Abstract
To molecularly define high-risk disease, we performed microarray analysis on tumor cells from 532 newly diagnosed patients with multiple myeloma (MM) treated on 2 separate protocols. Using log-rank tests of expression quartiles, 70 genes, 30% mapping to chromosome 1 (P < .001), were linked to early disease-related death. Importantly, most up-regulated genes mapped to chromosome 1q, and down-regulated genes mapped to chromosome 1p. The ratio of mean expression levels of up-regulated to down-regulated genes defined a high-risk score present in 13% of patients with shorter durations of complete remission, event-free survival, and overall survival (training set: hazard ratio [HR], 5.16; P < .001; test cohort: HR, 4.75; P < .001). The high-risk score also was an independent predictor of outcome endpoints in multivariate analysis (P < .001) that included the International Staging System and high-risk translocations. In a comparison of paired baseline and relapse samples, the high-risk score frequency rose to 76% at relapse and predicted short postrelapse survival (P < .05). Multivariate discriminant analysis revealed that a 17-gene subset could predict outcome as well as the 70-gene model. Our data suggest that altered transcriptional regulation of genes mapping to chromosome 1 may contribute to disease progression, and that expression profiling can be used to identify high-risk disease and guide therapeutic interventions.
Introduction
Multiple myeloma (MM), a malignancy of terminally differentiated plasma cells homing to and expanding in the bone marrow, is characterized by a tremendous heterogeneity in outcome following standard and high-dose therapies. Although many of the genetic and molecular lesions associated with disease initiation are known, the lesions that promote an aggressive clinical course have remained elusive.
All myelomas can be broadly divided into hyperdiploid and nonhyperdiploid disease.1–4 Hyperdiploidy, typically associated with trisomies of chromosomes 3, 5, 9, 11, 15, 19, and 21, is present in approximately 60% of patients.5 Unsupervised clustering and nonnegative matrix factorization of high-resolution ologonucelotide array comparative genomic hybridization (aCGH) data has revealed hyperdiploid myeloma can be further segregated into 2 groups, one exhibiting trisomies of the odd chromosomes listed here and another exhibiting, in addition, gains of chromosomes 1q and 7, deletion of chromosome 13, and absence of trisomy 11.6 Nonhyperdiploid myeloma can also be divided into 2 groups, one characterized by high-level amplification of chromosome 1q and deletions of chromosomes 1p and 13, and another characterized by the absence of chromosome 1 abnormalities but which harbors deletions of chromosomes 8 and 13.6 Furthermore, transcriptional activation of CCND1, CCND3, MAF, MAFB, or FGFR3/MMSET (resulting from translocations involving the immunoglobulin heavy chain locus on chromosome 14q32) is typical of nonhyperdiploid myeloma and is present in approximately 40% of patients.5,7,8
Using unsupervised hierarchical clustering of global gene expression patterns, we recently defined and validated the existence of 7 myeloma subgroups exhibiting strong correlations with hyperdiploidy and recurrent translocations.9 In this study, 2 high-risk entities were identified, one revealing overexpression of proliferation genes and derived from cases evolving from the other 6 classes, while the other was defined by the t(4;14)(p16;q32) translocation.9
Gains of the long arm of chromosome 1 (1q) are one of the most common genetic abnormalities in myeloma.10 Tandem duplications and jumping segmental duplications of the chromosome 1q band, resulting from decondensation of pericentromeric heterochromatin, are frequently associated with disease progression.11–13 Using aCGH on DNA isolated from plasma cells derived from patients with smoldering myeloma, Rosinol and colleagues showed that the risk of conversion to overt disease was linked to gains of 1q21 and loss of chromosome 13.14 Using interphase fluorescence in situ hybridization (FISH) analysis, we confirmed these findings. In addition, we showed that gains of 1q21 acquired in symptomatic myeloma were linked to inferior survival and were further amplified at disease relapse.15
We now report on gene expression profiling (GEP) of purified myeloma plasma cells obtained prior to initiation of therapy in 2 large, principally similarly treated, cohorts of patients with myeloma to identify a signature associated with short survival. Elevated expression levels of genes mapping to chromosome 1q and reduced expression levels of genes mapping to 1p constituted a high-risk score present in a small group of 13% of patients with very short survival.
Materials and methods
Patients
Purified plasma cells were obtained from normal healthy subjects and from patients with monoclonal gammopathy of undetermined significance (MGUS) and with overt myeloma requiring therapy. Patient characteristics of training (n = 351) and validation (n = 181) groups have been previously described.9 Of 351 patients in the training group, 51 also had samples taken at relapse. Both training and validation groups were treated on National Institutes of Health (NIH)–sponsored clinical trials UARK 98-026 and UARK 03-033, respectively. Both protocols used induction regimens followed by melphalan-based tandem autotransplantations, consolidation chemotherapy, and maintenance treatment. The Institutional Review Board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent approving use of their samples for research purposes.
GEP
Plasma cell purifications and GEP, using the Affymetrix U133Plus2.0 microarray (Santa Clara, CA), were performed as previously described.9,16 Microarray data and outcome data on the 532 patients used in this study have been deposited in the NIH Gene Expression Omnibus17 under accession number GSE2658.
Statistical and microarray analyses
Affymetrix U133Plus2.0 microarrays were preprocessed using GCOS1.1 software (Affymetrix, Santa Clara, CA) and normalized using conventional GCOS1.1 scaling. Log-rank tests for univariate association with disease-related survival were performed for each of the 54 675 “signal” summaries. Specifically, log-rank tests were performed for quartile 1 (Q1) versus Q2 through Q4 and Q4 versus Q1 through Q3 in order to identify under- and overexpressed prognostic genes, respectively. A false discovery rate cutoff of 2.5% was applied to each list of log-rank P values18 yielding 19 underexpressed and 51 overexpressed probe sets. Heat map–column dendrograms were computed with hierarchical clustering using Pearson correlation distances between patient pairs' log2-scale expression. Column-dendrogram branches were sorted left to right based upon each patient's difference between the average log2-scale expression of the 51 up-regulated and the 19 down-regulated genes: this difference is interpreted as an up-/down-regulated mean ratio (ie, geometric mean) on the log2 scale. This simple, univariate summary of the 70-gene expression profile for each patient may enhance robustness to residual array effects (ie, after MAS5.0 processing) that increase or decrease all 70 genes multiplicatively, and is also independent of the MAS5.0 scale factor. Weighting expression by hazard ratios, unstandardized or standardized (ie, Wald statistics), does not improve this score, and our design was to use no supervision by overall survival (OS) or event-free survival (EFS) beyond the gene-by-gene log-rank tests. We then clustered the log2 up-/down-regulated mean ratio using K-means into 3 groups to separate out the small extreme right mode in the histogram: the 2 groups with lower up/down mean ratios were combined. The single extreme mode in the up/down mean expression ratio is consistent with the extreme quartile log-rank tests used in the differential expression analysis, though the histograms and the right-hand side of the heat maps suggest that the extreme patient group is smaller than 25% (closer to 13%). Note that different clustering algorithms and numbers of groups generate high mean ratio groups between 12% and 29% of patients: we chose K-means (with K = 3) since it was best (ie, among simple algorithms for the univariate log2 ratio) at separating the small right-hand mode from the larger distribution. Any univariate cutoff capturing between 10% and 30% patients is significant for OS in the 351 patient training set. In the 181 patient validation set, K-means clustering was performed independently to produce an independent cutoff for high versus low log2 ratios. Application of the training set cutoff in the validation set provides an independently validated classification error of 1.7% (ie, 3 patients in the low-risk validation set are classified as high risk). We present an early validation based upon an independent cohort treated under a newer protocol in order to illustrate and provide strong supporting evidence for the association of the 70 gene up-/down-regulated mean ratio with OS. We expect the high-risk cutoff for the mean ratio to be associated with survival broadly in newly diagnosed patients, regardless of protocol, so that the difference in protocol for the validation set strengthens the evidence rather than weakening it. The mean ratio may also be associated with outcome in previously treated patients; however, new cutoffs for the ratio would be required to define a high-risk group. An important caveat is that the 70 genes are not particularly suited to explaining outcome among the lower two-thirds of patients (ranked by the mean ratio): this is consistent with the original log-rank screens, which lumped 75% of the patients into a single group for the Q1 and Q4 log-rank tests: these genes identify the most aggressive myeloma plasma cells, by design.
To determine the exact genome map location and order of the probe sets on the Affymetrix U133Plus2.0 microarray, software was developed to automatically query the National Center for Biotechnology Information (NCBI) search engine (http://www.ncbi.nlm.nih.gov/entrez) for all gene start and end sites. The location of each probe set was then compared with its corresponding gene or transcript start point and aligned from the p-arm telomere to q-arm telomere. In this manner, more than 98% (53 581 of 54 675) probe sets were given an exact chromosome position.
Distributions of EFS, OS, and duration of complete remission (dated from onset of complete response) were estimated using the Kaplan-Meier method,19 and log-rank statistics were used to test for their equality across groups.20 Chi-square tests and Fisher exact tests were used to test for the independence of categories. Multivariate proportional hazards analyses, adjusted the effects of predictors and the proportions of observed heterogeneity explained by the combined predictors (ie, R2), were computed.21 Table 5 summarizes a multivariate linear-regression analysis of the log2-scale up-/down-regulation ratio. The statistical package R version 2.0.122 was used for this analysis.
A stepwise multiple linear discriminant analysis (MSDA) with the Wilk lambda criterion23 was used to select a subset of the 70 genes equally capable of differentiating high-risk and low-risk MM. The MSDA selected the following equation: Discriminant score = 200 638_s_at × 0.283 − 1 557 277_a_at × 0.296 × 200 850_s_at × 0.208 + 201 897_s_at × 0.314 × 202 729_s_at × 0.287 + 203 432_at × 0.251 + 204 016_at × 0.193 + 205 235_s_at × 0.269 + 206 364_at × 0.375 + 206 513_at × 0.158 + 211 576_s_at × 0.316 + 213 607_at × 0.232 − 213 628_at × 0.251 − 218 924_s_at × 0.230 − 219 918_s_at × 0.402 + 220 789_s_at × 0.191 + 242 488_at × 0.148 (where the variables represent the Affymetrix value for the particular probe). The cutoff value was 1.5, such that values less than 1.5 indicated the sample belonged to the low-risk group, and values more than 1.5 indicated the sample belonged to the high-risk MM group. Both forward and backward variable selections were performed. The choice to enter or remove variables was based on minimizing the within group variability with respect to the total variability across all the samples.
Results
Gene expression patterns are an independent predictor of survival in myeloma
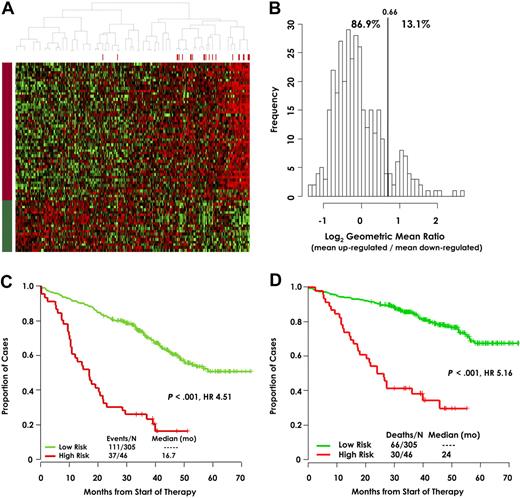
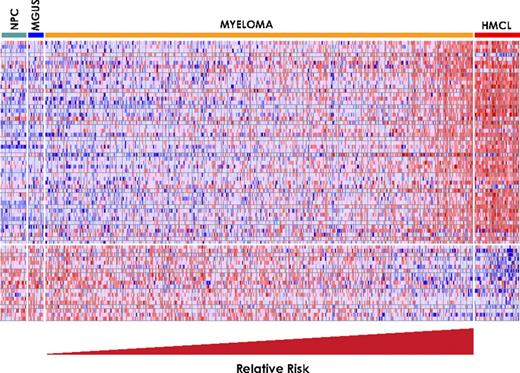
To identify a distinctive molecular signature of high-risk myeloma, we correlated early disease-related death with gene expression extremes. Gene expression levels from microarray data on CD138-selected plasma cells from 351 newly diagnosed patients were divided into quartiles, and log-rank tests were used to identify 70 genes that were linked to short survival: 51 had high expression (Q4) and 19 had low expression (Q1) (Table 1), the expression levels of which are depicted in a colorgram (Figure 1A). Noteworthy is the simultaneous up-regulation of the 51 genes and down-regulation of the 19 genes among the patients on the right-hand side. We therefore calculated the difference between the averages of Q4 and Q1 log2-scale expression for each patient. This unsupervised expression summary is interpretable as a log2-scale up- versus down-regulated mean expression ratio (referred to as a risk score). Its frequency distribution reveals a distinct group having high log2 up-/down-regulation ratios (Figure 1B). This is precisely the kind of extreme-expression group that Q1 and Q4 log-rank tests were designed to screen for, though both the frequency plot and heat map suggest that the group's size is smaller than 25%. Unsupervised K-means clustering of the log2 ratio estimated its proportion at 13.4%. This group exhibited significantly poorer EFS (Figure 1C; P < .001), with an unadjusted HR of 4.51, and also inferior OS (Figure 1D; P < 0.001), with an unadjusted HR of 5.16. Significant associations are expected for the training cohort, in whom the 70 genes were discovered, and they are reported for illustration. The early disease-related death outcome was chosen specifically for the purpose of identifying target genes in aggressive myeloma and, consequently, only 24 deaths were available for the log-rank tests used for gene discovery in the original cohort of 351 patients. Supervised clustering with the 70 genes was applied to plasma cells from 22 healthy donors, 14 patients with MGUS, 351 patients of the training cohort, and 38 human myeloma cell lines. Results revealed that the low-risk myeloma group had a pattern similar to that of MGUS and normal plasma cells, while the high-risk group exhibited a pattern similar to that of human myeloma cell lines (Figure 2).
List of genes comprising the 70-gene high-risk signature
| Rank . | Chromosome . | Affymetrix probe set . | Gene symbol . |
|---|---|---|---|
| Q4 | |||
| 1 | 8q21.13 | 202345_s_at | FABP5 |
| 2 | Xp22.12 | 1555864_s_at | PDHA1 |
| 3 | 5p15.33 | 204033_at | TRIP13 |
| 4 | 1q22 | 206513_at | AIM2 |
| 5 | 2p24.1 | 1555274_a_at | SELI |
| 6 | 21q22.3 | 211576_s_at | SLCI19A1 |
| 7 | 3p21.3 | 204016_at | LARS2 |
| 8 | 1q43 | 1565951_s_at | OPN3 |
| 9 | 1q31.3 | 219918_s_at | ASPM |
| 10 | 12q15 | 201947_s_at | CCT2 |
| 11 | 16p13.3 | 213535_s_at | UBE2I |
| 12 | 20q13.31 | 204092_s_at | STK6 |
| 13 | 1p36.33 | 213607_x_at | FLJ13052 |
| 14 | Xq12 | 208117_s_at | LAS1L |
| 15 | 17q25 | 210334_x_at | BIRC5 |
| 16 | 3q27 | 204023_at | RFC4 |
| 17 | 1q21.2 | 201897_s_at | CKS1B |
| 18 | 19q13.12 | 216194_s_at | CKAP1 |
| 19 | 1p11 | 225834_at | MGC57827 |
| 20 | 19q13.12 | 238952_x_at | DKFZp779O175 |
| 21 | 17p13.3 | 200634_at | PFN1 |
| 22 | 19p13.2 | 208931_s_at | ILF3 |
| 23 | 1q22 | 206332_s_at | IFI16 |
| 24 | 7p13 | 220789_s_at | TBRG4 |
| 25 | 10p11.23 | 218947_s_at | PAPD1 |
| 26 | 8q24 | 213310_at | EIF2C2 |
| 27 | 3q12.1 | 224523_s_at | MGC4308 |
| 28 | 1p36.13 | 201231_s_at | ENO1 |
| 29 | 18q12.1 | 217901_at | DSG2 |
| 30 | 6q22 | 226936_at | C6orf173 |
| 31 | 8q24.3 | 58696_at | EXOSC4 |
| 32 | 1q23.3 | 200916_at | TAGLN2 |
| 33 | 3q21 | 201614_s_at | RUVBL1 |
| 34 | 16p11.2 | 200966_x_at | ALDOA |
| 35 | 2p25.1 | 225082_at | CPSF3 |
| 36 | 1q43 | 242488_at | NA |
| 37 | 3q12.3 | 243011_at | MGC15606 |
| 38 | 22q13.1 | 201105_at | LGALS1 |
| 39 | 3p25.3 | 224200_s_at | RAD18 |
| 40 | 20p11 | 222417_s_at | SNX5 |
| 41 | 1q21.2 | 210460_s_at | PSMD4 |
| 42 | 12q24.3 | 200750_s_at | RAN |
| 43 | 1q32.1 | 206364_at | KIF14 |
| 44 | 7p15.2 | 201091_s_at | CBX3 |
| 45 | 12q22 | 203432_at | TMPO |
| 46 | 17q24.2 | 221970_s_at | DKFZP586L0724 |
| 47 | 11p15.4 | 212533_at | WEE1 |
| 48 | 3p12 | 213194_at | ROBO1 |
| 49 | 5q33.1 | 244686_at | TCOF1 |
| 50 | 8q23.1 | 200638_s_at | YWHAZ |
| 51 | 10q23.31 | 205235_s_at | MPHOSPH1 |
| Q1 | |||
| 1 | 9q31.3 | 201921_at | GNG10 |
| 2 | 1p13 | 227278_at | NA |
| 3 | Xp22.3 | 209740_s_at | PNPLA4 |
| 4 | 20q11.21 | 227547_at | NA |
| 5 | 10q25.1 | 225582_at | KIAA1754 |
| 6 | 1p13.2 | 200850_s_at | AHCYL1 |
| 7 | 1p13.3 | 213628_at | MCLC |
| 8 | 1p22 | 209717_at | EVI5 |
| 9 | 1p13.3 | 222495_at | AD-020 |
| 10 | 6p21.31 | 1557277_a_at | NA |
| 11 | 1p22.1 | 1554736_at | PARG1 |
| 12 | 1p22 | 218924_s_at | CTBS |
| 13 | 9p13.2 | 226954_at | UBE2R2 |
| 14 | 1p34 | 202838_at | FUCA1 |
| 15 | 13q14 | 230192_at | RFP2 |
| 16 | 12q13.11 | 48106_at | FLJ20489 |
| 17 | 11q13.1 | 237964_at | NA |
| 18 | 2p22.3 | 202729_s_at | LTBP1 |
| 19 | 1p13.1 | 212435_at | TRIM33 |
| Rank . | Chromosome . | Affymetrix probe set . | Gene symbol . |
|---|---|---|---|
| Q4 | |||
| 1 | 8q21.13 | 202345_s_at | FABP5 |
| 2 | Xp22.12 | 1555864_s_at | PDHA1 |
| 3 | 5p15.33 | 204033_at | TRIP13 |
| 4 | 1q22 | 206513_at | AIM2 |
| 5 | 2p24.1 | 1555274_a_at | SELI |
| 6 | 21q22.3 | 211576_s_at | SLCI19A1 |
| 7 | 3p21.3 | 204016_at | LARS2 |
| 8 | 1q43 | 1565951_s_at | OPN3 |
| 9 | 1q31.3 | 219918_s_at | ASPM |
| 10 | 12q15 | 201947_s_at | CCT2 |
| 11 | 16p13.3 | 213535_s_at | UBE2I |
| 12 | 20q13.31 | 204092_s_at | STK6 |
| 13 | 1p36.33 | 213607_x_at | FLJ13052 |
| 14 | Xq12 | 208117_s_at | LAS1L |
| 15 | 17q25 | 210334_x_at | BIRC5 |
| 16 | 3q27 | 204023_at | RFC4 |
| 17 | 1q21.2 | 201897_s_at | CKS1B |
| 18 | 19q13.12 | 216194_s_at | CKAP1 |
| 19 | 1p11 | 225834_at | MGC57827 |
| 20 | 19q13.12 | 238952_x_at | DKFZp779O175 |
| 21 | 17p13.3 | 200634_at | PFN1 |
| 22 | 19p13.2 | 208931_s_at | ILF3 |
| 23 | 1q22 | 206332_s_at | IFI16 |
| 24 | 7p13 | 220789_s_at | TBRG4 |
| 25 | 10p11.23 | 218947_s_at | PAPD1 |
| 26 | 8q24 | 213310_at | EIF2C2 |
| 27 | 3q12.1 | 224523_s_at | MGC4308 |
| 28 | 1p36.13 | 201231_s_at | ENO1 |
| 29 | 18q12.1 | 217901_at | DSG2 |
| 30 | 6q22 | 226936_at | C6orf173 |
| 31 | 8q24.3 | 58696_at | EXOSC4 |
| 32 | 1q23.3 | 200916_at | TAGLN2 |
| 33 | 3q21 | 201614_s_at | RUVBL1 |
| 34 | 16p11.2 | 200966_x_at | ALDOA |
| 35 | 2p25.1 | 225082_at | CPSF3 |
| 36 | 1q43 | 242488_at | NA |
| 37 | 3q12.3 | 243011_at | MGC15606 |
| 38 | 22q13.1 | 201105_at | LGALS1 |
| 39 | 3p25.3 | 224200_s_at | RAD18 |
| 40 | 20p11 | 222417_s_at | SNX5 |
| 41 | 1q21.2 | 210460_s_at | PSMD4 |
| 42 | 12q24.3 | 200750_s_at | RAN |
| 43 | 1q32.1 | 206364_at | KIF14 |
| 44 | 7p15.2 | 201091_s_at | CBX3 |
| 45 | 12q22 | 203432_at | TMPO |
| 46 | 17q24.2 | 221970_s_at | DKFZP586L0724 |
| 47 | 11p15.4 | 212533_at | WEE1 |
| 48 | 3p12 | 213194_at | ROBO1 |
| 49 | 5q33.1 | 244686_at | TCOF1 |
| 50 | 8q23.1 | 200638_s_at | YWHAZ |
| 51 | 10q23.31 | 205235_s_at | MPHOSPH1 |
| Q1 | |||
| 1 | 9q31.3 | 201921_at | GNG10 |
| 2 | 1p13 | 227278_at | NA |
| 3 | Xp22.3 | 209740_s_at | PNPLA4 |
| 4 | 20q11.21 | 227547_at | NA |
| 5 | 10q25.1 | 225582_at | KIAA1754 |
| 6 | 1p13.2 | 200850_s_at | AHCYL1 |
| 7 | 1p13.3 | 213628_at | MCLC |
| 8 | 1p22 | 209717_at | EVI5 |
| 9 | 1p13.3 | 222495_at | AD-020 |
| 10 | 6p21.31 | 1557277_a_at | NA |
| 11 | 1p22.1 | 1554736_at | PARG1 |
| 12 | 1p22 | 218924_s_at | CTBS |
| 13 | 9p13.2 | 226954_at | UBE2R2 |
| 14 | 1p34 | 202838_at | FUCA1 |
| 15 | 13q14 | 230192_at | RFP2 |
| 16 | 12q13.11 | 48106_at | FLJ20489 |
| 17 | 11q13.1 | 237964_at | NA |
| 18 | 2p22.3 | 202729_s_at | LTBP1 |
| 19 | 1p13.1 | 212435_at | TRIM33 |
Gene expression patterns can distinguish risk groups in training cohort. (A) Heat maps of the 70 genes illustrate remarkably similar expression patterns among 351 newly diagnosed patients used to identify the 70 genes. Red bars above the patient columns denote patients with disease-related deaths. The 51 genes in rows designated by the red bar on the left (top rows; up-regulated) identified patients in the upper quartile of expression at high risk for early disease-related death. The 19 gene rows designated by the green bar (down-regulated), identified patients in the lower quartile of expression at high risk of early disease-related death. (B) Training cohort frequencies for sample differences between ratios of the mean of log2 expression of the 51 up-regulated genes/19 down-regulated genes. This self-normalizing expression ratio has a marked bimodal distribution, consistent with the upper/lower quartile log-rank differential expression analysis, which was designed to detect genes that define a single high-risk group (13.1%) with an extreme expression distribution. Interpreted as an up/down-regulation ratio on the log2 scale, higher values are associated with poor outcome. The vertical line shows the high-risk versus low-risk cutoff for the log2-scale ratio determined by K-means clustering: the percentage of samples below and above the cutoff is also shown. Kaplan-Meier estimates of EFS (C) and OS (D) in low-risk myeloma (green) and high-risk myeloma (red) showed inferior 5-year actuarial probabilities of EFS (18% vs 60%, P < .001; HR = 4.51) and OS (28% vs 78%, P < .001; HR = 5.16) in the 13.1% patients with a high-risk signature.
Gene expression patterns can distinguish risk groups in training cohort. (A) Heat maps of the 70 genes illustrate remarkably similar expression patterns among 351 newly diagnosed patients used to identify the 70 genes. Red bars above the patient columns denote patients with disease-related deaths. The 51 genes in rows designated by the red bar on the left (top rows; up-regulated) identified patients in the upper quartile of expression at high risk for early disease-related death. The 19 gene rows designated by the green bar (down-regulated), identified patients in the lower quartile of expression at high risk of early disease-related death. (B) Training cohort frequencies for sample differences between ratios of the mean of log2 expression of the 51 up-regulated genes/19 down-regulated genes. This self-normalizing expression ratio has a marked bimodal distribution, consistent with the upper/lower quartile log-rank differential expression analysis, which was designed to detect genes that define a single high-risk group (13.1%) with an extreme expression distribution. Interpreted as an up/down-regulation ratio on the log2 scale, higher values are associated with poor outcome. The vertical line shows the high-risk versus low-risk cutoff for the log2-scale ratio determined by K-means clustering: the percentage of samples below and above the cutoff is also shown. Kaplan-Meier estimates of EFS (C) and OS (D) in low-risk myeloma (green) and high-risk myeloma (red) showed inferior 5-year actuarial probabilities of EFS (18% vs 60%, P < .001; HR = 4.51) and OS (28% vs 78%, P < .001; HR = 5.16) in the 13.1% patients with a high-risk signature.
Gene expression clustergram of 70 high-risk genes in plasma cells from 22 healthy subjects (NPC), 14 subjects with MGUS, 351 patients with newly diagnosed MM, and 42 human MM cell lines (HMCL). Each row represents a gene and each column represents a sample. The genes are ordered from top to bottom based on the rank in Table 1. Red color for a gene indicates expression above the median and blue color indicates expression below the median. Samples within myeloma risk groups were ordered so that the predicted risk increases continuously from left to right.
Gene expression clustergram of 70 high-risk genes in plasma cells from 22 healthy subjects (NPC), 14 subjects with MGUS, 351 patients with newly diagnosed MM, and 42 human MM cell lines (HMCL). Each row represents a gene and each column represents a sample. The genes are ordered from top to bottom based on the rank in Table 1. Red color for a gene indicates expression above the median and blue color indicates expression below the median. Samples within myeloma risk groups were ordered so that the predicted risk increases continuously from left to right.
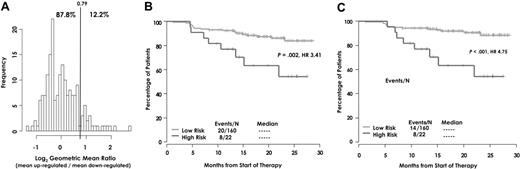
Next, we sought to confirm the association of the expression signature with OS in an independent test cohort of 181 patients. Indeed, an independent, unsupervised clustering of the log2-scale up-/down-regulated expression ratio identified a proportionally similar subset of patients exhibiting extreme dysregulation (12.2%; Figure 3A). A similar result of survival distribution and HR was found in both EFS (HR = 3.41, P = .002; Figure 3B) and OS(HR = 4.75, P < .001; Figure 3C) as seen in the training cohort. Absence of a high-risk score identified a favorable subset of patients with a 5-year continuous complete remission of 60% as opposed to a 3-year rate of only 20% in those with a high-risk score (data not shown).
Risk group distribution and survival analyses in the test cohort. (A) Test cohort frequencies for the ratio of the mean of the log2 up-/down-regulated genes. The cutoff for high risk was determined by independent clustering of the log2 ratio. The training and validation sets have a similar distribution for this expression summary of the 70 genes, including similar cutoffs for high risk and similar proportions clustered into the high-risk group. Kaplan-Meier estimates of (B) EFS and (C) OS between molecular risk groups in the test cohort.
Risk group distribution and survival analyses in the test cohort. (A) Test cohort frequencies for the ratio of the mean of the log2 up-/down-regulated genes. The cutoff for high risk was determined by independent clustering of the log2 ratio. The training and validation sets have a similar distribution for this expression summary of the 70 genes, including similar cutoffs for high risk and similar proportions clustered into the high-risk group. Kaplan-Meier estimates of (B) EFS and (C) OS between molecular risk groups in the test cohort.
To further assess the validity of the clusters with respect to clinical features, correlations of various clinical parameters were analyzed between the low- and high-risk subgroups in both training (Table 2) and test sets (Table 3). A remarkable similarity of clinical feature distribution in risk groups was observed in both training and test cohorts: higher serum levels of β2-microglobulin, C-reactive protein, creatinine, and lactate dehydrogenase (LDH), as well as FISH-defined chromosome 13 deletion and metaphase cytogenetic abnormalities, all were significantly more common in the high-risk group of both training and test sets (P < .05). Similarly, the clinically more benign CCND1 subgroup predominated in the low-risk and the MMSET/FGFR3 subgroup in the high-risk cohort, as depicted for the training set in Table 2 and for the test set in Table 3.
Correlation of clinical parameters with risk groups in the training cohort (n = 351)
| Characteristic . | Low-risk, % . | High-risk, % . | P . |
|---|---|---|---|
| Age, 65 y or older | 20 | 20 | .856 |
| Albumin, less than 35 g/L | 13 | 35 | .001 |
| β2-microglobulin | |||
| Less than 297.5 nM | 62 | 42 | .005 |
| 297.5 nM or more to less than 467.5 nM | 20 | 20 | |
| 467.5 nM or more | 19 | 40 | |
| C-reactive protein, 4 mg/L or more | 51 | 62 | .235 |
| LDH, 190 IU/L or more | 30 | 59 | < .001 |
| Interphase FISH-defined del13 | 31 | 49 | .031 |
| Cytogenetic abnormalities | 26 | 70 | < .001 |
| GEP-based translocations | |||
| CCND1 | 20 | 0 | < .001 |
| MMSET | 12 | 28 | |
| MAF/MAFB | 3 | 9 | |
| No spike | 65 | 63 | |
| Characteristic . | Low-risk, % . | High-risk, % . | P . |
|---|---|---|---|
| Age, 65 y or older | 20 | 20 | .856 |
| Albumin, less than 35 g/L | 13 | 35 | .001 |
| β2-microglobulin | |||
| Less than 297.5 nM | 62 | 42 | .005 |
| 297.5 nM or more to less than 467.5 nM | 20 | 20 | |
| 467.5 nM or more | 19 | 40 | |
| C-reactive protein, 4 mg/L or more | 51 | 62 | .235 |
| LDH, 190 IU/L or more | 30 | 59 | < .001 |
| Interphase FISH-defined del13 | 31 | 49 | .031 |
| Cytogenetic abnormalities | 26 | 70 | < .001 |
| GEP-based translocations | |||
| CCND1 | 20 | 0 | < .001 |
| MMSET | 12 | 28 | |
| MAF/MAFB | 3 | 9 | |
| No spike | 65 | 63 | |
Correlation of clinical parameters with risk groups in the test cohort (n = 181)
| Characteristic . | Low risk, % . | High risk, % . | P . |
|---|---|---|---|
| Age, 65 y or older | 30 | 23 | .692 |
| Albumin, less than 35 g/L | 17 | 32 | .163 |
| β2-microglobulin | |||
| Less than 297.5 nM | 57 | 32 | .005 |
| 297.5 nM to less than 467.5 nM | 23 | 18 | |
| 467.5 nM or more | 19 | 50 | |
| C-reactive protein, 4 mg/L or above | 44 | 59 | .271 |
| LDH, 190 IU/L or above | 18 | 59 | < .001 |
| Cytogenetic abnormalities | 27 | 77 | < .001 |
| GEP-based translocations | |||
| CCND1 | 14 | 0 | < .001 |
| MMSET | 12 | 23 | |
| MAF/MAFB | 7 | 36 | |
| No spike | 67 | 41 | |
| Characteristic . | Low risk, % . | High risk, % . | P . |
|---|---|---|---|
| Age, 65 y or older | 30 | 23 | .692 |
| Albumin, less than 35 g/L | 17 | 32 | .163 |
| β2-microglobulin | |||
| Less than 297.5 nM | 57 | 32 | .005 |
| 297.5 nM to less than 467.5 nM | 23 | 18 | |
| 467.5 nM or more | 19 | 50 | |
| C-reactive protein, 4 mg/L or above | 44 | 59 | .271 |
| LDH, 190 IU/L or above | 18 | 59 | < .001 |
| Cytogenetic abnormalities | 27 | 77 | < .001 |
| GEP-based translocations | |||
| CCND1 | 14 | 0 | < .001 |
| MMSET | 12 | 23 | |
| MAF/MAFB | 7 | 36 | |
| No spike | 67 | 41 | |
In a multivariate analysis of variables associated with OS and EFS, the high up-/down-regulation ratio predictor (high-risk score) retained its significance after adjustment for competing genetic and clinical variables (even including the International Staging System [ISS]) in both the training set (Table 4: HR = 4.1, P < .001) and the test set (data not shown; P = .025). Importantly, the high-risk score also was the only independent baseline parameter that affected complete response duration adversely (HR = 3.07; P < .001). This strong prognostic performance of the GEP-derived risk score can be partly explained by its strong association with known clinical prognostic variables, as shown by a multivariate analysis with the up-/down-regulation ratio as the outcome (Table 5). While the variables in Table 5 may serve as temporary, partial substitutes for a broadly available GEP assay, Table 4 suggests that such an assay, combined with high-risk translocations (also measurable via GEP), has the potential to provide a powerful simple prognostic test for myeloma.
Multivariate analysis of EFS and OS in the training cohort
| Significant predictors* . | % of cases . | EFS . | OS . | ||
|---|---|---|---|---|---|
| HR . | P . | HR . | P . | ||
| High-risk up-/down-regulated expression ratio (log2-scale)† | 13 | 3.24 | < .001 | 4.09 | < .001 |
| β2-microglobulin | |||||
| 297.5 nM to less than 467.5 nM | 20 | 1.72 | .001 | — | — |
| 467.5 nM or greater | 21 | 2.01 | — | — | |
| LDH, 190 IU/L or above | 34 | — | — | 1.92 | .004 |
| Interphase FISH, defined del13 | 33 | 1.63 | .007 | — | — |
| GEP-defined high-risk translocations | 18 | 1.97 | .001 | 1.85 | .012 |
| Significant predictors* . | % of cases . | EFS . | OS . | ||
|---|---|---|---|---|---|
| HR . | P . | HR . | P . | ||
| High-risk up-/down-regulated expression ratio (log2-scale)† | 13 | 3.24 | < .001 | 4.09 | < .001 |
| β2-microglobulin | |||||
| 297.5 nM to less than 467.5 nM | 20 | 1.72 | .001 | — | — |
| 467.5 nM or greater | 21 | 2.01 | — | — | |
| LDH, 190 IU/L or above | 34 | — | — | 1.92 | .004 |
| Interphase FISH, defined del13 | 33 | 1.63 | .007 | — | — |
| GEP-defined high-risk translocations | 18 | 1.97 | .001 | 1.85 | .012 |
N = 325; 26 of 351 patients were missing FISH-defined del13. MMSET/FGFR3 spikes (14.1%) are combined with MAF/MAFB spikes (3.7%). Low risk includes CCND1 spikes (16.9%) and no spike (65.3%). The collapsed categories perform better as prognostic categories due to the similarity in outcome distribution for the subgroups within high-risk and low-risk categories and the small size of the MAF/MAFB subgroup. For EFS, the number of events or deaths was 138; for OS, it was 87; the R2 values were 0.324 and 0.288, respectively.
—indicates insignificance for one or the other outcome.
Predictors with P >.05 for both outcomes: aged 65 years or older, metaphase cytogenetic abnormalities, albumin of 35 g/L or less, and C-reactive protein of 4 mg/L or more.
The average log2 (expression) of the 51 Q4 genes minus the average log2 (expression) of the 19 Q1 genes (ie, the log2 scale ratio of geometric mean up-regulated vs down-regulated genes). High risk by K-means clustering is 0.66 or greater (ie, a ratio of 1.58).
Multivariate analysis of fold-change in the up-/down-regulated expression ratio
| Significant predictors* . | % of cases . | Fold change . | P . |
|---|---|---|---|
| Interphase FISH-defined amp1q21 | 43 | 0.316 | < .001 |
| Cytogenetic abnormalities | 30 | 0.353 | < .001 |
| CCND1 or CCND3 spike | 20 | −0.248 | .008 |
| MAF/MAFB spike | 4 | 0.430 | .030 |
| MMSET/FGFR3 spike | 14 | 0.297 | .005 |
| LDH, 190 U/L or more | 31 | 0.332 | < .001 |
| Albumin, 35 g/L or less | 18 | 0.249 | .014 |
| Significant predictors* . | % of cases . | Fold change . | P . |
|---|---|---|---|
| Interphase FISH-defined amp1q21 | 43 | 0.316 | < .001 |
| Cytogenetic abnormalities | 30 | 0.353 | < .001 |
| CCND1 or CCND3 spike | 20 | −0.248 | .008 |
| MAF/MAFB spike | 4 | 0.430 | .030 |
| MMSET/FGFR3 spike | 14 | 0.297 | .005 |
| LDH, 190 U/L or more | 31 | 0.332 | < .001 |
| Albumin, 35 g/L or less | 18 | 0.249 | .014 |
N = 250; R2 = 0.324. Of the 351 patients, 98 were missing amp1q21 by FISH, and an additional 3 were missing albumin.
Predictors with P > .05 for both outcomes: age ≥ 65 years, β2-microglobulin (≥ 297.5 nM to ≥ 467.5 nM), C-reactive protein (≥ 4 mg/L).
Gene-expression model predicts postrelapse risk and survival
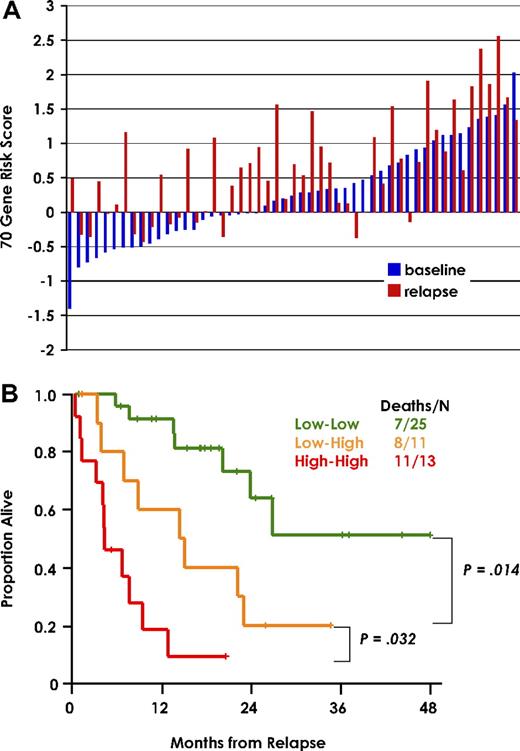
When the 70-gene risk model was applied to relapse samples from 51 of the 351 patients of the training set, 39 (76%) exhibited a high-risk score (Figure 4A). In a paired analysis of baseline and relapse samples, the 25 patients with low-risk designation at both diagnosis and relapse had a superior postrelapse survival, followed by 11 patients with low-risk designation at diagnosis and high-risk at relapse and 13 patients exhibiting a high-risk designation at both observation times (Figure 4B). There were only 2 patients with high risk at diagnosis and low risk at relapse.
70-gene risk score at diagnosis and relapse predicts postrelapse survival. (A) 70-gene risk score in paired diagnostic (blue) and relapse (red) samples of 51 patients from the training cohort. The gene expression risk score is indicated to the left. Sample pairs are order from left to right based on lowest baseline score. (B) Kaplan-Meier plots of postrelapse survival of the 3 groups defined by low risk both at diagnosis and relapse (Low-Low), low risk at diagnosis and high risk at relapse (Low-High), and high risk at both time points (High-High).
70-gene risk score at diagnosis and relapse predicts postrelapse survival. (A) 70-gene risk score in paired diagnostic (blue) and relapse (red) samples of 51 patients from the training cohort. The gene expression risk score is indicated to the left. Sample pairs are order from left to right based on lowest baseline score. (B) Kaplan-Meier plots of postrelapse survival of the 3 groups defined by low risk both at diagnosis and relapse (Low-Low), low risk at diagnosis and high risk at relapse (Low-High), and high risk at both time points (High-High).
Chromosome 1 genes are overrepresented in high-risk model
To determine whether the 70-gene high-risk signature may reflect specific gains or losses of genomic DNA in high-risk MM, the map positions of the 70 genes comprising the gene expression risk signature were compared (Table 6). While representing only 10% of genes on the microarray, 21 (30%) of the 70 high-risk genes mapped to chromosome 1 (P < .001): 9 (47%) of 19 Q1 genes mapped to 1p, with 5 mapping to 1p13; among 12 (24%) of 51 Q4 genes mapping to chromosome 1, 9 resided on 1q, while the 4 on 1p mapped to the extreme telomeric and centromeric regions of the p arm. These data suggest that gain of DNA material on 1q and loss of 1p are significant determinants of high risk in MM.
Chromosome distribution of all mapped probe sets on U133Plus2.0 microarray and the 70 genes of the high-risk signature
| Chromosome . | U133Plus2.0 . | Q1 . | Q4 . | Combined . |
|---|---|---|---|---|
| Gene no. (%) . | Gene no. (%) . | Gene no. (%) . | Gene no. (%) . | |
| 1 | 5379 (10) | 9 (47.4) | 12 (23.5) | 21 (30)* |
| 2 | 3958 (7.3) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 3 | 3275 (6.1) | 0 (0) | 7 (13.7) | 7 (10) |
| 4 | 2314 (4.3) | 0 (0) | 0 (0) | 0 (0) |
| 5 | 2615 (4.8) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 6 | 2956 (5.5) | 1 (5.3) | 1 (2) | 2 (2.9) |
| 7 | 2769 (5.1) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 8 | 2014 (3.7) | 0 (0) | 4 (7.8) | 4 (5.7) |
| 9 | 2139 (4) | 2 (10.5) | 0 (0) | 2 (2.9) |
| 10 | 2192 (4.1) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 11 | 2889 (5.4) | 1 (5.3) | 1 (2) | 2 (2.9) |
| 12 | 2739 (5.1) | 1 (5.3) | 3 (5.9) | 4 (5.7) |
| 13 | 1250 (2.3) | 1 (5.3) | 0 (0) | 1 (1.4) |
| 14 | 1793 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| 15 | 1805 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| 16 | 2084 (3.9) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 17 | 2843 (5.3) | 0 (0) | 3 (5.9) | 3 (4.3) |
| 18 | 966 (1.8) | 0 (0) | 1 (2) | 1 (1.4) |
| 19 | 2839 (5.3) | 0 (0) | 3 (5.9) | 3 (4.3) |
| 20 | 1487 (2.8) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 21 | 662 (1.2) | 0 (0) | 1 (2) | 1 (1.4) |
| 22 | 1225 (2.3) | 0 (0) | 1 (2) | 1 (1.4) |
| X | 1691 (3.1) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| Y | 107 (0.2) | 0 (0) | 0 (0) | 0 (0) |
| Total | 53991 | 19 (100) | 51 (100) | 70 (100) |
| Chromosome . | U133Plus2.0 . | Q1 . | Q4 . | Combined . |
|---|---|---|---|---|
| Gene no. (%) . | Gene no. (%) . | Gene no. (%) . | Gene no. (%) . | |
| 1 | 5379 (10) | 9 (47.4) | 12 (23.5) | 21 (30)* |
| 2 | 3958 (7.3) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 3 | 3275 (6.1) | 0 (0) | 7 (13.7) | 7 (10) |
| 4 | 2314 (4.3) | 0 (0) | 0 (0) | 0 (0) |
| 5 | 2615 (4.8) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 6 | 2956 (5.5) | 1 (5.3) | 1 (2) | 2 (2.9) |
| 7 | 2769 (5.1) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 8 | 2014 (3.7) | 0 (0) | 4 (7.8) | 4 (5.7) |
| 9 | 2139 (4) | 2 (10.5) | 0 (0) | 2 (2.9) |
| 10 | 2192 (4.1) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 11 | 2889 (5.4) | 1 (5.3) | 1 (2) | 2 (2.9) |
| 12 | 2739 (5.1) | 1 (5.3) | 3 (5.9) | 4 (5.7) |
| 13 | 1250 (2.3) | 1 (5.3) | 0 (0) | 1 (1.4) |
| 14 | 1793 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| 15 | 1805 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| 16 | 2084 (3.9) | 0 (0) | 2 (3.9) | 2 (2.9) |
| 17 | 2843 (5.3) | 0 (0) | 3 (5.9) | 3 (4.3) |
| 18 | 966 (1.8) | 0 (0) | 1 (2) | 1 (1.4) |
| 19 | 2839 (5.3) | 0 (0) | 3 (5.9) | 3 (4.3) |
| 20 | 1487 (2.8) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| 21 | 662 (1.2) | 0 (0) | 1 (2) | 1 (1.4) |
| 22 | 1225 (2.3) | 0 (0) | 1 (2) | 1 (1.4) |
| X | 1691 (3.1) | 1 (5.3) | 2 (3.9) | 3 (4.3) |
| Y | 107 (0.2) | 0 (0) | 0 (0) | 0 (0) |
| Total | 53991 | 19 (100) | 51 (100) | 70 (100) |
P < .001. An exact test for binomial proportions was used to compare the proportion of retained probe sets mapping to chromosome 1 to the proportion for the entire array. The Affymetrix control gene number was 62; unknown, 622, and total, 54 675.
A 17-gene model can substitute for 70-gene model
Having shown that high risk is likely related to genomic alterations of chromosome 1, we next wanted to identify a minimum set of genes capable of discriminating high-risk and low-risk myeloma. Applying a MSDA of the 70 high-risk–associated genes across the high-risk (n = 46) and low-risk (n = 305) patients defined by the 70-gene model in the training set, we identified 17 genes in the resultant linear discriminant function (Table 7). It is noteworthy that 3 (60%) of the 5 Q1 genes and 5 (45%) of the 12 Q4 genes in the model map to 1p and 1q, respectively. The 17-gene model was then applied to the training group and predicted, with 97.7% accuracy, the correct class based on the high-risk/low-risk classification of the 70-gene model (Table 8). A cross-validation analysis was performed where samples were removed one at a time from the sample set, and the predictive model was recalculated without that sample. Then the model was used to classify the removed observation. In this cross-validation approach, the prediction accuracy was 96.9%. The 17-gene model was then applied to the test set of 181 newly diagnosed patients receiving the second protocol UARK 03-033. The MSDA model again correctly classified 150 (94.3%) of 159 low-risk samples and 21 (95.5%) of 22 high-risk samples (Table 9). The Kaplan-Meier estimates of OS of the high-risk and low-risk groups were similar whether defined by the 17-gene model (Figure 5) or the 70-gene model (Figure 3D).
17 genes defined by MDSA ordered by their score
| Affymetrix probe set . | Gene symbol . | Chromosome location . | MSDA score . | 70-gene quartile . |
|---|---|---|---|---|
| 206364_at | KIF14 | 1q32.1 | 0.38 | Q4 |
| 211576_s_at | SLC19A1 | 21q22.3 | 0.32 | Q4 |
| 201897_s_at | CKS1B | 1q21.2 | 0.31 | Q4 |
| 200638_s_at | YWHAZ | 8q23.1 | 0.28 | Q4 |
| 205235_s_at | MPHOSPH1 | 10q23.31 | 0.27 | Q4 |
| 203432_at | TMPO | 12q22 | 0.25 | Q4 |
| 213607_x_at | NADK | 1p36.21 | 0.23 | Q4 |
| 204016_at | LARS2 | 3p21.3 | 0.19 | Q4 |
| 220789_s_at | TBRG4 | 7p14-p13 | 0.19 | Q4 |
| 206513_at | AIM2 | 1q22 | 0.16 | Q4 |
| 242488_at | NA | 1q43 | 0.15 | Q4 |
| 219918_s_at | ASPM | 1q31 | −0.40 | Q4 |
| 200850_s_at | AHCYL1 | 1p13.2 | −0.21 | Q1 |
| 218924_s_at | CTBS | 1p22 | −0.23 | Q1 |
| 213628_at | MCLC | 1p13.3 | −0.25 | Q1 |
| 202729_s_at | LTBP1 | 2p22-p21 | −0.29 | Q1 |
| 1557277_a_at | NA | 6p21 | −0.30 | Q1 |
| Affymetrix probe set . | Gene symbol . | Chromosome location . | MSDA score . | 70-gene quartile . |
|---|---|---|---|---|
| 206364_at | KIF14 | 1q32.1 | 0.38 | Q4 |
| 211576_s_at | SLC19A1 | 21q22.3 | 0.32 | Q4 |
| 201897_s_at | CKS1B | 1q21.2 | 0.31 | Q4 |
| 200638_s_at | YWHAZ | 8q23.1 | 0.28 | Q4 |
| 205235_s_at | MPHOSPH1 | 10q23.31 | 0.27 | Q4 |
| 203432_at | TMPO | 12q22 | 0.25 | Q4 |
| 213607_x_at | NADK | 1p36.21 | 0.23 | Q4 |
| 204016_at | LARS2 | 3p21.3 | 0.19 | Q4 |
| 220789_s_at | TBRG4 | 7p14-p13 | 0.19 | Q4 |
| 206513_at | AIM2 | 1q22 | 0.16 | Q4 |
| 242488_at | NA | 1q43 | 0.15 | Q4 |
| 219918_s_at | ASPM | 1q31 | −0.40 | Q4 |
| 200850_s_at | AHCYL1 | 1p13.2 | −0.21 | Q1 |
| 218924_s_at | CTBS | 1p22 | −0.23 | Q1 |
| 213628_at | MCLC | 1p13.3 | −0.25 | Q1 |
| 202729_s_at | LTBP1 | 2p22-p21 | −0.29 | Q1 |
| 1557277_a_at | NA | 6p21 | −0.30 | Q1 |
Confusion matrix of risk prediction in training set using 17-gene model
| 70-gene risk group . | Total . | 17-gene risk group . | |
|---|---|---|---|
| Low . | High . | ||
| Low | 305 | 298 | 7 |
| High | 46 | 1 | 45 |
| 70-gene risk group . | Total . | 17-gene risk group . | |
|---|---|---|---|
| Low . | High . | ||
| Low | 305 | 298 | 7 |
| High | 46 | 1 | 45 |
Confusion matrix of risk prediction in test set using 17-gene model
| 70-gene risk group . | Total . | 17-gene risk group . | |
|---|---|---|---|
| Low-risk . | High-risk . | ||
| Low | 159 | 150 | 9 |
| High | 22 | 1 | 21 |
| 70-gene risk group . | Total . | 17-gene risk group . | |
|---|---|---|---|
| Low-risk . | High-risk . | ||
| Low | 159 | 150 | 9 |
| High | 22 | 1 | 21 |
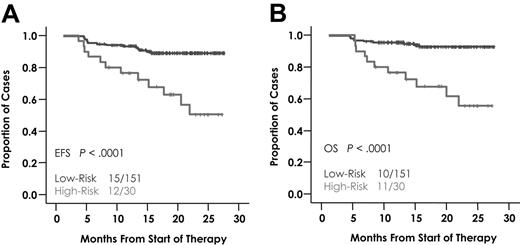
EFS and OS in risk groups defined by the 17-gene model in the test set. The 181 newly diagnosed patients with MM were predicted into high-risk (16.6%) and low-risk (83.4%) groups as described. Kaplan-Meier estimates of survival in low-risk and high-risk myeloma showed 2-year actuarial probabilities of EFS (A) of 88% for the high risk (red) versus 50% for low risk (blue) (P < .001) and OS (B) of 91% for the high-risk (red) versus 54% for the low-risk (blue) (P < .001).
EFS and OS in risk groups defined by the 17-gene model in the test set. The 181 newly diagnosed patients with MM were predicted into high-risk (16.6%) and low-risk (83.4%) groups as described. Kaplan-Meier estimates of survival in low-risk and high-risk myeloma showed 2-year actuarial probabilities of EFS (A) of 88% for the high risk (red) versus 50% for low risk (blue) (P < .001) and OS (B) of 91% for the high-risk (red) versus 54% for the low-risk (blue) (P < .001).
Relating 70-gene model–defined high-risk myeloma with molecular subgroups defined by unsupervised hierarchical cluster analysis
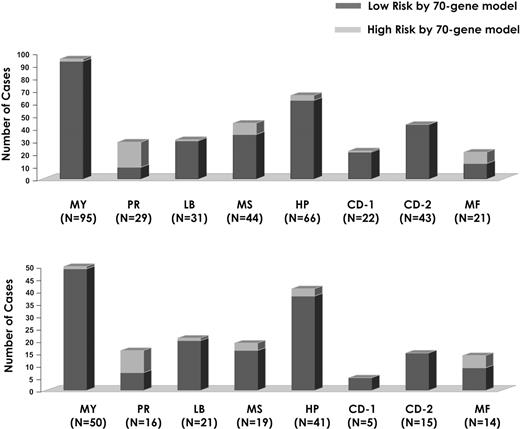
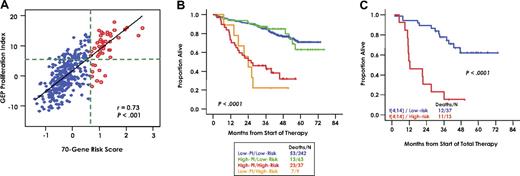
The high-risk model identified here was examined in the context of a previously defined molecular classification.9 High-risk disease designation pertained to all myeloma classes except for the CD-2 type characterized by CCND1 or CCND3 spikes and CD20 and VPREB3 expression (Figure 6). Despite a strong correlation between the high-risk signature and the proliferation (PR) subgroup (Figure 6), the presence of outlier cases suggests that the high-risk signature not only reflects tumor cell proliferation but may encompass also other features of disease conferring short survival, such as drug resistance. Analysis of the 351 training patients according to a 70-gene high-risk cut point of .66 and a proliferation index (PI) of 5 (Figure 7A) revealed that high and low PI designations failed to identify subgroups with different survival among low-risk and high-risk groups (Figure 7B). When applied to the 50 patients with t(4;14)(p16;q32), the 70-gene risk score again separated low-risk and high-risk subgroups (P < .001; Figure 7C).
Relationship between high risk and low risk defined by the 70-gene supervised model and the 7-subgroup unsupervised classifier.9 Data are presented as a stacked bar-view of the number of high-risk (red) and low-risk patients (blue) in each of the 7 subtypes, including the group of patients with the so-called myeloid signature (MY; far left).
Relationship between high risk and low risk defined by the 70-gene supervised model and the 7-subgroup unsupervised classifier.9 Data are presented as a stacked bar-view of the number of high-risk (red) and low-risk patients (blue) in each of the 7 subtypes, including the group of patients with the so-called myeloid signature (MY; far left).
Relating 70 gene model-defined high risk with molecular features. (A) Scatterplot of gene expression–based proliferation index (x-axis) by 70-gene risk score in 351 patients of the training cohort. Low-risk patients (blue) and high-risk patients (red) defined by the 70-gene model are indicated. The 2 variables show a substantial degree of correlation (r = 0.73; P < .001). To evaluate the influence of the 2 variables on outcome, we divided the population into 4 subgroups using a PI cut-point of 5 and a high-risk cut-point of .66. The groups are defined by the intersection of the 2 green dotted lines. The top left quadrant contains patients with high PI/low risk, the top right quadrant contains patients with high PI/high risk, the bottom left quadrant contains patients with low PI/low risk, and the bottom right quadrant contains patients with low PI/high risk. The line represents the linear trend in the data. (B) Kaplan-Meier plots of overall survival estimates of the 4 groups defined in panel A, revealing no impact of PI within risk groups. (C) Kaplan-Meier plots of overall survival estimates of t(4;14)-positive myeloma in relationship to the 70-gene high-risk score designation of the given sample, showing the profound impact of high- and low-risk scores.
Relating 70 gene model-defined high risk with molecular features. (A) Scatterplot of gene expression–based proliferation index (x-axis) by 70-gene risk score in 351 patients of the training cohort. Low-risk patients (blue) and high-risk patients (red) defined by the 70-gene model are indicated. The 2 variables show a substantial degree of correlation (r = 0.73; P < .001). To evaluate the influence of the 2 variables on outcome, we divided the population into 4 subgroups using a PI cut-point of 5 and a high-risk cut-point of .66. The groups are defined by the intersection of the 2 green dotted lines. The top left quadrant contains patients with high PI/low risk, the top right quadrant contains patients with high PI/high risk, the bottom left quadrant contains patients with low PI/low risk, and the bottom right quadrant contains patients with low PI/high risk. The line represents the linear trend in the data. (B) Kaplan-Meier plots of overall survival estimates of the 4 groups defined in panel A, revealing no impact of PI within risk groups. (C) Kaplan-Meier plots of overall survival estimates of t(4;14)-positive myeloma in relationship to the 70-gene high-risk score designation of the given sample, showing the profound impact of high- and low-risk scores.
Discussion
The survival variability of patients with MM is not well accounted for with current laboratory parameters, such as β2-microglobulin and albumin levels used in the ISS staging system.24 De novo high-risk disease may be fundamentally different from myeloma acquiring drug resistance and an aggressive clinical course after recurrent relapses.
A central hypothesis of the work presented in this paper was that expression extremes of a subset of genes correlating with survival might be representative of the effects of DNA copy changes in myeloma disease progression. We were thus able to identify a set of 70 genes, the expression levels of which permitted the identification of a small cohort of 13% to 14% of patients at high risk for early disease-related death. High-risk disease defined by this model was an independent and highly significant prognostic variable to be validated in the context of other treatment approaches.
The marked increase in the frequency of high-risk designation from 13% at diagnosis to 76% at relapse provides molecular evidence of disease evolution that influences postrelapse outcome. An aggressive myeloma phenotype, whether de novo or acquired, may develop through a similar mechanism. With further refinement of our model, we expect to develop tools for quantitative risk assessment during the entire course of therapeutic management.
In addition to its clinical relevance, our findings may also shed important light on the underlying molecular mechanisms that drive disease progression. A striking feature of the high-risk signature was the significant overrepresentation of genes from chromosome 1: nearly 50% of 19 underexpressed genes and 30% of 51 overexpressed genes were derived from chromosomes 1p and 1q, respectively. The predominance of chromosome 1q–derived genes in the high-risk score is in agreement with our recent report showing that disease progression is associated not only with an increase in copy number but also the percentage of cells with 1q21 amplification.15 The gene expression–based high-risk signature defined here is also remarkably consistent with a class of disease defined by high-resolution aCGH profiling and characterized by high-level amplification of 1q21 and deletion of 1p13.6 Taken together, these data suggest that alterations in this chromosome, either through genetic and/or epigenetic modifications, may play a significant role in disease evolution by providing a growth and/or survival advantage.
Using a combination of high-resolution aCGH and microarray profiling, we recently identified 47 minimal common regions (MCRs) of genomic gain across the myeloma genome and 207 genes mapping within these MCRs whose expression increases with increased in copy number.6 When the expression of these copy number–sensitive genes was compared between the high- and low-risk classes defined by the 70-gene model, we found that only genes mapping to MCRs at 1q21, 1q22, and 1q43-q44 were significantly overexpressed in high-risk disease (J.S., unpublished data, July 2006).
Although this report implicates chromosome 1 genes as key players in disease progression, the residence of 4 other genes, FABP5, YWHAZ, EXOSC4, and EIFC2, in the 8q21-8q24 region implies that gains of 8q may also contribute to high-risk disease. These genes encompass recently defined MCRs of gain/amplification at 8q24.12-8q24.13 and 8q24.2-8q24.3.6 Interestingly, expression of MYC, mapping to an MCR at 8q24, was not linked to survival in the current study.
Chromosome 13q14 deletion is an important predictor of survival in patients with myeloma treated on tandem transplantation trials.25 It is noteworthy that loss of expression of a single gene mapping to chromosome 13q14, RFP2, which was previously identified as a candidate tumor-suppressor gene in B-cell chronic lymhocytic leukemia (B-CLL) with significant homology to BRCA1,26 was again linked to poor survival in this analysis. RFP2 was also found to exhibit copy number-sensitive expression in myeloma.6
The frequent alteration of chromosome 1 in many late-stage cancers, including 1q21 amplification in non-Hodgkin lymphoma, Wilms tumor, Ewing sarcoma, and breast and ovarian cancer,12,27–31 warrants studies to determine whether the gene expression model described here has prognostic relevance in other cancers.
Through multivariate discriminant analyses, we found that of the original 70 genes, 17 probe sets could be used to detect high-risk myeloma. Future work will be aimed at developing and validating a quantitative RT-PCR–based assay that combines these staging/risk-associated genes with molecular subtype/etiology–linked genes identified in our unsupervised molecular classification.9 Assessment of the expression levels of these genes may provide a simple and powerful molecular-based prognostic test that would eliminate the need for testing many of the standard variables currently in use with limited prognostic implications devoid also on drug-able targets. Use of a PCR-based methodology would not only dramatically reduce time and effort expended in FISH-based analyses but also reduce markedly the quantity of tissue required for analysis. If these gene signatures are unique to myeloma tumor cells, such a test may be useful after treatment to assess minimal residual disease, possibly using peripheral blood as a sample source.
Authorship
Author contribution: J.D.S. and B.B. conceptualized work, supervised studies, analyzed data, and wrote the paper; F.Z. and B.E.B. analyzed data and wrote the paper; Y.H. analyzed data; S.C., I.H., J.P.S., B.K., C.R., D.R.W., Y.X., H.X., and O.S. performed essential laboratory research; E.A., Y.A., M.C.-F., S.G.K., K.H., A.M., M.P.R., F.V.R., G.T., R.W., M.Z., and B.B. enrolled patients to this study and/or performed other essential clinical research; and J.E. and J.C. provided critical evaluation of the work.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: John D. Shaughnessy Jr, University of Arkansas for Medical Sciences, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu; or Bart Barlogie, University of Arkansas for Medical Sciences, Little Rock, AR 72205; e-mail: barthelbarlogie@uams.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Clyde Bailey and Jennifer Gurley for database management and the nurses and administrative staff of the Myeloma Institute for their supportive roles. We are indebted to Kahla Hebert for assistance with manuscript preparation.
Supported by National Institutes of Health grants CA55819 (J.D.S., J.C., F.Z., G.T., F.V.R., and B.B.) and CA97513 (J.D.S.), and by the Fund to Cure Myeloma and Peninsula Community Foundation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal