Abstract
Micro(mi)RNAs are small noncoding RNAs that orchestrate many key aspects of cell physiology and their deregulation is often linked to distinct diseases including cancer. Here, we studied the contribution of miRNAs in a well-characterized human myeloid leukemia, acute promyelocytic leukemia (APL), targeted by retinoic acid and trioxide arsenic therapy. We identified several miRNAs transcriptionally repressed by the APL-associated PML-RAR oncogene which are released after treatment with all-trans retinoic acid. These coregulated miRNAs were found to control, in a coordinated manner, crucial pathways linked to leukemogenesis, such as HOX proteins and cell adhesion molecules whose expressions are thereby repressed by the chemotherapy. Thus, APL appears linked to transcriptional perturbation of miRNA genes, and clinical protocols able to successfully eradicate cancer cells may do so by restoring miRNA expression. The identification of abnormal miRNA biogenesis in cancer may therefore provide novel biomarkers and therapeutic targets in myeloid leukemias.
Introduction
The micro(mi)RNAs are approximately 22 nt-long RNAs that orchestrate the expression of genes involved in many aspects of cell biology (reviewed in Kim and Nam1 and Filipowicz et al2 ). The human genome encodes more than 500 miRNAs located in introns, exons, or intergenic regions and it is now admitted that each cell type produces a specific miRNA repertoire. Most miRNA genes are transcribed in a manner similar to coding genes but differ in their mode of posttranscriptional processing.1 As a consequence of the fundamental functions of miRNAs, their deregulations are implicated in diverse human pathologies, in particular cancers (reviewed in Calin and Croce3 ). MiRNAs are now considered as oncogenes or tumor suppressors and represent promising diagnostic and prognostic markers.3 miRNAs are also envisaged as novel targets of therapeutic strategies.4 The identification of miRNAs implicated in cancer might therefore not only help in getting a better understanding of the molecular basis of human diseases, but also characterize novel biomarkers and therapeutic targets of cancers. Within this context, we studied the potential contribution of miRNAs in acute promyelocytic leukemia (APL). APL is identified as the M3 subtype of acute myeloid leukemia (AML) by the French-American-British (FAB) classification and is characterized by a differentiation arrest of granulopoiesis at the promyelocytic stage (Wang and Chen5 and references herein). APL is associated with chromosomal translocations that invariably implicate the gene encoding the retinoic acid receptor alpha (RARA). The resulting fusion proteins exert dominant and negative effect on RARA and hence, retinoic acid (RA)–regulated genes become insensitive to physiologic doses of RA. The most frequent translocation fuses the RARA with the promyelocytic leukemia protein (PML) gene.6 APL is considered the most malignant form of acute leukemia with a severe bleeding tendency and a fatal course of only weeks. However, pharmacologic doses of different RAR agonists, such as all-trans retinoic acid (ATRA), overcome the PML-RARA–mediated repression and restore normal transcription and granulocytic differentiation.5 Providing our knowledge of APL leukemogenesis and therapy, this disease is considered an excellent model of cancer therapies. APL therefore appeared appropriate to study the exact contribution of miRNAs in the initiating events of leukemia and in associated chemotherapies.
In this study, using bioinformatic and transcriptomic approaches followed by experimental validations, we identified a group of miRNAs targeted by PML-RARA. We further show that the repression exerted by PML-RARA/RXR complexes can be stopped by ATRA even in primary blast cells. Some of the identified miRNAs were also shown to be regulated by RARA in non-APL cells. Finally, transcriptomic analyses of mRNAs modulated by ATRA in distinct APL cell lines allowed the identification of targeted messengers that are implicated in crucial pathways linked to leukemogenesis, such as HOX proteins and cell adhesion molecules (notably the urokinase receptor, uPAR).
Methods
Cells and treatment
The NB4, NB4-LR1, and NB4-LR2 cells were cultured in RPMI (Invitrogen, Carlsbad, CA) as previously described7 and treated with 1 μM ATRA (Sigma-Aldrich, St Louis, MO). 293T cells were maintained in Dulbecco modified Eagle medium (DMEM; Invitrogen) supplemented with 2 mM l-glutamine, 100 μg/mL penicillin, 50 μg/mL streptomycin, and 10% heat-inactivated fetal calf serum. Primary blast cells extracted from the bone marrow of APL patients were treated for 6 days with ATRA (0.1 μM). The diagnosis of APL was established according to clinical presentation and morphologic criteria of the FAB classification and was subsequently confirmed by cytogenetic assays and reverse transcriptase–polymerase chain reaction (RT-PCR) analyses for detection of the t(15;17) translocation and PML-RARA transcripts. All patients had consented to the use of their medical records. The study was approved by the ethics committees of participating institutions and informed consent was obtained in accordance with the Declaration of Helsinki.
miRNA microarrays
Small RNA fraction was purified with the miRvana miRNA isolation kit (Ambion, Austin, TX), labeled with Alexa fluor dyes and hybridized in a dye-swap experiment (Agilent) on specific chips (http://www.microarray.fr/microRNA). TIF images containing the data from each fluorescence channel were quantified with the Genepix pro 6.0 program (Molecular Devices, Sunnyvale, CA) using a ‘circular features’ quantification method. Normalizations were performed using the limma package available from Bioconductor (http://www.bioconductor.org). Intraslide and interslide normalization were performed using the Print Tip Loess and the quantile methods, respectively, and means of ratios from ATRA-treated/control were calculated for each cell line. Experimental data and associated microarray designs have been deposited in the NCBI Gene Expression Omnibus (accession no. GSE11379).
cDNA microarrays
Total RNA samples were amplified using the Amino Allyl MessageAmp II aRNA Amplification kit (Ambion) and coupled to Cy3 or Cy5 dyes (GE Healthcare, Little Chalfont, United Kingdom). Whole human genome arrays (Human Operon version 2, University Medical Center of Utrecht) were used. For each condition, 3 independent hybridizations including one dye-swap hybridization were realized. For statistical analyses using the significance analysis of microarrays (SAM) method, the 2 classes unpaired response type was used. Potentially interesting genes resulting from the SAM analysis were selected at a false discovery rate of 5%.
DNA contructs and luciferase assays
The miR-23a/24-2 promoter8 was cloned into the BglII/HindIII restriction sites of pGL3b vector (Promega, Madison, WI). All sets of primers are indicated in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The predicted PML-RARA response element was mutagenized using the QuickChange Mutagenesis procedure (Stratagene, Cambridge, United Kingdom). The 3′ untranslated regions (UTRs) of uPAR and EBF-3 mRNAs were fused to the renilla gene using the XhoI/NotI restrictions sites of the psiCHECK2 vector (Promega). The pcDNA3 vector encoding PML-RARA is described in Duprez et al.9 Transfections of 293T cells were performed using Lipofectamine 2000 (Invitrogen). When indicated, ATRA treatment (1 μM, 16 hours) was realized 24 hours after transfection. Luciferase assays were performed using Dual-Luciferase (Promega). The pRLTK vector was used to normalize the experiments except for those conducted with psiCHECK2 which contains a firefly gene internal control. LNAs (Table S1) were provided by Sigma-Proligo (Evry, France). Indicated results are means of at least 3 independent experiments.
Quantitative RT-PCR
Total RNAs were extracted using Trizol (Invitrogen) for NB4, HL-60, and 293T cell lines or RNAplus (QBiogene, Irvine, CA) for primary blast cells. RTs were realized using oligodT(N) for RARB, uPAR, HOXB8, and GAPDH or specific stem-loop oligonucleotides indicated in Table S1 for miRNAs. The miRNA-specific RT-qPCR protocol was adapted from Chen et al10 to SYBR Green PCR (Roche, Indianapolis, IN). Indicated results are means of at least 3 independent experiments.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed using a standard protocol. Briefly, the cross-linked chromatin was sonicated and immunoprecipitated overnight at 4°C by using antibodies against PML, RARA, or RXRA (10 mg; Santa Cruz Biotechnologies, Santa Cruz, CA). Immunoprecipitated DNA was used as template for PCR and qPCR using sets of primers indicated in Table S1.
Results
Prediction of PML-RARA response elements in miRNA genes
To identify potential transcription factor binding sites (TFBS) for the PML-RARA fusion protein, microRNA regulatory regions were analyzed with the NHR-Scan system.11 The most common TFBS prediction methods, position weight matrices, are inappropriate for the analysis of nuclear receptor target sites due to the tolerance of these transcription factors for paired half-sites with variable spacing and orientation. NHR-Scan uses a flexible Hidden Markov Model (HMM) framework to capture those characteristics. We constructed a specific HMM model for the PML-RARA fusion protein using the results from a selection/amplification experiment.12 The PML-RARA–specified form of NHR-Scan was used to predict binding sites in the promoter regions of 247 human intergenic microRNAs (from a miRNA promoter dataset provided by Mahony et al13 ). We limited the search space to the 2 kb upstream of the transcription start site (TSS) or, in the event of a proximal upstream gene, to the intergenic region. For adjacent microRNAs separated by less than 250 bp, the 5′-most TSS was considered. As shown in Table 1, 65 of 247 microRNA promoters contain a PML-RARA predicted site.
List of microRNAs carrying a predicted PML-RARA binding site in their promoter
| MicroRNA name . | Predicted site . | Site type . |
|---|---|---|
| hsa-let-7a-2 | TGGCCTTCTTGAACT | DR3 |
| hsa-let-7a-3 | TGGCCTGAGCTGACCG | DR4 |
| hsa-let-7c | TGAACTATTGTTGAACG | DR5 |
| hsa-let-7d | TAACCTGTTAAATTAGGTCA | ER8 |
| hsa-mir-100 | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-10b | TGTTCAGACAGGTCA | DR3 |
| hsa-mir-130a | TGACCCAGTGAACT | DR2 |
| hsa-mir-133b | AGGCCAAGAAGTTCA | DR3 |
| hsa-mir-135a-1 | AGTTCATGACCT | IR0 |
| hsa-mir-146a | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-154 | TGACCTCAGAATCATGGCCT | DR8 |
| hsa-mir-183 mir-96 | TGACCTTCTGGCCT | DR2 |
| hsa-mir-194-2 mir-192 | TAACCTCTCTGAACC | DR3 |
| hsa-mir-196a-1 | TGAACTCCTGACCT | DR2 |
| hsa-mir-200c | AGGTCAGCGAGGGGTCA | DR5 |
| hsa-mir-205 | TGACCTGCTCTGGACT | DR4 |
| hsa-mir-210 | TGACCCCTTGACCC | DR2 |
| hsa-mir-217 | TGAACTCCTGACCT | DR2 |
| hsa-mir-22 | TGAACTGGCCCTGACCC | DR5 |
| hsa-mir-23a mir-27a mir-24-2 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-29b-2 | TGACCCCATGAACC | DR2 |
| hsa-mir-323 mir-758 | TGACCTGCACTGCACC | DR4 |
| hsa-mir-331 | TGAACTCCTGGCCT | DR2 |
| hsa-mir-345 | TACCCTGGTGACCC | DR2 |
| hsa-mir-34a | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-365-2 | AGGGCAGTAGAGGGTCA | DR5 |
| hsa-mir-377 | TGAACCATGTGACCG | DR3 |
| hsa-mir-379 | GGGGCATGAACT | IR0 |
| hsa-mir-380 | AGGTCAGTGAAGAGGCCA | DR6 |
| hsa-mir-383 | TGAACTCCTGACCT | DR2 |
| hsa-mir-422a | TGGCCTTGTTGACCT | DR3 |
| hsa-mir-455 | GGGTCACCCAGGGCCA | DR4 |
| hsa-mir-497 mir-195 | GGGCCAGGAGGTCA | DR2 |
| hsa-mir-500 | AGGTCAACAAAGTTCA | DR4 |
| hsa-mir-507 mir-506 | TGAACTCCTGGCCT | DR2 |
| hsa-mir-513-2 | AGGCCAGGTGCAGTGGGTCA | DR8 |
| hsa-mir-516-1 | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-516-3 | TGAACTCCTGACCT | DR2 |
| hsa-mir-517a | TGACCTGGTCATGCACC | DR5 |
| hsa-mir-518a-2 | GGGTCACCTGAGGTCA | DR4 |
| hsa-mir-518d | TGAACTCCTGACCT | DR2 |
| hsa-mir-519e | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-520b | AGTTCAGGAGTTCA | DR2 |
| hsa-mir-520d | TGGACTCCTGACCT | DR2 |
| hsa-mir-520e | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-520f | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-521-2 | TGAACTCCTGACCT | DR2 |
| hsa-mir-524 | AGTTCAGGAGTTCA | DR2 |
| hsa-mir-527 | TGCCCTCCAGCCTGGGTCA | ER7 |
| hsa-mir-539 | TGCACCAAGTTTGACCT | DR5 |
| hsa-mir-548a-2 | TGACCTCCTGGCCT | DR2 |
| hsa-mir-551b | TGACCTTCATTTTAACCT | DR6 |
| hsa-mir-563 | TGACCTTTCCCTGCACT | DR5 |
| hsa-mir-573 | AGGTTAGGAGTTCA | DR2 |
| hsa-mir-583 | TGGCCCCATGACCT | DR2 |
| hsa-mir-607 | TGAACTCCTGACCT | DR2 |
| hsa-mir-612 | TGAACTCCTGACCT | DR2 |
| hsa-mir-613 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-622 | AGGCCAGGCGGTCA | DR2 |
| hsa-mir-626 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-645 | TGAACCCTATTGTGAACT | DR6 |
| hsa-mir-801 | AGGTCACAAGGTCA | DR2 |
| hsa-mir-9-2 | TGAACCTTATGAACT | DR3 |
| hsa-mir-92b | TGAACCCCTGACCT | DR2 |
| hsa-mir-9-3 | AGGCCAGCCACGGTTCA | DR5 |
| MicroRNA name . | Predicted site . | Site type . |
|---|---|---|
| hsa-let-7a-2 | TGGCCTTCTTGAACT | DR3 |
| hsa-let-7a-3 | TGGCCTGAGCTGACCG | DR4 |
| hsa-let-7c | TGAACTATTGTTGAACG | DR5 |
| hsa-let-7d | TAACCTGTTAAATTAGGTCA | ER8 |
| hsa-mir-100 | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-10b | TGTTCAGACAGGTCA | DR3 |
| hsa-mir-130a | TGACCCAGTGAACT | DR2 |
| hsa-mir-133b | AGGCCAAGAAGTTCA | DR3 |
| hsa-mir-135a-1 | AGTTCATGACCT | IR0 |
| hsa-mir-146a | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-154 | TGACCTCAGAATCATGGCCT | DR8 |
| hsa-mir-183 mir-96 | TGACCTTCTGGCCT | DR2 |
| hsa-mir-194-2 mir-192 | TAACCTCTCTGAACC | DR3 |
| hsa-mir-196a-1 | TGAACTCCTGACCT | DR2 |
| hsa-mir-200c | AGGTCAGCGAGGGGTCA | DR5 |
| hsa-mir-205 | TGACCTGCTCTGGACT | DR4 |
| hsa-mir-210 | TGACCCCTTGACCC | DR2 |
| hsa-mir-217 | TGAACTCCTGACCT | DR2 |
| hsa-mir-22 | TGAACTGGCCCTGACCC | DR5 |
| hsa-mir-23a mir-27a mir-24-2 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-29b-2 | TGACCCCATGAACC | DR2 |
| hsa-mir-323 mir-758 | TGACCTGCACTGCACC | DR4 |
| hsa-mir-331 | TGAACTCCTGGCCT | DR2 |
| hsa-mir-345 | TACCCTGGTGACCC | DR2 |
| hsa-mir-34a | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-365-2 | AGGGCAGTAGAGGGTCA | DR5 |
| hsa-mir-377 | TGAACCATGTGACCG | DR3 |
| hsa-mir-379 | GGGGCATGAACT | IR0 |
| hsa-mir-380 | AGGTCAGTGAAGAGGCCA | DR6 |
| hsa-mir-383 | TGAACTCCTGACCT | DR2 |
| hsa-mir-422a | TGGCCTTGTTGACCT | DR3 |
| hsa-mir-455 | GGGTCACCCAGGGCCA | DR4 |
| hsa-mir-497 mir-195 | GGGCCAGGAGGTCA | DR2 |
| hsa-mir-500 | AGGTCAACAAAGTTCA | DR4 |
| hsa-mir-507 mir-506 | TGAACTCCTGGCCT | DR2 |
| hsa-mir-513-2 | AGGCCAGGTGCAGTGGGTCA | DR8 |
| hsa-mir-516-1 | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-516-3 | TGAACTCCTGACCT | DR2 |
| hsa-mir-517a | TGACCTGGTCATGCACC | DR5 |
| hsa-mir-518a-2 | GGGTCACCTGAGGTCA | DR4 |
| hsa-mir-518d | TGAACTCCTGACCT | DR2 |
| hsa-mir-519e | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-520b | AGTTCAGGAGTTCA | DR2 |
| hsa-mir-520d | TGGACTCCTGACCT | DR2 |
| hsa-mir-520e | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-520f | AGGTCAGGAGTTCA | DR2 |
| hsa-mir-521-2 | TGAACTCCTGACCT | DR2 |
| hsa-mir-524 | AGTTCAGGAGTTCA | DR2 |
| hsa-mir-527 | TGCCCTCCAGCCTGGGTCA | ER7 |
| hsa-mir-539 | TGCACCAAGTTTGACCT | DR5 |
| hsa-mir-548a-2 | TGACCTCCTGGCCT | DR2 |
| hsa-mir-551b | TGACCTTCATTTTAACCT | DR6 |
| hsa-mir-563 | TGACCTTTCCCTGCACT | DR5 |
| hsa-mir-573 | AGGTTAGGAGTTCA | DR2 |
| hsa-mir-583 | TGGCCCCATGACCT | DR2 |
| hsa-mir-607 | TGAACTCCTGACCT | DR2 |
| hsa-mir-612 | TGAACTCCTGACCT | DR2 |
| hsa-mir-613 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-622 | AGGCCAGGCGGTCA | DR2 |
| hsa-mir-626 | AGGCCAGGAGTTCA | DR2 |
| hsa-mir-645 | TGAACCCTATTGTGAACT | DR6 |
| hsa-mir-801 | AGGTCACAAGGTCA | DR2 |
| hsa-mir-9-2 | TGAACCTTATGAACT | DR3 |
| hsa-mir-92b | TGAACCCCTGACCT | DR2 |
| hsa-mir-9-3 | AGGCCAGCCACGGTTCA | DR5 |
The site type describes the half-site combination (IR indicates inverted repeat; DR, direct repeat; ER, everted repeat) followed by the spacer size (eg, DR2 refers to a direct repeat with 2 bp between the half-sites).
Transcriptomic analyses of the miRNA modulations induced by ATRA in NB4 cells
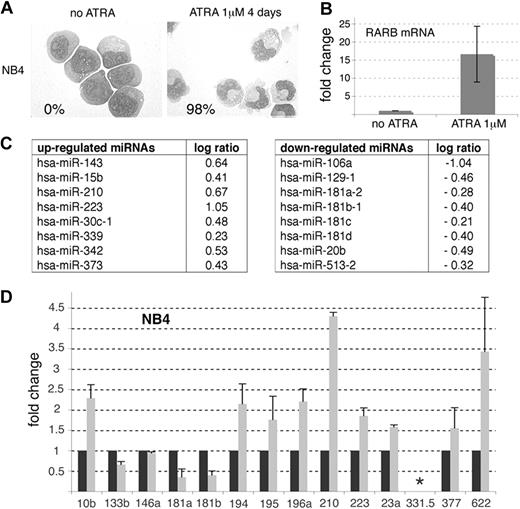
Next, we reasoned that if some miRNAs are repressed by PML-RARA, then pharmacologic doses of RA should abolish this repression and lead to an increase in the level of expression of the corresponding miRNAs. This expression pattern should in fact be similar to those observed for known RA-regulated genes, such as RARB (or RARb2), a well-characterized target of the RARA and PML-RARA proteins.14 We treated NB4 cells7 with 1 μM ATRA for 16 hours, as this concentration induces NB4 maturation in 4 days (Figure 1A) and this time point was previously shown to be appropriate to observe gene expression fluctuations.15 First, using RT-qPCR, we confirmed that ATRA induced an up-regulation of the RARB mRNA (Figure 1B), indicating that our settings were suitable to detect transcriptional modulations induced by ATRA. The same RNA samples were then analyzed using miRNA microarrays (Figure 1C). These analyses revealed that ATRA readily modulated several miRNAs, among those the miR-181b, which was previously shown to be down-regulated by ATRA in NB4 cells16 ; and the miR-15b, the miR-223, and the miR-342, 3 miRNAs previously found up-regulated in similar settings16,17 (Figure 1C). The miR-106a, miR-129-1, and miR-20b were also found down-regulated by ATRA while the miR-143, miR-30c, and miR-378 were up-regulated (Figure 1C). Only 2 miRNAs, among the 65 bioinformatically predicted to be repressed by PML-RARA, were detected by the arrays: the miR-210 and the miR-513-2 (compare Figure 1C with Table 1). While the miR-210 was clearly up-regulated by ATRA, suggesting that this miRNA could be an authentic transcriptional target of PML-RARA, the miR-513-2 was down-regulated, excluding this miRNA as a candidate.
miRNA profiles of NB4 cell line upon ATRA treatment. (A) Morphology of May-Grünwald-Giemsa (MGG)–stained NB4 cells treated or not with ATRA (1 μM) for 4 days. Maturation was monitored by the Nitroblue Tetrazolium (NBT) dye reduction assay and the percentage of NBT-positive cells obtained at day 4 of treatment is indicated (bottom left). (B-D) NB4 cells were treated with 1 μM ATRA for 16 hours. (B) RT-qPCR analysis of RARB mRNA expression in NB4 cells upon ATRA treatment. Results are indicated as fold change, 1 being the cycle threshold (Ct) obtained in the absence of treatment. (C) Table of miRNAs modulated by ATRA. See “Methods.” (D) miRNA-specific RT-qPCRs performed in untreated (black histograms) and ATRA-treated NB4 cells (gray histograms). The asterisk (*) indicates an absence of amplification. Results are indicated as fold change, 1 being the value obtained in the absence of treatment.
miRNA profiles of NB4 cell line upon ATRA treatment. (A) Morphology of May-Grünwald-Giemsa (MGG)–stained NB4 cells treated or not with ATRA (1 μM) for 4 days. Maturation was monitored by the Nitroblue Tetrazolium (NBT) dye reduction assay and the percentage of NBT-positive cells obtained at day 4 of treatment is indicated (bottom left). (B-D) NB4 cells were treated with 1 μM ATRA for 16 hours. (B) RT-qPCR analysis of RARB mRNA expression in NB4 cells upon ATRA treatment. Results are indicated as fold change, 1 being the cycle threshold (Ct) obtained in the absence of treatment. (C) Table of miRNAs modulated by ATRA. See “Methods.” (D) miRNA-specific RT-qPCRs performed in untreated (black histograms) and ATRA-treated NB4 cells (gray histograms). The asterisk (*) indicates an absence of amplification. Results are indicated as fold change, 1 being the value obtained in the absence of treatment.
RT-qPCR analyses of the miRNA modulations induced by ATRA in NB4 cells
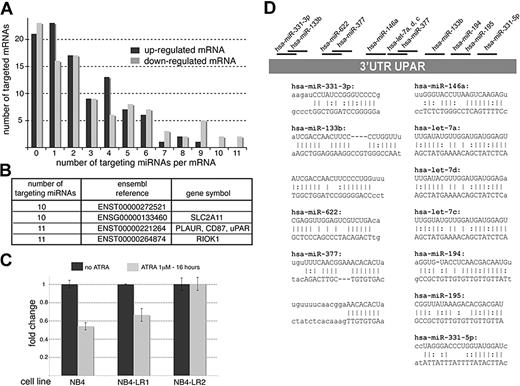
In order to identify additional miRNA candidates, we performed miRNA-specific RT-qPCRs directed against 11 of the 65 miRNAs presented in Table 1 (Figure 1D). The miR-223, miR-181a, and miR-181b, detected by microarrays, were also amplified (Figure 1D). Whereas RT-qPCRs confirmed the results obtained by microarrays for the miR-223, miR-181a, miR-181b, and miR-210, this approach revealed that the miR-10b, miR-194, miR-195, miR-196a, miR-23a, miR-377, and miR-622 were also up-regulated by ATRA in NB4 cells (Figure 1D). Some miRNAs (miR-331-5p) could not be detected and the expression of others (miR-133b and miR-146a) did not follow the expected pattern of PML-RARA–repressed genes (Figure 1D). Overall, the RT-qPCR analyses corroborated 75% of our predictions and suggested that the miR-10b, miR-194, miR-195, miR-196a, miR-210, miR-23a, miR-377, and miR-622 could represent authentic targets of PML-RARA. To validate our findings, investigations were further focused on the miR-210 and miR23a (belonging to a cluster of miRNAs, namely miR-23a, miR-27a, and miR-24-2). Of note, since the miR-23a, miR-27a, and miR-24-2 are generated from the same primary miRNA8 and because clustered miRNAs exhibit similar expression pattern,18 we considered that the miR-23a was representative of the expression of the entire miRNA cluster.
Transcriptional repression of the miR-210 and the miR23a/24-2 by PML-RARA
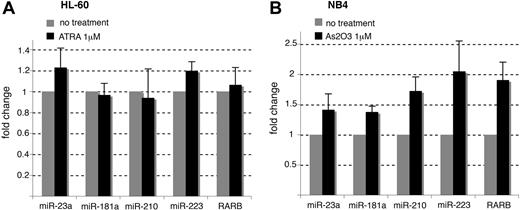
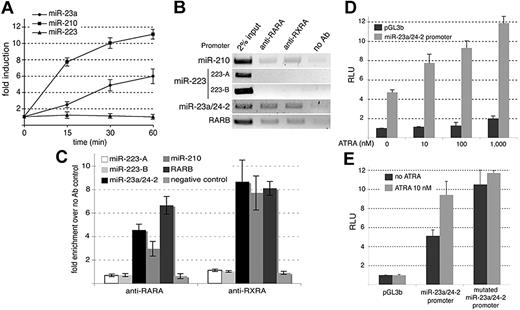
To directly confirm the binding of PML-RARA to the miR-210 and miR-23a/24-2 promoters in NB4 cells, we performed ChIP experiments using anti-PML, anti-RARA, and anti-RXRA antibodies. Anti-RXRA immunoprecipitations were performed because, although PML-RARA is able to bind DNA in the absence of RXR, the fusion protein has a greater DNA-binding affinity when complexed with RXRA12,14 The RARB promoter was used as a positive control19 while the miR-223 promoter was used as a negative control (Table 1). Whereas the miR-223 promoters17,20 were not associated with PML, RARA, or RXRA, these 3 proteins were physically bound to the RARB as well as the miR-210 and miR-23a/24-2 promoters (Figure 2A,B), suggesting that these miRNAs are repressed by PML-RARA/RXRA complexes in APL cells, similarly to the RARB promoter. As RXRA was recently shown to be an essential factor in APL pathogenesis,14 these findings might draw attention to the importance of the transcriptional repression of miRNAs in the development of APL. The sequence located directly upstream the miRNA precursor does not necessarily define the miRNA promoter20 and the formal characterization of the transcriptional start site of the primary miRNA is required before cloning miRNA promoters. We took advantage of the study performed by Lee et al21 and cloned the miR-23a/24-2 promoter to drive the expression of the firefly luciferase reporter gene. The reporter was transfected in 293T cells together with a PML-RARA expression vector.9 We observed that PML-RARA readily reduced the transcriptional activity of the miR-23a/24-2 promoters even in non-APL cells (Figure 2C). This reduction was limited by the addition of 1 μM ATRA (Figure 2C). In accordance with our bioinformatics predictions, a mutant of the miR-23a/24-2 promoter corresponding to the predicted PML-RARA response element was not sensitive to PML-RARA expression (Figure 2C). These results indicated that PML-RARA repressed the miR-23a/24-2 promoter through the response element identified in silico. Furthermore, RA did not induce noticeable changes in the expression of the miR-23a and miR-210 in PML-RARA–lacking HL-60 cells treated with ATRA for 16 hours (Figure 3A).22 On the other hand, degrading PML-RARA with arsenic trioxide in NB4 cells increased the expression of the miR-23a, miR-210, and the RARB (Figure 3B).23 These results confirmed that the expression of the miR-23a and miR-210 is dependent on PML-RARA.
The miR-23a/24-2 and miR-210 promoters are repressed by PML-RARA. (A) ChIP experiments performed in NB4 cells. Chromatin was immunoprecipitated using the indicated antibodies and the enriched genomic fragments were PCR amplified using specific primers. 223-A and 223-B correspond to the 2 miR-223 promoters described in Fukao et al20 and Fazi et al,17 respectively. The RARB promoter was used as a positive control. (B) qPCR analyses performed on DNA immunoprecipitated in panel A. The negative control corresponds to a sequence located 3.9 kb downstream the miR-23a/24-2 precursor. Fold enrichment over the negative control (no antibody) was calculated using the following formula: 2[(Ct input − Ct IP) − (Ct input − Ct no Ab)]. (C) LUC assays performed in 293T cells transfected with a firefly luciferase reporter gene driven by the miR-23a/24-2 promoter together with the control empty pcDNA3 plasmid (black histograms) or the PML-RARA–expressing vector (gray histograms). A promoterless vector was used as a negative control (pGL3b). A miR-23a/24-2 promoter mutated in the predicted PML-RARA response element was also tested (mutated miR-23a/24-2 promoter). At 24 hours after transfection, 293T cells were treated or not with ATRA (1 μM) for 16 hours. Results are expressed as relative light units (RLUs), 1 representing the value obtained with the promoterless vector pGL3b in absence of PML-RARA and ATRA treatments.
The miR-23a/24-2 and miR-210 promoters are repressed by PML-RARA. (A) ChIP experiments performed in NB4 cells. Chromatin was immunoprecipitated using the indicated antibodies and the enriched genomic fragments were PCR amplified using specific primers. 223-A and 223-B correspond to the 2 miR-223 promoters described in Fukao et al20 and Fazi et al,17 respectively. The RARB promoter was used as a positive control. (B) qPCR analyses performed on DNA immunoprecipitated in panel A. The negative control corresponds to a sequence located 3.9 kb downstream the miR-23a/24-2 precursor. Fold enrichment over the negative control (no antibody) was calculated using the following formula: 2[(Ct input − Ct IP) − (Ct input − Ct no Ab)]. (C) LUC assays performed in 293T cells transfected with a firefly luciferase reporter gene driven by the miR-23a/24-2 promoter together with the control empty pcDNA3 plasmid (black histograms) or the PML-RARA–expressing vector (gray histograms). A promoterless vector was used as a negative control (pGL3b). A miR-23a/24-2 promoter mutated in the predicted PML-RARA response element was also tested (mutated miR-23a/24-2 promoter). At 24 hours after transfection, 293T cells were treated or not with ATRA (1 μM) for 16 hours. Results are expressed as relative light units (RLUs), 1 representing the value obtained with the promoterless vector pGL3b in absence of PML-RARA and ATRA treatments.
miRNA modulations in ATRA-treated HL-60 and arsenic-treated NB4 cells. (A) miRNAs and RARB mRNA modulations in HL-60 cells treated with 1 μM ATRA for 16 hours. (B) miRNAs and RARB mRNA modulations in NB4 cells treated with 1 μM arsenic trioxide (As2O3) for 16 hours. RT-qPCR was performed as in Figure 1.
miRNA modulations in ATRA-treated HL-60 and arsenic-treated NB4 cells. (A) miRNAs and RARB mRNA modulations in HL-60 cells treated with 1 μM ATRA for 16 hours. (B) miRNAs and RARB mRNA modulations in NB4 cells treated with 1 μM arsenic trioxide (As2O3) for 16 hours. RT-qPCR was performed as in Figure 1.
Evaluation of the miRNA modulations induced by ATRA in primary APL blast cells
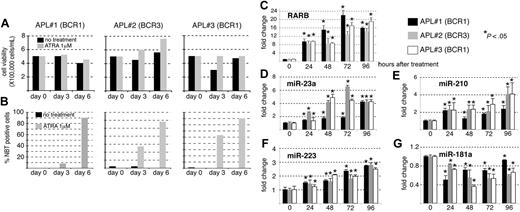
Next, we determined whether the miRNA modulations observed in the NB4 cell line were biologically relevant and tested miRNA expression levels in primary APL blast cells extracted from the bone marrow of 3 different patients. RT-PCR analyses performed during diagnosis revealed that patient 2 expressed the short BCR3 isoform of PML-RARA (11 000 copies) whereas patients 1 and 3 expressed the long BCR1 isoform (33 900 and 18 900 copies, respectively). The blast cells were treated with 100 nM ATRA for 6 days and RNA extraction was performed each day from day 0 to day 4 of treatment.24,25 Cell viability and maturation of APL cells was assessed by trypan blue (Figure 4A) and NitroBlueTetrazolium (NBT) dye reduction assays (Figure 4B), respectively, at days 0, 3, and 6 of ATRA treament. We also controlled the induction of the prototypic RARB by ATRA (Figure 4C). RT-qPCRs specific for the miR-23a, miR-210, miR-223, and miR-181a were then performed (Figure 4D-G, respectively). The miR-181a was chosen because the expression of this miRNA was recently shown to correlate with morphologic subclass of AML.26 We observed that the expression of the miR-23a, miR-210, and miR-223 increased throughout the ATRA treatment although the fold changes were not exactly comparable, the miR-223 being the least-induced miRNA (Figure 4D-F). The expression of the miR-181a was generally diminished by ATRA (Figure 4G). Hence, the modulations of the miR-23a, miR-210, miR-223, and miR-181a induced by ATRA in the NB4 cell line were confirmed in primary blast cells.
miRNA profiles of APL primary blast upon ATRA treatment. Blast cells extracted from bone marrow of 3 different patients with APL were cultured for 6 days with ATRA (0.1 μM). (A) Viable cell counts during ATRA treatment were determined using Trypan blue. (B) Granulocytic differentiation was assessed by NBT dye reduction assay. (C-G) RT-qPCR analyses specific for RARB (C), miR-23a (D), miR-210 (E), miR-223 (F), miR-181a (G). Total RNAs were extracted each day of ATRA treatment for 4 days. Results are indicated as fold change, 1 being the value obtained in the absence of ATRA treatment (0 hour after treatment) for each patient (*P < .05).
miRNA profiles of APL primary blast upon ATRA treatment. Blast cells extracted from bone marrow of 3 different patients with APL were cultured for 6 days with ATRA (0.1 μM). (A) Viable cell counts during ATRA treatment were determined using Trypan blue. (B) Granulocytic differentiation was assessed by NBT dye reduction assay. (C-G) RT-qPCR analyses specific for RARB (C), miR-23a (D), miR-210 (E), miR-223 (F), miR-181a (G). Total RNAs were extracted each day of ATRA treatment for 4 days. Results are indicated as fold change, 1 being the value obtained in the absence of ATRA treatment (0 hour after treatment) for each patient (*P < .05).
Transcriptional regulation of the miR-210 and the miR23a/24-2 by RARA in non-APL cells
PML-RARA is thought to interfere with the functions of both parental proteins in a dominant-negative manner. However, although PML-RARA binds to canonical RARA binding sites, it also recognizes a wider range of DNA-target sequences that are not regulated by RARA.12,27 Therefore, we tested whether the miR-210 and the miR-23a/24-2 were also regulated by RARA in non-APL cells. The promoters of the miR-210 and miR-23a/24-2 exhibited response elements compatible with the binding of RA-integrating complexes (ie, RARA/RXRA heterodimers; Table 1).28 We verified, using RT-qPCRs, the effect of physiologic doses (10 nM) of RA on the expression of the miR-210 and the miR-23a/24-2 in non-APL 293T cells. As shown in Figure 5A, the miR-210 and miR-23a were rapidly up-regulated by physiologic doses of ATRA while no modulation of the miR-223 expression could be detected in the same time frame. These observations suggested that the miR-210 and the miR-23a/24-2 could be transcriptionally modulated by RA in non-APL cells. ChIP experiments directed against both RARA and RXRA in 293T cells revealed that RARA/RXRA heterodimers could bind the miR-210 as well as the miR-23a/24-2 promoters but not the miR-223 promoters (Figure 5B,C). Luciferase assays were also conducted to substantiate these regulations. We transfected the firefly reporter containing the miR-23a/24-2 promoter in 293T cells and treated the cells with increasing doses of ATRA (from physiologic [10 nM] to pharmacologic doses [1 μM]) for 16 hours (Figure 5D). We observed that 10 nM ATRA was sufficient to induce a significant increase in the luciferase activity (Figure 5D), indicating that the miR-23a/24-2 promoter was sensitive to physiologic doses of ATRA. Moreover, we also tested the mutant described in Figure 2C and observed that this mutant was not responsive to 10 nM ATRA (Figure 5E). This result showed that, in the case of the miR-23a/24-2 promoter, the PML-RARA response element is similar to the RARA response element. Together, these results revealed that the miR-210 and the miR-23a/24-2 are directly repressed by the PML-RARA oncogene in APL cells and are regulated by RARA/RXRA heterodimers in non-APL cells.
The miR-23a/24-2 and miR-210 promoters are regulated by RARA. (A) RT-qPCRs directed against the miR-23a, miR-210, and miR-223 were performed in 293T cells treated with ATRA (10 nM). Results are indicated as fold change, 1 being the value obtained in the absence of treatment. (B) ChIPs performed in 293T cells using the indicated antibodies. The enriched genomic fragments were amplified using specific primers. The RARB promoter was used as a positive control. 223-A and 223-B as in Figure 2A. (C) qPCR analyses performed on DNA immunoprecipitated in panel B. (D) LUC assays performed in 293T cells transfected with promoterless pGL3b plasmid (black histograms) or the firefly luciferase reporter driven by the miR-23a/24-2 promoter (gray histograms). Cells were treated with increased doses of ATRA for 16 hours. Results are expressed as RLUs, 1 representing the value obtained with the pGL3b vector in absence of treatment. (E) LUC assays performed in 293T cells transfected with the firefly luciferase reporter driven by the miR-23a/24-2 promoter or the miR-23a/24-2 promoter mutated in the predicted PML-RARA response element. Cells were treated (gray histograms) or not (black histograms) with ATRA (10 nM) for 16 hours. Results are expressed as RLU, 1 representing the value obtained with the pGL3b vector in absence of treatment.
The miR-23a/24-2 and miR-210 promoters are regulated by RARA. (A) RT-qPCRs directed against the miR-23a, miR-210, and miR-223 were performed in 293T cells treated with ATRA (10 nM). Results are indicated as fold change, 1 being the value obtained in the absence of treatment. (B) ChIPs performed in 293T cells using the indicated antibodies. The enriched genomic fragments were amplified using specific primers. The RARB promoter was used as a positive control. 223-A and 223-B as in Figure 2A. (C) qPCR analyses performed on DNA immunoprecipitated in panel B. (D) LUC assays performed in 293T cells transfected with promoterless pGL3b plasmid (black histograms) or the firefly luciferase reporter driven by the miR-23a/24-2 promoter (gray histograms). Cells were treated with increased doses of ATRA for 16 hours. Results are expressed as RLUs, 1 representing the value obtained with the pGL3b vector in absence of treatment. (E) LUC assays performed in 293T cells transfected with the firefly luciferase reporter driven by the miR-23a/24-2 promoter or the miR-23a/24-2 promoter mutated in the predicted PML-RARA response element. Cells were treated (gray histograms) or not (black histograms) with ATRA (10 nM) for 16 hours. Results are expressed as RLU, 1 representing the value obtained with the pGL3b vector in absence of treatment.
Functional consequences of the PML-RARA–mediated miRNA repression
Next, we evaluated the functional consequences of PML-RARA–mediated miRNA repression in NB4 cells and used transcriptomic approaches to identify mRNAs potentially regulated by the group of miRNAs repressed by PML-RARA. In fact, simultaneous profiling of miRNA and mRNA expression is an appropriate strategy to identify functional miRNA targets.29 We reasoned that the up-regulation of miRNAs induced by ATRA should be accompanied by the down-regulation of the corresponding mRNA targets. We analyzed the RNA samples used in Figure 1 using pan-genomic cDNA microarrays. In order to focus only on genes that are both sensitive to ATRA and PML-RARA repression (and, hence, possibly to PML-RARA–repressed miRNAs), we compared these results to those obtained with RNA samples extracted from ATRA-resistant APL cells, NB4-LR1 and NB4-LR2.9,30 The NB4-LR1 cells do transcriptionally respond to ATRA but do not maturate.30 In contrast, the NB4-LR2 cells show a clear defect in RA signaling.9 We reasoned that, in contrast to genes modulated by other pathways, RA-sensitive genes should be up-regulated by ATRA in both NB4 and NB4-LR1 cells but remain unchanged in NB4-LR2 cells. This expression pattern was indeed observed in the case of the PML-RARA–targeted RARB gene (Figure 1B and Figure S1). Of note, the increase of the RARB mRNA was less pronounced in NB4-LR1 than in NB4 cells, likely reflecting the activation of additional pathways in NB4 cells, such as the PKA pathway, that might synergize with ATRA.31 Genes that were found up- or down-regulated in NB4 and NB4-LR1 cells but unchanged in NB4-LR2 are recapitulated in Tables S2 and S3, respectively. We noticed that global changes of the transcriptome were less pronounced in NB4-LR1 than in NB4 cells, again likely reflecting the activation of other cellular pathways by ATRA in NB4 cells.31 Overall, our microarray analyses were consistent with those previously published.25,32 The lists of the up- and down-regulated genes were then compared with the list of miRNAs bioinformatically predicted to be repressed by PML-RARA using the miRBase targets (http://microrna.sanger.ac.uk/targets/v5/). Among the first hundred genes analyzed in each case, we observed that 77% of the down-regulated genes and 79% of the up-regulated genes could potentially be targeted by at least one miRNA predicted to be repressed by PML-RARA (Figure 6A). Hence, no significant enrichment in down-regulated genes could be observed. However, translational repression orchestrated by miRNAs generally requires several targets of the same miRNA and/or distinct targeting miRNAs.2 As our studies identified a subset of miRNAs that are coordinately induced by ATRA, it is likely that these miRNAs do not exert their action independently but rather synergize and target the same mRNAs. Thus, we counted the number of miRNAs potentially repressed by PML-RARA targeting each mRNA indicated in Tables S2 and S3 (Figure 6A). These analyses revealed that 9 of the 100 down-regulated mRNAs could be targeted by more than 9 PML-RARA–repressed miRNA candidates as opposed to only 1 of the 100 up-regulated mRNAs. In order to confirm that these genes were genuine miRNA targets, we considered the 4 down-regulated mRNAs targeted by 10 or more miRNA candidates (Figure 6B). While only few data were available for 3 of those, the uPAR gene was particularly appealing because this gene has already been studied in the context of AML,33,34 NB4, and ATRA response.32,35
Functional consequences of miRNA inactivation by PML-RARA. (A) cDNA profiles obtained by microarrays (recapitulated in Tables S2 and S3) were compared with the data indicated in Table 1 in order to determine the number of targeted mRNAs and the number of targeting miRNAs per mRNA. (B) List of 4 down-regulated mRNAs targeted by 10 or more miRNA candidates. (C) RT-qPCR directed against the uPAR mRNA performed on RNAs used in panel A. (D) Schematic representation of the uPAR 3′ UTR and the targeting miRNAs. The miRNA::mRNA hybrids are indicated.
Functional consequences of miRNA inactivation by PML-RARA. (A) cDNA profiles obtained by microarrays (recapitulated in Tables S2 and S3) were compared with the data indicated in Table 1 in order to determine the number of targeted mRNAs and the number of targeting miRNAs per mRNA. (B) List of 4 down-regulated mRNAs targeted by 10 or more miRNA candidates. (C) RT-qPCR directed against the uPAR mRNA performed on RNAs used in panel A. (D) Schematic representation of the uPAR 3′ UTR and the targeting miRNAs. The miRNA::mRNA hybrids are indicated.
miRNAs repressed by PML-RARA control key cancer genes
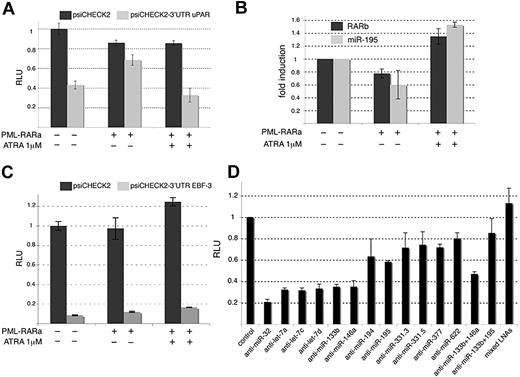
The uPAR protein plays essential roles in a variety of cell functions that exploit extracellular proteolysis, adhesion, and chemotaxis (reviewed in Pillay et al36 ). The uPAR expression level has been strongly correlated with poor prognosis in a variety of malignant tumors36 and patients suffering with AML have high expression of uPAR and high relapse risk after therapy.33,34 In addition, the uPAR mRNA was specifically shown to be down-regulated by ATRA in NB4.32,35 We first validated microarray analyses by RT-qPCRs and confirmed that the uPAR mRNA was down-regulated by ATRA in NB4 and NB4-LR1 but not in NB4-LR2 cells (Figure 6C). The miRBase targets revealed that the 3′ UTR of uPAR harbored 13 sequences that might be targeted by several miRNAs predicted to be repressed by PML-RARA (Figure 6D). To test these predictions, we cloned the 3′ UTR of uPAR downstream the renilla luciferase reporter gene (psiCHECK2-uPAR 3′ UTR) and transfected this construct into 293T cells (Figure 7A). We observed that the renilla expression was drastically reduced when fused to uPAR 3′ UTR (Figure 7A). We additionally observed that the introduction of PML-RARA in 293T cells restored the expression of the renilla reporter containing the uPAR 3′ UTR, whereas the fusion protein had little effect on the expression of the parental renilla (Figure 7A). Moreover, treatment of the transfected cells with 1 μM ATRA abolished the effect of PML-RARA on psiCHECK2-uPAR 3′ UTR (Figure 7A). In parallel, we verified that PML-RARA repressed the expression of the RARB mRNA whereas the addition of 1 μM ATRA stopped this repression (Figure 7B). A similar pattern was obtained for the miR-195 (Figure 7B), one of the 11 miRNAs potentially targeting uPAR mRNA and suspected to be repressed by PML-RARA (Figures 1D, 6D). It is noteworthy that, in both cases, the addition of 1 μM ATRA slightly increased the RNA expression (Figure 7B), suggesting that the miR-195, similarly to the RARB, is sensitive to RA. These observations indicate that the modulation exerted by the uPAR 3′ UTR is inversely correlated to the capacity of PML-RARA to repress transcription. We also tested a candidate gene targeted by less than 11 miRNAs and evaluated the effect of the 3′ UTR of the EBF-3 mRNA, targeted by only 2 miRNAs of the 65 indicated in Table 1 (Table S3 and Figure 7C). While the 3′ UTR of EBF-3 clearly decreased renilla expression, potentially due to negative posttranscriptional regulations, this effect was insensitive to PML-RARA expression (Figure 7C). These results suggested that EBF-3 is not a target of the miRNAs repressed by PML-RARA and underscored the importance of the number of miRNA binding sites per target. Next, we verified that the regulation elicited by the 3′ UTR of uPAR was orchestrated by miRNAs. For that purpose, we transfected 293T cells with the psiCHECK2-uPAR 3′ UTR vector and specific LNA miRNA inhibitors (Figure 7D). A functional LNA anti–miR-32 was used as a negative control. The inhibition of the miRNAs let-7a, let-7c, let-7d, miR-133b, and miR-146a had a modest effect on the expression of the reporter harboring the 3′ UTR of uPAR whereas the inhibition of the miR-194, miR-195, miR-331-5p, miR-331-3p, miR-377, and miR-622 significantly restored the expression of the Renilla (Figure 7D). Mixing LNAs directed against the miR-133b and the miR-195 had a stronger effect than each LNA transfected alone (Figure 7D). Likewise, the strongest effect on Renilla expression was observed when all LNAs were mixed (Figure 7D). This might suggest that this subset of miRNAs act in a synergistic manner. The similar effects of the LNAs directed against the miR-377 and the miR-622 might also indicate that these miRNAs act in a redundant manner. Overall, these observations showed that the miRNAs repressed by PML-RARA do not exert their action independently but are rather coordinated to regulate the same mRNAs. Finally, some of these miRNAs (ie, miR-331-5p) could not be detected in NB4 cells but seemed efficient in regulating uPAR 3′ UTR in 293T cells. This indicated that distinct miRNAs could regulate uPAR expression depending on the cell type, a proposal consistent with the idea that each cell type harbors a particular miRNA repertoire.
miRNAs repressed by PML-RAR control crucial cancer genes. (A) Luciferase assay performed in 293T cells transfected with the empty renilla luciferase reporter gene psiCHECK2 (dark histograms) or with the reporter fused to the uPAR 3′ UTR (psiCHECK2–3′ UTR uPAR; gray histograms). The cells were also transfected with the pcDNA3 plasmid or the PML-RARA–expressing vector and treated or not with ATRA (1 μM) for 16 hours as indicated. Results are expressed as RLUs, 1 representing the value obtained with the empty psiCHECK2 plasmid in absence of PML-RARA and treatment. (B) RT-qPCR analyses directed against the RARB mRNA and the miR-195 in 293T cells transfected and treated as in panel A. (C) Luciferase assay performed in 293T cells transfected with the empty renilla luciferase reporter gene psiCHECK2 (dark histograms) or with the reporter fused to the EBF-3 3′ UTR (psiCHECK2-3′ UTR EBF-3; gray histograms). Cells were treated and results analyzed as in panel A. (D) LUC assay performed in 293T cells transfected with the psiCHECK2 or psiCHECK2-3′ UTR uPAR together with specific LNA miRNA inhibitors as indicated. LNA anti–miR-32 was used as a negative control. The results are expressed as RLU, 1 being the value obtained with the empty psiCHECK2 plasmid for each LNA treatment (control).
miRNAs repressed by PML-RAR control crucial cancer genes. (A) Luciferase assay performed in 293T cells transfected with the empty renilla luciferase reporter gene psiCHECK2 (dark histograms) or with the reporter fused to the uPAR 3′ UTR (psiCHECK2–3′ UTR uPAR; gray histograms). The cells were also transfected with the pcDNA3 plasmid or the PML-RARA–expressing vector and treated or not with ATRA (1 μM) for 16 hours as indicated. Results are expressed as RLUs, 1 representing the value obtained with the empty psiCHECK2 plasmid in absence of PML-RARA and treatment. (B) RT-qPCR analyses directed against the RARB mRNA and the miR-195 in 293T cells transfected and treated as in panel A. (C) Luciferase assay performed in 293T cells transfected with the empty renilla luciferase reporter gene psiCHECK2 (dark histograms) or with the reporter fused to the EBF-3 3′ UTR (psiCHECK2-3′ UTR EBF-3; gray histograms). Cells were treated and results analyzed as in panel A. (D) LUC assay performed in 293T cells transfected with the psiCHECK2 or psiCHECK2-3′ UTR uPAR together with specific LNA miRNA inhibitors as indicated. LNA anti–miR-32 was used as a negative control. The results are expressed as RLU, 1 being the value obtained with the empty psiCHECK2 plasmid for each LNA treatment (control).
Finally, we also inspected validated miRNA targets whose expression was shown to correlate with some miRNAs of our subset.26 Notably, our bioinformatics and RT-qPCR analyses have revealed that the miR-196a might be repressed by PML-RARA (Table 1 and Figure 1D). This miRNA is known to regulate the expression of the HOXB8 mRNA.37 HOXB8 is also predicted to be repressed by the miR-27a, miR-377, miR-520d, and miR-524, which are validated or potential targets of PML-RARA (Table 1 and Figure 2). Using specific RT-qPCR, we found that the level of this messenger was down-regulated by ATRA in NB4 and NB4-LR1 while remaining unchanged in NB4-LR2 (Figure S5). We also found using specific RT-qPCR that the level of HoxB8 messenger was systematically increased in 293T cells when blocking the miR-377 (Figure S5). This result suggested that HOXB8 expression could be coordinately regulated by several miRNAs, in particular the miR-377 and the miR-196a.37
Discussion
We revealed that PML-RARA is able to transcriptionally repress several miRNA genes. Because the expression of these miRNAs is restored by ATRA and As2O3, our results suggest that clinical protocols, able to successfully eradicate cancer cells, may do so at least in part by impacting miRNA expression. We also showed that the miR-23a/24-2 and miR-210 are physiologically regulated by RA. These findings indicate that, in addition to its canonical properties of transcription regulation, RA, through miRNAs, can indirectly affect posttranscriptional processes such as translation.
Our miRNA microarray analyses and those performed by Garzon et al16 converged to the findings that the miR-15b, miR-223, and miR-342 are up-regulated by ATRA whereas the miR-181a and miR-181b are down-regulated in NB4 cell lines. The diminution of the expression of miR-181a induced by ATRA was confirmed in primary APL blasts (Figure 4E). The expression of this miRNA was recently correlated with a particular subclass of AML as the miR-181a is highly expressed in AML-M1 or AML-M2 compared with AML-M4 or AML-M5.26 No AML-M3 sample was tested in this study.26 As the miR-181a is down-regulated by ATRA (Figures 1D, 3E), our observations might suggest that the expression of the miR-181a is also relevant in the case of AML-M3. The up-regulation of the miR-223 in ATRA-treated NB4 cells was also observed by Fazi et al and was implicated in NB4 cell maturation.17 The miR-223 has also been implicated in AML-M2 leukemia wherein it is transcriptionally silenced by the AML1/ETO oncogene associated with the t(8;21) translocation.38 Interestingly, the let-7a, let-7c, and let-7d miRNAs were found up-regulated by ATRA in APL cells16 and we predicted those miRNAs to be repressed by PML-RARA (Table 1). We decided not to include these miRNAs in our RT-qPCR analyses because we suspected that their high degree of homology could make difficult their discrimination by PCR. However, given the essential functions of the let-7 targets,16,39 it may be interesting to further test whether the let-7a, let-7c, and let-7d are repressed by PML-RARA.
The findings that PML-RARA represses miRNA genes, including miRNAs located in intergenic regions, reveal that the fusion protein affects unsuspected regions of the chromatin. Our results in fact extend previous findings obtained by Hoemme et al who showed, using ChIP to chip approach, that PML-RARA regulates key cancer coding genes.40 The miRNAs repressed by PML-RARA are also implicated in the control of crucial pathways linked to leukemogenesis. For instance, the expression of HOXB8 seemed to be controlled by miRNAs potentially repressed by PML-RARA (Figures 1, S2). RA is known to induce a temporal program of HOX gene expression and this expression is often perturbed in leukemias.25,41-44 Of note, the miR-10b, which is potentially repressed by PML-RARA (Table 1 and Figure 1D), also regulates HOXD10.45 The up-regulation of HOX genes in AML due to the down-regulation of miRNAs has already been suspected26,46,47 but we revealed here a potential link with oncogene-mediated transcriptional repression at least in the case of APL. Likewise, we identified the uPAR-coding mRNA as a target of miRNAs repressed by PML-RARA. High expression of uPAR reflects a significant lower remission rate after chemotherapy and a higher risk for relapse.34 Curiously, Mustjoki et al have shown that 13-trans RA increased uPAR mRNA and protein levels in NB4 cells.48 However, the authors showed that the global effect of RA was a decrease in proteolytic activity due to the activation of plasminogen activator inhibitors (PAIs).48,49 Although we and others32,35 have found that ATRA rather decreases uPAR mRNA in NB4 (Figure 6C), our results are consistent with the observation that the overall outcome of ATRA is a decrease in uPA activity.49 But, in addition to PAIs,49 we showed that the miRNA pathway could also be implicated in this process. This regulation appeared orchestrated by several miRNAs organized in a complex but synchronized network. Although further investigations are required to evaluate the extent of this type of process, our results suggested that coordination may play central role(s) in the action of coregulated miRNAs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to the patients for their contributions. We thank Michel Lanotte for cells and reagents. We are grateful to Valérie Courgnaud, François Bernardin, and Mikalail Yatskou for technical advice and help.
This work was supported by Inserm, CNRS, and the National Research Fund of Luxembourg.
Authorship
Contribution: A.S., G.V., M.B., and C.-H.L. performed molecular analyses; E.P.-C. and W.W. performed bioinformatics; T.M., B.M., and P.B. performed miRNA microarrays; G.V., L.V., and E.F. performed cDNA microarrays; K.A. provided miRNA inhibitors; B.C. and C.C. diagnosed and prepared APL blasts; and A.S., C.C., and C.-H.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charles-Henri Lecellier, Institut de Génétique Moléculaire de Montpellier, CNRS UMR 5535, IFR 122, Université de Montpellier, 1919 Route de Mende, F-34293 Montpellier Cedex 5 France; e-mail: charles.lecellier@igmm.cnrs.fr.

![Figure 2. The miR-23a/24-2 and miR-210 promoters are repressed by PML-RARA. (A) ChIP experiments performed in NB4 cells. Chromatin was immunoprecipitated using the indicated antibodies and the enriched genomic fragments were PCR amplified using specific primers. 223-A and 223-B correspond to the 2 miR-223 promoters described in Fukao et al20 and Fazi et al,17 respectively. The RARB promoter was used as a positive control. (B) qPCR analyses performed on DNA immunoprecipitated in panel A. The negative control corresponds to a sequence located 3.9 kb downstream the miR-23a/24-2 precursor. Fold enrichment over the negative control (no antibody) was calculated using the following formula: 2[(Ct input − Ct IP) − (Ct input − Ct no Ab)]. (C) LUC assays performed in 293T cells transfected with a firefly luciferase reporter gene driven by the miR-23a/24-2 promoter together with the control empty pcDNA3 plasmid (black histograms) or the PML-RARA–expressing vector (gray histograms). A promoterless vector was used as a negative control (pGL3b). A miR-23a/24-2 promoter mutated in the predicted PML-RARA response element was also tested (mutated miR-23a/24-2 promoter). At 24 hours after transfection, 293T cells were treated or not with ATRA (1 μM) for 16 hours. Results are expressed as relative light units (RLUs), 1 representing the value obtained with the promoterless vector pGL3b in absence of PML-RARA and ATRA treatments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/2/10.1182_blood-2008-05-158139/6/m_zh80020929120002.jpeg?Expires=1767704827&Signature=ifqCXTpgtQCaIlSDsYoSzE2ez~e5yKTyIJRXKI9otw6asX385TpdyosbdW1BcI7hI9c35CY~q-3ZJmr6LNVa956~J8P00fYMyNcS-G0CccR9HDb2L6PHxSeRE-B7iULSLqsEcv~Bhfaatk8mF7VyNR-eMqx2VKfB4yIMhIEDUxDwaj2oZjMFjWe5fPtUdzcoE5bozAw4kBrmWIoxXsRcLPgeqHA2iq-9e4xQJdPlE-kdmRG54utSQGo~4lFJKJjhmMy6EuAqeLydmh9Fr~GFI6XmeerUBirjTmJCoNGt4wq5ioc8XAku-nZM6IvLPJ8U7QU5YTy42XYMh-DiHzwyww__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal