Abstract
NPM1-mutated acute myeloid leukemia (AML) is a provisional entity in the 2008 World Health Organization (WHO) classification of myeloid neoplasms. The significance of multilineage dysplasia (MLD) in NPM1-mutated AML is unclear. Thus, in the 2008 WHO classification, NPM1-mutated AML with MLD is classified as AML with myelodysplasia (MD)–related changes (MRCs). We evaluated morphologically 318 NPM1-mutated AML patients and found MLD in 23.3%. Except for a male predominance and a lower fms-related tyrosine kinase 3–internal tandem duplication (FLT3-ITD) incidence in the MLD+ group, no differences were observed in age, sex, cytogenetics, and FLT3-–tyrosine kinase domain between NPM1-mutated AML with and without MLD. NPM1-mutated AML with and without MLD showed overlapping immunophenotype (CD34 negativity) and gene expression profile (CD34 down-regulation, HOX genes up-regulation). Moreover, overall and event-free survival did not differ among NPM1-mutated AML patients independently of whether they were MLD+ or MLD−, the NPM1-mutated/FLT3-ITD negative genotype showing the better prognosis. Lack of MLD impact on survival was confirmed by multivariate analysis that highlighted FLT3-ITD as the only significant prognostic parameter in NPM1-mutated AML. Our findings indicate that NPM1 mutations rather than MLD dictate the distinctive features of NPM1-mutated AML. Thus, irrespective of MLD, NPM1-mutated AML represents one disease entity clearly distinct from AML with MRCs.
Introduction
Acute myeloid leukemia (AML) harboring an NPM1 mutation causing aberrant cytoplasmic expression of nucleophosmin (NPMc+ AML)1,2 accounts for approximately 30% of adult AML and exhibits distinctive biologic and clinicopathologic features.3-5 Moreover, AML with mutated NPM1 shows a favorable prognosis, in the absence of fms-related tyrosine kinase 3–internal tandem duplication (FLT3-ITD) mutation.6-11 For these reasons, AML with mutated NPM1 is listed as a provisional entity in the 2008 World Health Organization (WHO) classification of myeloid neoplasms.12
At the time of preparation of the 2008 WHO classification, one argument for considering NPM1-mutated AML as a provisional rather than a distinct entity was that it showed overlapping features with AML with myelodysplasia (MD)–related changes, whose significance was unclear. According to the 2008 WHO,13 one case can be assigned to AML with MD-related changes, because of the following reasons: (1) AML arising from previous myelodysplastic syndrome (MDS) or MDS/myeloproliferative neoplasm (MPN); and/or (2) AML with an MDS-related cytogenetic abnormality; and/or (3) AML with multilineage dysplasia (MLD).
Previous history of MDS or MDS/MPN is not usually a matter of controversy in distinguishing AML with MD-related changes from NPM1-mutated AML, because the latter is mostly a de novo leukemia.1,3,14 Presence of MDS-related cytogenetic abnormalities is also rarely confounding, because approximately 85% of NPM1-mutated AMLs show a normal karyotype.1 Moreover, if a cytogenetic abnormality is present (∼ 15% of cases), this only exceptionally involves gain or loss of major segments of certain chromosomes, for example, complex karyotypes, −7/del(7q), or −5/del(5q),15 as is frequently observed in AML with MD-related changes.13 Even more importantly, we recently demonstrated that the presence of cytogenetic aberrations has no impact on the biologic and prognostic features of NPM1-mutated AML.15 Thus, the major controversy in the 2008 WHO classification mainly consists in how to classify AML cases presenting with MLD (defined according to WHO morphologic criteria13 ) and a concomitant NPM1 mutation.
According to the 2008 WHO,13 cases that are recognized as distinct entities in the group of “AML with recurrent genetic abnormalities” but show MD-related changes of any type, are not classified as “AML with MD-related changes.” In fact, the recurrent genetic lesion takes predominance over the MD-related changes. This criterion, however, does not apply to NPM1-mutated AML, because the WHO classification states that “It is currently unclear whether NPM1-positive/FLT3-negative genotype in AML with multilineage dysplasia confers the same good prognosis as in other AML with NPM1 mutations.”13 Thus, the WHO classification presently recommends that cases with overlapping features should be diagnosed as AML with MD-related changes, additionally annotating the presence of NPM1 mutation.13 p126
To better clarify the significance of MLD in NPM1-mutated AML, we investigated 318 patients with the following aims: (1) to determine the frequency and type of MLD in NPM1-mutated AML; (2) to assess the gene expression profile in NPM1-mutated AML with and without MLD; and (3) to compare the clinicopathologic, cytogenetic, FLT3 mutation status, and prognostic features of NPM1-mutated AML with and without MLD.
Our results indicate that MLD has no impact on biologic and prognostic features of NPM1-mutated AML. Taken together with previous observations that features of NPM1-mutated AML are not influenced by concomitant chromosomal aberrations,15 our findings reinforce the concept that NPM1 mutation is a founder genetic lesion and clearly point to AML with mutated NPM1 and AML with MD-related changes as distinct disease entities.
Methods
Patients with leukemia
We investigated a total of 318 patients with AML with de novo cytoplasmic/mutated NPM1: 110 were enrolled in the Gruppo Italiano Malattie Ematologiche Maligne dell'Adulto (GIMEMA) LAM99P and GIMEMA/European Organisation for Research and Treatment of Cancer (EORTC) AML12 trials, and 208 were from the Munich Leukemia Laboratory (MLL). Only patients with NPM1-mutated AML with information on cytomorphologic features, cytogenetics, and prognosis were included in this study. Cytomorphologic evaluation in the 208 cases from the MLL and in the 110 patients from GIMEMA/EORTC was performed on bone marrow and peripheral blood smears.
The 110 patients from the LAM99P and AML12 (registration phase only) protocols received the same therapy, that consisted in a 3-drug induction cycle with daunorubicin, etoposide, and arabinosylcytosine. In case of complete remission (CR), a single course of consolidation therapy was administered: intermediate-dose arabinosylcytosine (500 mg/m2 every 12 hours in a 2-hour infusion on days 1-6) plus daunorubicin, given on days 4 to 6. After consolidation, younger patients with a sibling donor were assigned to undergo allogeneic stem cell transplantation. Those without donor (as well as older patients) received unpurged autologous stem cell transplantation. Approval for the above studies was obtained from the institutional board of each participating center with consent obtained in accordance with the Declaration of Helsinki.
The 208 NPM1-mutated AML cases from the MLL received the following therapy: 94 patients were treated within the AML Cooperative Group (AMLCG) trials16 (see information on study centers in “Acknowledgments”), the remaining 114 patients received treatment according to AMLCG protocols but were not enrolled in the AMLCG trial (n = 17) or other intensive AML therapy protocols. As a control group, 55 patients with NPM1 wild-type AML without recurrent cytogenetic abnormalities excluding therapy-related AML were analyzed for MLD. These patients also received therapy according to the AMLCG trial.
Defining criteria for AML with mutated NPM1
The 208 AML cases from MLL were defined by mutational analysis of the NPM1 gene, as previously reported.6
Presence of the NPM1 mutation in the 110 patients with AML from GIMEMA LAM99P and GIMEMA/EORTC AML12 trials was defined by immunohistochemical criteria, that is, demonstration of aberrant cytoplasmic expression of NPM (NPMc+.17,18 NPMc+) was detected in paraffin sections from bone marrow biopsies fixed in B5/decalcified in EDTA with the use of an anti-NPM monoclonal antibody (clone 376), as previously described.17 Immunostaining with monoclonal antibody against nucleolin/C23 (clone 4E2; Santa Cruz Biotechnology) served as negative control (nucleus-restricted positivity). When fresh cells were available for molecular or biochemical studies, the presence of NPM1 gene mutations or a mutated NPM1 protein was confirmed by mutational analysis1 or Western blotting19 with antibodies specifically directed against the NPM1 mutants.
Morphologic studies
Morphologic analysis of all 318 AML cases was performed by 3 separate investigators in Munich (K.M. or U.B., validation by T.H.) on Pappenheim-stained or May-Grünwald–stained smears obtained from peripheral blood and bone marrow. Evaluation of dysplastic features was done searching for the presence of dysplasia in at least 50% of the cells in at least 2 cell lineages in bone marrow smears, according to WHO criteria.13,20
In 110 patients from GIMEMA/EORTC, both smears and bone marrow trephines were available for analysis. This offered the opportunity to assess blindly whether the bone marrow biopsy could be used for detecting MLD in NPM1-mutated AML. The bone marrow biopsy is a well-accepted procedure for detecting dysmegakaryopoiesis because clusters and dysplastic changes of megakaryocytes, such as absence of lobulation or multinuclearity, are usually well appreciated in tissue sections.13,21 In contrast, dysplastic changes affecting the myeloid and erythroid cell lineages are hard to assess by histology,22 and this is the reason why the WHO guidelines require that MLD must be evaluated in smears.13 To overcome this problem, we used our GIMEMA cohort to evaluate whether aberrant cytoplasmic expression in hemopoietic precursors of different lineages in bone marrow biopsies23 could be used as surrogate for predicting MLD. Multilineage involvement was considered to be present when 2 or more myeloid (but not lymphoid24 ) cell lineages showed at immunohistochemistry aberrant cytoplasmic expression of NPM.23 Immunohistochemical detection of NPM, which also included double staining for glycophorin and NPM, was carried out as previously described.23 Morphologic and immunohistochemical patterns in the bone marrow biopsies were compared with smear analysis that we used as the “gold standard” procedure for assessing MLD.13 Parallel analysis of smears and bone marrow trephines was performed blindly. Smears were analyzed in the MLL by K.M. (validation by T.H.) whereas bone marrow biopsies were evaluated by 2 independent investigators, in Perugia (B.F.) and in Bologna (S.P.).
Cytogenetics and mutational analysis of the FLT3 gene
Karyotypes were analyzed after G-banding and described according to the International System for Human Cytogenetic nomenclature.25
Gene expression profiling
The sample preparation assay was performed as previously reported (Affymetrix HG-U133 Plus 2.0 microarrays).28-30 Gene expression raw data were processed according to the manufacturer's recommendations. After quality control, raw data were normalized with the use of the robust multiarray average normalization algorithm as implemented in the R-package affy Version 1.18.0.31 For supervised statistical analyses, samples were grouped accordingly, and for each disease entity differentially expressed genes were calculated by t statistics. To visualize the similarity of gene expression patterns, principal component analyses were applied. Transformed gene expression data were analyzed with the use of Partek Genomics Suite Version 6.4 (Partek Inc). Microarray data can be downloaded at Gene Expression Omnibus, under accession no. GSE18018.32
Statistical analysis
We compared distribution of variables such age, sex, white blood cell (WBC) count, CD34 expression, cytogenetics, and FLT3-ITD status in the 2 NPM1-mutated AML groups (with and without MLD). Mean differences were analyzed with the t test, and the chi-square test was applied in case of contingency tables. In case of 2 × 2 contingency tables, the Fisher exact test was applied.
For prognostic analysis, characteristics of patients were summarized by cross-tabulations (categorical variables), quantiles (median for ordinal factors) or by standard positional and variation parameters (mean and SD for continuous variables). Differences in the distributions of prognostic factors in subgroups were analyzed by the χ2 or Fisher exact test and by the Wilcoxon test. Survival was defined as the time from diagnosis to death or date of the last follow-up. Event-free survival (EFS) was defined as the time from diagnosis to date of failure (no CR, relapse, death) or date of the last follow-up. The probabilities of overall survival (OS) and EFS were estimated with the use of the Kaplan-Meier method. The log-rank test was used to compare risk factor categories in survival analysis, confidence intervals when estimated (95% confidence intervals) with the Simon and Lee method. Cox proportional hazard regression models were performed to examine and check for treatment results and the risk factors affecting time to event.
All tests were 2-sided, accepting P values less than or equal to .05 as indicating a statistically significant difference. Statistical analysis of patients from the MLL were performed with SPSS Version 14.0.1 software (SPSS Inc); data of patients with AML from the GIMEMA LAM99P and GIMEMA/EORTC AML12 studies were analyzed with SAS 9.1.3 software (SAS Institute Inc).
Results
Frequency and type of MLD in AML with mutated NPM1
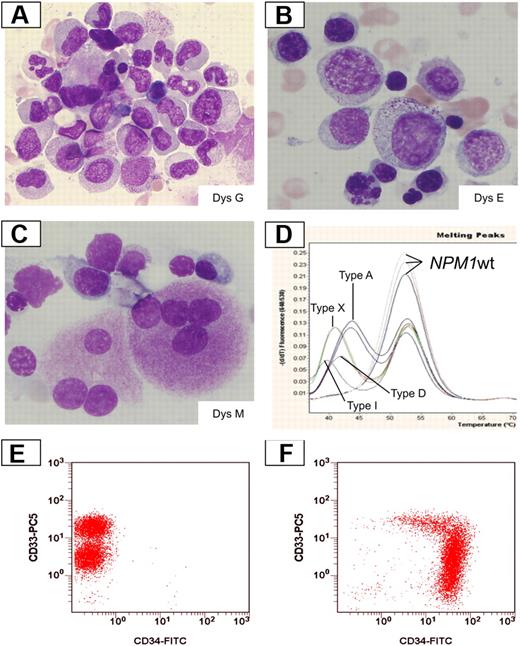
Frequency and type of morphologic MLD changes were evaluated in smears from 318 patients with AML with mutated/cytoplasmic NPM1, according to the 2008 WHO criteria.13 MLD was detected in 74 of 318 cases (23.3%). In particular, 57 of 318 cases showed dysplastic changes of 2 marrow lineages (erythroid and myeloid, n = 3; erythroid and megakaryocytic, n = 19; myeloid and megakaryocytic, n = 35). Dysplasia of 3 lineages was observed in 17 of 318 cases. Representative examples of NPM1-mutated AML with MLD are shown in Figure 1.
Dysplastic changes in smears from NPM1-mutated AML. (A) Dysgranulopoiesis (DysG) in a case of NPM1-mutated AML showing myeloid cells with hypogranulated cytoplasm and pseudo-Pelger cells. Bone marrow; Pappenheim staining. (B) Dyserythropoiesis (DysE) in a case of NPM1-mutated AML showing nuclear irregularity with fragmentation and multinucleation of red precursors. Bone marrow; Pappenheim staining. (C) Dysmegakaryopoiesis (DysM) in a case of NPM1-mutated AML showing 2 dysplastic megakaryocytes with multiple nuclei; Pappenheim staining. All images were collected using a Zeiss Axio Imager A1, 63×/1.4 oil objective Plan-Apochromat; 10×/23 eyepiece Sony camera 3CCD HD, and Model MC-HD 1/3 Horn imaging DHS solution. (D) LightCycler-based melting curve analyses showing different NPM1 mutation types in AML with MLD changes: type A (nt959insTCTG), type D (nt959insCCTG), type I (nt959insCTTG), type X (nt959insTTCC), and wild-type patients. (E-F) Expression of CD34 by multiparameter flow cytometry. A case with NPM1 mutation and MLD changes shows a lack of expression of CD34 (panel E, note the different levels of CD33 expression between myeloblasts and monoblasts). A different AML MLD+ case without NPM1 mutation shows a strong expression of CD34 with a part of the population lacking CD33 expression (F).
Dysplastic changes in smears from NPM1-mutated AML. (A) Dysgranulopoiesis (DysG) in a case of NPM1-mutated AML showing myeloid cells with hypogranulated cytoplasm and pseudo-Pelger cells. Bone marrow; Pappenheim staining. (B) Dyserythropoiesis (DysE) in a case of NPM1-mutated AML showing nuclear irregularity with fragmentation and multinucleation of red precursors. Bone marrow; Pappenheim staining. (C) Dysmegakaryopoiesis (DysM) in a case of NPM1-mutated AML showing 2 dysplastic megakaryocytes with multiple nuclei; Pappenheim staining. All images were collected using a Zeiss Axio Imager A1, 63×/1.4 oil objective Plan-Apochromat; 10×/23 eyepiece Sony camera 3CCD HD, and Model MC-HD 1/3 Horn imaging DHS solution. (D) LightCycler-based melting curve analyses showing different NPM1 mutation types in AML with MLD changes: type A (nt959insTCTG), type D (nt959insCCTG), type I (nt959insCTTG), type X (nt959insTTCC), and wild-type patients. (E-F) Expression of CD34 by multiparameter flow cytometry. A case with NPM1 mutation and MLD changes shows a lack of expression of CD34 (panel E, note the different levels of CD33 expression between myeloblasts and monoblasts). A different AML MLD+ case without NPM1 mutation shows a strong expression of CD34 with a part of the population lacking CD33 expression (F).
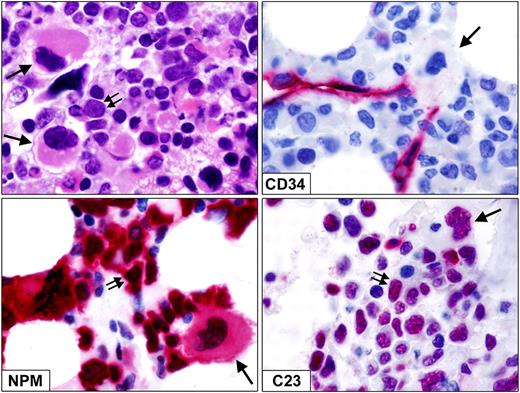
In the 110 patients from GIMEMA/EORTC, the presence of MLD in smears and the aberrant NPMc+ in hemopoietic precursors of different lineages in bone marrow biopsies were correlated. MLD was detected in smears from 20 of 110 cases (18.2%), whereas the aberrant NPMc+ staining pattern in hemopoietic precursors of different lineages was found in bone marrow biopsies from 23 of 110 patients (20.9%). Correlation between these 2 parameters was poor. In fact, only 10 of the 20 MLD+ cases in the smears showed the aberrant NPMc+ staining pattern in hemopoietic precursors of different lineages in tissue sections; conversely, 13 of 90 MLD− cases in the smears were NPMc+ in various lineages in the bone marrow biopsy. These findings clearly indicate that multilineage involvement, as detected in trephine biopsies by NPMc+ immunohistochemistry, cannot be used as surrogate for dysplastic changes in smears to predict MLD in the erythroid and myeloid cell lineages. In addition to smears, morphologic examination of bone marrow biopsy helped to identify dysplastic megakaryocytes (Figure 2), which is in keeping with the data from the literature. Like myeloid blasts, morphologically dysplastic megakaryocytes frequently showed aberrant cytoplasmic expression of NPM and nucleus-restricted expression of C23/nucleolin (Figure 2), clearly indicating that they belonged to the leukemic clone.
Dysplastic megakaryocytes in bone marrow biopsy from NPM1-mutated AML. (Top left) Infiltration by myeloid blasts (double arrows) admixed with dysplastic, monolobated megakaryocytes (single arrows; paraffin sections; hematoxylin-eosin. (Top right) Myeloid blasts as well as a monolobated megakaryocyte (single arrow) are CD34−. CD34+ small vessels serve as positive control. (Bottom left) Myeloid blasts (double arrows) and a dysplastic monolobated megakaryocyte (single arrow) show aberrant cytoplasmic expression of NPM. (Bottom right) Myeloid blasts (double arrows) and a dysplastic monolobated megakaryocyte (single arrow) show nucleus-restricted expression of nucleolin/C23. Immunostainings for CD34, NPM, and C23 were shown with the APAAP technique (hematoxylin counterstaining). All images were collected using an Olympus B61 microscope and a Plan Fl 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0.
Dysplastic megakaryocytes in bone marrow biopsy from NPM1-mutated AML. (Top left) Infiltration by myeloid blasts (double arrows) admixed with dysplastic, monolobated megakaryocytes (single arrows; paraffin sections; hematoxylin-eosin. (Top right) Myeloid blasts as well as a monolobated megakaryocyte (single arrow) are CD34−. CD34+ small vessels serve as positive control. (Bottom left) Myeloid blasts (double arrows) and a dysplastic monolobated megakaryocyte (single arrow) show aberrant cytoplasmic expression of NPM. (Bottom right) Myeloid blasts (double arrows) and a dysplastic monolobated megakaryocyte (single arrow) show nucleus-restricted expression of nucleolin/C23. Immunostainings for CD34, NPM, and C23 were shown with the APAAP technique (hematoxylin counterstaining). All images were collected using an Olympus B61 microscope and a Plan Fl 100×/1.3 NA oil objective; Camedia 4040, Dp_soft Version 3.2; and Adobe Photoshop 7.0.
The type of NPM1 mutation was correlated with the presence/absence of MLD in 208 patients. No differences in the type and frequency of MLD changes were observed in AML carrying NPM1 mutation A (n = 153), B (n = 22), D (n = 7), I (n = 7), or other rare mutation variants, that is, K, R, M, H, J, U, V, ZA, ZD, ZG, ZI ZJ, or ZK (n = 19).
In conclusion, analysis of smears from 318 patients with NPM1-mutated AML showed MLD in approximately 23% of cases and showed that dysplastic changes in smears did not correlate significantly with multilineage involvement in bone marrow biopsies, as defined by the NPMc+ staining pattern in hemopoietic precursors of different lineages.
Gene expression profiling in MLD+ versus MLD− AML and comparison to NPM1-mutation status
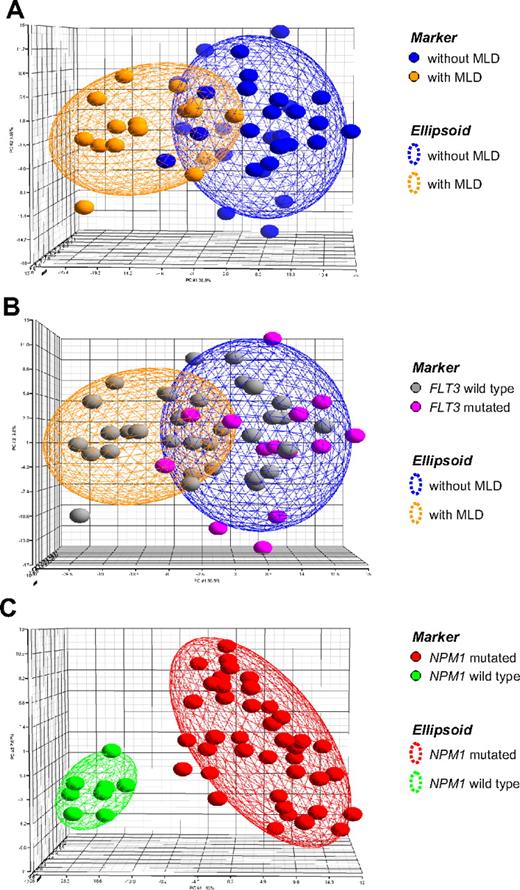
To further investigate the biologic significance of MLD, an analysis of 48 AML cases according to MLD status was performed with gene expression microarrays (Figure 3). First, a group of 16 MLD+ cases was compared against 32 MLD− cases. As given in the principal component analysis, the top 400 genes were not able to separate the corresponding groups (Figure 3A), but different expression patterns were mainly caused by the respective known FLT3-ITD status of these cases (Figure 3B). In contrast, when the respective cases were analyzed according to their NPM1 mutation status, 40 cases were NPM1 mutated, 8 cases were NPM1 wild-type, respectively; the resulting signature of the top 400 differentially expressed probe sets clearly separated these 2 AML subgroups (Figure 3C), indicating they are biologically distinct. As observed previously by similar analyses restricted to AML cases with a normal karyotype,3,4,8 the genes overexpressed in AML with mutated NPM1 included characteristic signature candidates such as HOX cluster members HOXA3, HOXA6, HOXA7, HOXB2, HOXB3, and HOXB6.
Principal component analysis. In this supervised analysis each patient (n = 48) is represented by a colored sphere. Ellipsoids are drawn with 2-fold standard deviations. (A) The gene expression signature is given for the top 400 probe sets differentially expressed between 32 AML cases without MLD and 16 AML cases displaying MLD. (B) The same signature is now annotated according to known FLT3 mutation status of the cases. (C) The gene expression signature is given for the top 400 probe sets differentially expressed between 40 NPM1-mutated and 8 NPM1 wild-type AML cases. All 8 cases with NPM1 wild-type AML displayed MLD. Detailed information on the probe sets is available online.
Principal component analysis. In this supervised analysis each patient (n = 48) is represented by a colored sphere. Ellipsoids are drawn with 2-fold standard deviations. (A) The gene expression signature is given for the top 400 probe sets differentially expressed between 32 AML cases without MLD and 16 AML cases displaying MLD. (B) The same signature is now annotated according to known FLT3 mutation status of the cases. (C) The gene expression signature is given for the top 400 probe sets differentially expressed between 40 NPM1-mutated and 8 NPM1 wild-type AML cases. All 8 cases with NPM1 wild-type AML displayed MLD. Detailed information on the probe sets is available online.
Thus, a strong underlying gene expression pattern separating biologic subgroups was observable according to mutation status of NPM1, but not according to MLD. The corresponding gene lists are provided in the supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
AML with mutated NPM1 with or without MLD show overlapping features
NPM1-mutated AML with and without MLD were then compared in terms of age, sex, WBC count, morphologic appearance according to French-American-British/WHO, expression of CD34, cytogenetics, FLT3-ITD status, and FLT3–tyrosine kinase domain (TKD) status.
In the full cohort of 318 patients with NPM1-mutated AML (74 with MLD; 244 without MLD) median age was 53 years (range, 19-85 years) in the group with MLD and 57.9 years (range, 16-87 years) in the group without MLD (P = .14). No significant difference in age distribution of the 2 groups emerged when they were stratified in decades (< 21 years, 21-30 years, 31-40 years, 41-50 years, 51-60 years, and > 60 years). Males were 41 of 74 (55.4%) in the group with MLD versus 102 of 244 (41.8%) in the group without MLD (P = .046). The median WBC count was 25.7 × 109/L (range, 0.9-196 × 109/L) in the group with MLD and 33.7 × 109/L (range, 0.5-600.0 × 109/L) in the group without MLD (P = .26). Morphology according to French-American-British/WHO was available in 316 of 318 patients, and no significant differences were observed in the 2 groups of NPM1-mutated AML with and without MLD.
Because down-regulation of CD34 is a distinguishing immunophenotypic feature of AML with cytoplasmic/mutated NPM1,1 we assessed this parameter in NPM1-mutated AML with and without MLD. Data on CD34 expression in cell suspensions or bone marrow sections were available for 228 of 318 cases of AML with cytoplasmic/mutated NPM1 (55 with MLD; 173 without MLD). Cases were regarded as negative if the percentage of CD34+ cells was 10% or less. Negativity for CD34 was found in 42 of 55 (76.4%) and 137 of 173 (79.2%) cases of the groups with and without MLD, respectively (P = .70). CD34 expression in representative cases is shown at flow cytometry (Figure 1) and in bone marrow tissue sections (Figure 2).
Cytogenetics was available in all 318 cases. Abnormal karyotype was present in 43 of 318 cases (13.5%): 12 of 74 (16.2%) cases of NPM1-mutated AML with MLD and 31 of 244 (12.7%) cases of NPM1-mutated AML without MLD (P = .44). Twelve cases had cytogenetic features (eg, del(9), −7, others) that define AML with MD-related changes (Tables 1 and 2). ITD analysis of the FLT3 gene was available in 312 of 318 NPM1-mutated AML cases (72 with MLD; 240 without MLD). FLT3-ITD was detected in 14 of 72 (19.4%) cases of the group with MLD and 92 of 240 (38.3%) cases of the group without MLD, the difference being statistically significant (P = .003). Analysis of FLT3-TKD mutation was available in 254 of 318 (63 with MLD; 191 without MLD). FLT3-TKD mutations were detected in 8 of 63 (12.7%) cases of the group with MLD and 16 of 191 (8.4%) cases of the group without MLD, respectively (P = .32).
Abnormal karyotypes in patients with NPM1-mutated AML from the Munich Leukemia Laboratory
| Patient . | Karyotype . | MRC . |
|---|---|---|
| MLD− | ||
| Aberrant 1 | 45,X,−Y[18]/46,XY[4] | − |
| Aberrant 2 | 45,X,−Y[20] | − |
| Aberrant 3 | 45,X,−Y[7]/44,X,−Y,dic(12;13)(p11;p11)[13] | + |
| Aberrant 4 | 45,X,−Y[20] | − |
| Aberrant 5 | 45,X,−Y[20] | − |
| Aberrant 6 | 46,XX,del(12)(p12)[8]/46,XX[15] | + |
| Aberrant 7 | 46,XX,del(9)(q22q34)[20] | + |
| Aberrant 8 | 46,XX,der(1)t(1;1)(p36;q12)[14]/46,XX[1] | − |
| Aberrant 9 | 47,XX,+21[4]/46,XX[16] | − |
| Aberrant 10 | 47,XX,+21[17]/46,XX[3] | − |
| Aberrant 11 | 47,XX,+21[3]/46,XX[17] | − |
| Aberrant 12 | 47,XX,+4[20] | − |
| Aberrant 13 | 47,XX,+8[6]/46,XX[4] | − |
| Aberrant 14 | 47,XX,+8[20]/46,XX[1] | − |
| Aberrant 15 | 47,XY,+4[4]/46,XY[16] | − |
| Aberrant 16 | 47,XY,+4[4]/46,XY[18] | − |
| Aberrant 17 | 47,XY,+8[14]/46,XY[6] | − |
| Aberrant 18 | 47,XY,+8[6]/46,XY[14] | − |
| Aberrant 19 | 55,XY,+X,+4,+5,+8,+10,+13,+14,+17,+18[4]/47, XY,+X[8] | + |
| Aberrant 20 | 90,XXXX,−3,+8,+8,−10,−11,−17[5]/46,XX[15] | + |
| MLD+ | ||
| Aberrant 21 | 45,X,−Y[18]/46,XY[2] | − |
| Aberrant 22 | 45,XY,−17,der(18)t(17;18)(q11;q23),del(20)(q12)[16]/ 46,XY[4] | − |
| Aberrant 23 | 46,X,−Y,+8[17]/47,X,−Y,+8,+8[3] | − |
| Aberrant 24 | 46,XX,del(9)(q22)[7]/46,XX[13] | + |
| Aberrant 25 | 46,XX,der(19)t(3;19)(q21;p13)[6]/47,XX,+8[2]/47,XX, der(19)t(3;19)(q21;p13),+8[2]/46,XX[1] | − |
| Aberrant 26 | 46,XY,del(20)(q11)[11]/46,XY[9] | − |
| Aberrant 27 | 46,XY,der(18)t(18;21)(p11;q11)[16]46,XY[4] | − |
| Aberrant 28 | 46,XY,t(3;10)(q12;q21)*[20] | − |
| Aberrant 29 | 47,XX,inv(2)(p12q21)*,+8[4]/46,XX,inv(2)(p12q21)*[16] | − |
| Aberrant 30 | 47,XY,+4[15]/46,XY[5] | − |
| Aberrant 31 | 47,XY,+8[14]/46,XY[6] | − |
| Patient . | Karyotype . | MRC . |
|---|---|---|
| MLD− | ||
| Aberrant 1 | 45,X,−Y[18]/46,XY[4] | − |
| Aberrant 2 | 45,X,−Y[20] | − |
| Aberrant 3 | 45,X,−Y[7]/44,X,−Y,dic(12;13)(p11;p11)[13] | + |
| Aberrant 4 | 45,X,−Y[20] | − |
| Aberrant 5 | 45,X,−Y[20] | − |
| Aberrant 6 | 46,XX,del(12)(p12)[8]/46,XX[15] | + |
| Aberrant 7 | 46,XX,del(9)(q22q34)[20] | + |
| Aberrant 8 | 46,XX,der(1)t(1;1)(p36;q12)[14]/46,XX[1] | − |
| Aberrant 9 | 47,XX,+21[4]/46,XX[16] | − |
| Aberrant 10 | 47,XX,+21[17]/46,XX[3] | − |
| Aberrant 11 | 47,XX,+21[3]/46,XX[17] | − |
| Aberrant 12 | 47,XX,+4[20] | − |
| Aberrant 13 | 47,XX,+8[6]/46,XX[4] | − |
| Aberrant 14 | 47,XX,+8[20]/46,XX[1] | − |
| Aberrant 15 | 47,XY,+4[4]/46,XY[16] | − |
| Aberrant 16 | 47,XY,+4[4]/46,XY[18] | − |
| Aberrant 17 | 47,XY,+8[14]/46,XY[6] | − |
| Aberrant 18 | 47,XY,+8[6]/46,XY[14] | − |
| Aberrant 19 | 55,XY,+X,+4,+5,+8,+10,+13,+14,+17,+18[4]/47, XY,+X[8] | + |
| Aberrant 20 | 90,XXXX,−3,+8,+8,−10,−11,−17[5]/46,XX[15] | + |
| MLD+ | ||
| Aberrant 21 | 45,X,−Y[18]/46,XY[2] | − |
| Aberrant 22 | 45,XY,−17,der(18)t(17;18)(q11;q23),del(20)(q12)[16]/ 46,XY[4] | − |
| Aberrant 23 | 46,X,−Y,+8[17]/47,X,−Y,+8,+8[3] | − |
| Aberrant 24 | 46,XX,del(9)(q22)[7]/46,XX[13] | + |
| Aberrant 25 | 46,XX,der(19)t(3;19)(q21;p13)[6]/47,XX,+8[2]/47,XX, der(19)t(3;19)(q21;p13),+8[2]/46,XX[1] | − |
| Aberrant 26 | 46,XY,del(20)(q11)[11]/46,XY[9] | − |
| Aberrant 27 | 46,XY,der(18)t(18;21)(p11;q11)[16]46,XY[4] | − |
| Aberrant 28 | 46,XY,t(3;10)(q12;q21)*[20] | − |
| Aberrant 29 | 47,XX,inv(2)(p12q21)*,+8[4]/46,XX,inv(2)(p12q21)*[16] | − |
| Aberrant 30 | 47,XY,+4[15]/46,XY[5] | − |
| Aberrant 31 | 47,XY,+8[14]/46,XY[6] | − |
MRC (myelodysplasia-related change) indicates which cases show cytogenetic abnormalities sufficient to diagnose acute myeloid leukemia (AML) with myelodysplasia-related features (according to World Health Organization [WHO] 200813 ); MLD+, cases with multilineage dysplasia (according to WHO 200813 ); and MLD−, cases without multilineage dysplasia (according to WHO 200813 ).
Possible constitutional chromosomal aberration.
Abnormal karyotypes in patients with NPM1-mutated from GIMEMA/EORTC
| . | Code Pt . | Karyotype . | MRC . | MLD . |
|---|---|---|---|---|
| 1 | 479A/09 | 46,XX,del(9)(q22)[3]/46,XX[17] | + | + |
| 2 | 382A/04 | 44–45,X,−Y,−22[cp7]/46,XY[12] | − | − |
| 3 | 04A31 | 43–45,XY,−7/46,XY,del(7)(q?)/46,XY | + | − |
| 4 | 19 158 | 46,XX,inv(3)(q21q26)*[2]/46,XX[19] | − | − |
| 5 | 548A/19 | 35–44,−X,−6,−8,−19[7]/46,XY[7] | + | − |
| 6 | 19909 | 79–163<3n>XXXYYY[14]/ 46,XY[6] | + | − |
| 7 | 248A/24 | 45,XY,−21[3]/46,XY[17] | − | − |
| 8 | 24922 | 47,XX,+8,add(18)(q23)[26]/46,XX[1] | − | − |
| 9 | 210A/28 | 47,XX,+8[10] | − | − |
| 10 | 742A/28 | 46,XX,t(10;17)(q26;q11),del(13)(q12q31)[5]/ 46,XX[10] | + | − |
| 11 | 276A/30 | 45,XY,del(11)(q13q23),−15[12]/46,XY[13] | + | − |
| 12 | 623A/85 | 45,X,−X[15] | − | − |
| . | Code Pt . | Karyotype . | MRC . | MLD . |
|---|---|---|---|---|
| 1 | 479A/09 | 46,XX,del(9)(q22)[3]/46,XX[17] | + | + |
| 2 | 382A/04 | 44–45,X,−Y,−22[cp7]/46,XY[12] | − | − |
| 3 | 04A31 | 43–45,XY,−7/46,XY,del(7)(q?)/46,XY | + | − |
| 4 | 19 158 | 46,XX,inv(3)(q21q26)*[2]/46,XX[19] | − | − |
| 5 | 548A/19 | 35–44,−X,−6,−8,−19[7]/46,XY[7] | + | − |
| 6 | 19909 | 79–163<3n>XXXYYY[14]/ 46,XY[6] | + | − |
| 7 | 248A/24 | 45,XY,−21[3]/46,XY[17] | − | − |
| 8 | 24922 | 47,XX,+8,add(18)(q23)[26]/46,XX[1] | − | − |
| 9 | 210A/28 | 47,XX,+8[10] | − | − |
| 10 | 742A/28 | 46,XX,t(10;17)(q26;q11),del(13)(q12q31)[5]/ 46,XX[10] | + | − |
| 11 | 276A/30 | 45,XY,del(11)(q13q23),−15[12]/46,XY[13] | + | − |
| 12 | 623A/85 | 45,X,−X[15] | − | − |
MRC (myelodysplasia-related change) indicates which cases show cytogenetic abnormalities sufficient to diagnose acute myeloid leukemia (AML) with myelodysplasia-related features (according to World Health Organization [WHO] 200813 ); and MLD, cases with multilineage dysplasia (according to WHO 200813 ).
Probable inv(3) (difficult interpretation because of too few suboptimal metaphases).
Thus, with the exception of a slight male predominance and a lower incidence of FLT3-ITD mutations in the MLD+ group, NPM1-mutated AML with and without MLD showed overlapping features.
NPM1-mutated AML with and without MLD show a similar outcome
The effect of MLD on prognosis was assessed in a total of 318 patients with AML with mutated NPM1. Survival was analyzed separately in the GIMEMA/EORTC and German series.
The 110 patients from GIMEMA LAM99P and GIMEMA/EORTC AML12 received the same treatment (although enrolled in 2 consecutive studies). The CR rate difference between NPM1-mutated AML with or without MLD was not statistically significant (83.3% vs 80.6%, respectively; P = .999). No differences were observed for both OS and EFS in the whole population according to the presence or absence of MLD (20 and 90 cases, respectively; Figure 4A-B). Similar results were obtained when the analysis was restricted only to NPM1-mutated AML cases with normal karyotype (19 with MLD; 79 without MLD; Figure 4C-D).
Survival curves of patients with NPM1-mutated AML (MLD+ vs MLD−) from GIMEMA LAM99P and GIMEMA/EORTC AML12 trials. (A) No significant differences in OS are observed between NPM1-mutated AML with (red line) and without (blue line) MLD (P = .608). (B) No significant differences in EFS are observed between NPM1-mutated AML with (red line) and without (blue line) MLD (P = .367). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated AML with (red line) and without (blue line) MLD (P = .862). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated AML with and without MLD (P = .541).
Survival curves of patients with NPM1-mutated AML (MLD+ vs MLD−) from GIMEMA LAM99P and GIMEMA/EORTC AML12 trials. (A) No significant differences in OS are observed between NPM1-mutated AML with (red line) and without (blue line) MLD (P = .608). (B) No significant differences in EFS are observed between NPM1-mutated AML with (red line) and without (blue line) MLD (P = .367). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated AML with (red line) and without (blue line) MLD (P = .862). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated AML with and without MLD (P = .541).
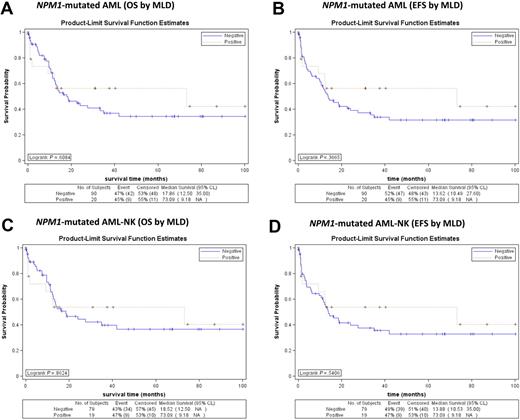
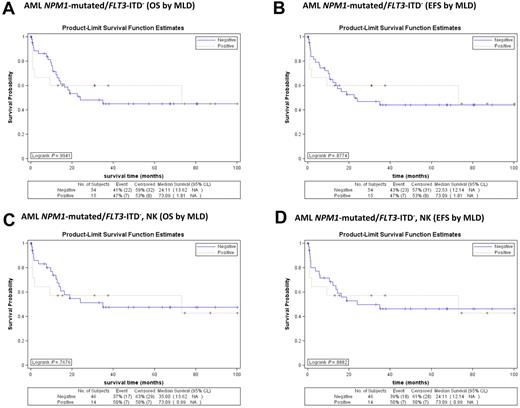
In addition, considering separately the NPM1-mutated/FLT3-ITD− cases, either as whole population (Figure 5A-B) or as normal karyotype (Figure 5C-D), again, the outcome in terms of OS and EFS was not influenced by MLD. Similar results were obtained when FLT3-ITD+ cases were analyzed separately (not shown).
Survival curves of patients with NPM1-mutated/FLT3-ITD− AML (MLD+ vs MLD−) from GIMEMA LAM99P and GIMEMA/EORTC AML12 trials. (A) No significant differences in OS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .994). (B) No significant differences in EFS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .877). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .768). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .888).
Survival curves of patients with NPM1-mutated/FLT3-ITD− AML (MLD+ vs MLD−) from GIMEMA LAM99P and GIMEMA/EORTC AML12 trials. (A) No significant differences in OS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .994). (B) No significant differences in EFS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .877). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .768). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (blue line) MLD (P = .888).
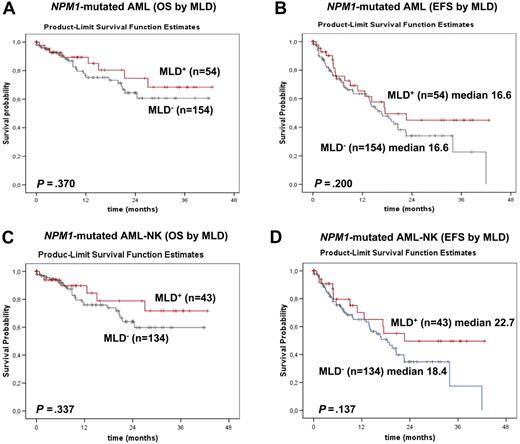
Overlapping results emerged from the analysis of the 208 NPM1-mutated patients from the MLL. OS and EFS did not significantly differ between NPM1-mutated AML with MLD (n = 54) and NPM1-mutated AML without MLD (n = 154; alive at 2 years: 74.6% vs 64.4%; P = .370; median EFS: 17.5 months vs 16.6 months; P = .200; Figure 6A-B). The same was true when this analysis was restricted to cases with normal karyotype (Figure 6C-D).
Survival curves of patients with NPM1-mutated AML (MLD+ vs MLD−) from the MLL. (A) No significant differences in OS are observed between NPM1-mutated AML with (red line) and without (gray line) MLD (P = .370). (B) No significant differences in EFS are observed between NPM1-mutated AML with (red line) and without (gray line) MLD (P = .200). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated AML with (red line) and without (gray line) MLD (P = .337). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated AML with (red line) and without (gray line) MLD (P = .137).
Survival curves of patients with NPM1-mutated AML (MLD+ vs MLD−) from the MLL. (A) No significant differences in OS are observed between NPM1-mutated AML with (red line) and without (gray line) MLD (P = .370). (B) No significant differences in EFS are observed between NPM1-mutated AML with (red line) and without (gray line) MLD (P = .200). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated AML with (red line) and without (gray line) MLD (P = .337). (D) No significant differences in EFS are observed between normal karyotype NPM1-mutated AML with (red line) and without (gray line) MLD (P = .137).
OS was significantly shorter in NPM1-mutated cases with additional FLT3-ITD (n = 71) than in NPM1-mutated cases without FLT3-ITD (n = 137; P = .044; median OS not reached vs 21.5 months; supplemental Figure 1A). There was a tendency for EFS to be shorter in the NPM1-mutated/FLT3-ITD+ subgroup than in the NPM1-mutated/FLT3-ITD− group (14.2 vs 19.6 months), although this was not statistically significant (P = .226; supplemental Figure 1B).
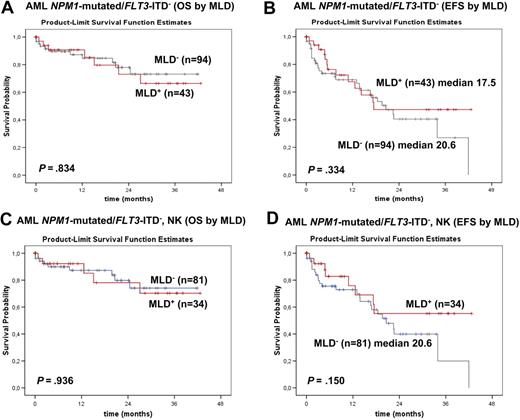
Considering only FLT3-ITD− cases, no statistically significant difference emerged in OS and EFS of NPM1-mutated AML with and without MLD (alive at 2 years: 73.0% vs 78.0%; P = .834; median EFS: 17.5 vs 20.6 months; P = .334; Figure 7A-B). The same was true when this analysis was restricted to FLT3-ITD− cases with normal karyotype (Figure 7C-D). Also in the subgroups of NPM1-mutated/FLT3-ITD+ cases, OS and EFS was not influenced by MLD (median OS, not reached with MLD+ vs 20.8 months with MLD−; alive at 2 years: 85.7% with MLD+ vs 43.3% with MLD−; P = .195; supplemental Figure 1C); and median EFS was 22.7 months for MLD+ versus 14.0 months for MLD− (P = .547; supplemental Figure 1D).
Survival curves of patients with NPM1-mutated/FLT3-ITD− AML (MLD+ vs MLD−) from the MLL. (A) No significant differences in OS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .834). (B) No significant differences in EFS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .334). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .936). (D) No significant differences in EFS are observed between normal karyotypeNPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .150).
Survival curves of patients with NPM1-mutated/FLT3-ITD− AML (MLD+ vs MLD−) from the MLL. (A) No significant differences in OS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .834). (B) No significant differences in EFS are observed between NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .334). (C) No significant differences in OS are observed between normal karyotype NPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .936). (D) No significant differences in EFS are observed between normal karyotypeNPM1-mutated/FLT3-ITD− AML with (red line) and without (gray line) MLD (P = .150).
As a comparison group, we used 55 patients who were randomly selected to include only de novo AML cases (excluding therapy-related AML) with unmutated NPM1 and normal karyotype; all were diagnosed at the MLL. MLD was identified in 20 of 55 (36.4%) cases: 35 MLD− patients were classified as AML–not otherwise specified and 20 MLD+ patients were classified as AML with MD-related changes, according to the 2008 WHO classification. Neither OS (median not reached vs 25.5 months; P = .230) nor EFS (median of 16.9 vs 11.9 months; P = .679) differed significantly between MLD+ and MLD− patients within this subgroup (supplemental Figure 2A-B).
Multivariate analysis for OS and EFS carried out in all 318 patients with NPM1-mutated AML (Tables 3–4) indicated that MLD had no significant effect on OS and EFS but showed a significant effect of FLT3 status (mutated vs unmutated) on both OS (P = .016) and EFS (P = .016).
Multivariate data regression (analysis of maximum likelihood estimates) of OS (n = 318 NPM1-mutated AML cases)
| Variable . | Pr > χ2 test . | Hazard ratio (95% confidence limits) . |
|---|---|---|
| Age as continuous variable | 0.5615 | 1.005 (0.987-1.024) |
| FLT3: mutated vs unmutated | 0.0156 | 1.767 (1.114-2.802) |
| Karyotype: aberrant vs normal | 0.5535 | 1.217 (0.636-2.326) |
| MLD: positive vs negative | 0.4242 | 0.796 (0.455-1.393) |
| Variable . | Pr > χ2 test . | Hazard ratio (95% confidence limits) . |
|---|---|---|
| Age as continuous variable | 0.5615 | 1.005 (0.987-1.024) |
| FLT3: mutated vs unmutated | 0.0156 | 1.767 (1.114-2.802) |
| Karyotype: aberrant vs normal | 0.5535 | 1.217 (0.636-2.326) |
| MLD: positive vs negative | 0.4242 | 0.796 (0.455-1.393) |
OS indicates overall survival; AML, acute myeloid leukemia; Pr, probability; and MLD, multilineage dysplasia.
Multivariate data regression (analysis of maximum likelihood estimates) of EFS (n = 318 NPM1-mutated AML cases)
| Variable . | Pr > χ2 test . | Hazard ratio (95% confidence limits) . |
|---|---|---|
| Age as continuous variable | 0.1697 | 1.010 (0.996-1.025) |
| FLT3: Mutated vs unmutated | 0.0161 | 1.597 (1.091-2.337) |
| Karyotype: aberrant vs normal | 0.1335 | 1.482 (0.886-2.479) |
| MLD: positive vs negative | 0.4084 | 0.827 (0.526-1.298) |
| Variable . | Pr > χ2 test . | Hazard ratio (95% confidence limits) . |
|---|---|---|
| Age as continuous variable | 0.1697 | 1.010 (0.996-1.025) |
| FLT3: Mutated vs unmutated | 0.0161 | 1.597 (1.091-2.337) |
| Karyotype: aberrant vs normal | 0.1335 | 1.482 (0.886-2.479) |
| MLD: positive vs negative | 0.4084 | 0.827 (0.526-1.298) |
EFS indicates event-free survival; AML, acute myeloid leukemia; Pr, probability; and MLD, multilineage dysplasia.
Discussion
This study provides evidence that NPM1-mutated AML with and without MLD changes have an overlapping gene expression profile and show similar immunophenotypic and prognostic features. These findings, taken together with previous observations that the presence of additional chromosomal abnormalities have no effect on the biologic and clinical characteristics features of NPM1-mutated AML,15 support the view that this leukemia, irrespective of concomitant MD-related changes (MLD or chromosomal abnormalities), represents a single distinct entity whose molecularly defining feature is the presence of a mutated NPM1 gene. Thus, for classification purposes, the detection of an NPM1 mutation should take diagnostic precedence over the presence of MD-related changes, as is now accepted for cytogenetic alterations listed in the group of AML with recurrent genetic abnormalities.13 Our results are not only diagnostically relevant because they allow one to better define the border between NPM1-mutated AML and AML with MD-related changes, but they also have important prognostic implications.
This study and previous findings from our group1,15 strongly suggest that MD-related changes as defined by the 2008 WHO classification,13 that is, previous history of MDS or MDS/MPN, presence of MDS-related cytogenetic abnormalities, and MLD, play a scarce relevance in defining biologically and clinically NPM1-mutated AML. Notably, AML with mutated NPM1 is a de novo leukemia.1,3 In our experience, none of the AML occurring in patients with well-documented clinical history of MDS showed cytoplasmic-mutated NPM.1 Moreover, we found that only 1 of 46 cases of AML secondary to chronic MPN harbored a mutated NPM1 gene.14 More in-depth studies of this single case, however, suggested that it might have represented a de novo AML arising from a normal residual hemopoietic stem cell rather than a clonally related evolution of the original MPN.14
We previously found that approximately 15% of patients with NPM1-mutated AML harbor chromosomal aberrations.1,15 These aberrations appear to be secondary genetic events that do not appear to significantly effect distinctive biologic, clinicopathologic, and prognostic features of AML with mutated NPM1.15 The most frequent chromosomal aberrations in NPM1-mutated AML were +8, +4, −Y, del(9q), and +21.15 However, with the exception of del(9q) that, in the 2008 WHO classification, is accepted as an MDS-related cytogenetic abnormality,13 trisomy 8 or the loss of chromosome Y should not be considered, by themselves, as sufficient evidence for assigning a case to AML with MD-related changes. Other typical MD-related cytogenetic abnormalities are rarely detected in NPM1-mutated AML. Recently, we found −7 or a complex karyotype (≥ 3 clonal chromosome aberrations according to the Southwest Oncology Group) in only 3 of 631 and 4 of 631 NPM1-mutated AML cases, respectively.15 Interestingly, the complex karyotypes did not show the typical pattern of chromosomal gains and losses, such as loss of 5q, 7q 12p, 16q, and 17p and gain of 8q, 11q, and 21q, usually observed in MDS.15
The results of this study further expand the analysis of correlation between NPM1-mutated AML and AML with MD-related changes. In particular, they provide evidence that MLD has less effect than the genetic lesion (NPM1 mutation) in defining the leukemia entity. In fact, NPM1-mutated AML with and without MLD exhibited similar immunophenotype and gene expression profile: down-regulation of the CD34 and CD133 genes and overexpression of HOXA and HOXB cluster genes, respectively.4,8,33 These findings differ from AML with MD-related changes without NPM1 mutation. Thus, when present, MLD should be regarded the equivalent of a morphologic variant of AML with mutated NPM1. The reason why that in NPM1-mutated AML multilineage dysplasia appears to associate with a lower frequency of FLT3-ITD remains to be explored.
The de novo origin of NPM1-mutated AML and our findings pointing to MD-related changes in NPM1-mutated AML as secondary events add to the growing body of evidence that NPM1 mutation is a driver genetic lesion in AML, as supported by the following observations: (1) cytoplasmic mutated NPM is specific for AML1,34,35 ; (2) NPM1 mutations are mutually exclusive of recurrent cytogenetic abnormalities in AML36 ; (3) NPM1-mutated AML exhibits a distinctive gene expression signature4,8,33 and microRNA profile5,37 ; (4) all NPM1 mutations result in common changes at the C-terminal portion of NPM that maximize the export of this protein from the nucleus to cytoplasm,1,38-40 pointing to aberrant cytoplasmic expression of NPM1 as a key event for leukemogenesis39,41 ; (5) NPM1 mutation is one of the few recurrent genetic lesions so far identified at whole genome sequencing of AML with normal karyotype42,43 ; and (6) with few exceptions,44 which could represent clonally unrelated secondary leukemias,45 NPM1 mutations are stable during the course of the disease.46-49
Based on our findings, we propose that AML cases with an NPM1 mutation and MLD should be classified as AML with mutated NPM1 rather than AML with MD-related changes. Indeed, there are several clinical and immunophenotypic differences that call for a separation of the 2 entities. AML with MD-related dysplasia usually presents with pancytopenia13 that is quite uncommon in NPM1-mutated AML.3 Moreover, in cases with aberrations of chromosomes 5 and 7, a high incidence of CD34 and CD7 expression has been reported.50 In contrast, most NPM1-mutated AML, including rare cases carrying −7 or a complex karyotype, are CD34−.15
When using immunohistochemistry as first screening for NPM1-mutated AML, care should be taken in avoiding confusion with AML carrying the t(3;5) translocation because this rare leukemia subtype may show MD-related changes and aberrant cytoplasmic expression of NPM51 (because of the presence of the NPM-MLF1 fusion protein). Further molecular and cytogenetic/fluorescent in situ hybridization analyses clarify the issue showing an NPM1 mutation, or the typical t(3;5) or the NPM/MLF1 fusion gene/transcript.51
Clinically, we showed no significant effect of MLD in response to induction therapy of patients with NPM1-mutated AML (∼ 80%). The most clinically relevant finding in this study was, however, that MLD had no effect in the prognosis of NPM1-mutated AML. In particular, patients with NPM1-mutated FLT3-ITD− AML with or with MLD showed the same relatively favorable prognosis.
In conclusion, this study and our previous findings15 strongly support the view that NPM1-mutated AML represents a distinct entity, irrespective of MD-related changes. We suggest that the current criteria of 2008 WHO classification should be reconsidered accordingly and that AML showing concomitant NPM1 mutation and MD-related changes of any type be classified as AML with mutated NPM1 rather than AML with MD-related changes. On the basis of our results, the term AML with MD-related changes should be restricted to cases with MD-related changes without accompanying recurrent genetic abnormalities, including NPM1 mutation. AML with MD-related changes was recently claimed to differ prognostically from AML–not otherwise specified,52 although morphologic dysplasia alone was not found to significantly affect prognosis of AML in general in other large studies.20,53 The above concepts have important implications in the classification and treatment of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mrs Claudia Tibidò for secretarial assistance and Dr Geraldine Boyd for assistance in editing the manuscript. We are indebted to Prof Martin Dugas, University of Münster, for support in microarray analyses; and to Profs Giuseppe Saglio, Francesco Lo Coco, Daniela Diverio, and Fabrizio Pane for performing molecular analyses of the FLT3 gene.
The authors from the Munich Leukemia Laboratory (MLL) thank all physicians and especially participants of the AMLCG group for sending bone marrow or blood samples to our laboratory for reference diagnosis and for submitting clinical data. For those patients (n = 94) that were referred to our laboratory from centers of the AMLCG group, we listed the centers and investigators in order of the number of cases provided in detail: Universitätsklinikum Köln (M. Hallek); Vivantes Klinikum Neukölln, Berlin (A. Grüneisen); Universitätsklinikum Mannheim (E. Lengfelder); Krankenhaus Düren (M. Flaßhove); St Antonius Krankenhaus, Eschweiler (P. Staib, F. Schlegel); St Johannes Krankenhaus, Dortmund (H. Pielken); Städtisches Klinikum, Gütersloh (C. Gropp); Katholisches Krankenhaus, Hagen (H.-W. Lindemann); Evangelisches Krankenhaus, Hamm (L. Balleisen); Paracelsus Klinik, Osnabrück (O. Koch); Universitätsklnikum Regensburg (A. Reichle); Evangelisches Waldkrankenhaus Spandau, Berlin (U. Wahnschaffe); Helios Klinikum, Berlin (W.-D. Ludwig); Vivantes Klinikum Spandau, Berlin (E. Späth-Schwalbe); St Franziskus Krankenhaus, Bielefeld (H.-J. Weh); Knappschaftskrankenhaus, Bottrop (G. Trenn); Klinikum Bremen-Mitte, Bremen (B. Hertenstein); Kreisklinik Fürstenfeldbruck (R. Eissele); Klinikum Krefeld (Th. Frieling); Maria-Hilf-Krankenhaus, Mönchengladbach (D. Graeven); Städtisches Krankenhaus München-Neuperlach, München (D. Fleckenstein); Vinzenz Palotti Hospital, Bergisch-Gladbach (S. Korsten); Vivantes Klinikum am Urban, Berlin (J. Beyer); Städtisches Krankenhaus Martha-Maria, Halle/Saale (U. Haak); Klinik für Knochenmarktransplantation und Hämatologie/Onkologie, Idar-Oberstein (A. Fauser); Städtische Kliniken, Kassel (M. Wolf); Bundeswehrzentralkrankenhaus, Koblenz (G. Herzog); Klinikum Landshut (B. Kempf), St Walburga Krankenhaus, Meschede (M. Schwonzen); Klinikum Passau (Th. Südhoff); Krankenhaus Barmherzige Brüder, Regensburg (E.-D. Kreuser); Praxis Dr Pihusch, Rosenheim, St Marien Krankenhaus, Siegen (W. Gassmann).
The authors from the GIMEMA group thank all centers and investigators contributing to the GIMEMA LAM99P and GIMEMA/EORTC AML12 studies. The centers and investigators are listed in order of the number of cases provided: Istituto di Ematologia, Universita La Sapienza, Roma (G. Meloni); Divisione di Ematologia, Ospedale V. Cervello, Palermo (F. Fabbiano); Divisione di Ematologia, Azienda USL di Pescara, Pescara (M. Sborgia); Cattedra di Ematologia, Bari, (V. Liso); Az. Osp. S. G. Moscati, Avellino (N. Cantore); Dipartimento di Emato-Oncologia, Azienda Ospedaliera Bianchi-Melacrino-Morelli Reggio Calabria (F. Nobile); Ospedale Ferrarotto-S. Bambino, Catania (F. Di Raimondo); Divisione di Ematologia e Oncologia Clinica, Catanzaro (D. Magro); Istituto di Ematologia, Università di Ancona, Ancona (P. Leoni); Istituto di Ematologia, Policlinico Monteluce, Perugia (A. Tabilio); Cattedra di Ematologia, Ospedale S. Chiara, Pisa (M. Petrini); Sezione di Medicina Interna, Oncologia ed Ematologia, Dipartimento Scienze Mediche, Oncologiche e Radiologiche, Modena (G. Torelli); Azienda Sanitaria Locale Salerno 1, Medicina Interna, Ematologia–Oncologia, Nocera Inferiore (A. M. D'Arco); Istituto di Ematologia, Universita Federico II, Napoli (B. Rotoli); Divisione di Ematologia, Ospedale S. Carlo, Potenza (F. Ricciuti); Istituto di Ematologia, Policlinico Gemelli, Roma (S. Sica); Divisione di Ematologia, Ospedale Casa Sollievo della Sofferenza, S. Giovanni Rotondo (L. Melillo); Istituto di Ematologia, Ospedale A. Businco, Cagliari (E. Angelucci); Divisione di Ematologia, Ospedale S. Giovanni Bosco, Napoli (E. Miraglia); Divisione di Ematologia, Fondazione Centro S. Raffaele del Monte Tabor, Milano (L. Camba); Divisione di Ematologia, Azienda Ospedaliera Cremona, Cremona (S. Moranti); Divisione di Ematologia, Ospedale S. Eugenio, Roma (A. Venditti); Divisione Medica, Ospedale Maggiore, Lodi (G. Nalli); Divisione di Ematologia, Ospedale S. Francesco, Nuoro (A. Gabbas); Cattedra di Ematologia-CTMO, Università di Parma, Parma (V. Rizzoli); Divisione di Ematologia, Universita di Sassari (F. Dore); Divisione di Ematologia, Ospedale SS. Antonio e Biagio, Alessandria (A. Levis); Azienda Ospedaliera A. Di Summa, Brindisi (G. Quarta); Divisione di Ematologia, Ospedale S. Croce, Cuneo (A. Gallamini); Sezione Ematologia, Dipartimento Scienze Biomediche, Arcispedale S. Anna, Ferrara (P-L Castaldi); Divisione di Ematologia, Ospedale di Messina (M. Brugiatelli).
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC).
Authorship
Contribution: B.F. had the original idea for the study and wrote the paper; K.M., U.B., and M.M. analyzed the smears and contributed to writing the paper; A.V. is responsible for the centralization and analysis of smears in the GIMEMA trial and contributed to writing the paper; S.S. and C.H. performed, respectively, the cytogenetic and molecular studies on patients from the Munich Leukemia Laboratory and contributed to writing the paper; T.W. and W.K. carried out the prognostic analysis of patients from the Munich Leukemia Laboratory and contributed to writing the paper; A.K. investigated the gene expression profile of cases from the Munich Leukemia Laboratory and contributed to writing the paper; H.-U.K. carried out statistical analyses on gene expression data; M.P.M. and S.P. characterized by immunohistochemistry/WB the samples from the GIMEMA/EORTC patients; A.S. performed the statistical analyses on patients enrolled in the GIMEMA/EORTC trials; M.V., A.P., and P.F. carried out the prognostic analysis of the GIMEMA/EORTC patients; F.M. was the coordinator of the GIMEMA-EORTC clinical trial; T.H. validated cytomorphology of the Munich and the GIMEMA cases and contributed to the design of the study and to writing the paper.
Conflict-of-interest disclosure: B.F. applied for a patent on clinical use of NPM1 mutants. T.H., C.H., S.S., and W.K. own the Munich Leukemia Laboratory GmbH. K.M., T.W., and A.K. are employed by the Munich Leukemia Laboratory. The remaining authors declare no competing financial interests.
Correspondence: Brunangelo Falini, Institute of Hematology, University of Perugia, Ospedale Santa Maria della Misericordia, Perugia, Italy; e-mail: faliniem@unipg.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal