JAK mutational status, and outcome. As in previous reports,30,31 we observed a significant association between the presence of CRLF2 alteration and JAK mutations. Indeed, JAK-mutated B-progenitor ALL cases lacking CRLF2 alteration are rare. Importantly, although JAK mutations are most commonly observed at or near JAK2 R683 in B-progenitor ALL, additional residues in the JAK2 kinase domain and JAK1 pseudokinase domain are mutated in CRLF2-rearranged ALL cases. We also observed that CRLF2 alterations and JAK mutations are highly correlated with the presence of IKZF1 alterations. Together, these observations suggest that multiple genetic alterations cooperate in the pathogenesis of high-risk ALL, including perturbations in normal lymphoid development (mediated by disruption of IKZF1, which encodes a zinc finger transcription factor required for normal lymphoid development), and aberrant signaling through the heterodimeric CRLF2 receptor, mediated by CRLF2 rearrangement and JAK mutations.
Recent data indicate that CRLF2 overexpression and JAK mutations contribute to lymphoid transformation. B-progenitor ALL cell lines harboring both lesions exhibit constitutive JAK-STAT activation,31 and enforced expression of CRLF2 in primary murine hematopoietic progenitors results in expansion of B-lymphoid cells. Using the murine pro-B Ba/F3 cell line expressing IL7RA (the heterodimeric partner of CRLF2), coexpression of both P2RY8-CRLF2 and JAK mutations was required for efficient transformation as assessed by cytokine-independent cell growth and JAK-STAT activation.30 Only JAK1 V658F and the homologous mutation JAK2 V617F (which is commonly observed in myeloproliferative disease)39 transformed Ba/F3 cells in the absence of enforced expression of IL7RA and/or CRLF2, suggesting that different JAK mutations interact with different cytokine receptors and mediate signaling through different cellular pathways.30
The association between CRLF2 alteration and poor outcome (in univariate analysis) is in stark contrast to DS-ALL, in which no association with outcome was observed.30 Although our study contained only 8 DS cases, 2 were among the highest CRLF2-expressing samples; both had JAK2 R683G mutations and both relapsed. Although CRLF2 was not significantly associated with outcome in multivariable analyses independently of alteration of IKZF1, a notable finding from stratified analyses was that CRLF2 was associated with poor outcome irrespective of JAK mutational status. This finding, and the presence of a BCR-ABL1–like gene expression profile in CRLF2-rearranged cases lacking JAK mutations, suggests that additional kinase-activating mutations remain to be identified in high-risk ALL. Furthermore, IKZF1 alteration was associated with poor outcome in both CRLF2-rearranged and -nonrearranged cases, suggesting that additional high-risk cases with IKZF1 alterations harbor other genetic alterations that promote resistance to therapy and treatment failure.
These findings have important clinical implications. There are currently few tests to rapidly identify patients who lack known chromosomal alterations that are at high risk of relapse. Our data indicate that detection of IKZF1 alteration, CRLF2 alteration (by FISH, flow cytometry, or PCR/RT-PCR analysis), and JAK mutation should be considered at diagnosis in childhood ALL, particularly in those with clinical high-risk features (eg, older age and high leukocyte count). In addition, as CRLF2/JAK-mediated transformation is abrogated by JAK inhibitors in vitro30 and as therapeutic inhibitors of JAK-STAT signaling are entering clinical trials for hematopoietic malignancies,42 clinical trials of JAK inhibitors in high-risk ALL are warranted. As those trials are designed, it will be critical to determine whether they should be limited to patients with documented JAK mutations or should also include patients with JAK wild-type ALLs with genomic lesions leading to CRLF2 overexpression.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Audrey Phillips for technical assistance.
This work was supported by National Institutes of Health Department of Health and Human Services: National Cancer Institute Strategic Partnerships to Evaluate Cancer Gene Signatures Program National Cancer Institute (U01 CA114762; principal investigator, C.L.W.) and National Cancer Institute (U10CA98543, supporting the COG and Statistical Center, principal investigator, G.H.R.), the American Lebanese Syrian Associated Charities (J.R.D.), the National Childhood Cancer Foundation, COG (cell banking grant U24 CA114766, G.H.R.), and a Leukemia & Lymphoma Society Specialized Center of Research (program grant 7388-06, principal investigator, C.L.W.). This work was supported in part by grants to the COG (CA114766 and a supplement to that grant for the Therapeutically Applicable Research to Generate Effective Targets project) and a Pew Scholar award (C.G.M.). S.P.H. is the Ergen Family Chair in Pediatric Cancer. Optimization of FISH probes was performed at the University of New Mexico Cancer Center Fluorescence Microscopy Facility (supported as detailed on the webpage: http://hsc.unm.edu/crtc/microscopy/Facility.html).
National Institutes of Health
Authorship
Contribution: R.C.H., C.G.M., and I.-M.C. performed experiments, analyzed data, and wrote the manuscript; W.W. performed experiments and wrote the manuscript; F.M.M. performed FISH experiments and analyzed data; A.J.C. analyzed FISH data; H.K. and W.L. performed statistical analysis; K.K.D. performed experiments and analyzed data; M.A.S., W.L.C., and M.D. analyzed data; W.P.B., B.M.C., G.H.R., and J.R.D. designed the study; and S.P.H. and C.L.W. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cheryl L. Willman, University of New Mexico Cancer Center, MSC08 4630 1, University of New Mexico, Albuquerque, NM 87131; e-mail: cwillman@salud.unm.edu; or Charles G. Mullighan, MS342, Rm D4047E, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105-3678; e-mail: charles.mullighan@stjude.org; or Stephen P. Hunger, Children's Hospital, 13123 East 16th Ave, B115, Aurora, CO 80045; e-mail: hunger.stephen@tchden.org.
References
Author notes
R.C.H. and C.G.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal