Abstract
The CBL ubiquitin ligase targets a variety of activated tyrosine kinases (TKs) for degradation. Many TKs are mutationally or autocrine activated and/or often overexpressed at the mRNA and protein levels in acute leukemias. We hypothesized that CBL is mutated in patients with acute myeloid leukemia (AML). Four of 12 patients and the MOLM-13 cell line harbored c-CBL mutations, either RNA splicing mutations, missense mutations, or a nucleotide insertion. Additionally, 1 of the 12 patients harbored a missense mutation in the related CBL-b gene. Each c-CBL mutation involves the structurally important α-helix within the linker region, while the mutation in CBL-b was located in the Ub-E2 protein-binding RING finger. Short-interfering RNA knockdown of mutant c-CBL present in MOLM-13 cells was growth inhibitory. In summary, novel mutations in c-CBL and CBL-b have been identified in human AML and may represent potential targets for novel therapeutics.
Introduction
The Casitas B-cell lymphoma (CBL) gene gives rise to a protein with multiadaptor function and E3 ubiquitin ligase activity that targets a variety of tyrosine kinases (TKs) for degradation.1-3 c-CBL, and the related family member, CBL-b, contain several functional domains, including a tyrosine kinase-binding domain (TKB), a RING finger (RF) domain, a conserved linker region between the TKB and RF, and a C-terminal domain with ubiquitin ligase activity.
The first described oncogenic mutant of c-CBL, the viral homolog, v-cbl, retains the TKB domain but lacks the RF and the C-terminal domains and induces B-cell lymphoma and myeloid leukemia in mice.4,5 The transforming mutant isolated from a murine pre-B lymphoma cell line (70Z), is an in-frame 17-amino acid deletion arising from a splice site mutation, and leads to partial deletion that includes a linker region conserved in c-CBL and CBL-b that lies between the TKB and RF.6 The murine p95cbl transforming mutant is an in-frame deletion of 111 amino acids due to a splicing mutation and results in loss of the entire RF and a portion of the linker region.7 In the human HUT78 T-cell lymphoma cell line, c-CBL is truncated but keeps an intact linker and RF, and is nontransforming.6,8,9 Lastly, MLL-CBL gene fusions have been reported for 2 acute myeloid leukemia (AML) patients.10,11 To our knowledge, transforming mutations in CBL-b have not been described. Herein, we report several novel monoallelic c-CBL and CBL-b mutations in human AML that involve the linker region and the RF, respectively.
Patients, materials, and methods
MOLM-13, THP-1, EOL-1, MV4-11, and RS4-11 cells were screened for c-CBL mutations. Cryopreserved leukopheresis samples from 12 newly diagnosed AML patients were selected regardless of karyotype after informed consent in accordance with the Declaration of Helsinki and with Ohio State University institutional review board approval. All had 80% to 100% circulating blasts. Highly enriched CD34+ BM cells from consented healthy volunteers were obtained using magnetic bead separation.
Reverse-transcription-polymerase chain reaction (RT-PCR) was performed with primer pairs for c-CBL: e2F (5′-TCAGCCTAGGCGAAACCTAA) and e9R (5′-TCCCTCTAGGATCAAACGGA), and for CBL-b: e10F (5′-CCGGTTAAGTTGCACTCGAT) and e13R (5′-CAAAGGGGTCCACGATTATG). For c-CBL, PCR conditions were 95°C for 5 minutes; 27 cycles of 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 1 minute; one cycle of 72°C for 10 minutes. For CBL-b, PCR conditions were 95°C for 5 minutes; 27 cycles of 95°C for 30 seconds, 59°C for 30 seconds, and 72°C for 30 seconds; one cycle at 72°C for 7 minutes. Cloned sequences were analyzed using the Basic Local Alignment Search Tool at http://www.ncbi.nlm.nih.gov/blast/ (National Center for Biotechnology Information).
The MolD8CBL siRNA targeting the exon 7-exon 9 junction of the MOLM-13 mutant c-CBL and scrambled (Scr) oligonucleotides were designed using tools at http://www.ambion.com (Ambion).
Total proteins were separated through a 4% to 15% SDS-PA gel, transferred to nitrocellulose, and incubated with anti-c-CBL (C-15; Santa Cruz Biotechnology, Santa Cruz, CA).
MTS cell proliferation assays were carried out 48 hours after siRNA transfection of MOLM-13 cells (CellTiter96 Proliferation Assay; Promega, Madison, WI). Analysis of variance and Tukey pairwise comparison tests were performed with a 2-sided α-level = .05 (GraphPad Prism 4.0; GraphPad Software, San Diego, CA).
Results and discussion
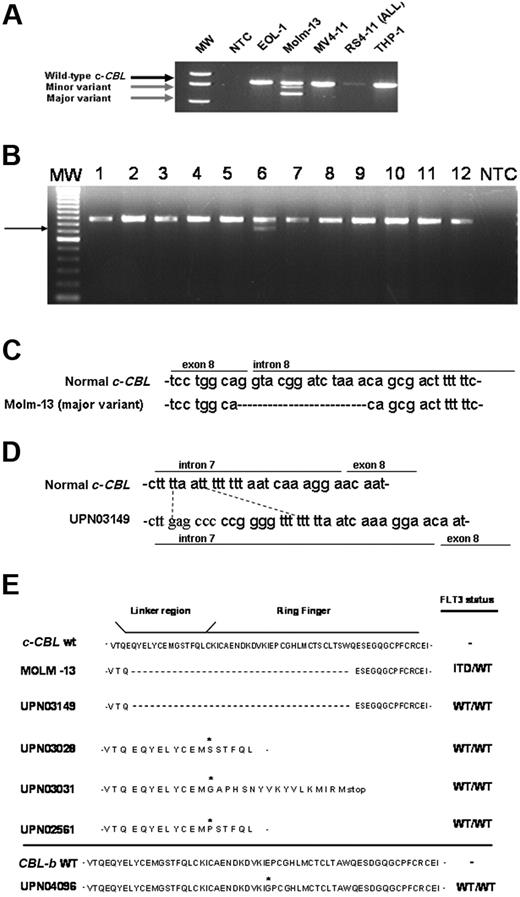
Of the cell lines evaluated, only MOLM-13 cells expressed c-CBL transcript size variants (Figure 1A). The shortest transcript lacks exon 8 and the minor variant resulted from an alternative donor-splice site (not shown). One of 12 AML patient samples screened expressed a WT c-CBL transcript and a smaller transcript lacking exon 8 (Figure 1B, UPN03149, lane 6). No mutations were found in the linker region of the related family member, CBL-b. However, one patient having normal c-CBL harbored a missense mutation in the CBL-b RF, changing a glutamate to a glycine residue (Figure 1E). No mutations in either gene were found in 5 normal CD34+ BM samples (not shown).
Aberrant CBL transcripts in AML. (A) RT-PCR screening of AML cell lines and the RS(4;11) acute lymphoblastic leukemia (ALL) cell line was carried out as described in “Patients, materials, and methods.” Gel electrophoresis of resulting amplification products using a primer set covering human c-CBL exons 2 through 9 revealed major and minor transcript variants (gray arrows) in the MOLM-13 cell line. MW indicates 1 kb + DNA size ladder; NTC, no template control reaction. (B) RT-PCR screening for c-CBL transcript was performed using AML patient samples (lanes 1-12). A representative agarose gel is shown. (Lane 6) UPN03149 expresses a smaller transcript variant (arrow) and the wild-type mRNA for c-CBL. MW indicates 1 kb + DNA size marker; NTC, no template control reaction. (C) Partial sequence alignment of gDNA PCR amplification products in MOLM-13 compared with the normal c-CBL gene sequence. MOLM-13 is missing 14 bp at the exon 8-intron 8 boundary, including the normal “G” donor site. (D) Partial sequence alignment of gDNA PCR amplification products in UPN03149 relative to wild-type c-CBL DNA. UPN03419 is missing 4 bp that is replaced with a 12-bp sequence. (E) The predicted amino acid sequences derived from the region of interest of mutant CBL transcripts are shown in comparison with the corresponding CBL WT sequence. FLT3 TK allelic status, that is, absence (WT/WT) or presence of internal tandem duplication (FLT3-ITD) or activating domain mutations (FLT3-TKD), was determined for each case of AML and is shown on the right. Asterisk indicates location of missense mutation or nucleotide insertion that gives rise to an amino acid change.
Aberrant CBL transcripts in AML. (A) RT-PCR screening of AML cell lines and the RS(4;11) acute lymphoblastic leukemia (ALL) cell line was carried out as described in “Patients, materials, and methods.” Gel electrophoresis of resulting amplification products using a primer set covering human c-CBL exons 2 through 9 revealed major and minor transcript variants (gray arrows) in the MOLM-13 cell line. MW indicates 1 kb + DNA size ladder; NTC, no template control reaction. (B) RT-PCR screening for c-CBL transcript was performed using AML patient samples (lanes 1-12). A representative agarose gel is shown. (Lane 6) UPN03149 expresses a smaller transcript variant (arrow) and the wild-type mRNA for c-CBL. MW indicates 1 kb + DNA size marker; NTC, no template control reaction. (C) Partial sequence alignment of gDNA PCR amplification products in MOLM-13 compared with the normal c-CBL gene sequence. MOLM-13 is missing 14 bp at the exon 8-intron 8 boundary, including the normal “G” donor site. (D) Partial sequence alignment of gDNA PCR amplification products in UPN03149 relative to wild-type c-CBL DNA. UPN03419 is missing 4 bp that is replaced with a 12-bp sequence. (E) The predicted amino acid sequences derived from the region of interest of mutant CBL transcripts are shown in comparison with the corresponding CBL WT sequence. FLT3 TK allelic status, that is, absence (WT/WT) or presence of internal tandem duplication (FLT3-ITD) or activating domain mutations (FLT3-TKD), was determined for each case of AML and is shown on the right. Asterisk indicates location of missense mutation or nucleotide insertion that gives rise to an amino acid change.
A splice site mutation in one c-CBL allele was observed in MOLM-13 (Figure 1C). In UPN03149, one allele carried a deletion-insertion near the intron 7-exon 8 junction (Figure 1D). This mutation may function as an “intronic splicing silencer” element, an entity described previously for other genes.12,13 Three additional patient samples (UPN03028, UPN03031, and UPN02561) each had single missense mutations involving the same codon within an α-helical structure of the linker region (Figure 1E). Two important tyrosine residues lie within this linker region.14 Phosphorylation of one or both tyrosine residues occurs in response to activated TKs.15-18 This phosphorylation can lead to CBL binding to the activated TK and subsequent E3 Ub ligase activity followed by receptor internalization and degradation.15,19 The deletion of one or both tyrosines induces a conformational change that alters one or more c-CBL-associated events.6,8,19 It remains to be formally established whether the mutations identified in the 3 additional patients disrupt the α-helical structure and alter the function of c-CBL.
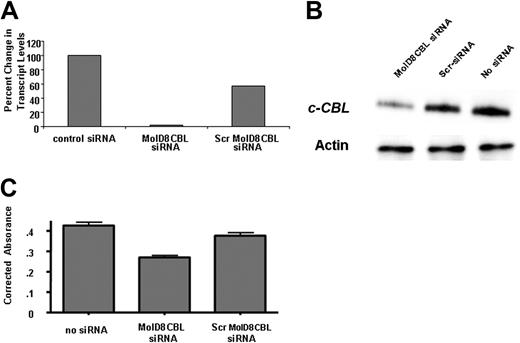
siRNA oligonucleotides targeting the mutant c-CBL (MolD8CBL-siRNA) in MOLM-13 cells reduced mutant mRNA and protein levels (Figure 2A-B). This reduction in mutant CBL had a modest effect on proliferation when comparing cells transfected with MolD8CBL-siRNA, scrambled RNA (Scr-siRNA), or control siRNA (ANOVA, P < .001). An approximately 36% reduction in number of cells exposed to MolD8CBL-siRNA was observed when compared with cells exposed to Scr-siRNA (P < .01) (Figure 2C). No significant difference in viable cell number between no siRNA control and Scr-siRNA-transfected cells was observed (P > .05).
siRNA-mediated reduction in MOLM-13-expressed mutant c-CBL transcript and protein is associated with reduced cell proliferation. (A) MOLM-13 cells were transfected (Nucleofector; Amaxa, Gaithersburg, MD) with siRNAs targeting the MOLM-13 c-CBL mutant transcript fusing exon 7 and exon 9 (siMolD8CBL) or a scrambled sequence (scrMolD8CBL) or transfected with “control siRNA” that has been validated to not affect mRNA levels in mammalian cells (Ambion, Austin, TX) and incubated at 37°C for 24 hours. Relative real-time RT-PCR was carried out using total RNA and mutant-specific primers and primers common to mutant and wild-type c-CBL transcripts (sequences available upon written request). 18S rRNA was used to normalize for starting template amounts. Expression of c-CBL WT or mutant transcript in each transfected sample was measured. Data are depicted as percent change in the mutant transcript level relative to the level of mutant in the control siRNA-transfected cells. No difference in c-CBL WT mRNA levels was observed (not shown). siRNA sequences are available upon request. (B) Immunoblot analysis of c-CBL protein in MOLM-13 cells transfected with no siRNA or either Scr or MolD8CBL siRNAs. Detection of actin was used to control for well loading. (C) MTS cell proliferation assay was carried out at 48 hours after transfection of MOLM-13 cells without (no siRNA) or with 1 nmol MolD8CBL or Scr siRNA. Viability is indicated as the average absorbance at 492 nm of triplicate wells corrected for background and relative to the 0-hour time point.
siRNA-mediated reduction in MOLM-13-expressed mutant c-CBL transcript and protein is associated with reduced cell proliferation. (A) MOLM-13 cells were transfected (Nucleofector; Amaxa, Gaithersburg, MD) with siRNAs targeting the MOLM-13 c-CBL mutant transcript fusing exon 7 and exon 9 (siMolD8CBL) or a scrambled sequence (scrMolD8CBL) or transfected with “control siRNA” that has been validated to not affect mRNA levels in mammalian cells (Ambion, Austin, TX) and incubated at 37°C for 24 hours. Relative real-time RT-PCR was carried out using total RNA and mutant-specific primers and primers common to mutant and wild-type c-CBL transcripts (sequences available upon written request). 18S rRNA was used to normalize for starting template amounts. Expression of c-CBL WT or mutant transcript in each transfected sample was measured. Data are depicted as percent change in the mutant transcript level relative to the level of mutant in the control siRNA-transfected cells. No difference in c-CBL WT mRNA levels was observed (not shown). siRNA sequences are available upon request. (B) Immunoblot analysis of c-CBL protein in MOLM-13 cells transfected with no siRNA or either Scr or MolD8CBL siRNAs. Detection of actin was used to control for well loading. (C) MTS cell proliferation assay was carried out at 48 hours after transfection of MOLM-13 cells without (no siRNA) or with 1 nmol MolD8CBL or Scr siRNA. Viability is indicated as the average absorbance at 492 nm of triplicate wells corrected for background and relative to the 0-hour time point.
In summary, we have discovered novel c-CBL linker region gene mutations and a novel mutation in the RF of CBL-b in human AML. The linker sequence is critical to maintaining the interaction between the TKB and RF domain that is necessary for CBL's negative regulatory function.8,20 Disruption of this linker sequence renders CBL unable to degrade RTKs and leads to enhanced proliferation and survival signaling (eg, the murine p70Z mutant renders 32D cells growth-factor independent and induces sustained levels of tyrosine kinases).21 Ectopic expression of p70Z CBL inhibits endogenous CBL function, suggesting a dominant-negative effect by the mutant CBL.22 An amino acid sequence comparison of wild-type (WT) CBL with that of v-cbl, murine p70Z CBL, and the predicted amino acid sequences encoded by the human MOLM-13 and (UPN03149) AML mutant CBL mRNAs indicates a loss of the majority of the linker region sequence, including the 2 critical tyrosine residues, in these 2 samples (Figure 1E).
Thus, one would predict that the CBL mutations present in the human AML blasts would inhibit down-regulation of activated TKs. Several TKs (eg, FLT3 and KIT) have critical functions in normal hematopoiesis and are overexpressed and/or constitutively active without mutational activation in AML.8,20 While c-CBL and CBL-b regulate KIT,23 it was not clear if they modulate FLT3 in AML. Concurrent with this report, Sargin et al used the murine 32D model to show that c-CBL WT binds to FLT3, resulting in its degradation and a diminution in downstream signaling.24 A novel transforming c-CBL RF mutant identified in an AML patient24 does bind FLT3 yet sustains activated FLT3 levels as well as the downstream signaling mediators, ERK1/2 and AKT. The oncogenic role of the mutations we detected in MOLM-13 and UPN03149 remains unproven, as does the functional relevance of the mutations we detected in the remaining AML samples. Moreover, an investigation of mutant CBL on other activated TKs in AML is warranted. The full elucidation of the role of mutant CBL proteins in the Ub-mediated proteolytic pathway should provide additional insight into the molecular heterogeneity of AML.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded in part by NCI grant funding R01 CA89341 and P30 CA16058 (M.A.C.) and K01 CA96887 (S.P.W.).
We express our gratitude to Dr Hamid Band, who provided us with the pAlterMax plasmid expressing c-CBL WT. We thank The Ohio State University Comprehensive Cancer Center's Nucleic Acid Shared Resource for sequencing and real-time RT-PCR support.
National Institutes of Health
Authorship
Contribution: S.P.W. designed and supervised research, analyzed data, and cowrote the paper; M.A.C. contributed human samples and cowrote the paper; C.D.B. provided patient AML samples; R.B. provided technical and intellectual expertise; L.W., J.Y., M.W., K.J.A., T.B.M., and J.W. performed research; and D.P. contributed intellectual expertise.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: S. P. Whitman, 2001 Polaris Pkwy, The Ohio State University Comprehensive Cancer Center, Columbus, OH 43240; e-mail: susan.whitman@osumc.edu.