GATA-1 and PU.1 are essential hematopoietic transcription factors that control erythromegakaryocytic and myelolymphoid differentiation, respectively. These proteins antagonize each other through direct physical interaction to repress alternate lineage programs. We used immortalized Gata1− erythromegakaryocytic progenitor cells to study how PU.1/Sfpi1 expression is regulated by GATA-1 and GATA-2, a related factor that is normally expressed at earlier stages of hematopoiesis. Both GATA factors bind the PU.1/Sfpi1 gene at 2 highly conserved regions. In the absence of GATA-1, GATA-2 binding is associated with an undifferentiated state, intermediate level PU.1/Sfpi1 expression, and low-level expression of its downstream myeloid target genes. Restoration of GATA-1 function induces erythromegakaryocytic differentiation. Concomitantly, GATA-1 replaces GATA-2 at the PU.1/Sfpi1 locus and PU.1/Sfpi1 expression is extinguished. In contrast, when GATA-1 is not present, shRNA knockdown of GATA-2 increases PU.1/Sfpi1 expression by 3-fold and reprograms the cells to become macrophages. Our findings indicate that GATA factors act sequentially to regulate lineage determination during hematopoiesis, in part by exerting variable repressive effects at the PU.1/Sfpi1 locus.
Introduction
Hematopoiesis is largely regulated by transcription factors that drive differentiation by activating cell type–specific programs of gene expression, and concurrently, by repressing alternate lineage programs (reviewed in Laiosa et al1 and Cantor and Orkin2 ).3,,–6 It is believed that hematopoietic progenitors maintain their plasticity by coexpressing relatively low levels of multiple key lineage-determining transcription factors.4,7,,,–11 In some instances, these factors positively regulate their own expression and, simultaneously, cross-antagonize the expression and/or function of other nuclear proteins that promote development of different lineages. This creates reinforcing regulatory loops in which one or several transcriptional programs can eventually predominate to guide differentiation down a specific lineage pathway.
The nuclear protein GATA-1 serves as a prototype for the actions of lineage-restricted transcription factors in hematopoietic development (reviewed in Crispino12 and Ferreira et al13 ). GATA-1 is the founding member of a small family of zinc finger transcription factors that recognize the DNA motif (T/A(GATA)A/G). GATA-1 promotes erythroid, megakaryocytic, and mast and eosinophil development. In humans, germline GATA1 gene mutations cause congenital anemia and thrombocytopenia, whereas somatic mutations collaborate with constitutional trisomy 21 (Down syndrome) to cause acute megakaryoblastic leukemia. GATA-1 promotes hematopoiesis by activating transcription of genes that define the phenotypes and functions of mature blood cells. In addition, GATA-1 facilitates cellular differentiation by repressing gene expression.14 For example, GATA-1 inhibits the transcription of Kit and Gata2, genes that are expressed predominantly at relatively early stages of hematopoiesis.15,–17 The Kit and Gata2 loci contain GATA motifs that are bound by GATA-2 in immature progenitors, where these genes are transcriptionally active. During the later stages of erythroid (and possibly megakaryocytic) development, GATA-1 expression predominates and replaces GATA-2 at the same motifs to inhibit transcription. Both GATA-1 and GATA-2 participate in numerous functionally important protein interactions. For example, binding to the cofactor Friend of GATA-1 (FOG-1) is essential for most aspects of GATA-1 and GATA-2 function during erythromegakaryocytic development.18,,,–22 In turn, FOG recruits additional proteins, including TACC3,18 CtBP2,23 and the NuRD complex,24,25 to influence target gene activity.
GATA-1 also binds the master myeloid transcription factor, PU.1, an Ets family member that is essential for monocytic, granulocytic, and lymphoid lineages (reviewed in Koschmieder et al26 ). The physical interaction between GATA-1 and PU.1 is mutually antagonistic through several possible mechanisms.27,,,,,–33 For example, it is proposed that GATA-1 inhibits PU.1 by preventing its interaction with the essential coactivator c-Jun,29 whereas PU.1 blocks GATA-1 function by inhibiting its ability to bind to DNA30 and/or by recruiting a complex that contains the retinoblastoma protein and histone modifying enzymes.31,32 In multipotential progenitors, GATA-1 and PU.1 are expressed together at low levels and their relative stoichiometry influences subsequent hematopoietic development and differentiation in avian,34 mammalian,35,–37 and zebrafish38,39 cells (reviewed in Laiosa et al1 ). Insertional activation of PU.1 by the spleen focus-forming virus arrests erythroid development and promotes erythroleukemia, in part by antagonizing GATA-1 activity.28,40 Thus, normal and pathologic hematopoiesis are influenced by cross-antagonism between GATA-1 and PU.1.
Studies of GATA-1, GATA-2, and PU.1 illustrate how cooperative and antagonistic relationships between transcription factors, as well as their timing of expression, exert critical roles in hematopoietic development. However, the regulatory networks through which these important nuclear proteins function are not entirely defined. We studied interactions between GATA-1, GATA-2, and PU.1 in genetically modified bipotential erythromegakaryocytic progenitor cells. Our findings illustrate 2 new facets of developmental hematopoiesis. First, GATA-1 inhibits PU.1 not only by physical protein interaction, but also through direct transcriptional repression. Second, in addition to having opposing effects on transcription of the same target gene as described previously, GATA-2 and GATA-1 act cooperatively and successively to exert repressive effects of differing magnitudes that gradually restrict gene expression during hematopoietic development.
Methods
Cell culture
GATA-1− megakaryocyte-erythroid (G1ME) cells were cultured and genetically manipulated as described.41 G1ME cells were maintained in α-MEM supplemented with 20% fetal bovine serum, 1% penicillin/streptomycin, 1% glutamine, and 20 ng/mL thrombopoietin (TPO). Cytokines used in various experiments include erythropoietin (EPO; 2 U/mL), interleukin-3 (IL-3; 20 ng/mL), kit ligand (stem cell factor; 50 ng/mL), macrophage colony-stimulating factor (MCSF; 5 ng/mL), and granulocyte-macrophage colony-stimulating factor (GMCSF; 5 ng/mL; R&D Systems). For some experiments, we used TPO-conditioned media (CM) prepared from cells engineered to express murine TPO.42 The concentration of TPO CM was optimized for G1ME cell culture using Celltiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). Based on comparison with recombinant TPO, we used approximately 20 ng/mL TPO CM.
Retroviral transduction
The MSCV-based retroviral vector MIGR1-GFP was used to express wild-type murine GATA-1 in G1ME cells.41 For GATA-2 knockdown in G1ME cells and Gata-1− embryonic stem (ES) cell–derived embryoid body (EB) cells, cells were infected with Banshee retroviruses harboring GFP alone or a modified version that contained GFP and shRNA against GATA2. The following sequence was cloned into the Banshee plasmid to create a shRNA against GATA2: 5′-CGCCGCCATTACTGTGAATATTTAGTGAAGCCACAGATGTAAATATTCACAGTAATGGCGGCA-3′. Retroviral particles were generated via transient transfection of the Plat-E retrovirus packaging cell line.43 For retroviral transduction, 4 × 106 G1ME cells or ES cell–derived EB cells were placed in 1 well of a 6-well plate containing 5 mL retroviral supernatant with 8 mg/mL polybrene and 10 mM HEPES. TPO CM was added to G1ME cell transductions and for GATA-1–null ES cell–derived EB cell transduction, 100 ng/mL murine SCF, 20 ng/mL murine TPO, 5 ng/mL human VEGF, 50 ng/mL murine FLT-3 ligand, 1 ng/mL murine IL-3, 10 ng/mL murine IL-6, and 5 ng/mL murine IL-11 were added. The cells were spinoculated at 2400g for 90 minutes at 20°C, incubated at 37°C in 5% CO2, and then 4 hours later the infection was repeated.
Western blot
Nuclear extracts were prepared from untransduced and transduced G1ME and murine erythroleukemia (MEL) cells according to standard methods, fractionated on sodium dodecyl sulfate–2% polyacrylamide gels, and transferred to nitrocellulose membranes by electroblotting. Antibodies for immunoblot included rat anti–GATA-1 (N6), rabbit anti-PU.1 (T-21), rabbit anti–GATA-2 (H-116), and rabbit anti-HDAC2 (H-54; Santa Cruz Biotechnology).
Flow cytometry
Cells were stained in PBS with 1% FCS at 4°C for 20 minutes with the following antibodies: anti–Ter119-APC, anti–CD41-PE, anti–CD45R-PE (B220), anti–CD11b-PE (Mac1), anti–CD11b-APC (Mac1), anti–CD11b-PE-Cy7 (Mac1), anti–Ly6C-PE (Gr1), anti–CD80-APC, anti–CD86-APC, and anti–CD69-PE (BD Biosciences), as well as anti–GPIb-PE (Emfret Analytics), and anti–F4/80-PE (Invitrogen). Flow cytometry was performed on either FACSCalibur or FACSCanto flow cytometers (BD Biosciences) and analyzed with FlowJo Software (TreeStar). GFP+ cells were sorted on a FACSDiva (BD Biosciences).
Microarray experiments
G1ME cells were transduced with MIGR1 or MIGR1-GATA-1 and sorted for GFP expression 18 and 42 hours later. Total RNA was extracted using Trizol and the Qiagen RNeasy kit (Qiagen). RNA was hybridized to Affymetrix GeneChip Genome 430 2.0. The studies were performed on biologic triplicate samples. Microarray data reported here were submitted to Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/44 ), accession number GSE14980. Details on the analysis of microarray experiments are provided as supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Real-time polymerase chain reaction
Total cellular RNA was isolated with Trizol and cDNA was prepared by the oligo(dT) method (Invitrogen) from 2 mg total RNA. Polymerase chain reaction (PCR) was performed using SYBR green dye on an ABI 7900 real-time machine (PE Applied Biosystems). mRNA levels were normalized to those of glyceraldehyde phosphate dehydrogenase (Gapdh). Primer sequences are described in supplemental Table 2.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described previously.45,46 Antibodies used include anti–GATA-1 (N6), anti–GATA-2 (H-116), anti-PU.1 (E-19), anti-FOG (M-20), anti-MTA (C-20; Santa Cruz Biotechnology), and acetyl-H3 (Upstate Biotechnology Inc). Immunoprecipitated DNA was analyzed by real-time PCR. Oligonucleotide primer pairs were designed by Primer Express software (Applied Biosystems) to amplify 50- to 150-bp amplicons and are described in supplemental Table 3. PCR products were quantified using SYBR green dye on an ABI 7900 real-time machine (PE Applied Biosystems). The signals were referenced to a dilution series of the relevant input genomic DNA.
Morphologic analysis
Cells were centrifuged onto a glass slide and stained with May-Grünwald-Giemsa (Sigma-Aldrich). Light microscopy images were obtained with a Zeiss Axioskope 2 microscope, Zeiss Axiocam camera, and Zeiss AxioVision 3.1 software (Carl Zeiss Microimaging) at room temperature.
Macrophage cytokine stimulation assays
Six days after infection, sorted GFP+ G1ME cells transduced with MIGR1 vector or MIGR1-shGATA2 were resuspended at 105 cells/well in 96-well flat-bottom plates in G1ME medium containing TPO, IL-3, MCSF, GMCSF as well as 100 ng/mL lipopolysaccharide (Sigma-Aldrich) and/or 10 ng/mL interferon-γ (R&D Systems). Culture supernatants were harvested after 18 hours. TNF-α concentrations were determined by enzyme-linked immunosorbent assay according to protocols provided by the manufacturer (R&D Systems). Nitric oxide (NO) content was measured using the Greiss reagent (Invitrogen Corporation). Cells were also stained and examined for up-regulation of surface marker expression as outlined in “Flow cytometry.”
Murine ES cell differentiation and shRNA-mediated GATA-2 knockdown
Results
GATA-1 gene regulation in a bipotential erythromegakaryocytic precursor
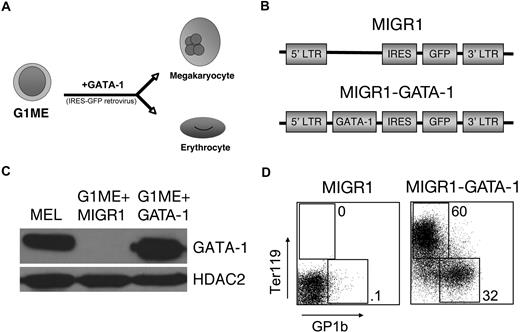
We studied the actions of GATA-1 in G1ME (for Gata1− megakaryocyte-erythroid) cells, a hematopoietic line derived from in vitro differentiation of murine Gata1− embryonic stem (ES) cells.41 G1ME cells self-renew in culture as thrombopoietin (TPO)–dependent, undifferentiated blasts. Restoration of GATA-1 by retroviral transfer induces erythroid and megakaryocytic differentiation (Figure 1). Of note, G1ME cells cultured in multiple cytokines (IL-3, GMCSF, GCSF, MCSF, and kit ligand; alone and in combination) exhibit no obvious signs of myeloid differentiation in short-term cultures, based on analysis of cell surface markers and cell morphology, either at baseline or after GATA-1 rescue. According to these findings, we speculated that G1ME cells approximate a bipotential megakaryocyte-erythroid progenitor (MEP) whose development is arrested by loss of GATA-1.41
GATA-1 induces erythroid and megakaryocytic differentiation in G1ME cells. (A) Schematic of G1ME cell differentiation into megakaryocytes and erythrocytes after GATA-1 is retrovirally restored. (B) Retroviral constructs used for gene rescue. The MIGR1 vector encodes green fluorescent protein (GFP) linked to an internal ribosome entry site (IRES). MIGR1-GATA-1 also contains the full-length coding region of the murine GATA-1 cDNA. (C) GATA-1 protein expression in transduced G1ME cells determined by Western blotting. GATA-1 expression in transduced G1ME cells approximated endogenous expression in murine erythroleukemia (MEL) cells. (D) Expression of the erythroid-specific marker Ter119 and terminal megakaryocytic marker GP1b of G1ME cells 4 days after transduction with MIGR1 or MIGR1-GATA-1. Percentages refer to fraction of GFP+ cells expressing Ter119 or GP1b.
GATA-1 induces erythroid and megakaryocytic differentiation in G1ME cells. (A) Schematic of G1ME cell differentiation into megakaryocytes and erythrocytes after GATA-1 is retrovirally restored. (B) Retroviral constructs used for gene rescue. The MIGR1 vector encodes green fluorescent protein (GFP) linked to an internal ribosome entry site (IRES). MIGR1-GATA-1 also contains the full-length coding region of the murine GATA-1 cDNA. (C) GATA-1 protein expression in transduced G1ME cells determined by Western blotting. GATA-1 expression in transduced G1ME cells approximated endogenous expression in murine erythroleukemia (MEL) cells. (D) Expression of the erythroid-specific marker Ter119 and terminal megakaryocytic marker GP1b of G1ME cells 4 days after transduction with MIGR1 or MIGR1-GATA-1. Percentages refer to fraction of GFP+ cells expressing Ter119 or GP1b.
GATA-1 is strongly up-regulated during the transition of multipotential granulocyte/erythroid/macrophage/megakaryocyte progenitors to MEPs49 and is believed to play an important functional role in this process. To better understand the role of GATA-1 in MEPs, we performed transcriptome analysis to define the GATA-1–regulated program of gene expression in G1ME cells. We used the MIGR1 retroviral vector in which GATA-1 cDNA is linked to green fluorescent protein (GFP) via an internal ribosome entry site (IRES). Cells were transduced with virus encoding vector alone or MIGR1-GATA1 and cultured with TPO and EPO to support erythromegakaryocytic differentiation. GFP+ cells were purified by fluorescence activated cell sorting (FACS) at 18 and 42 hours after transduction, and mRNA transcripts were compared using an Affymetrix Gene-Chip microarray (Genome 430 2.0, which interrogates 39 000 mouse genes and expression sequence tags). As predicted (Figure 1 and Stachura et al41 ), GATA-1 induced numerous erythroid and megakaryocytic marker genes including known GATA-1 targets (supplemental Table 1). These include genes encoding erythroid Kruppel-like factor (EKLF), globins, alpha hemoglobin stabilizing protein (AHSP, ERAF), and specialized membrane proteins. GATA-1 also activated numerous genes encoding megakaryocytic proteins such as von Willebrand factor (VWF), thrombospondin 1, and proplatelet basic protein. These observations validate further the use of G1ME cells as a model system to study gene expression during the maturation of bipotential MEPs. Of note, in G1ME cells, GATA-1 induces β-globin chains of both embryonic (Hbb-y, Hbb-bh1) and adult (Hbb-b1) origin. Concurrently, adult α-globin (Hba-a1) is significantly up-regulated with minimal increase of embryonic zeta chain (Hba-x) expression. These patterns differ from globin expression profiles observed in circulating erythrocytes at various stages of mouse embryogenesis.50 Thus, it is unknown whether G1ME cells most closely approximate adult-type definitive MEPs or primitive MEPs, which were recently shown to arise in early embryonic yolk sac.51
GATA-1 represses a myeloid program by inhibiting PU.1 expression
In addition to its established role as a transcriptional activator, GATA-1 also represses gene expression.14,–16,24,25,52,–54 This property is important for normal hematopoiesis and may contribute to leukemia, as a subset of GATA-1 target genes is inadequately repressed in Down syndrome-associated acute megakaryoblastic leukemia, which expresses the amino-truncated mutant protein GATA-1s.14,55,56 To study the GATA-1–regulated program of gene repression in MEPs, we used the binomial test for significance to examine overlap between the 300 genes most strongly down-regulated by GATA-1 in G1ME cells and Gene Ontology functional classifications (http://www.geneontology.org).57 We found that GATA-1–repressed transcripts were significantly enriched for those involved in immune responses, specifically myeloid and lymphoid-expressed mRNAs (Table 1). Based on this finding, we examined the expression of mRNAs encoding the key myeloid and lymphoid regulators PU.1, CEBPa, and Ikaros. Among these, only PU.1, which positively regulates many myeloid and lymphoid genes, was significantly expressed in G1ME cells. Microarray analysis demonstrated that PU.1 mRNA was decreased by 30% and 70% at 18 and 42 hours after GATA-1 restoration, respectively (not shown). Real-time reverse-transcription (RT)–PCR and Western blot analysis confirmed that PU.1 mRNA and protein were strongly down-regulated by GATA-1 in G1ME cells (Figure 2A-B). In the absence of GATA-1, G1ME cells express PU.1 protein at a similar level to that observed in murine erythroleukemia (MEL) cells where aberrant activation of the PU.1/Sfpi1 gene contributes to maturation arrest and malignant transformation58,–60 (Figure 2B). In contrast, G1ME cells express PU.1/Sfpi1 mRNA at about one-third the level observed in the myeloid cell line 416B (Figure 2C).
Function of genes down-regulated by GATA1 in G1ME cells
| GO biologic process list . | Matches . | Fold excess . | P . |
|---|---|---|---|
| Immune response | 34 | 4.16 | 8.78 × 10−12 |
| Immune system process | 41 | 3.43 | 2.80 × 10−11 |
| Cell activation | 15 | 3.94 | .000010 |
| Leukocyte activation | 14 | 4.05 | .000014 |
| Lymphocyte activation | 13 | 4.27 | .000016 |
| Regulation of mononuclear cell proliferation | 6 | 9.83 | .000035 |
| Regulation of lymphocyte proliferation | 6 | 9.83 | .000035 |
| Regulation of lymphocyte activation | 8 | 6.50 | .000037 |
| Response to stimulus | 58 | 1.77 | .000042 |
| Mononuclear cell proliferation | 7 | 7.24 | .000057 |
| GO biologic process list . | Matches . | Fold excess . | P . |
|---|---|---|---|
| Immune response | 34 | 4.16 | 8.78 × 10−12 |
| Immune system process | 41 | 3.43 | 2.80 × 10−11 |
| Cell activation | 15 | 3.94 | .000010 |
| Leukocyte activation | 14 | 4.05 | .000014 |
| Lymphocyte activation | 13 | 4.27 | .000016 |
| Regulation of mononuclear cell proliferation | 6 | 9.83 | .000035 |
| Regulation of lymphocyte proliferation | 6 | 9.83 | .000035 |
| Regulation of lymphocyte activation | 8 | 6.50 | .000037 |
| Response to stimulus | 58 | 1.77 | .000042 |
| Mononuclear cell proliferation | 7 | 7.24 | .000057 |
The Gene Ontology biologic processes most overrepresented among the 300 genes whose expression in G1ME was most decreased by GATA-1 transduction are shown. For each category, the number of down-regulated genes falling into that category is given, the factor by which this number of matches exceeds the number expected by chance, and the probability that the result is nonrandom. All categories with P < .00005 are shown.
GATA-1 inhibits PU.1 expression. (A) PU.1/Sfpi1 mRNA expression by real-time reverse-transcribed polymerase chain reaction (RT-PCR) after GATA-1 rescue in G1ME cells relative to untransduced cells. Bars represent the mean of 3 independent experiments ± SD. (B) PU.1 protein expression by Western blotting of sorted GFP+ G1ME cells before and after GATA-1 restoration. PU.1 expression in untransduced G1ME cells approximates endogenous murine erythroleukemia (MEL) cells. Two bands are visualized for PU.1, most likely due to posttranslational modifications. HDAC2 indicates histone deacetylase 2. (C) PU.1/Sfpi1 mRNA expression by RT-PCR in G1ME cells relative to the myeloid cell line 416B. Bars represent the mean of 3 independent experiments ± SD.
GATA-1 inhibits PU.1 expression. (A) PU.1/Sfpi1 mRNA expression by real-time reverse-transcribed polymerase chain reaction (RT-PCR) after GATA-1 rescue in G1ME cells relative to untransduced cells. Bars represent the mean of 3 independent experiments ± SD. (B) PU.1 protein expression by Western blotting of sorted GFP+ G1ME cells before and after GATA-1 restoration. PU.1 expression in untransduced G1ME cells approximates endogenous murine erythroleukemia (MEL) cells. Two bands are visualized for PU.1, most likely due to posttranslational modifications. HDAC2 indicates histone deacetylase 2. (C) PU.1/Sfpi1 mRNA expression by RT-PCR in G1ME cells relative to the myeloid cell line 416B. Bars represent the mean of 3 independent experiments ± SD.
We next examined the levels of known PU.1-activated myeloid genes in G1ME cells. In a prior study, Laslo et al identified a cohort of mRNAs that is induced after restoration of PU.1 expression in PU.1/Sfpi1−/− hematopoietic progenitors.61 We examined how GATA-1 regulates these same mRNAs in G1ME cells. Among the top 150 PU.1-induced transcripts, 77 were deemed by GeneSpring software (Agilent Technologies) to be present in G1ME cells. Among those, 45 (58%) were down-regulated by GATA-1, 23 (30%) were unchanged, and 9 (12%) were up-regulated (Figure 3). Hence, the majority of genes activated by PU.1 in hematopoietic progenitors is repressed by GATA-1 in G1ME cells. These data support the hypothesis that GATA-1 represses a myeloid program in MEPs by inhibiting PU.1/Sfpi1 expression, although failure of GATA-1–rescued G1ME cells to down-regulate all PU.1 targets identified by Laslo et al is expected, since the effects of both GATA-1 and PU.1 are dependent on their expression levels and cellular contexts (“Discussion”). Thus, G1ME cells may not simply represent bipotent MEPs, but rather, they may be poised toward myeloid differentiation due to loss of GATA-1. G1ME cells express relatively low levels of myeloid genes and exhibit an undifferentiated phenotype, probably because PU.1 expression is not sufficient to support overt granulocyte or macrophage maturation (“Discussion”).
Repression of PU.1 targets by GATA-1 in G1ME cells. The columns represent triplicate MIGR1 or MIGR1-GATA-1 transduced G1ME cells sorted by GFP positivity 42 hours after infection. The color scale ranges from green to red, corresponding to decreases and increases, respectively, in expression level. Transcripts shown are a cohort of PU.1 targets, with approximately two-thirds down-regulated by GATA-1 restoration.
Repression of PU.1 targets by GATA-1 in G1ME cells. The columns represent triplicate MIGR1 or MIGR1-GATA-1 transduced G1ME cells sorted by GFP positivity 42 hours after infection. The color scale ranges from green to red, corresponding to decreases and increases, respectively, in expression level. Transcripts shown are a cohort of PU.1 targets, with approximately two-thirds down-regulated by GATA-1 restoration.
GATA-1 directly regulates the PU.1/Sfpi1 gene
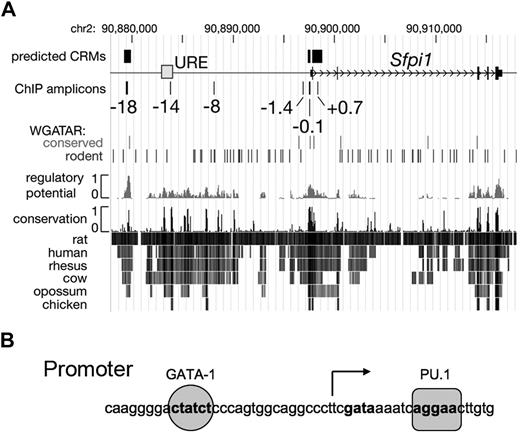
To investigate whether GATA-1 represses transcription of PU.1/Sfpi1 directly, we investigated whether functionally important GATA binding motifs are present in the PU.1/Sfpi1 gene. GATA-regulated cis-regulatory modules (CRMs), such as enhancers, can be predicted in aligned mammalian genomic DNA sequences by the presence of one or more conserved GATA consensus binding motifs within regions whose alignment patterns are similar to those found in a training set of known regulatory regions.62,63 Using this model, we found 2 predicted GATA-1 binding CRMs within the PU.1/Sfpi1 gene, one in the proximal promoter and a second 18 kb upstream of the transcriptional start site (Figure 4). Both of these regions are deeply conserved in evolution and contain canonical GATA-1 DNA binding sites (T/A(GATA)A/G). The proximal promoter region is known to be important for PU.1/Sfpi1 expression64,–66 ; a functional role for the −18-kb region has not been appreciated. The PU.1/Sfpi1 gene contains an upstream regulatory enhancer (URE) at −14 kb that is required for normal gene expression67,68 ; this region does not contain conserved GATA binding motifs (Figure 4). Both the URE and the proximal promoter contain essential positive autoregulatory PU.1 binding motifs.64,68 In the promoter region, the canonical GATA-1 site is 17-bp upstream of the transcriptional start and 27-bp upstream of the autoregulatory PU.1 site (Figure 4B). The presence of a conserved GATA motif in the PU.1/Sfpi1 proximal promoter was noted previously64 and has been shown to bind GATA-2 in chromatin immunoprecipitation (ChIP) experiments69 (“Discussion”).
The Sfpi1/PU.1 locus. (A) A 35-kb segment of mouse chromosome 2 is represented with the DNA encoding Sfpi1/PU.1 (thin black rectangles), the transcription start site (open arrow), and the −14-kb upstream regulatory element (URE) as a gray box. Predicted erythroid cis-regulatory modules (CRMs)63 and amplicons used in chromatin immunoprecipitation (ChIP) assays are shown as rectangles above and below the line, respectively, with the positions of the amplicons relative to the transcription start given in kilobases. The positions of GATA consensus binding motifs (WGATAR), both conserved in mammals and present only in rodents, are indicated as vertical lines. The track labeled “regulatory potential” plots sequence similarity to alignment patterns in known regulatory regions.62 Rows labeled “conservation” show interspecies alignments with the mouse genome; darker lines indicate greater sequence similarity. (B) The sequence of the PU.1/Sfpi1 promoter region. A PU.1 binding site (AGGAA) is located just downstream of the transcription start site, which is indicated by the arrow. A GATA-1 binding site (CTATCT) is located 27-bp upstream of the PU.1 binding site.
The Sfpi1/PU.1 locus. (A) A 35-kb segment of mouse chromosome 2 is represented with the DNA encoding Sfpi1/PU.1 (thin black rectangles), the transcription start site (open arrow), and the −14-kb upstream regulatory element (URE) as a gray box. Predicted erythroid cis-regulatory modules (CRMs)63 and amplicons used in chromatin immunoprecipitation (ChIP) assays are shown as rectangles above and below the line, respectively, with the positions of the amplicons relative to the transcription start given in kilobases. The positions of GATA consensus binding motifs (WGATAR), both conserved in mammals and present only in rodents, are indicated as vertical lines. The track labeled “regulatory potential” plots sequence similarity to alignment patterns in known regulatory regions.62 Rows labeled “conservation” show interspecies alignments with the mouse genome; darker lines indicate greater sequence similarity. (B) The sequence of the PU.1/Sfpi1 promoter region. A PU.1 binding site (AGGAA) is located just downstream of the transcription start site, which is indicated by the arrow. A GATA-1 binding site (CTATCT) is located 27-bp upstream of the PU.1 binding site.
We used ChIP to analyze transcription factor binding and histone modifications at the PU.1/Sfpi1 locus. In G1ME cells, GATA-2 physically occupies both the PU.1/Sfpi1 promoter and −18-kb region when the gene is active (Figure 5A). During induced erythromegakaryocytic maturation, GATA-1 replaces GATA-2 at the locus, coincident with down-regulated gene expression (Figure 5A-B). These findings resemble the “GATA-factor switch” described at other loci such as Gata2 and Kit where GATA-2 and GATA-1 are proposed to compete for the same cis elements to activate and repress transcription, respectively.15,16
Quantitative chromatin immunoprecipitation (ChIP) analysis of the PU.1/Sfpi1 locus in G1ME cells untransduced, or transduced with MIGR1-GATA-1, examined at 24 hours and 48 hours after infection. The relative occupancy of GATA-2 (A), GATA-1 (B), PU.1 (C), Friend of GATA-1 (FOG-1, D), metastasis associated protein-2 (MTA-2, E), and acetylase H3 (acH3, F) are indicated as vertical bars. As a negative control, ChIP experiments were performed with isotype-matched preimmune IgG. The hypersensitivity 3 (HS3) region of the β-globin locus and GPIIb promoter region are controls. * denotes not done. The results represent the average of 3 independent ChIP experiments. Error bars represent SD.
Quantitative chromatin immunoprecipitation (ChIP) analysis of the PU.1/Sfpi1 locus in G1ME cells untransduced, or transduced with MIGR1-GATA-1, examined at 24 hours and 48 hours after infection. The relative occupancy of GATA-2 (A), GATA-1 (B), PU.1 (C), Friend of GATA-1 (FOG-1, D), metastasis associated protein-2 (MTA-2, E), and acetylase H3 (acH3, F) are indicated as vertical bars. As a negative control, ChIP experiments were performed with isotype-matched preimmune IgG. The hypersensitivity 3 (HS3) region of the β-globin locus and GPIIb promoter region are controls. * denotes not done. The results represent the average of 3 independent ChIP experiments. Error bars represent SD.
In the absence of GATA-1, PU.1 occupies the −14-kb URE and promoter of the PU.1/Sfpi1 locus (Figure 5C), as previously reported.68 Relatively weak PU.1 binding was also detected at the −18-kb region, consistent with the presence of a predicted PU.1 binding motif (not shown). Retroviral expression of GATA-1 reduced PU.1 binding at all 3 regions, most likely because PU.1 protein expression was decreased. Together, these studies indicate that both GATA-1 and GATA-2 bind the PU.1/Sfpi1 gene in MEP-like cells. We also detected GATA-1 binding to the PU.1 locus in several other cellular contexts including the erythroblast cell line G1E after GATA-1 induction and the megakaryoblastic cell line Y10/L8057 (supplemental Figure 1A-B). In embryonic day 13.5 murine fetal liver, which contains mainly differentiated erythroid precursors, low-level GATA-1 occupancy was detected at the PU.1/Sfpi1 promoter, but not at the −18-kb region (supplemental Figure 1C).
Most activities of GATA-2 and GATA-1 depend on physical interaction with FOG-1. ChIP analysis demonstrated that FOG-1 occupies the PU.1/Sfpi1 gene at the same regions bound by GATA factors (Figure 5D). FOG-1 occupancy of PU.1/Sfpi1 was unaffected after GATA-1 rescue, indicating that both GATA-1 and GATA-2 recruit FOG-1 to the locus. We also detected the FOG-1–associated NuRD component MTA-224,25 at the −18-kb and promoter regions of PU.1/Sfpi1 in G1ME cells, both before and after GATA-1 rescue (Figure 5E). Of note, FOG-1 and MTA-2 also bound the −14-kb URE region, where we did not detect GATA factors. Thus, NuRD is probably recruited to the locus by GATA-dependent and -independent mechanisms. Together, our ChIP studies indicate that GATA factor–associated multisubunit transcription factor complexes assemble at the PU.1/Sfpi1 promoter and also at the −18-kb upstream region.
Next, we examined histone H3K9/K14 acetylation across the PU.1/Sfpi1 locus in G1ME cells (Figure 5F). This histone mark is associated with an open chromatin conformation and is generally observed at promoters and enhancers of active genes. As expected, histone H3K9/K14 was hyperacetylated at the promoter and −14-kb URE.70 We also observed a peak of hyperacetylation around −18 kb where GATA factors bind. During GATA-1–induced gene repression, acetylation decreased significantly at all regions. These data are consistent with known roles for the promoter and −14-kb URE in gene expression and also support the possibility that an additional functional element(s) resides around −18 kb. The ChIP findings also indicate that GATA-1 represses PU.1/Sfpi1 transcription by modifying chromatin, presumably by directly or indirectly recruiting histone deacetylases and/or interfering with acetylases.
GATA-2 deficiency induces PU.1/Sfpi1 and reprograms G1ME cells to macrophages
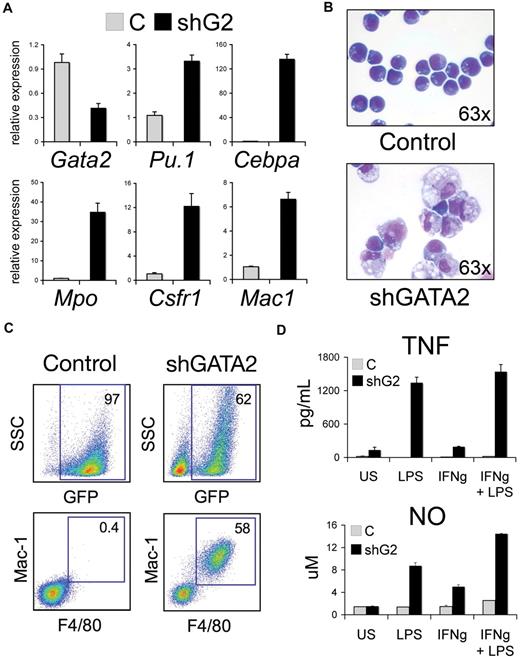
In G1ME cells, GATA-2 binds PU.1/Sfpi1 when the gene is active. To examine how GATA-2 occupancy regulates PU.1/Sfpi1 expression, we infected parental (Gata1−) G1ME cells with a bicistronic retrovirus expressing GFP and an anti-Gata2 shRNA. Infected GFP+ cells were purified by FACS and cultured in a multilineage cytokine mixture (EPO, TPO, MCSF, GMCSF, SCF). Expression of the shRNA specifically reduced Gata2 mRNA and protein by about 60% (Figure 6A and supplemental Figure 2) by 48 hours. Simultaneously, PU.1/Sfpi1 mRNA increased approximately 3-fold and there was a profound increase in several PU.1-regulated myeloid genes including Cebpa, Mpo, Csf1r, and Mac1. The myeloid cytokines MCSF and GMCSF were required to support the growth and survival of cells infected with Gata2 shRNA but not with control virus (not shown). By 5 days, the shRNA-expressing cells became adherent to plastic (not shown), acquired a macrophage morphology (Figure 6B), and expressed the macrophage cell surface markers Mac-1 and F4/80 (Figure 6C). There was no expression of granulocytic, lymphoid, erythroid, or late megakaryocytic lineage markers (Gr1, B220, Ter119, and Gp1b, respectively; not shown). The Gata2 shRNA-expressing cells also exhibited functional properties of macrophages including secretion of the proinflammatory cytokine tumor necrosis factor-α (TNF-α) at baseline and after stimulation with lipopolysaccharide (LPS) and interferon-γ (IFN-γ; Figure 6D). Stimulation of Gata2 knockdown G1ME cells with LPS/IFN-γ also induced nitric oxide production (Figure 6D) and up-regulated several macrophage activation markers including the early activation molecule CD69, and the costimulatory molecules B7/CD80 and B72/CD86 (supplemental Figure 3).71 Together, these data indicate that Gata2 knockdown induces PU.1/Sfpi1 expression and reprograms G1ME cells toward the macrophage lineage.
GATA-2 deficiency induces PU-1/Sfpi1 and reprograms G1ME cells to macrophages. (A) Relative expression of Gata2 and selected myeloid and macrophage-specific target genes in G1ME cells transduced with Banshee control (C) or Banshee shGATA2 retrovirus and flow-purified by GFP positivity. Mean plus or minus standard deviation values are shown for one representative experiment performed in triplicate. CCAAT/enhancer binding protein alpha (Cebpa), myeloperoxidase (Mpo), colony-stimulating factor receptor 1 (Csfr1), and macrophage 1 (Mac1). (B) Morphology of G1ME cells transduced with control or shGATA2 virus 6 days after infection. The shG2-infected cells are larger with abundant cytoplasm-containing granules and vacuoles. May-Grünwald Giemsa stain. Original magnification, ×63. Photographs were obtained using an Axioskope 2 microscope equipped with an AxioCam camera and AxioVision acquisition software (Carl Zeiss Microimaging) at room temperature. (C) Representative flow cytometric analysis of Banshee control– and Banshee shGATA2–infected G1ME cells. The numbers indicate the percentage of GFP+ cells within the live population in the top panels and the percentage of Mac-1+ F4/80+ within the GFP+ population in the bottom panels. (D) Macrophage stimulation assays of Banshee control (C)– and Banshee shGATA2–transduced G1ME cells. Tumor necrosis factor (TNF) and nitric oxide (NO) levels were measured from the supernatant of unstimulated (US) cells or 18 hours after stimulation with 100 ng/mL lipopolysaccharide (LPS) and/or interferon-γ (IFN-γ). GATA-2 knockdown G1ME cells induced TNF secretion and produced nitric oxide at baseline and after stimulation with LPS and IFN-γ. Mean ± SD values are shown for one representative experiment performed in triplicate.
GATA-2 deficiency induces PU-1/Sfpi1 and reprograms G1ME cells to macrophages. (A) Relative expression of Gata2 and selected myeloid and macrophage-specific target genes in G1ME cells transduced with Banshee control (C) or Banshee shGATA2 retrovirus and flow-purified by GFP positivity. Mean plus or minus standard deviation values are shown for one representative experiment performed in triplicate. CCAAT/enhancer binding protein alpha (Cebpa), myeloperoxidase (Mpo), colony-stimulating factor receptor 1 (Csfr1), and macrophage 1 (Mac1). (B) Morphology of G1ME cells transduced with control or shGATA2 virus 6 days after infection. The shG2-infected cells are larger with abundant cytoplasm-containing granules and vacuoles. May-Grünwald Giemsa stain. Original magnification, ×63. Photographs were obtained using an Axioskope 2 microscope equipped with an AxioCam camera and AxioVision acquisition software (Carl Zeiss Microimaging) at room temperature. (C) Representative flow cytometric analysis of Banshee control– and Banshee shGATA2–infected G1ME cells. The numbers indicate the percentage of GFP+ cells within the live population in the top panels and the percentage of Mac-1+ F4/80+ within the GFP+ population in the bottom panels. (D) Macrophage stimulation assays of Banshee control (C)– and Banshee shGATA2–transduced G1ME cells. Tumor necrosis factor (TNF) and nitric oxide (NO) levels were measured from the supernatant of unstimulated (US) cells or 18 hours after stimulation with 100 ng/mL lipopolysaccharide (LPS) and/or interferon-γ (IFN-γ). GATA-2 knockdown G1ME cells induced TNF secretion and produced nitric oxide at baseline and after stimulation with LPS and IFN-γ. Mean ± SD values are shown for one representative experiment performed in triplicate.
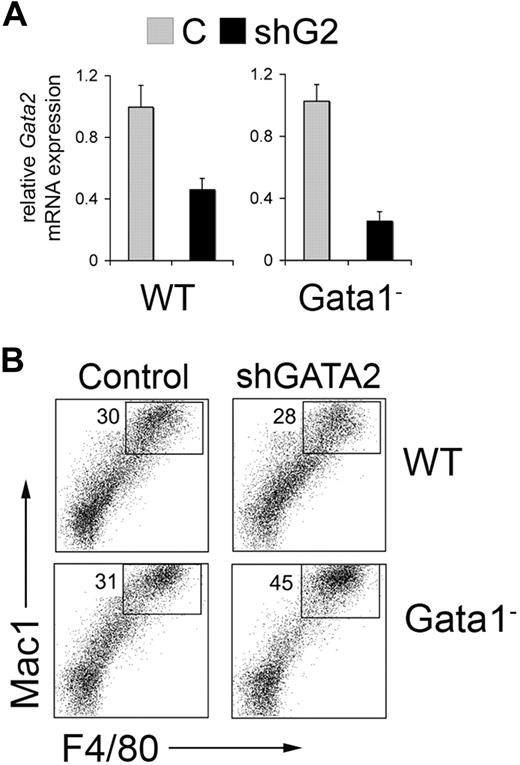
Our results indicate that combined GATA-1 and GATA-2 deficiency in MEP-like progenitors induces PU.1/Sfpi1 expression to favor macrophage differentiation. To test this further in a different model, we differentiated wild-type and Gata-1− murine embryonic stem (ES) cells into embryoid bodies (EBs) using defined serum-free culture conditions that enrich for hematopoietic progenitors (see supplemental methods). After 6 days, approximately 30% and 70% of cells within the wt and Gata-1− EBs, respectively, expressed CD41 (not shown), which marks multipotential hematopoietic progenitors in this experimental model and in early embryos.72,73 This finding is consistent with our previous finding that loss of GATA-1 expands CD41+ hematopoietic progenitors in ES cell differentiation cultures.41 We disaggregated the EBs with trypsin, infected single cell suspensions with retrovirus containing GFP alone or GFP plus shRNA against Gata2, and FACS-purified the resultant GFP+ cells. The shRNA specifically reduced Gata2 expression by 60% and 75% in wt and Gata-1− cells, respectively (Figure 7A).
Gata2 knockdown in wild-type (WT) and Gata1− cells derived from in vitro differentiation of embryonic stem (ES) cells. (A) Five-day-old embryoid bodies were disrupted and the cells were transduced with Banshee control (C) or Banshee shGATA2 retrovirus. Twenty-four hours later, GFP+ cells were purified by flow cytometry and analyzed for Gata2 expression by RT-PCR. Gata2 mRNA expression normalized to Gapdh mRNA is assigned a value of 1.0 in control virus–infected cells from WT and Gata1− embryoid bodies. Error bars represent SD. (B) Representative flow cytometric analysis of Banshee control– and Banshee shGATA2–infected wild-type (WT) and Gata1− hematopoietic progenitors derived from embryoid bodies. The numbers indicate the percentage of high Mac-1+ F4/80+ cells within the GFP+ population.
Gata2 knockdown in wild-type (WT) and Gata1− cells derived from in vitro differentiation of embryonic stem (ES) cells. (A) Five-day-old embryoid bodies were disrupted and the cells were transduced with Banshee control (C) or Banshee shGATA2 retrovirus. Twenty-four hours later, GFP+ cells were purified by flow cytometry and analyzed for Gata2 expression by RT-PCR. Gata2 mRNA expression normalized to Gapdh mRNA is assigned a value of 1.0 in control virus–infected cells from WT and Gata1− embryoid bodies. Error bars represent SD. (B) Representative flow cytometric analysis of Banshee control– and Banshee shGATA2–infected wild-type (WT) and Gata1− hematopoietic progenitors derived from embryoid bodies. The numbers indicate the percentage of high Mac-1+ F4/80+ cells within the GFP+ population.
Next, we cultured GFP+ cells in a multilineage cytokine cocktail containing SCF, EPO, TPO, IL-3, IL-6, IL-11, MCSF, and GMCSF for 5 to 6 days and assessed the expression of hematopoietic lineage–specific cell surface markers. In 2 separate experiments, Gata2 knockdown in Gata1− progenitors increased the numbers of macrophages by approximately 50%, as indicated by Mac-1 and F4/80 expression (Figure 7B). In contrast, reduced Gata2 expression had no detectable effect on macrophage differentiation of wild type EB-derived hematopoietic progenitors. We did not identify a difference in Gr1 or B220 expression after Gata2 knockdown in wt or Gata1− progenitors (not shown). Hence, GATA-1 and GATA-2 cooperate to restrain myelopoiesis, particularly macrophage differentiation. This effect occurs at least in part via repression of PU.1/Sfpi1.
Discussion
Understanding how lineage-determining transcription factors intercombine to control specific blood cell fates is a central problem for developmental hematology. GATA-1 is required for erythroid and megakaryocytic development, whereas PU.1 is necessary for proper lymphoid and myeloid differentiation. Both of these transcription factors activate genes associated with mature hematopoietic lineages. In addition, GATA-1 and PU.1 proteins physically interact to inhibit each other's activities, thereby repressing alternate lineage gene programs.27,,–30,74 Our current data indicate that GATA-1 inhibits PU.1 not only via protein-protein interactions, but also at the level of transcription. Specifically, we show that restoration of GATA-1 function in the Gata1− erythromegakaryocytic cell line G1ME induces down-regulation of PU.1/Sfpi1 mRNA and protein coincident with physical binding of GATA-1 to the PU.1/Sfpi1 gene at the promoter and −18-kb region.
G1ME cells resemble MEPs, multipotent precursors that give rise to lineage-committed erythroid and megakaryocytic cells.41 We detected GATA-1 at the PU.1/Sfpi1 promoter and −18-kb region in induced G1E erythroid cells75 and Y10/L8057 megakaryocytic cells76 (supplemental Figure 1). However, in these lineage-committed cell types, GATA-1 binding to the PU.1/Sfpi1 locus was less prominent compared with that observed in G1ME cells, which represent an earlier stage of development. Moreover, in primary murine fetal liver, which contains approximately 90% late-stage erythroid precursors, GATA-1 binding was observed at the PU.1/Sfpi1 promoter, but not at the −18-kb region. Hence it is possible that during normal hematopoiesis, GATA-1–mediated transcriptional repression of PU.1/Sfpi1 plays a more predominant role in multilineage progenitors that are facing differentiation decisions. This interpretation is consistent with findings that transcription factor binding to the PU.1/Sfpi1 locus is dynamic and cell-context dependent. For example, physical binding of RUNX1 to the PU.1/Sfpi1 gene is strongest in progenitor cells and down-regulated during subsequent maturation.70 Moreover, the functional effects of this protein-gene interaction vary according to hematopoietic stage.77 Our ability to expand G1ME cells in culture and rescue erythromegakaryocytic development by genetic complementation provides a convenient model system to study the biochemical and genetic actions of GATA-1 at a specific, synchronized stage of hematopoiesis that is difficult to access in primary tissues. Here we use this system to show that GATA factors regulate PU.1/Sfpi1 transcription in MEP-like cells. Currently, it would be problematic to purify sufficient quantities of primary Gata1− or wt MEPs for the experiments described here.
GATA-1 directly represses numerous genes including Kit, Gata2, Myc, Myb, and others.14,–16,24,25,52,–54 In multipotential progenitors, the Kit and Gata2 loci are relatively highly expressed and are occupied by GATA-2. During erythroid maturation, GATA-1 replaces GATA-2 at these loci, coincident with down-regulated expression of these genes. This “GATA factor switch” suggests a model whereby GATA-2 and GATA-1 sequentially bind the same cis elements with activating and repressive effects, respectively. The model is supported by our findings that a GATA-2 to GATA-1 switch at PU.1/Sfpi1 coincides with histone deacetylation and gene repression. However, in the absence of GATA-1, GATA-2 knockdown in G1ME cells increases PU.1/Sfpi1 expression, an effect opposite to that expected if GATA-2 were activating. There are several potential explanations for this finding. First, GATA-2 knockdown may activate PU.1/Sfpi1 expression indirectly by modulating additional unidentified transcription factors. Second, GATA-2 and GATA-1 could be equally potent direct inhibitors of PU.1/Sfpi1 transcription. In this case, PU.1/Sfpi1 expression would be regulated by the overall extent to which either GATA factor saturates the locus. However, this “quantitative” model is not supported by the observation that the GATA factor–associated proteins FOG-1 and MTA-2 occupy PU.1/Sfpi1 at similar levels, irrespective of whether GATA-1 or GATA-2 is bound (Figure 5). This suggests a similar degree of GATA-factor binding to PU.1/Sfpi1, yet expression is significantly higher when GATA-2 is bound. A third possibility, which we favor, is that both GATA-1 and GATA-2 inhibit PU.1/Sfpi1 expression, but to different extents, with GATA-1 being a stronger repressor.
The notion that GATA-1 and GATA-2 exert graded repressive effects suggests a new model for their sequential activities during hematopoiesis. Thus, in early hematopoietic development GATA-2 predominates to help maintain expression of PU.1/Sfpi1 at an intermediate level. This is compatible with the “lineage priming” concept whereby multipotent progenitors maintain their plasticity through relatively low-level expression of both myeloid and erythroid genes.4,7,,,–11 Upon erythromegakaryocytic differentiation, GATA-1 predominates, replaces GATA-2 at the PU.1/Sfpi1 locus, and shuts down expression to help ensure terminal maturation. Conversely, during myeloid differentiation, both GATA-1 and GATA-2 are extinguished, which allows for higher-level PU.1 expression and increased activation of its downstream targets. These pathways are intricately interconnected. For example, there is evidence to suggest that the PU.1/Sfpi1, Gata1, and Gata2 genes are all autoregulatory and GATA-1 inhibits Gata2 expression.15,64,67,68,78,–80 In addition, PU.1 can inhibit Gata2 expression.74 The differentiation of hematopoietic progenitors is highly dependent on the precise timing and levels of GATA factors and PU.1.34,81,–83 Moreover, alterations in the levels of activities of these transcription factors mark some of the earliest lineage fate decisions during normal hematopoiesis84,85 and are also associated with malignant transformation.59,86,–88 Our finding that GATA factors exert graded repression on PU.1/Sfpi1 transcription illustrates a new facet of the complex regulatory network that controls and balances the effects of these nuclear proteins.
The ability of GATA-2 knockdown to reprogram erythromegakaryocytic progenitors into macrophage underscores the importance of the GATA factor–PU.1 regulatory axis in lineage determination. We propose that loss of GATA-1 endows hematopoietic progenitors with increased lineage plasticity, in part by increasing PU.1/Sfpi1 transcription. In this setting, reduced GATA-2 derepresses PU.1/Sfpi1 further, allowing PU.1 protein level to reach a critical threshold for myeloid reprogramming. This hypothesis is consistent with several prior studies. First, altered balance of PU.1 and GATA-1 determines lineage choice in multipotential progenitors and can reprogram more committed precursors.34,38,39,81,89 Second, loss of GATA-1 can arrest hematopoietic maturation and increase lineage plasticity.41,73,90,–92 Third, in ES cell–derived hematopoietic progenitors, enforced expression of GATA-2 inhibits PU.1/Sfpi1 expression and reduces macrophage differentiation.69 It is important to note that the combinatorial effects of GATA factors and PU.1 are highly cell-context dependent. For example, GATA factors and PU.1 synergize in eosinophil and mast cell development.74,93 These lineages lack FOG-1, which may facilitate GATA factor–PU.1 cross-antagonism.94,95
In this study, Gata2 knockdown enhanced myelopoiesis specifically in cells intrinsically lacking Gata1. Gata2-targeted mice with intact Gata1 alleles were not noted to have increased myelopoiesis, probably because GATA-2 has important actions on hematopoietic progenitor proliferation/survival that are independent of the effects shown here.96,97 Knockdown of gata1 in zebrafish with intact gata2 alleles increased myeloid to erythroid ratios,38 but similar findings have not been reported in Gata1-deficient mice. Overall, our current findings, combined with these previous ones, indicate that Pu.1/Sfpi1 gene transcription is dependent on GATA-1 and GATA-2 expression levels and is likely to vary according to hematopoietic stage and species.
One unanswered question relates to the mechanisms by which GATA factors regulate PU.1/Sfpi1. Our ChIP studies suggest direct effects since, coincident with repression, GATA-1 binds the locus in at least 2 conserved regions: the promoter and the 18-kb upstream regions. In a prior study, GATA-2 was noted to bind the PU.1/Sfpi1 promoter in ES cell–derived hematopoietic progenitors and overexpressed GATA-2 antagonized PU.1-induced activation of the PU.1/Sfpi1 promoter in luciferase reporter assays.69 Both GATA-1 and GATA-2 recruit FOG-1-NuRD complexes to the PU.1/Sfpi1 locus. Although these complexes are associated with both activation and repression of gene expression,25,98 their role in regulating the PU.1/Sfpi1 locus through GATA factors is unknown. It will be interesting to study this further by determining the effects of Gata-2 shRNA knockdown on NuRD occupancy at PU.1/Sfpi1.
GATA factors may repress PU.1/Sfpi1 via binding at −18 kb, although a regulatory role for this region has not been appreciated previously. Transgenic reporter constructs lacking the −18-kb region largely recapitulate PU.1 expression in myeloid cells.68 Moreover, the normal −18-kb region or one containing a mutated GATA site had no effects on PU.1/Sfpi1 promoter activity in transient reporter assays using G1ME cells (not shown). However, several lines of evidence indicate that this region may be important for gene regulation, perhaps only in the context of normally chromatinized DNA. First, the −18-kb region and the GATA binding element are highly conserved in evolution (Figure 4). Second, GATA-FOG-NuRD complexes assemble at this region in hematopoietic cells (Figure 5). Third, the region marks a peak of histone H3K9/K14 acetylation (Figure 5), which commonly signifies the presence of positive regulatory elements. Furthermore, this DNA segment is marked by high levels of monomethylation and trimethylation on histone H3K4 along with trimethylation of histone H3K27 in mouse ES cells (published data viewed as the “Broad H3 ChIPseq” track at the UCSC Genome Browser99 ), characteristic of enhancers that are poised for lineage-specific activation or repression.100 Taken together, our current studies indicate that this region could participate in fine-tuning gene expression during myelopoiesis or play a more predominant role at specific stages of hematopoietic development, particularly within the MEP.
How GATA-1 inhibits transcription is incompletely understood and likely involves varied mechanisms at individual genes. Repression can be either FOG-1 dependent or independent.25,54 At the Kit locus, GATA-1 reconfigures the long-range spatial arrangement of chromatinized DNA to disrupt promoter-enhancer interactions.16 GATA factors may also inhibit gene expression by recruiting chromatin-modifying enzymes,15,16 which is consistent with our finding that GATA-1 binding is associated with widespread H3K9/K14 deacetylation across the PU.1/Sfpi1 gene. In the future, we are interested in better defining how these potential mechanisms apply to regulation of PU.1/Sfpi1 transcription by GATA factors during normal and malignant hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr John J. Rossi of the Beckman Research Institute of City of Hope for the Banshee vector. We thank Peter Laslo and Harinder Singh of The University of Chicago for PUER expression data. We thank David Stachura for helpful comments on the manuscript.
This study was supported by National Institutes of Health (NIH) grant K08 HL093290 (S.T.C.), R01DK58044 (G.A.B.), and R01 DK065806 (R.C.H., M.J.W., and G.A.B.). E.K. is supported by NIH training grant T32 GM008216-22. M.J.W. is a Leukemia & Lymphoma Society Scholar.
National Institutes of Health
Authorship
Contribution: S.T.C. designed and conducted research, analyzed data, and wrote the paper; E.K., K.E.N., C.V., Y.Y., and Z.H. designed and conducted research, and analyzed data; L.C.B., J.D.C., R.C.H., and G.A.B. designed research and analyzed data; and M.J.W. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mitchell J. Weiss, 3615 Civic Center Blvd, 316B ARC, Philadelphia, PA 19104; e-mail: weissmi@email.chop.edu.