Abstract
We analyzed somatic hypermutation (SHM) patterns and secondary rearrangements involving the immunoglobulin (IG) light chain (LC) gene loci in 725 patients with chronic lymphocytic leukemia (CLL). Important differences regarding mutational load and targeting were identified in groups of sequences defined by IGKV/IGLV gene usage and/or K/LCDR3 features. Recurrent amino acid (AA) changes in the IGKV/IGLV sequences were observed in subsets of CLL cases with stereotyped B-cell receptors (BCRs), especially those expressing IGHV3-21/IGLV3-21 and IGHV4-34/IGKV2-30 BCRs. Comparison with CLL LC sequences carrying heterogeneous K/LCDR3s or non-CLL LC sequences revealed that distinct amino acid changes appear to be “CLL-biased.” Finally, a significant proportion of CLL cases with monotypic LC expression were found to carry multiple potentially functional LC rearrangements, alluding to active, (auto)antigen-driven receptor editing. In conclusion, SHM targeting in CLL LCs is just as precise and, likely, functionally driven as in heavy chains. Secondary LC gene rearrangements and subset-biased mutations in CLL LC genes are strong indications that LCs are crucial in shaping the specificity of leukemic BCRs, in association with defined heavy chains. Therefore, CLL is characterized not only by stereotyped HCDR3 and heavy chains but, rather, by stereotyped BCRs involving both chains, which generate distinctive antigen-binding grooves.
Introduction
During the past decade, several groups, including ours, have revealed the unique molecular features of the B-cell receptors (BCRs) expressed by chronic lymphocytic leukemia (CLL)-malignant B cells. In particular, the immunoglobulin (Ig) gene repertoire of CLL is biased1-4 and uniquely characterized by the existence of subsets of cases carrying closely homologous (“stereotyped”) heavy chain complementarity-determining region 3 (HCDR3) sequences5-11 and often showing restricted light chain gene rearrangements. This remarkable BCR similarity implies the recognition of individual, discrete antigens or classes of structurally similar epitopes, likely selecting the leukemic clones. In addition, as recently reported from our group, certain subsets of CLL patients with stereotyped BCRs exhibit very precise targeting and distinctive features of somatic hypermutation (SHM) in IG heavy chain–variable (IGHV) genes, providing further evidence for selection by specific antigenic element(s).12
That notwithstanding, several lines of evidence from normal, autoreactive, or malignant B cells indicate that light chains also can play a critical role in antigen recognition. For instance, a defect in light chain gene usage may lead to impaired immune responses, as witnessed by a lack of a specific allelic variant of the IGKV2D-29 gene in the Navajo populations,13 which predisposes them to infection by Haemophilus influenzae. In addition, some light chain germline-encoded specificities are inherently dangerous and may predispose to serious pathology, as exemplified by the IGKV1-17 gene, which has major influences on the cationic charge of anti-DNA antibodies associated with lupus nephritis14 regardless IGHV gene usage. The marked light chain gene repertoire biases shown by us and others in various types of B-cell malignancies, including CLL, seem to suggest a similar critical role also in B-cell neoplasias15-22 that still requires further elucidation.
Studies in healthy humans as well as transgenic mouse models have shown that secondary rearrangements involving the Ig light chain loci represent a major somatic diversification mechanism which, along with SHM, has the capacity to drastically alter BCR specificity (“receptor editing”).23 In fact, B cells with autoreactive specificities may be rescued by such light chain editing processes, which have the potential to modify a BCR with a “dangerous” or nondesirable specificity.24-27 However, in some cases, antigen-driven receptor editing can promote a breach of allelic exclusion, leading to the appearance of cells with functional IgK and IgL rearrangements coexisting28 that are believed to dilute the autoreactive specificity.29 Previous studies from our group have indicated that receptor editing may also be active in CLL progenitors and critically affect the specificity of the clonotypic BCRs.30,31 This argument was based on the finding that many CLL cases, especially those expressing λ light chains, carried potentially functional IGKV-J joints inactivated by secondary rearrangements involving the κ-deleting element, very likely occurring as part of receptor editing.30,31
In the present study, we performed a systematic analysis of SHM and secondary light chain rearrangements in a series of 725 patients with CLL. Our study aims were to explore the putative role of light chains as editors to heavy chains expressed by the malignant CLL cells and to identify disease-biased features of SHM in the variable region of κ and λ genes. The results reported here emphasize that IG light chains complement heavy chains in shaping BCR specificity in CLL. Furthermore, they reinforce the concept that certain subsets of CLL cases expressing stereotyped BCRs are characterized by stereotyped IG heavy and light chain SHM patterns, further support for the role of antigen in CLL development.
Methods
Patient group
The patient group included 725 patients with CLL from collaborating institutions in France (n = 70), Greece (n = 488), Italy (n = 71), and Sweden (n = 96). All patients displayed the typical CLL immunophenotype as described previously and met the diagnostic criteria of the National Cancer Institute Working Group (NCI-WG).32 The light chain isotype as determined by flow cytometry was known in 709 of 725 patients. A case was considered to be κ- or λ-expressing if the ratio of κ to λ expression on CD19+ cells was greater than 3 or less than 0.3, respectively. On the basis of the aforementioned definitions, 425 of 709 patients and 284 of 709 patients with available data expressed κ and λ light chains, respectively.
No form of selection was applied in the Greek cohort, as all patients were analyzed in parallel for Ig heavy and light chain gene rearrangements. In the other national cohorts, patients were selected for Ig light chain analysis based on IGHV usage and HCDR3 features, in particular the expression of stereotyped HCDR3s. The gene repertoires, mutational status, and CDR3 features of 288 IGKV and 97 IGLV sequences of unselected cases from the Greek cohort have been reported previously.3,31 Written informed consent was obtained in accordance with the Declaration of Helsinki, and the study was approved by the local Ethics Review Committee of each institution.
PCR amplification of IGK-J/IGLV-J rearrangements and sequence analysis
In the vast majority of patients (686 of 725, 94.6%), peripheral blood samples were analyzed; bone marrow (23 patients), lymph nodes (11 patients), and spleen specimens (5 patients) also were analyzed. Amplification and sequence analysis of IGKV-J and IGLV-J rearrangements was performed on either DNA or complementary (c)DNA as previously described.3,5,9-11,30,31 Sequence data were analyzed with the IMGT database and tools.33,34 Any partial sequences that did not include the entire KCDR1/LCDR1 were not evaluated further.
PCR amplification of IGLV3-21 germline sequences
Germline sequence analysis was performed on the DNA of positively selected CD4+ T cells or granulocytes, which do not harbor rearranged IG genes. For germline analysis of the IGLV3-21 gene, peripheral blood from a patient belonging to subset no. 211 (stereotyped IGHV3-21/IGLV3-21 BCRs) was first subjected to CD19-positive selection to remove the B-cell population, followed by CD4-positive selection of T cells (Dynal CD4/CD19 Positive Isolation Kit, Invitrogen, Oslo, Sweden). With this approach, the resulting CD4+ T-cell fraction contained less than 0.5% of contaminating B cells. In a second case from subset no. 2, granulocytes obtained after Ficoll separation were analyzed. The upstream primer for amplification of IGLV3-21 sequences was complementary to the LCDR2 of the IGLV3-21 germline (5′-ccctgtgctggtcatctattat-3′), whereas the downstream primer was complementary to the IGLV3-21 intronic region (5′-GTGTTTTTGTCTCACTTCCTCATC-3′).
Definitions and groupings
All IGKV-J and IGLV-J rearrangements involving functional genes were considered potentially functional (PF) if the corresponding junctions were in-frame without stop codons. In-frame rearrangements of functional V genes carrying stop codons outside the junctions also were considered PF, at least in the primary repertoire; in such cases, the possibility that an initially functional rearrangement may have acquired stop codons in the context of the SHM process cannot be a priori excluded. In many cases, several PF rearrangements were found to coexist in the same clone. In such cases, the expressed rearrangement could not be identified with certainty, particularly if both rearrangements involved the same locus as the expressed light chain (for instance, 2 in-frame IGKV-J rearrangements in a κ-expressing case). For this reason, we refrained from making a distinction on the basis of the expression status of each rearrangement. Finally, sequences were considered nonfunctional (NF) if they involved pseudogenes, were rearranged out-of-frame, or carried stop codons created by the junctions.
Collection of sequence data from public databases
IGKV-J/IGLV-J sequences were retrieved from the IMGT/LIGM-DB database. Redundant, poorly annotated, out-of-frame, incomplete, or clonally related sequences were excluded from the analysis. A total of 4709 sequences (2346 IGKV-J and 2363 IGLV-J rearrangements) were included in the analysis. The final collection consisted of (1) 1591 IgK and 1501 IgL sequences from normal B cells; (2) 332 IgK and 610 IgL sequences from autoreactive cells; (3) 42 IgK and 37 IgL sequences from immune dysregulation disorders (allergy, asthma, various types of immunodeficiency); (4) 261 IgK and 175 IgL sequences from B-cell lymphoproliferative disorders, excluding CLL; and (5) 120 IgK and 40 IgL sequences from patients with CLL (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sequence analysis and data mining
All sequences were submitted to the IMGT V-QUEST tool34 and the following information was extracted:
IGKV, IGLV, IGKJ, and IGLJ gene and allele usage; percentage of identity to germline; and CDR3 length.
SHM characteristics for the part of the V region extending from CDR1 to CDR3: FR1 was excluded from the analysis to avoid misidentification of mutations caused by the use of FR1 consensus primers in the amplification reactions. A change in the IGKV/IGLV part of KCDR3/LCDR3 was counted as mutation only if followed by a stretch of at least 3 germline nucleotides. To account for the fact that a mutation is more likely to occur in a K/LFR than a K/LCDR simply because of its greater length, each mutation was normalized as previously described.12
Each nucleotide mutation was first recorded as replacement (R) or silent (S). The amino acids (AAs) were grouped in 5 AA categories according to standardized biochemical criteria.35 If a somatically introduced AA belonged to the same biochemical category as the mutated AA, the change was considered as “conservative,” if not the change was considered as “nonconservative.”35
Statistical analysis
Descriptive statistics for discrete parameters included counts and frequency distributions. For quantitative variables, statistical measures included means, medians, standard deviation, and min–max values. Significance of bivariate relationships between factors was assessed with the use of χ2 and Fisher exact tests. For all comparisons, a significance level of P at .05 was set and all statistical analyses were performed with the use of the Statistical Package SPSS version 12.0 (SPSS, Chicago, IL).
Results
Ig repertoires
A total of 612 IGKV-J and 279 IGLV-J sequences obtained from 725 patients with CLL were included in the analysis. Two or 3 light chain gene rearrangements were amplified in 124 of 725 (17%) and 21 of 725 (2.7%) patients, respectively. A total of 63 of 145 patients with multiple amplified light chain sequences carried at least 2 in-frame rearrangements. All cases with multiple light chain rearrangements carried a single in-frame heavy chain rearrangement, effectively ruling out the possibility of biclonal populations. Following the definitions detailed in “Methods,” we considered the 497/612 IGKV-J and 266/279 IGLV-J sequences to be PF; we considered the remaining 115/612 IGKV-J and 13/279 IGLV-J sequences to be NF. The Ig subgroup and individual gene usage was generally similar to previous studies3,4,31,40 and is reported in detail in Tables S2Table S3. IGLV repertoire in the Potentially Functional and Non Functional subgroups (XLS, 16.5 KB)Table S4. IGKJ repertoires in the Potentially Functional and Non Functional subgroups (XLS, 13.5 KB)–S5. The only exception concerns an overrepresentation of the IGLV3-21 gene, which is attributable to the enrichment for cases belonging to the major subset with stereotyped IGHV3-21 heavy chains (subset no. 2: 37 of 725 patients, 5.1%; subset numbering follows our previous reports11,12 ). As previously demonstrated by several groups, including ours,5,9-11,30 cases of this subset show restricted usage of IGLV3-21 light chains with long (12 AA) LCDR3s (Figure 1). In keeping with our previous report,3 the patterns of association of individual rearranged IGKV or IGLV genes with certain IGKJ or IGLJ genes, respectively, varied significantly (Tables S6Table S7. IGKJ12 vs IGKV3-5 gene usage among individual rearranged IGKV genes in the IGK-"non-functional" subgroup (XLS, 17 KB)Table S8. IGLJ1 vs IGLV2-3 gene usage among individual rearranged IGLV genes in the IGL-“potentially functional” subgroup (XLS, 16.5 KB)–S9).
Distribution of LCDR3 lengths in different IGLV subgroups. The peak at codon length 12 is composed predominantly of IGLV3-21 rearrangements in cases belonging to subset no. 2.11,12
Restricted light chain gene usage was observed for the vast majority of CLL cases in subsets with stereotyped HCDR3s, for example, IGKV1-39/1D-39 in 30 of 31 cases of subset no. 1 (stereotyped IGHV1/5/7/IGKV1-39/1D-39 BCRs), IGLV3-21 in 36 of 37 cases of subset no. 2 (stereotyped IGHV3-21/IGLV3-21 BCRs), and IGKV2-30 in all 15 cases of subset no. 4 (stereotyped IGHV4-24/IGLK2-30 BCRs) (Table S10). Furthermore, we identified subset-biased light chain CDR3 motifs among sequences by using the same IGKV or IGLV gene (Table S11). For instance, all 30 IGKV1-39/1D-39 light chains of subset no. 1 carried long KCDR3s (10-11 AAs) generated by significant N region addition, with a junctional proline in 26 of 30 cases; in contrast, all 9 IGKV1-39/1D-39 light chains of subset no. 8 carried 9 amino-acid-long KCDR3s with a junctional arginine appearing in 5 of 9 patients (similar to a previous report6 ). Interestingly, evidence for charge balancing emerged by examination of the median CDR3 isoelectric point (pI) values in heavy/light pairs of several subsets (Table S11). Thus, in some cases, the light chain CDR3 was found to have an opposite median pI charge to that of the heavy chain (eg, subset no. 13).
IGKV and IGLV mutational status: an overview
Given that even a low level of mutations can be functionally relevant and following our recent analysis on SHM features of IGH sequences in CLL,12 we investigated SHM features in all sequences of the present series with less than 100% identity to germline. Sequences were subdivided into a “truly unmutated” subgroup with IGKV/IGLV genes in germline configuration (100% identity), a “minimally mutated” subgroup, with 99.0% to 99.9% germline identity; a “borderline mutated” subgroup, with 98.0% to 98.9% identity; and a “mutated” subgroup, with less than 98% identity (Table 1). The IGKV/IGLV repertoires of the “truly unmutated,” “minimally mutated,” “borderline mutated,” and “mutated” subgroups differed (Tables S12,S13).
Mutational status of IGKV-J and IGLV-J rearrangements
| Mutational status . | IGKV-J, PF (n = 497) . | IGKV-J, NF (n = 115) . | IGLV-J, PF (n = 266) . | IGLV-J, NF (n = 13) . | ||||
|---|---|---|---|---|---|---|---|---|
| No. . | % . | No. . | % . | No. . | % . | No. . | % . | |
| Truly unmutated (100% identity to germline) | 232 | 46.7 | 76 | 66.1 | 72 | 27.1 | 5 | 38.5 |
| Minimally mutated (99.0%-99.9% identity to germline) | 32 | 6.4 | 16 | 13.9 | 32 | 12.0 | 0 | 0.0 |
| Borderline mutated (98.0%-98.9% identity to germline) | 27 | 5.4 | 4 | 3.5 | 57 | 21.4 | 2 | 15.4 |
| Mutated (< 98% identity to germline) | 206 | 41.5 | 19 | 16.5 | 105 | 39.5 | 6 | 46.1 |
| Mutational status . | IGKV-J, PF (n = 497) . | IGKV-J, NF (n = 115) . | IGLV-J, PF (n = 266) . | IGLV-J, NF (n = 13) . | ||||
|---|---|---|---|---|---|---|---|---|
| No. . | % . | No. . | % . | No. . | % . | No. . | % . | |
| Truly unmutated (100% identity to germline) | 232 | 46.7 | 76 | 66.1 | 72 | 27.1 | 5 | 38.5 |
| Minimally mutated (99.0%-99.9% identity to germline) | 32 | 6.4 | 16 | 13.9 | 32 | 12.0 | 0 | 0.0 |
| Borderline mutated (98.0%-98.9% identity to germline) | 27 | 5.4 | 4 | 3.5 | 57 | 21.4 | 2 | 15.4 |
| Mutated (< 98% identity to germline) | 206 | 41.5 | 19 | 16.5 | 105 | 39.5 | 6 | 46.1 |
PF indicates potentially functional; and NF, nonfunctional.
At the individual gene level, the distribution of rearrangements of IGKV/IGLV genes according to mutation status varied significantly (Figure 2; Tables S14,S15). Furthermore, significant differences also were observed with regard to mutational status among subgroups of sequences using different alleles of the same IGK/LV gene, in particular the IGKV1-5, IGLV1-51, and IGLV3-21 genes (Table S16).
Ig gene usage and mutational status. (A) Distribution of rearrangements of the most frequent IGKV genes of the present series according to mutational status. (B) Distribution of rearrangements of the 9 most frequent IGLV genes of the present series according to mutational status.
Ig gene usage and mutational status. (A) Distribution of rearrangements of the most frequent IGKV genes of the present series according to mutational status. (B) Distribution of rearrangements of the 9 most frequent IGLV genes of the present series according to mutational status.
Targeting of somatic hypermutation
Nucleotide substitution analysis in CLL sequences of our series with more than 100% identity to germline revealed that transitions predominated (2273/3948, 57.6%) over transversions, in keeping with a canonical SHM process.41 A noncanonical distribution of transitions-transversions was observed only in the CDRs of NF IgK or IgL rearrangements.
At the individual gene level, the IGKV2-30 gene (preferentially pairing with IGHV4-34 in subset no. 411,12 ) and IGLV3-21 gene (in subset no. 211,12 ) were characterized by distinctive nucleotide substitution spectra. In particular, the IGKV2-30 gene carried significantly more transitions compared with all other IGKV2 genes (66% vs 54%; P = .03); comparison with non-CLL IGKV2-30 sequences revealed that this overrepresentation was “CLL-biased.” In addition, compared with all other IGLV3 subgroup genes, IGLV3-21 rearrangements showed (1) significantly fewer G-to-A substitutions (11.6% vs 21%; P < .01) and (2) significantly more C→T and A→G substitutions (30.4% vs 17.7% and 21.8% vs 10.8%, respectively; P < .01 for both substitutions). However, as described to follow in this subsection, the high frequency of the C→T substitution could be attributed to a novel germline variant of the IGLV3-21 gene.
SHM frequencies in the FRs and CDRs were calculated for all IGKV/IGLV subgroups. Here, as in all analyses, the normalized distribution percentages (as described in “Methods”) were used. Overall, there was a greater targeting of R mutations to the CDRs versus FRs for both IGKV and IGLV “PF” subgroups (Table S17). In contrast, the IGKV NF subgroup, as expected, exhibited a more “homogeneous” targeting of R mutations in CDRs and FRs (Table S17); the number of NF IGLV rearrangements was too low (n = 13) to allow for meaningful comparisons.
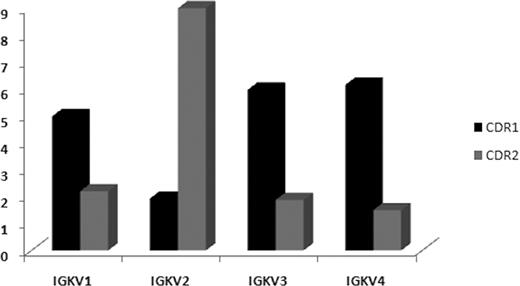
IGK sequences were distinguished by a clustering of R mutations in KCDR1; this was evident for all IGKV subgroups with the notable exception of the IGKV2 subgroup, which exhibited preferential targeting to the KCDR2, especially in IGKV2-30 rearrangements (Figure 3; Tables S18,S19). To explore whether the observed differences in mutation distributions might reflect differences in germline composition, we compared the KCDR2 sequences of IGKV genes from all IGKV subgroups. This comparison revealed no significant differences in the number of hotspot motifs for SHM between IGKV genes of different subgroups.
R/S normalized mutation ratios in the KCDR1 and KCDR2. A clustering of R mutations in KCDR1 was evident for all IGKV subgroups with the notable exception of the IGKV2 subgroup, which exhibited preferential targeting to the KCDR2, especially in IGKV2-30 rearrangements of cases with stereotyped IGHV4-34/IGKV2-30 BCRs (subset no. 4).11,12
R/S normalized mutation ratios in the KCDR1 and KCDR2. A clustering of R mutations in KCDR1 was evident for all IGKV subgroups with the notable exception of the IGKV2 subgroup, which exhibited preferential targeting to the KCDR2, especially in IGKV2-30 rearrangements of cases with stereotyped IGHV4-34/IGKV2-30 BCRs (subset no. 4).11,12
IGL sequences were mainly targeted for R mutations in LCDR2 (Tables S20,S21). Among IGL sequences, a distinctive pattern was observed in rearrangements of the IGLV3-21 gene, which were characterized by clustering of S mutations within LCDR3. In particular, a C→T silent mutation at codon IMGT/LCDR3-108 was observed in 60/92 IGLV3-21 rearrangements of the present series. However, as indicated by comparison of the germline and clonal sequences from 2 IGLV3-21–expressing cases with this mutation, a T in codon 108 appears to be a germline variant compared with all alleles of the IGLV3-21 gene included in the IMGT database, because T was present in germline in both cases.
Recurrent amino acid changes in subsets of CLL cases with stereotyped BCRs
The frequency of AA changes among mutated sequences that used the same IGKV or IGLV gene was recorded in CLL patients with stereotyped BCRs (Table S10). Recurrent AA changes (ie, the same AA replacement at the same position) across the whole IGKV or IGLV gene sequence were identified for subsets of CLL sequences with stereotyped BCRs. As revealed by comparison of the CLL versus non-CLL datasets, certain AA changes could be considered as “CLL-biased.” A comprehensive list of stereotyped AA changes is provided in Table S22. Furthermore, for certain IGKV or IGLV genes, many stereotyped AA changes occurred significantly more frequently in cases with stereotyped rather than heterogeneous BCRs, and, therefore, could be considered as “subset-biased.” The most striking “CLL-biased” hypermutations were observed in the following 2 subsets (Table 2):
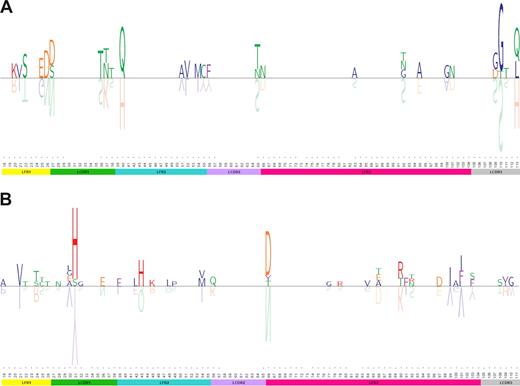
Seventeen of 37 (46%) IGLV3-21 sequences in cases with stereotyped IGHV3-21/IGLV3-21 BCRs (subset no. 2)11,12 carried an S-to-G change at IMGT/HCDR3-110 (Figure 4A). In 2 cases of this subset, germline sequence analysis of the IGLV3-21 gene confirmed that the S-to-G change was generated somatically and, thus, did not represent a polymorphism. Comparison of “subset” IGLV3-21 sequences to CLL IGLV3-21 sequences of cases with heterogeneous BCRs or non-CLL IGLV3-21 sequences demonstrated that this change was “CLL-biased” and “subset-biased.”
In a group of 15 IGKV2-30 sequences in cases with stereotyped IGHV4-34/IGKV2-30 BCRs (subset no. 4),11,12 3 recurrent mutations were observed at a frequency of 27% to 67%. In particular, 10 of 15 sequences (67%) carried a Y-to-H change at IMGT/HCDR1-31, 4/15 sequences (27%) carried a Q-to-H change at IMGT/HFR2-43 and, finally, 8 of 15 sequences (53%) an N-to-D change at IMGT/HFR3-66 (Figure 4B). As in subset no. 2, comparison with CLL IGKV2-30 sequences with heterogeneous KCDR3 or non-CLL IGKV2-30 sequences demonstrated that all 3 AA changes were “subset-biased” (Table 2).
“Stereotyped” amino acid changes
| Sequence (subset no.) . | Change* . | CLL-subset . | CLL-heterogeneous . | Non-CLL† . |
|---|---|---|---|---|
| IGLV3-21 (no. 2) | ||||
| IMGT-LCDR3, codon 110 | S→G | 17/37 | 14/55 | 9/197 |
| IGKV2-30 (no. 4) | ||||
| IMGT-KCDR1, codon 31 | Y→H | 10/15 | 1/17 | 6/62 |
| IMGT-KFR2, codon 43 | Q→H | 4/15 | 2/17 | 2/62 |
| IMGT-KFR3, codon 66 | N→D | 8/15 | 3/17 | 1/62 |
| Sequence (subset no.) . | Change* . | CLL-subset . | CLL-heterogeneous . | Non-CLL† . |
|---|---|---|---|---|
| IGLV3-21 (no. 2) | ||||
| IMGT-LCDR3, codon 110 | S→G | 17/37 | 14/55 | 9/197 |
| IGKV2-30 (no. 4) | ||||
| IMGT-KCDR1, codon 31 | Y→H | 10/15 | 1/17 | 6/62 |
| IMGT-KFR2, codon 43 | Q→H | 4/15 | 2/17 | 2/62 |
| IMGT-KFR3, codon 66 | N→D | 8/15 | 3/17 | 1/62 |
The frequency of changes among mutated sequences using the same IGKV or IGLV gene was recorded in CLL cases with stereotyped BCRs (cases belonging to subsets, numbered as in References 11,12), CLL cases with heterogeneous BCRs, and IGKV/IGLV sequences from normal or autoreactive clones (non-CLL).
BCR indicates B-cell receptor; and CLL, chronic lymphocytic leukemia.
All amino acid changes listed in Table 2 were significantly more frequent among cases with stereotyped BCRs and, therefore, could be considered “subset-biased.”
A complete list of the non-CLL (external) sequences carrying the indicated changes is given in Table S27.
Sequence logos for alignments of selected subsets. In these subfamily logos,50 each FR/CDR position is displayed as a stack of upright AA symbols. The height of each one-letter AA symbol is directly proportional to the relative frequency of that AA at a given FR/CDR position in the alignment among all sequences of a given subset. For clarity, the germline AAs of the IGKV or IGLV allele used by a given subset are shown upside down in gray color: the height of the inverted germline AA symbol is the sum of the heights of the upright AAs representing only the germline AA used by the cases that showed a change. Blank spaces represent AAs that are unchanged in the CLL IGKV or IGLV sequence compared with the germline sequence. For clarity, logos are vertically stretched so that the tallest upright stacks are of the same size, irrespective of its information content (ie, number of sequences). More information about number of sequences with a certain AA change of total number of sequences in each subset can be found in Table 2 and Table S21. AAs are divided in groups and color-coded as follows: GAPVLIM in blue, FYW in purple, STCNQ in green, KRH in red, and DE in orange. (A) Subset no. 2; (B) subset no. 4.
Sequence logos for alignments of selected subsets. In these subfamily logos,50 each FR/CDR position is displayed as a stack of upright AA symbols. The height of each one-letter AA symbol is directly proportional to the relative frequency of that AA at a given FR/CDR position in the alignment among all sequences of a given subset. For clarity, the germline AAs of the IGKV or IGLV allele used by a given subset are shown upside down in gray color: the height of the inverted germline AA symbol is the sum of the heights of the upright AAs representing only the germline AA used by the cases that showed a change. Blank spaces represent AAs that are unchanged in the CLL IGKV or IGLV sequence compared with the germline sequence. For clarity, logos are vertically stretched so that the tallest upright stacks are of the same size, irrespective of its information content (ie, number of sequences). More information about number of sequences with a certain AA change of total number of sequences in each subset can be found in Table 2 and Table S21. AAs are divided in groups and color-coded as follows: GAPVLIM in blue, FYW in purple, STCNQ in green, KRH in red, and DE in orange. (A) Subset no. 2; (B) subset no. 4.
Cases with multiple light chain rearrangements
As we have previously demonstrated,31 comprehensive polymerase chain reaction (PCR) analysis of rearrangements at the Ig light chain loci requires parallel assessment at the genomic DNA and cDNA level. With the caveat that this strategy was adopted for 488 (67%) of 725 cases of the present study, we identified 145 (20%) of 725 cases carrying multiple IgK and/or IgL gene rearrangements (Table 3). The frequency of detection of multiple rearrangements was significantly greater among λ- versus κ-expressing cases (98 [34.5%] of 284 vs 47 [11%] of 425 cases, respectively; P < .001)
Distribution of κ and λ CLL cases of the present series according to the number and type of light chain gene rearrangements
| No. of rearrangements . | No. κ-expressing . | No. λ-expressing . | Total . |
|---|---|---|---|
| One | |||
| IgK functional | 370 | 17 | 387 |
| IgL functional | 1 | 150 | 151 |
| IgK nonfunctional | 7 | 14 | 21 |
| IgL nonfunctional | 0 | 5 | 5 |
| Two | |||
| One IgK functional + one IGL functional | 6 | 27 | 33 |
| Two IgL functional | 0 | 3 | 3 |
| Two IgK nonfunctional | 0 | 1 | 1 |
| Two IgK functional | 17 | 0 | 17 |
| One IgK nonfunctional + one IgL functional | 0 | 45 | 45 |
| One IgK functional + one IgL nonfunctional | 1 | 0 | 1 |
| One IgK functional + one IgK nonfunctional | 20 | 0 | 20 |
| One IgL functional + one IgL nonfunctional | 0 | 4 | 4 |
| Three | |||
| Two IgK nonfunctional + one IgL functional | 0 | 10 | 10 |
| Two IgL functional + one IgK functional | 0 | 1 | 1 |
| Two IgK functional + one IgL functional | 0 | 1 | 1 |
| Two IgL functional + one IgK nonfunctional | 0 | 1 | 1 |
| Two IgK functional + one IgL nonfunctional | 1 | 0 | 1 |
| One IgK functional + one IgK nonfunctional + one IgL functional | 2 | 3 | 5 |
| One IgK nonfunctional + one IgL nonfunctional + one IGL functional | 0 | 1 | 1 |
| One IgK functional + one IgL nonfunctional + one IgL functional | 0 | 1 | 1 |
| Total | 425 | 284 | 709 |
| No. of rearrangements . | No. κ-expressing . | No. λ-expressing . | Total . |
|---|---|---|---|
| One | |||
| IgK functional | 370 | 17 | 387 |
| IgL functional | 1 | 150 | 151 |
| IgK nonfunctional | 7 | 14 | 21 |
| IgL nonfunctional | 0 | 5 | 5 |
| Two | |||
| One IgK functional + one IGL functional | 6 | 27 | 33 |
| Two IgL functional | 0 | 3 | 3 |
| Two IgK nonfunctional | 0 | 1 | 1 |
| Two IgK functional | 17 | 0 | 17 |
| One IgK nonfunctional + one IgL functional | 0 | 45 | 45 |
| One IgK functional + one IgL nonfunctional | 1 | 0 | 1 |
| One IgK functional + one IgK nonfunctional | 20 | 0 | 20 |
| One IgL functional + one IgL nonfunctional | 0 | 4 | 4 |
| Three | |||
| Two IgK nonfunctional + one IgL functional | 0 | 10 | 10 |
| Two IgL functional + one IgK functional | 0 | 1 | 1 |
| Two IgK functional + one IgL functional | 0 | 1 | 1 |
| Two IgL functional + one IgK nonfunctional | 0 | 1 | 1 |
| Two IgK functional + one IgL nonfunctional | 1 | 0 | 1 |
| One IgK functional + one IgK nonfunctional + one IgL functional | 2 | 3 | 5 |
| One IgK nonfunctional + one IgL nonfunctional + one IGL functional | 0 | 1 | 1 |
| One IgK functional + one IgL nonfunctional + one IgL functional | 0 | 1 | 1 |
| Total | 425 | 284 | 709 |
All calculations were performed on 709 of 725 patients with available information on the expressed light chain isotype.
IgK indicates immunoglobulin K; and IgL, immunoglobulin L. Other abbreviations are as explained in Table 2.
Overall, 63 patients from our series (26 κ- and 37 λ-expressing) carried at least 2 PF light-chain gene rearrangements; of note, 19 of these 63 patients (30%) belonged to subsets with stereotyped BCRs (Table S23). In all patients with multiple, transcribed, potentially functional light chain gene rearrangements, flow cytometry demonstrated that monotypic light chain expression was still maintained, indicating that allelic exclusion was probably regulated at the posttranscriptional level, at least for λ-expressing cases, but was not absolute at the transcriptional level.
Ig gene repertoires of cases with multiple rearrangements generally were similar to those at the cohort level. An interesting exception concerned the IGKV1-17 gene, usually present in anti-DNA antibodies,14 which was overrepresented among cases with multiple PF rearrangements. Interestingly, 4 of 12 IGKV1-17 PF rearrangements from our series were detected among λ-expressing cases (Table S24); of the remaining 8 IGKV1-17 rearrangements in κ-expressing cases, 2 were coamplified along with a second PF IGKV-J rearrangement (Table S24).
A second remarkable exception concerned IGKV3-15, the IGKV gene with the most electropositive germ line KCDR3 (KCDR3 isoelectric point value 10.9),42 which also was overrepresented among patients with multiple PF rearrangements compared with the cohort. Finally, it is noteworthy that 5 of 7 IGKV1-33/1D-33 rearrangements in patients with multiple PF light chain rearrangements used IGKJ4, a downstream IGKJ gene, suggesting that successive rearrangements may have occurred at the IGK locus.
Following the 98% cutoff for identity to germline as the criterion for considering a sequence as mutated or unmutated, we discovered that 57 (39%) of 145 patients carried multiple rearrangements with discordant mutational status (Table S25). To exclude the possibility that this discrepancy was caused by the presence of nonfunctional rearrangements, which are mainly unmutated, we analyzed separately all patients with multiple PF rearrangements and obtained a similar result, with 26 (41%) of 63 patients carrying rearrangements with discordant mutational status. Of particular interest was the finding of a truly mutated IGKV1-17 rearrangement coamplified along with a borderline mutated IGLV3-21 rearrangement in a case belonging to subset no. 2 (stereotyped IGHV3-21/IGLV3-21 BCRs), which was tantalizingly similar to what recently was reported for an unrelated case with stereotyped, borderline-mutated, IGHV3-21/IGLV3-21 BCR patient who also carried a truly mutated IGKV1-17 rearrangement.40
Several sequences in patients with multiple rearrangements exhibited deleterious changes introduced by SHM, for instance, mutation of “landmark” residues (eg, Cys at IMGT/FR3-104) or insertions/duplications/deletions (Table S26). Overall, sequence changes consistent with nucleotide insertions/duplications or deletions were identified in 11 rearrangements of the present series; 8 of 11 sequences were detected in cases with multiple rearrangements. Insertions/duplications or deletions occurred as multiples of 3 basepairs (therefore maintaining the original gene reading frame) in only 4 of 11 sequences. Interestingly, 1 of 4 sequences carried a large in-frame deletion of 30 nucleotides, starting at FR3-IMGT position 75; this rearrangement was coamplified by reverse transcription (RT)–PCR along with an in-frame IGKV3-20/IGKJ2 rearrangement. On these grounds, it would not be unreasonable to speculate that this large deletion probably led to structural impairment and thus precluded surface expression of the affected IG sequence.
Discussion
The CLL IG heavy chain repertoire demonstrates biases in the usage of certain IGHV genes,1,2 remarkable HCDR3 stereotypy5-11,30 and, as recently demonstrated by our group, stereotyped patterns of SHM in selected subgroups of patients.12 Although considerably less is known for light chain genes in CLL, the available data indicate that the pairing of Ig heavy and light chains in CLL-malignant B cells is nonstochastic and also that light chains may have a significant complementary role in antigen recognition by the clonotypic BCRs.3,4
In the present study, we showed that, in addition to restricted light chain gene usage in the vast majority of CLL patients with stereotyped HCDR3s, subset-biased light chain CDR3 motifs can be observed among sequences that use the same IGKV or IGLV gene. These findings strongly support the argument that CLL may develop from a limited set of B lymphocytes with a defined BCR structure and imply that selection of such cells by antigen may be critical for CLL pathogenesis.
Previous studies from our group have provided evidence to suggest that light chain gene biases in CLL may be driven by a series of BCR-mediated positive and negative selective processes after exposure to cognate, although mostly unknown, (auto)antigens.3,5,30,43 This argument is strengthened by the present finding that a significant proportion of patients with monotypic light chain expression carried multiple potentially functional light chain rearrangements, alluding to the possibility of secondary rearrangements likely occurring in the context of (auto)antigen-driven receptor editing.23-29,44
An illustrative example of receptor editing is provided by the small group of cases of the present study carrying potentially functional rearrangements of the IGKV1-17 gene. This gene has major influences on cationic charge of the autoantibodies in human lupus nephritis.14 Because IGKV1-17 is rarely used in the normal repertoire, it has been argued that normal B cells may edit IGKV1-17 rearrangements so as to avoid self-reactivity, whereas lupus B cells may have a defect in this mechanism.14 In the present series, 4 potentially functional IGKV1-17 rearrangements were detected among λ-expressing cases, strongly indicating the occurrence of receptor editing. Of the remaining 8 potentially functional IGKV1-17 rearrangements in κ-expressing cases, 2 were coamplified along with a second potentially functional IGKV-J rearrangement, perhaps attributed to lack of allelic exclusion, similar to what has been reported previously in CLL by analysis of IGH rearrangements.45 These results may imply that primary IGK rearrangements with autoreactive potential in CLL clonogenic cells were followed by a secondary light chain rearrangement in the context of a receptor “dilution” process.25,29 Alternatively, one might argue that the presence of dual receptors may allow the cell to escape tolerance mechanisms, in particular deletion, while maintaining the capacity to respond to autoantigen. This adaptation could provide the circumstances under which chronic autoantigenic stimulation could lead to proliferation, clonal expansion, and malignant transformation.
Another example of receptor editing is proposed by stereotyped IGHV3-21 CLL cases belonging to subset no. 2, which are characterized by a strikingly biased expression of λ light chains using the IGLV3-21 gene.5,10,11,30,40 We and others have previously demonstrated that the light-chain rearrangements in the cases of this subset have followed the hierarchical pattern of light chain recombination (IgK, IgK, IgL) and have gone through several rearranging attempts before producing a functional light chain.30,40 On the basis of this finding, we suggested that, although the reason for the multiple rearrangement events occurring in IGHV3-21 CLL cases is unknown, negative selection may have acted on BCRs with an IGHV3-21/IGKV rearrangement.30 This argument is strengthened by the findings reported here, with several subset no. 2 cases carrying potentially functional IGKV-J rearrangements though expressing λ light chains.
Analysis of SHM patterns in Ig genes is crucial to gain insight into the contribution of antigenic pressure in shaping the BCR structure. Recently this was demonstrated by our group in CLL by identifying marked differences in mutation patterns in subgroups of CLL cases using different IGHV genes.12 We here show that also in CLL light chains with less that 100% germline identity, the mutational load varied significantly according to IGKV/IGLV gene usage and K/LCDR3 features. Furthermore, in keeping with our recent findings in CLL IGH genes,12 differential mutation patterns were even observed between different alleles of the same IGKV or IGLV gene. These findings support the idea that SHM targeting in CLL light chains is just as precise and, perhaps, functionally driven as in heavy chains.
When we studied individual genes, the most striking SHM patterns were evident among IGLV3-21 and IGKV2-30 sequences. This mirrors the fact that these 2 light chain genes are used by the clonotypic stereotyped BCRs of subsets no. 2 and no. 4, respectively, which are also outstanding among CLL cases with regard to heavy chain SHM features.12 In particular, IGLV3-21 sequences of subset no. 2 cases showed an interesting analogy to their partner heavy chains, in that they mostly belonged to the borderline/minimally mutated categories. However, although minimally/borderline mutated, many IGLV3-21 sequences of this subset exhibit a stereotyped, “subset-biased” somatic hypermutation (S-to-G change at IMGT-LCDR3/110). Therefore, in keeping with our recent report on heavy chains,12 even very slight alterations in light chain sequences appear to be selected for and could very likely confer a clonal/functional advantage, at least for subsets of CLL cases.
In addition, we demonstrate that the IGKV2-30 light chains of subset no. 4 are also distinguished by stereotyped AA changes. Of note, 1 of the 3 “subset-biased” AA changes concerns the site-specific introduction of aspartic acid residues in the KFR3 (IMGT/KFR3-66). In transgenic mouse model systems, DNA binding antibodies carrying electropositive HCDR3s enriched in arginine residues can be edited by light chains with low isoelectric point values.46,47 In fact, the introduction of even a few strategically positioned aspartic acid residues in the κ chains of the transgenic animals was reported to be sufficient to negate DNA binding by 3H9 anti-DNA autoantibodies.46
These findings concur with our previous observation that all subset no. 4 IGHV4-34 CLL sequences have long HCDR3s, enriched in electropositive residues,11 reminiscent of many pathogenic anti-DNA antibodies48,49 and display distinctive SHM patterns, in particular stereotyped AA changes (consistent introduction of aspartic acid residues in HCDR1).12 Because this finding might suggest that the malignant clones belonging to subset no. 4 might have emerged from malignant transformation of cells with anti-DNA specificity,11,12 it would be tempting to speculate that the recurrent, site-specific introduction of aspartic acid residues in the KFR3 of subset no. 4 light chains may have represented a means of “charge balancing” to diminish responsiveness of the CLL progenitors against DNA. At this point, it is also worth underscoring the fact that a similar case of charge balancing, at least deduced by sequence analysis, emerged from examination of the median CDR3 pI values in heavy/light pairs of several subsets (Table S11). This finding may be considered as indirect evidence of light chains acting as editors to their paired heavy chain, by perhaps allowing an otherwise “inappropriate” heavy chain specificity to become acceptable by balancing the overall charge of the g molecule.
In conclusion, we here provide evidence that light chains may have a significant complementary role to heavy chains in the antigen recognition processes by the clonotypic BCRs expressed in CLL leukemic lymphocytes. Stereotyped CDR3 motifs and subset-biased mutations in the light chain genes are strong indications that light chains are crucial in shaping the antigenic recognition by leukemic BCRs, in association with defined heavy chains. On these grounds, we propose that not only stereotyped HCDR3 and heavy chains are present in CLL cells but rather stereotyped BCRs as a whole, involving both chains, which create highly distinctive antigen-binding grooves.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors sincerely thank Prof Marie-Paule Lefranc and Dr Veronique Giudicelli, Laboratoire d'Immunogenetique Moleculaire, LIGM, Universite Montpellier II, Montpellier, France, for their continuous support and invaluable help with the large scale immunoglobulin sequence analysis throughout this project. We also thank Prof Göran Roos, Department of Medical Biosciences, Umeå University, Sweden; Prof Christer Sundström, Department of Genetics and Pathology, Uppsala University, Sweden; Dr Karin Karlsson, Department of Hematology, Lund University Hospital, Lund, Sweden; Dr Mats Merup, Department of Medicine, Karolinska University Hospital, Huddinge, Sweden; and Prof Juhani Vilpo, Laboratory Center, Tampere University Hospital, Tampere, Finland, for providing samples and clinical data concerning Swedish and Finnish CLL patients. We also acknowledge the contribution of Dr Gerard Tobin, Dr Ulf Thunberg, and Dr Mia Thorsélius to the sequence analysis.
This work was supported by the General Secretariat for Research and Technology of Greece (Program INA-GENOME); the Swedish Cancer Society, the Swedish Research Council, the Medical Faculty of Uppsala University, Uppsala University Hospital, and the Lion's Cancer Research Foundation, Uppsala, Sweden; and the Associazione Italiana per la Ricerca sul Cancro-AIRC, Milano, Italy.
Authorship
Contribution: A.H., N.D., F.M., and T.S. performed research, analyzed data, and wrote the paper; E.A. performed research and wrote the paper; C.T., N.L., and A.A. provided samples and associated data; A.T. supervised research; and F.D., P.G., R.R., K.S., and C.B. designed and supervised the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paolo Ghia, Università Vita-Salute San Raffaele, Via Olgettina 58, 20132 Milano, Italy; e-mail: ghia.paolo@hsr.it.
References
Author notes
*A.H., N.D., and F.M. contributed equally to this work.