The Total Therapy 3 trial 2003-33 enrolled 303 newly diagnosed multiple myeloma patients and was noted to provide superior clinical outcomes compared with predecessor trial Total Therapy 2, especially in gene expression profiling (GEP)–defined low-risk disease. We report here on the results of successor trial 2006-66 with 177 patients, using bortezomib, lenalidomide, and dexamethasone maintenance for 3 years versus bortezomib, thalidomide, and dexamethasone in year 1 and thalidomide/dexamethasone in years 2 and 3 in the 2003-33 protocol. Overall survival (OS) and event-free survival (EFS) plots were super-imposable for the 2 trials, as were onset of complete response and complete response duration (CRD), regardless of GEP risk. GEP-defined high-risk designation, pertinent to 17% of patients, imparted inferior OS, EFS, and CRD in both protocols and, on multivariate analysis, was the sole adverse feature affecting OS, EFS, and CRD. Mathematical modeling of CRD in low-risk myeloma predicted a 55% cure fraction (P < .001). Despite more rapid onset and higher rate of CR than in other molecular subgroups, CRD was inferior in CCND1 without CD20 myeloma, resembling outcomes in MAF/MAFB and proliferation entities. The robustness of the GEP risk model should be exploited in clinical trials aimed at improving the notoriously poor outcome in high-risk disease.
Introduction
Total Therapy 3 (2003-33; TT3) used, for newly diagnosed multiple myeloma (MM), 2 cycles of VTD-PACE (bortezomib, thalidomide, and dexamethasone and 4-day continuous infusions of cisplatin, doxorubicin, cyclophosphamide, and etoposide) as induction before and consolidation therapy after melphalan-based tandem transplantation, which was followed by 3 years of intended maintenance with VTD in year 1 and thalidomide/dexamethasone in years 2 and 3.1,–3 Results revealed superior outcomes, especially in gene expression profiling (GEP)–defined low-risk MM, compared with predecessor phase 3 trial Total Therapy 2 (TT2), which randomized patients up-front to receive or not receive thalidomide during induction, consolidation, and maintenance phases.4 In further analyses examining the timeliness of completion of intended protocol steps, we concluded that the improved results in TT3 versus TT2 were attributable to the incorporation, up-front, of bortezomib in TT3.3 To validate these findings and bortezomib pharmacogenomic data,5 a successor trial 2006-66 enrolled another 177 patients. The trials were identical in design, except that the maintenance phase in 2006-66 applied 3 years rather than 1 year of bortezomib and used lenalidomide instead of thalidomide.
Methods
A total of 303 patients had been enrolled in the 2003-33 trial between February 2004 and July 2006 (median follow-up, 4.4 years), whereas 177 subjects had been accrued to 2006-66 between November 2006 and September 2008 (median follow-up, 2.0 years). Protocol details are as follows: both studies used 2 cycles each of VTD-PACE induction before and dose-reduced VTD-PACE consolidation after melphalan 200 mg/m2-based tandem transplantation. The 3-year maintenance phase differed in the 2 trials in that bortezomib in protocol 2003-33 was limited to year 1, whereas the 2006-66 study called for 3 years of proteasome inhibitor administration. Specifically, trial 2003-33 used VTD maintenance in year 1 with monthly cycles of bortezomib 1.0 mg/m2 on days 1, 4, 8, and 11, whereas thalidomide was given continuously at 100 mg/day and dexamethasone 20 mg on days 1 to 4 and 8 to 11; in years 2 and 3, thalidomide was continued at 100 mg/day and dexamethasone was limited to monthly pulses of 20 mg on days 1 to 4. In 2006-66, bortezomib, lenalidomide, and dexamethasone (VRD) was given for 3 years, composing monthly cycles of bortezomib 1.0 mg/m2 on days 1, 4, 8, and 11 in year 1 followed by weekly administration in years 2 and 3; lenalidomide was administered at 15 mg on days 1 to 20 followed by 5 mg on days 21 to 28 for all 3 years; dexamethasone was applied at 20 mg on days 1 to 4 and 8 to 11 in year 1 and was given weekly with bortezomib in years 2 and 3.
All 480 patients had signed a written informed consent in keeping with institutional and federal guidelines and in accordance with the Declaration of Helsinki. Both protocols and their revisions had been approved by the Institutional Review Board, and results were reviewed annually. A Data Safety and Monitoring Board performed annual reviews. A federally accredited team of independent reviewers audited nearly 80% of patient records for accuracy in reported toxicity, response, and response duration.
Clinical endpoints examined included overall survival (OS) and event-free survival (EFS) measured from initiation of therapy, whereas duration of complete response (CR) was counted from onset of documented CR. CR was defined according to International Myeloma Working Group criteria, as were relapse and EFS.6 International Staging System (ISS) criteria were applied to disease staging.7 Variables examined at baseline and at prespecified follow-up times included, in addition to standard variables of serum and urine myeloma-protein measurements, serum levels of free-light chains, β-2-microglobulin (B2M) and C-reactive protein; bone marrow aspirates and biopsies were examined for the presence of monoclonal plasma cells, whereas bone marrow aspirates were subjected to metaphase karyotyping to determine the presence of cytogenetic abnormalities. GEP of CD138-purified plasma cells was performed in 275 patients on 2003-33 and 166 on 2006-66 protocols to define the 70 gene-derived risk score,8 molecular subgroup,9 and delTP53 status.10 Molecular subgroup designations included CCND-1 without CD20 expression (CD-1) or with CD20 expression (CD-2), MAF/MAFB, MMSET/FGFR3, hyperdiploidy, low bone disease, myeloid, and proliferation.
Kaplan-Meier methods were used to generate survival distribution graphs,11 and comparisons were made through the log-rank test.12 The Pearson χ2 test was used for categorical comparisons.13 Stepwise selection and Cox proportional hazard regression modeling were applied to the multivariate analyses.14
A parametric mixture cure fraction model was used, using a logistic function to estimate the cured fraction and an exponential distribution to estimate the survival function of uncured persons. This was performed for EFS and the duration of CR. For CR duration, the fraction of patients achieving CR was multiplied by the cure fraction to calculate the overall cure rate.
Results
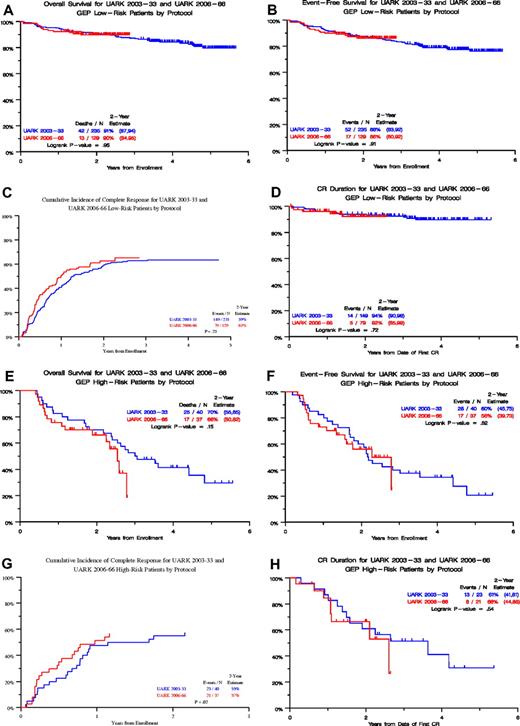
Clinical outcomes in the 2 studies were comparable (Table 1) except for a higher incidence of several adverse features in 2006-66: albumin less than 3.5 g/dL (45% vs 26%, P < .001) and B2M more than or equal to 3.5 mg/L (57% vs 45%, P = .012), resulting in higher incidence of ISS stages 2 and 3 (70% vs 55%, P < .001). GEP-defined high risk was also more frequent in 2006-66 (22% vs 15%, P = .038), and GEP-defined CD-1 molecular subgroup was also over-represented in 2006-66 (11% vs 5%, P = .037), whereas myeloid designation was underrepresented (3% vs 14%, P < .001). Figure 1 portrays clinical outcomes, including OS (Figure 1A) and EFS (Figure 1B) from initiation of protocol therapy, onset of CR timed from start of therapy (Figure 1C), and CR duration (CRD) timed from CR onset (Figure 1D). Despite the aforementioned differences in baseline prognostic variables in favor of 2003-33, results revealed, with the exception of faster CR onset in 2006-66, similar Kaplan-Meier plots for OS, EFS, and CRD. When examined in the context of GEP-defined risk, 2003-33 and 2006-66 results were again superimposable for both low-risk (Figure 2A-D) and high-risk disease (Figure 2E-H). For the 2003-33 trial with longer follow-up, the high 4-year CRD estimate in low-risk myeloma of 89% was consistent with a cure estimate of 55% (Figure 3).
Patient characteristics in 2003-33 and 2006-66 trials
| Factor . | Overall, n/N (%) . | 2003-33, n/N (%) . | 2006-66, n/N (%) . | P . |
|---|---|---|---|---|
| Age > 65 y | 130/480 (27) | 84/303 (28) | 46/177 (26) | .680 |
| Female | 179/480 (37) | 110/303 (36) | 69/177 (39) | .558 |
| White | 432/480 (90) | 268/303 (88) | 164/177 (93) | .138 |
| Albumin < 3.5 g/dL* | 160/479 (33) | 80/303 (26) | 80/176 (45) | < .001 |
| B2M > 3.5 mg/L* | 235/477 (49) | 136/303 (45) | 99/174 (57) | .012 |
| B2M > 5.5 mg/L | 116/477 (24) | 65/303 (21) | 51/174 (29) | .054 |
| ISS stage 1* | 189/477 (40) | 137/303 (45) | 52/174 (30) | < .001 |
| ISS stage 3 | 116/477 (24) | 65/303 (21) | 51/174 (29) | .054 |
| Creatinine > 2 mg/dL | 34/479 (7) | 23/303 (8) | 11/176 (6) | .582 |
| CRP > 8 mg/L | 159/477 (33) | 100/302 (33) | 59/175 (34) | .893 |
| Hb < 10 g/dL | 151/479 (32) | 94/303 (31) | 57/176 (32) | .757 |
| LDH > 190 U/L | 124/479 (26) | 81/303 (27) | 43/176 (24) | .579 |
| Platelet count < 150 × 109/L | 63/479 (13) | 38/303 (13) | 25/176 (14) | .604 |
| Cytogenetic abnormalities (anytime before enrollment) | 170/475 (36) | 100/302 (33) | 70/173 (40) | .108 |
| GEP high-risk* | 77/441 (17) | 40/275 (15) | 37/166 (22) | .038 |
| GEP CD-1 subgroup* | 33/441 (7) | 15/275 (5) | 18/166 (11) | .037 |
| GEP CD-2 subgroup | 52/441 (12) | 27/275 (10) | 25/166 (15) | .098 |
| GEP HY subgroup | 129/441 (29) | 80/275 (29) | 49/166 (30) | .924 |
| GEP LB subgroup | 45/441 (10) | 33/275 (12) | 12/166 (7) | .109 |
| GEP MF subgroup | 31/441 (7) | 22/275 (8) | 9/166 (5) | .305 |
| GEP MS subgroup | 55/441 (12) | 33/275 (12) | 22/166 (13) | .700 |
| GEP MY subgroup* | 43/441 (10) | 38/275 (14) | 5/166 (3) | < .001 |
| GEP PR subgroup | 53/441 (12) | 27/275 (10) | 26/166 (16) | .067 |
| GEP TP53 deletion | 46/441 (10) | 33/275 (12) | 13/166 (8) | .165 |
| Factor . | Overall, n/N (%) . | 2003-33, n/N (%) . | 2006-66, n/N (%) . | P . |
|---|---|---|---|---|
| Age > 65 y | 130/480 (27) | 84/303 (28) | 46/177 (26) | .680 |
| Female | 179/480 (37) | 110/303 (36) | 69/177 (39) | .558 |
| White | 432/480 (90) | 268/303 (88) | 164/177 (93) | .138 |
| Albumin < 3.5 g/dL* | 160/479 (33) | 80/303 (26) | 80/176 (45) | < .001 |
| B2M > 3.5 mg/L* | 235/477 (49) | 136/303 (45) | 99/174 (57) | .012 |
| B2M > 5.5 mg/L | 116/477 (24) | 65/303 (21) | 51/174 (29) | .054 |
| ISS stage 1* | 189/477 (40) | 137/303 (45) | 52/174 (30) | < .001 |
| ISS stage 3 | 116/477 (24) | 65/303 (21) | 51/174 (29) | .054 |
| Creatinine > 2 mg/dL | 34/479 (7) | 23/303 (8) | 11/176 (6) | .582 |
| CRP > 8 mg/L | 159/477 (33) | 100/302 (33) | 59/175 (34) | .893 |
| Hb < 10 g/dL | 151/479 (32) | 94/303 (31) | 57/176 (32) | .757 |
| LDH > 190 U/L | 124/479 (26) | 81/303 (27) | 43/176 (24) | .579 |
| Platelet count < 150 × 109/L | 63/479 (13) | 38/303 (13) | 25/176 (14) | .604 |
| Cytogenetic abnormalities (anytime before enrollment) | 170/475 (36) | 100/302 (33) | 70/173 (40) | .108 |
| GEP high-risk* | 77/441 (17) | 40/275 (15) | 37/166 (22) | .038 |
| GEP CD-1 subgroup* | 33/441 (7) | 15/275 (5) | 18/166 (11) | .037 |
| GEP CD-2 subgroup | 52/441 (12) | 27/275 (10) | 25/166 (15) | .098 |
| GEP HY subgroup | 129/441 (29) | 80/275 (29) | 49/166 (30) | .924 |
| GEP LB subgroup | 45/441 (10) | 33/275 (12) | 12/166 (7) | .109 |
| GEP MF subgroup | 31/441 (7) | 22/275 (8) | 9/166 (5) | .305 |
| GEP MS subgroup | 55/441 (12) | 33/275 (12) | 22/166 (13) | .700 |
| GEP MY subgroup* | 43/441 (10) | 38/275 (14) | 5/166 (3) | < .001 |
| GEP PR subgroup | 53/441 (12) | 27/275 (10) | 26/166 (16) | .067 |
| GEP TP53 deletion | 46/441 (10) | 33/275 (12) | 13/166 (8) | .165 |
Molecular subgroups included CD-1 (CCND1 without CD20), CD-2 (CCND1 with CD20 expression), MS (MMSET/FGFR3), MF (MAF/MAFB), HY (hyperdiploidy), LB (low bone disease), MY (myeloid), and PR (proliferation).
B2M indicates β-2-microglobulin; ISS, International Staging System; CRP, C-reactive protein; Hb, hemoglobin; LDH, lactate dehydrogenase; and GEP, gene expression profiling.
Statistically significant at P < .05.
Clinical outcomes in 2003-33 and 2006-66 trials. Overall survival (OS; A) and event-free survival (EFS; B) from initiation of therapy, as well as timing of onset of complete response (CR) from treatment start (C). Complete response duration (CRD) was measured from onset of CR (D).
Clinical outcomes in 2003-33 and 2006-66 trials. Overall survival (OS; A) and event-free survival (EFS; B) from initiation of therapy, as well as timing of onset of complete response (CR) from treatment start (C). Complete response duration (CRD) was measured from onset of CR (D).
Clinical outcomes in 2003-33 and 2006-66 trials according to gene expression profiling (GEP)–defined risk. (A-D) Low-risk disease: OS (A), EFS, (B), CR onset (C), and CRD (D). (E-H) High-risk disease: OS (E), EFS, (F), CR onset (G), and CRD (H).
Clinical outcomes in 2003-33 and 2006-66 trials according to gene expression profiling (GEP)–defined risk. (A-D) Low-risk disease: OS (A), EFS, (B), CR onset (C), and CRD (D). (E-H) High-risk disease: OS (E), EFS, (F), CR onset (G), and CRD (H).
Mathematical model for GEP-defined low-risk and high-risk myeloma, compatible with a cure fraction for low-risk disease of 55% (CR rate of 63% times 87.6%).
Mathematical model for GEP-defined low-risk and high-risk myeloma, compatible with a cure fraction for low-risk disease of 55% (CR rate of 63% times 87.6%).
We next examined clinical outcomes in the context of GEP-defined molecular subgroups (Figure 4). The comparability of outcomes in 2003-33 and 2006-66 trials overall (Figure 1) and within risk groups (Figure 2) justified data pooling to better convey the prognostic impact on clinical outcome of 8 molecular entities. The superior performance of CD-2, low bone disease, hyperdiploidy, MMSET/FGFR3, and myeloid subgroups is readily apparent for OS (Figure 4A), EFS (Figure 4B), and CRD (Figure 4D), whereas proliferation and MAF/MAFB were associated with distinctly inferior outcomes compared with the 5 superior subgroups (P < .001 for OS, EFS, and CRD). In the case of CD-1, CR onset was faster and its ultimate frequency higher compared with other subgroups (P < .001; Figure 4C), yet CRD tracked more closely with proliferation and MAF/MAFB (P = .91) than with the favorable subgroups (P = .005; Figure 4D).
Clinical outcomes in 2003-33 and 2006-66 trials combined for patients with GEP-derived molecular subgroups. OS (A), EFS, (B), CR onset (C), and CRD (D). Subgroup designations are CD-1, CD-2, MMSET/FGFR3, MAF/MAFB, hyperdiploidy, low bone disease, myeloid, and proliferation.9 Similar colors were used for similar curves (blue represents favorable; yellow, CD-1; and red, unfavorable). P values for individual colors, or 1 color versus the rest.
Clinical outcomes in 2003-33 and 2006-66 trials combined for patients with GEP-derived molecular subgroups. OS (A), EFS, (B), CR onset (C), and CRD (D). Subgroup designations are CD-1, CD-2, MMSET/FGFR3, MAF/MAFB, hyperdiploidy, low bone disease, myeloid, and proliferation.9 Similar colors were used for similar curves (blue represents favorable; yellow, CD-1; and red, unfavorable). P values for individual colors, or 1 color versus the rest.
Multivariate analyses were conducted to examine the adverse variables independently linked to clinical outcomes in 2003-33 and 2006-66 trials (Table 2). In both protocols, GEP-defined high-risk affected OS, EFS, and CRD adversely, whereas the CD-1 molecular subgroup was linked to higher CR rate. No other baseline parameter retained independent prognostic significance in 2006-66. In 2003-33, the presence of cytogenetic abnormalities and high LDH were additional negative features for OS and EFS; high B2M more than 5.5 mg/L was detrimental for OS and low albumin less than 3.5 g/dL for EFS. Both EFS and CRD were shorter in the presence of renal function impairment. Both low albumin and high creatinine more than or equal to 2 mg/dL were linked to lower CR rates.
Multivariate models in trials 2003-33 and 2006-66 linked to OS, EFS, CR rate, and CRD
| Variable . | 2003-33 . | 2006-66 . | ||||
|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| OS | ||||||
| GEP high-risk | 40/275 (15) | 2.43 (1.40-4.19) | .001 | 36/159 (23) | 3.00 (1.23-7.31) | .016* |
| Cytogenetic abnormalities | 95/275 (35) | 1.99 (1.19-3.34) | .009 | 66/159 (42) | 1.72 (0.73-4.02) | .213 |
| LDH > 190 U/L | 74/275 (27) | 1.78 (1.07-2.95) | .025 | 40/159 (25) | 1.25 (0.55-2.84) | .590 |
| B2M > 5.5 mg/L | 59/275 (21) | 2.24 (1.35-3.74) | .002 | 48/159 (30) | 1.28 (0.56-2.92) | .553 |
| EFS | ||||||
| GEP high-risk | 40/275 (15) | 2.57 (1.54-4.31) | < .001 | 36/159 (23) | 2.77 (1.18-6.45) | .019* |
| Cytogenetic abnormalities | 95/275 (35) | 1.80 (1.12-2.90) | .016 | 66/159 (42) | 1.17 (0.53-2.56) | .696 |
| LDH > 190 U/L | 74/275 (27) | 2.34 (1.46-3.74) | < .001 | 40/159 (25) | 1.54 (0.72-3.26) | .264 |
| Creatinine > 2 mg/dL | 21/275 (8) | 3.30 (1.86-5.85) | < .001 | 11/159 (7) | 0.61 (0.16-2.32) | .469 |
| Albumin < 3.5 g/dL | 76/275 (28) | 2.00 (1.25-3.22) | .004 | 71/159 (45) | 1.62 (0.74-3.55) | .226 |
| CR rate | ||||||
| GEP CD-1 subgroup | 15/275 (5) | 8.43 (1.08-65.84) | .042 | 18/165 (11) | 12.56 (1.62-97.30) | .015* |
| Creatinine > 2 mg/dL | 21/275 (8) | 0.28 (0.11-0.74) | .010 | 11/165 (7) | 1.76 (0.44-7.12) | .427 |
| Albumin < 3.5 g/dL | 76/275 (28) | 0.47 (0.27-0.82) | .008 | 74/165 (45) | 0.59 (0.31-1.13) | .110 |
| CRD | ||||||
| GEP high-risk | 23/172 (13) | 7.58 (3.28-17.50) | < .001 | 21/100 (21) | 5.87 (1.80-19.07) | .003* |
| GEP CD-1 subgroup | 14/172 (8) | 2.93 (0.81-10.56) | .100 | 17/100 (17) | 2.96 (0.92-9.59) | .070 |
| Creatinine > 2 mg/dL | 7/172 (4) | 4.23 (1.50-11.93) | .006 | 8/100 (8) | 0.61 (0.07-5.13) | .648 |
| Variable . | 2003-33 . | 2006-66 . | ||||
|---|---|---|---|---|---|---|
| n/N (%) . | HR (95% CI) . | P . | n/N (%) . | HR (95% CI) . | P . | |
| OS | ||||||
| GEP high-risk | 40/275 (15) | 2.43 (1.40-4.19) | .001 | 36/159 (23) | 3.00 (1.23-7.31) | .016* |
| Cytogenetic abnormalities | 95/275 (35) | 1.99 (1.19-3.34) | .009 | 66/159 (42) | 1.72 (0.73-4.02) | .213 |
| LDH > 190 U/L | 74/275 (27) | 1.78 (1.07-2.95) | .025 | 40/159 (25) | 1.25 (0.55-2.84) | .590 |
| B2M > 5.5 mg/L | 59/275 (21) | 2.24 (1.35-3.74) | .002 | 48/159 (30) | 1.28 (0.56-2.92) | .553 |
| EFS | ||||||
| GEP high-risk | 40/275 (15) | 2.57 (1.54-4.31) | < .001 | 36/159 (23) | 2.77 (1.18-6.45) | .019* |
| Cytogenetic abnormalities | 95/275 (35) | 1.80 (1.12-2.90) | .016 | 66/159 (42) | 1.17 (0.53-2.56) | .696 |
| LDH > 190 U/L | 74/275 (27) | 2.34 (1.46-3.74) | < .001 | 40/159 (25) | 1.54 (0.72-3.26) | .264 |
| Creatinine > 2 mg/dL | 21/275 (8) | 3.30 (1.86-5.85) | < .001 | 11/159 (7) | 0.61 (0.16-2.32) | .469 |
| Albumin < 3.5 g/dL | 76/275 (28) | 2.00 (1.25-3.22) | .004 | 71/159 (45) | 1.62 (0.74-3.55) | .226 |
| CR rate | ||||||
| GEP CD-1 subgroup | 15/275 (5) | 8.43 (1.08-65.84) | .042 | 18/165 (11) | 12.56 (1.62-97.30) | .015* |
| Creatinine > 2 mg/dL | 21/275 (8) | 0.28 (0.11-0.74) | .010 | 11/165 (7) | 1.76 (0.44-7.12) | .427 |
| Albumin < 3.5 g/dL | 76/275 (28) | 0.47 (0.27-0.82) | .008 | 74/165 (45) | 0.59 (0.31-1.13) | .110 |
| CRD | ||||||
| GEP high-risk | 23/172 (13) | 7.58 (3.28-17.50) | < .001 | 21/100 (21) | 5.87 (1.80-19.07) | .003* |
| GEP CD-1 subgroup | 14/172 (8) | 2.93 (0.81-10.56) | .100 | 17/100 (17) | 2.96 (0.92-9.59) | .070 |
| Creatinine > 2 mg/dL | 7/172 (4) | 4.23 (1.50-11.93) | .006 | 8/100 (8) | 0.61 (0.07-5.13) | .648 |
Multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level. A multivariate P value greater than .05 indicates variable forced into model with significant variables chosen using stepwise selection. P value for HR from the Wald χ2 test in Cox regression. Variables considered for the models included: age > 65 years, female, albumin < 3.5 g/dL, B2M > 3.5 mg/L, B2M > 5.5 mg/L, ISS stage 1, ISS stage 3, creatinine > 2 mg/dL, CRP > 8 mg/L, Hb < 10 g/dL, LDH > 190 U/L, cytogenetic abnormalities (any time before enrollment), GEP high-risk, GEP CD-1 subgroup, GEP CD-2 subgroup, GEP hyperdiploidy subgroup, GEP low bone disease subgroup, GEP MAF/MAFB subgroup, GEP MMSET/FGFR3 subgroup, GEP myeloid subgroup, GEP proliferation subgroup, and GEP TP53 deletion.
HR indicates hazard ratio; OS, overall survival; EFS, event-free survival; CI, confidence interval; LDH, lactate dehydrogenase; B2M, β-2-microglobulin; ISS, International Staging System; CR, complete response; CRD, complete response duration; and GEP, gene expression profiling.
Statistically significant at P < .05.
Discussion
The data presented here validate earlier reports of remarkable success of TT3 in GEP-defined low-risk myeloma.1,–3 Indeed, despite more adverse features in 2006-66, clinical endpoints (OS, EFS, and CRD) were superimposable in the 2 protocols, which may be attributable to a superior maintenance regimen in 2006-66 using 3 years of bortezomib and the purportedly more effective immunomodulatory agent, lenalidomide, instead of thalidomide in 2003-33. To address this issue, we are envisioning a pair-mate analysis once longer follow-up has been accomplished in 2006-66. For the more mature 2003-33 trial, the 4-year CRD estimate of 89% in GEP-defined low-risk myeloma was consistent with a cure estimate of more than 50%.15 Having again validated the robustness of the GEP risk model, we advocate its use for selecting the approximately 15% of patients with high-risk myeloma for innovative trials directed at improving currently dismal outcomes. Because the problem with high-risk myeloma is not achieving but sustaining CR, we are exploring, in Total Therapy 5, whether dose-dense and less dose-intense strategies can prevent disease relapse during treatment-free phases of TT3 (required for host recovery from high-dose therapy).
Having already discovered that bortezomib in TT3 overcomes the adverse roles in TT2 of the MMSET/FGFR3 subgroup3 and of delTP53 in the low-risk setting,16 we now draw attention to unique differences between CD-1 and CD-2 molecular subtypes, both of which share t(11;14). CD-1 myeloma was characterized by the steepest onset and highest level of CR but short CRD, similar to the proliferation subgroup, whereas, despite slowest timing of onset and lowest plateau of CR, the CRD of CD-2 was strikingly superior among all molecular entities analyzed. These data emphasize the potential pitfalls of using CR rather than CRD as a surrogate endpoint.17
The uniquely poor prognosis of the MAF/MAFB entity, affecting 6% of newly diagnosed patients, demands special attention. Although t(14;16)- and t(14;20)-positive myeloma cell lines exist, we have found that GEP signatures of primary MAF/MAFB disease and cell lines are not completely congruent, perhaps as a consequence of tumor cell–microenvironmental interactions. Therefore, scrutiny of primary disease samples is essential for identifying relevant targets, among which IGF1, IGF1R, and IL6R are noteworthy for their unique overexpression in MAF/MAFB disease (J.D.S., preliminary observations, 2009).9,18
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the National Cancer Institute, Bethesda, MD (grant CA 55813).
Authorship
Contribution: F.v.R., J.D.S., E.A., J.C., and B.B. conceived the work; B.N., F.v.R., E.A., Y.A., S.W., and B.B. participated in clinical research protocols; J.S., A.H., and J.C. performed statistical analyses; J.D.S. performed gene expression profiling; and B.N., F.v.R., and B.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bart Barlogie, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 West Markham, #816, Little Rock, AR 72205; e-mail: barlogiebart@uams.edu.