Abstract
Homozygous loss of function of Runx1 (Runt-related transcription factor 1 gene) during murine development results in an embryonic lethal phenotype characterized by a complete lack of definitive hematopoiesis. In light of recent reports of disparate requirements for hematopoietic transcription factors during development as opposed to adult hematopoiesis, we used a conditional gene-targeting strategy to effect the loss of Runx1 function in adult mice. In contrast with the critical role of Runx1 during development, Runx1 was not essential for hematopoiesis in the adult hematopoietic compartment, though a number of significant hematopoietic abnormalities were observed. Runx1 excision had lineage-specific effects on B- and T-cell maturation and pronounced inhibition of common lymphocyte progenitor production. Runx1 excision also resulted in inefficient platelet production. Of note, Runx1-deficient mice developed a mild myeloproliferative phenotype characterized by an increase in peripheral blood neutrophils, an increase in myeloid progenitor populations, and extramedullary hematopoiesis composed of maturing myeloid and erythroid elements. These findings indicate that Runx1 deficiency has markedly different consequences during development compared with adult hematopoiesis, and they provide insight into the phenotypic manifestations of Runx1 deficiency in hematopoietic malignancies.
Introduction
RUNX1 (also known as AML1, CBFA2, and PEPB2A) encodes a heterodimeric partner of CBFβ (core-binding factor beta gene), and together they constitute a transcription factor in the core-binding factor (CBF) family. Runx1-CBFβ is required for hematopoietic stem cell emergence,1,2 and it regulates a broad spectrum of genes in the myeloid and lymphoid lineages, including IL3 (interleukin 3 gene), CSF2 (colony-stimulating factor 2 [granulocyte-macrophage] gene), CSF1R (colony-stimulating factor 1 receptor gene), CD4 (CD4 antigen gene), and Tcrd (T-cell receptor delta chain gene).3-10 Mice that are deficient in either Runx1 or Cbfb lack definitive hematopoiesis and die during midgestation, emphasizing the important role that CBF plays in development.11-14 Recent expression studies using internal ribosomal entry site–green fluorescence protein (IRES-GFP) or lacZ knock-in mice further demonstrate the wide expression pattern of Runx1 throughout the mature hematopoietic system and suggest lineage-specific requirements for Runx1 expression in adult hematopoietic development.15,16
Several lines of evidence suggest that loss-of-function mutations in RUNX1 contribute to the pathogenesis of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). First, RUNX1 and its heterodimeric partner, CBFβ, are among the most common targets of chromosomal translocations in human leuke mia. Three examples—t(8;21)(q22;q22),17-21 inv16 (p13q22),22 and t(12;21)(p13;q22),23-25 giving rise to the RUNX1-ETO (eight to twenty-one), CBFB-MYH11, and ETV6-RUNX1 fusion proteins, respectively—account for approximately 25% of adult AML and 25% of pediatric acute lymphoblastic leukemia (ALL).26-34 Second, familial platelet disorder with propensity to develop acute myeloid leukemia (FPD/AML; MIM 301699) is an autosomal dominant disorder that is caused by loss-of-function mutations in RUNX135,36 and that has phenotypic similarities to MDS, including peripheral blood cytopenias, decreased colony-forming activity in myeloid progenitors, qualitative and quantitative platelet defects, and high likelihood of progression to AML during the lifetime of affected persons. Third, mutations in RUNX1 have been identified in sporadic leukemias at a frequency of approximately 3% to 5% and at a lower frequency in MDS. Mutations in RUNX1 are more common in undifferentiated myeloid leukemias (French-American-British [FAB] subtype M0), occurring at a frequency of approximately 25%, and in AML associated with trisomy 21.37 Most sporadic cases of AML with loss-of-function mutations in RUNX1 have biallelic mutations.38
Translocations that target CBF result in the expression of fusion proteins lacking the ability to transactivate expression of hematopoietic target genes.39-42 Furthermore, homologous recombination in which the RUNX1/ETO and CBFB/MYH11 alleles were “knocked-in” to the Runx1 or Cbfb loci, respectively, resulted in midgestation embryonic lethality with a phenotype that was nearly identical to that of mice with homozygous deficiency of either Runx1 or Cbfb, demonstrating that RUNX1/ETO and CBFB/MYH11 are dominant-negative Runx1 and Cbfb alleles.43-45 Collectively, these data indicate that loss of function of CBF, either because of chromosomal translocations or because of loss-of-function point mutations, contributes to the pathogenesis of AML in part by interfering with normal hematopoietic differentiation programs.
Thus, there is a paradox that RUNX1 function is required for definitive hematopoiesis, yet RUNX1 loss of function is associated with acute leukemias in which hematopoietic progenitors have self-renewal capacity. There are now a number of examples of disparate requirements for hematopoietic transcription factors during development compared with adult hematopoiesis. For example, Scl (Tal1) (the T-cell lymphocyte leukemia 1 gene) is essential for hematopoietic development during embryogenesis but is dispensable for adult hematopoiesis.46-48 To further characterize the role of Runx1 in adult hematopoiesis and in leukemia, we generated a conditional Runx1 allele that would allow for analysis of the role of Runx1 in the adult hematopoietic compartment.
Materials and methods
Generation of the conditional Runx1 mouse strain
A 4.3-kilobase (kb) XbaI-AvrII genomic fragment upstream of exon 4 of Runx1 and a 3.0-kb AflII-XhoI fragment downstream of exon 4 were introduced as the 5′ and the 3′ homologous region, respectively, of the targeting vector. A LoxP site was generated by oligo synthesis (Operon, Huntsville, AL) and was inserted into the junction between the 5′ homologous region and the AvrII-AflII genomic fragment containing exon 4. A second LoxP site was introduced from 1 of 2 LoxP sites in the Neo(LoxP) cassette (Figure 1A). The targeting vector was linearized with NotI and electroporated into J1 ES cells, and Runx1tm3Spe/+ (Runx1F/+) F1 mice were generated by standard protocols. The 5′ and 3′ targeting probes were used to screen for homologous recombination events and to confirm the correct targeting. The 5′ probe recognizes a 7.3-kb BamHI fragment in the wild-type Runx1 allele and a 4.7-kb BamHI fragment in the targeted allele. The 3′ probe recognizes an 11.5-kb SspI fragment in the wild-type allele and a 13.5-kb fragment in the targeted allele (Figure 1A).
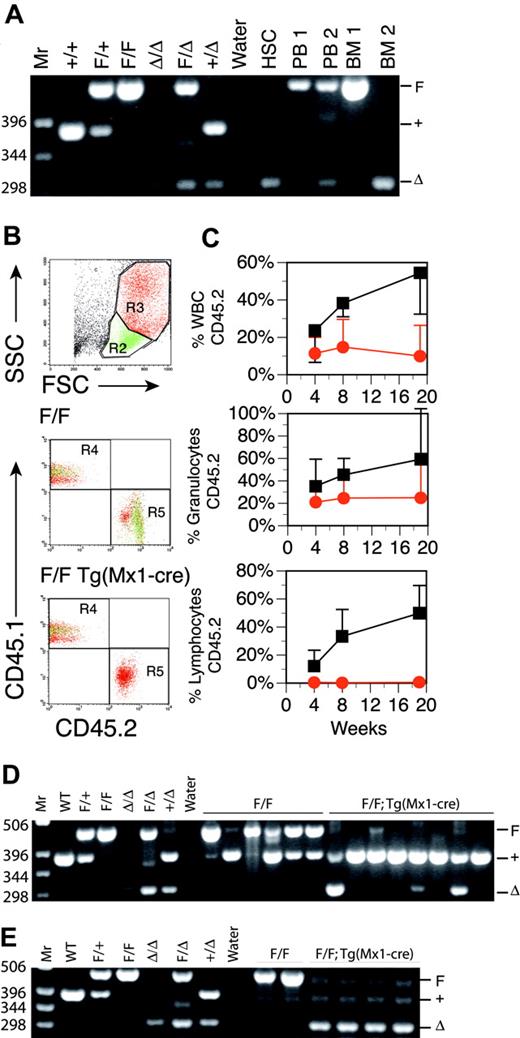
Inducible Runx1 excision in adult mice induces thrombocytopenia. (A) Schematic representation of Runx1 gene-targeting strategy used to flank exon 4 with LoxP-targeting sites. B indicates BamHI; S, SspI; E4, Runt domain exon 4. (filled boxes) 5′ and 3′ targeting probes. (dashed arrows) BamHI-digested genomic DNA fragments for wild-type (7.3-kb), Floxed (2.8-kb), and excised (2.5-kb) DNA detected with excision (Δ) probe (open boxes) located 3′ to the distal LoxP site. (B) Southern blot analysis of BamHI-digested genomic DNA from bone marrow [B] and spleen [S] cells of representative mice killed at 14, 35, and 91 days after pIpC injection. The dose of pIpC is indicated. The blots are probed with Δ probe, which detects the Floxed (2.8-kb) and excised (2.5-kb) Runx1 alleles. Numbers indicate percentage excision. (C) Mean ± SD of total WBC counts, total red blood cell (RBC) counts, total platelet counts, percentage lymphocytes, and percentage neutrophils in PB of pIpC-treated Runx1F/F—Tg(Mx1-Cre) (red symbols) and Runx1F/F (black symbols) mice. Mice were bled 12 days before pIpC injection and 14, 21, 28, 35, 49, 91, 119, and 154 days after pIpC injection. The number of animals evaluated for each genotype at the respective time points is N (Runx1F/F—Tg(Mx1-Cre) = 14, 14, 12, 8, 10, 9, 8, 7 and 6; N (Runx1F/F) = 7, 5, 7, 7, 5, 7, 6, 6, and 6.
Inducible Runx1 excision in adult mice induces thrombocytopenia. (A) Schematic representation of Runx1 gene-targeting strategy used to flank exon 4 with LoxP-targeting sites. B indicates BamHI; S, SspI; E4, Runt domain exon 4. (filled boxes) 5′ and 3′ targeting probes. (dashed arrows) BamHI-digested genomic DNA fragments for wild-type (7.3-kb), Floxed (2.8-kb), and excised (2.5-kb) DNA detected with excision (Δ) probe (open boxes) located 3′ to the distal LoxP site. (B) Southern blot analysis of BamHI-digested genomic DNA from bone marrow [B] and spleen [S] cells of representative mice killed at 14, 35, and 91 days after pIpC injection. The dose of pIpC is indicated. The blots are probed with Δ probe, which detects the Floxed (2.8-kb) and excised (2.5-kb) Runx1 alleles. Numbers indicate percentage excision. (C) Mean ± SD of total WBC counts, total red blood cell (RBC) counts, total platelet counts, percentage lymphocytes, and percentage neutrophils in PB of pIpC-treated Runx1F/F—Tg(Mx1-Cre) (red symbols) and Runx1F/F (black symbols) mice. Mice were bled 12 days before pIpC injection and 14, 21, 28, 35, 49, 91, 119, and 154 days after pIpC injection. The number of animals evaluated for each genotype at the respective time points is N (Runx1F/F—Tg(Mx1-Cre) = 14, 14, 12, 8, 10, 9, 8, 7 and 6; N (Runx1F/F) = 7, 5, 7, 7, 5, 7, 6, 6, and 6.
Runx1F/F and Runx1F/F—Tg(Mx1-Cre) colony generation
All mice were housed in a pathogen-free animal facility in microisolator cages. Runx1F/+ mice were backcrossed against C57BL/6 mice (Taconic, Germantown, NY) for 3 generations, then intercrossed to obtain Runx1F/F mice. Tg(Mx1 [myxovirus resistance 1 gene]–Cre) mice49 were similarly backcrossed onto C57BL/6. Runx1F/F mice were mated to Runx1+/+—Tg(Mx1-Cre) mice to generate Runx1F/+—Tg(Mx1-Cre) mice. Runx1F/+—Tg(Mx1-Cre) mice were mated to Runx1F/F mice to generate Runx1F/F—Tg(Mx1-Cre) mice. Runx1F/F—Tg(Mx1-Cre) mice were mated to Runx1F/F mice to generate Runx1F/F—Tg(Mx1-Cre) and Runx1F/F littermates for phenotypic analysis. To evaluate the Runx1F allele in the Runx1 null (designated Runx1tm1Spe or Runx1rd) background,11 Runx1F/F—Tg(Mx1-Cre) mice were crossed to Runx1+/rd mice backcrossed 7 generations onto BALB/c (Taconic) to generate CXB6F1 Runx1F/rd—Tg(Mx1-Cre) mice and littermate controls (Runx1F/rd, Runx1F/+—Tg(Mx1-Cre), and Runx1F/+). In Runx1F/rd—Tg(Mx1-Cre) mice, a single Runx1 Cre-mediated excision event results in a Runx1 null genotype.
Tail genomic DNA was obtained using a Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). Mice were genotyped for the Runx1F allele by polymerase chain reaction (PCR). Reactions (20 μL) were performed with 80 μM forward (Rdintron 5′, GAGTCCCAGCTGTCAATTCC 3′) and reverse (Rdexon4 5′, GGTGATGGTCAGAGTGAAGC 3′) primers, 250 μM dNTPs, 1.5 mM MgCl2, 2.5 U Taq (Invitrogen) and 5 to 50 ng DNA. DNA was denatured at 94°C for 3 minutes, then amplified by 40 cycles at 94°C for 30 seconds, 60°C for 1 minute, and 72°C for 1 minute. Runx1rd alleles were similarly genotyped with Rdintron and RD29 (5′) TCGCAGCGCATCGCCTTCTA 3′). The Mx1-Cre genotype was performed by Southern blot analysis of BamHI-digested genomic tail DNA.
Induction of Mx1-Cre expression and Runx1F/F excision in adult mice
Eight- to 12-week-old mice were injected intraperitoneally with sterile polyinosinic-polycytidylic acid (pIpC) (Sigma, St Louis, MO) dissolved at 2 mg/mL in phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA). One cohort of mice (3 Runx1F/F and 4 Runx1F/F—Tg(Mx1-Cre)) was injected every other day for 3 injections with 125 μL (250 μg/dose).49 Runx1F excision was variable with this dose (Figure 1B). Therefore, all studies were performed with mice receiving 7 injections, every other day, of 300 μL (600 μg/dose).46 All times after pIpC induction are counted from the first day of injection (day 0). Mice were anesthetized with methoxyflurane or isoflurane according to institutional guidelines (Harvard University, Boston, MA) and retro-orbital eye bleeds were performed. Automated cell counts were performed at Children's Hospital (Boston, MA) using murine-specific software.
Runx1F/F excision evaluation
Excision of floxed Runx1 alleles after Cre expression was evaluated using Southern blot or PCR analysis. Single-cell suspensions of bone marrow (BM), spleen, or thymus cells were prepared as described previously.50,51 Genomic DNA (10-20 μg) was digested with BamHI, separated in 0.7% agarose gel, and transferred to Hybond-N+ membranes (Amersham Biosciences, Piscataway, NJ). The excision (Δ) probe (Figure 1A) is located within Runx1 intron 4, 3′ to the most distal LoxP site, and hybridizes to Runx1+ (7.3 kb), Runx1F (2.8 kb), and Runx1F-Excised (designated Runx1Δ) (2.5 kb) genomic DNA (Figure 1A-B) but not to Runx1rd. Probe template DNA was generated in PCR with the primers RX1FldelF (5′) TGCGCTTACAGAATGTCAGG (3′) and RX1FldelR (5′) CATGACCATGATTGCAGGAG 3′. Alternatively, excision was evaluated by 3-primer PCR, performed as described earlier in this paragraph with 80 μM Rdintron and 40 μM each of Rdexon4 and LoxPrev113 (5′) CCAAGATAGTCCTTAACGGTCG 3′).
Histopathology
Histopathologic examination was conducted as previously described.51 Critical analyses were performed by a hematopathologist blinded to genotype (J.L.K.). Histologic images were obtained on a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) equipped with a SPOT RT color digital camera model 2.1.1 (Diagnostic Instruments, Sterling Heights, MI). The microscope was equipped with a 10×/22 ocular lens. Low power images (×100) were obtained with a 10×/0.25 objective lens. High power images (×600) were obtained with a 60×/1.4 objective lens with oil (Trak 300; Richard Allan Scientific, Kalamazoo, MI). Images were cropped in Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA) and composed in Adobe Illustrator for CS (Adobe Systems).
In vitro colony assays
Myeloid colony-plating assays were performed in methylcellulose-based medium (M3434) containing 3 U/mL erythropoietin (EPO), 10 ng/mL recombinant murine interleukin-3 (rmIL-3), 10 ng/mL rmIL-6, and 50 ng/mL recombinant murine stem cell factor (rmSCF), as per the manufacturer's protocols (StemCell Technologies, Vancouver, BC, Canada). Cells were plated in 3 dilutions (2 × 104, 1 × 104, 5 × 103 cells/dish) in duplicate and were incubated for 12 days at 37°C. Colonies were counted at dilutions in which 30 to 60 colonies per plate were observed. Similar plating efficiencies were observed with fresh and previously frozen BM cells. Colonies were scored by morphology and Wright-Giemsa–stained slides of cytospins. Individual colonies were washed and suspended in 200 μL PBS. One hundred microliters of this suspension was plated on slides with a Cytospin4 centrifuge (Thermo Shandon, Pittsburgh, PA) for 5 minutes at 36g. The remaining cells were suspended in 50 μL water and boiled for 5 minutes, and 5 μL was used in a PCR. Megakaryocyte plating assays were performed in collagen-containing medium according to the manufacturer's protocol (MegaCult-C; StemCell Technologies). Slides were fixed and stained for 6 hours with acetylcholine iodide (Sigma, St Louis, MO), as per the MegaCult-C protocol, and were counterstained with Harris hematoxylin solution (Sigma).
Bone marrow transplantation
Single-cell suspensions of BM cells were prepared as described previously.50,51 Cells were stored in 90% fetal calf serum and 10% dimethyl sulfoxide (DMSO; Sigma) in liquid nitrogen. Cells were thawed at 37°C and washed in PBS, and viable cells were counted by trypan blue dye exclusion (Sigma). Thawed cells consistently demonstrated viability of 40% to 50% after thawing (trypan blue dye exclusion). For competitive repopulation assays, competitor BM was obtained from wild-type B6.SJL (CD45.1+) mice (Jackson Laboratories, Bar Harbor, ME) on the day of transplantation. Total (6 × 105) viable BM cells (0.6 mL) at a test-competitor ratio of 4:1 were injected into the lateral tail veins of lethally irradiated (2 × 650 cGy) female recipient B6.SJL mice. Blood was obtained by retro-orbital bleeding at 4, 8, and 19 weeks after transplantation for flow cytometric analysis. For noncompetitive transplantations, 1 × 106 total BM cells in 0.6 mL were injected. Mice were housed in microisolator cages with autoclaved chow and acidified water.
Flow cytometric analysis
Sorting of myeloid progenitors was accomplished by staining BM cells from mice 17 weeks (fresh) or 27 and 39 weeks (frozen) after pIpC induction with purified rat anti–IL-7Rα chain monoclonal antibodies (A7R34) (e-Bioscience, San Diego, CA) and purified or phycoerythrin (PE)–Cy5-conjugated rat antibodies specific for the following lineage markers: CD3 (CT-CD3), CD4 (RM4-5), CD8 (5H10), B220 (6B2), Gr-1 (8C5), Ter119, and CD19 (6D5) (Caltag, Burlingame, CA). IL-7Rα+Lin+ cells were removed with sheep anti–rat immunoglobulin G (IgG)–conjugated magnetic beads (Dynabeads M-450; Dynal A.S., Oslo, Norway), and the remaining cells were stained with PE-Cy5–conjugated goat anti–rat IgG (Caltag). Cells were then stained with R-phycoerythrin (PE)–conjugated anti–FcγRII/III (2.4G2), fluorescein-isothiocyanate (FITC-)–conjugated anti–CD34 (RAM34), allophycocyanin (APC)–conjugated anti–c-Kit (2B8), and biotinylated anti–Sca-1 (E13-161-7) monoclonal antibodies (BD PharMingen, San Diego, CA), followed by avidin-APC-Cy7 (Caltag). Myeloid progenitors were sorted as IL-7Rα–Lin–Sca-1–c-Kit+CD34+FcγRII/IIIlo (common myeloid progenitor [CMP]), IL-7Rα–Lin–Sca-1–c-Kit+CD34+FcγRII/IIIhi (granulocyte-monocyte progenitor [GMP]), and as IL-7Rα–Lin–Sca-1–c-Kit+CD34–FcγRII/IIIlo megakaryocyte-erythrocyte progenitor [MEPs], as described previously.52 Hematopoietic stem cells (HSCs) and common lymphocyte progenitor cells (CLPs) were sorted as IL-7Rα–Lin–Sca-1hic-Kithi and IL-7Rα+Lin–Sca-1loc-Kitlo populations, respectively.53
Additional flow cytometric analysis was performed on a 4-color FACScalibur cytometer (Becton Dickinson, Mountain View, CA) and analyzed using CellQuest software. All antibodies were obtained from BD PharMingen. Cells were preincubated with 1 μg purified rat anti–mouse CD16/CD32 before staining. Competitive reconstitution was assayed by staining 250 μL fresh red blood cell–lysed peripheral blood (PB) with 1 μg fluorescein isothiocyanate (FITC)–mouse anti–mouse CD45.1 and 1 μg PerCP-Cy5.5 mouse anti–mouse CD45.2 of scatter-gated nucleated cells. Additional flow cytometric analyses were performed on previously frozen cell suspensions. Double-negative (DN) thymocytes were stained with FITC-rat anti–mouse CD8a and PerCP–Cy5.5-rat anti–mouse CD4 or with FITC-anti–CD45.1, PerCP–Cy5.5-rat anti–mouse CD4, PerCP–Cy5.5-rat anti–mouse CD8a, PE-rat anti–mouse CD44, and APC-rat anti–mouse CD25. CD25 and CD44 stainings were evaluated in CD45+CD4–CD8– gated cells. The percentage of myeloid cells in spleen and BM was determined by staining with FITC–anti-CD45.2, PE–rat anti–mouse CD11b (Mac-1), 7AAD-PerCP-Cy5.5 (for viability), and APC–rat anti–mouse Ly-6G and Ly-6C (Gr-1) or PE–rat anti–mouse c-KIT and APC–rat anti–mouse Ter119. Freshly derived BM cells from 2 femurs and 2 tibias were cultured in Dulbecco modified Eagle medium (DMEM) + 10% fetal calf serum with 10 ng/mL recombinant murine thrombopoietin (rmTPO; Sigma) and 50 ng/mL rhIL-11 (StemCell Technologies) for 4 days before flow cytometric analysis of DNA content. BM megakaryocytes were stained with FITC–rat anti–mouse CD41, followed by 50 μg/mL propidium iodide in 0.1% sodium citrate buffer and then incubated with 50 μg/mL RNAase (Qiagen, Valencia, CA), as previously described.54
Results
Excision of a conditional Runx1 allele in adult hematopoietic tissues
To test the effects of loss of Runx1 on adult hematopoietic tissues, we constructed a conditional knock-out allele of the Runx1 locus in mice that could be inactivated using the Cre-LoxP conditional targeting system (Figure 1A). This allows for tissue-specific deletion of a LoxP-flanked (floxed) exon of Runx1 after tissue-specific expression of Cre recombinase. Excision of the floxed Runx1 allele (Runx1tm3Spe, Runx1F) by the early embryonic Cre deleter strain Tg(EIIa-Cre)55 recapitulated the embryonic lethal phenotype associated with homozygosity for a previously derived Runx1 allele (Runx1rd), in which neo replaces exon 411 (data not shown). In transgenic (Tg) Mx1-Cre mice, the interferon-inducible promoter Mx1 allows for expression of Cre recombinase in the hematopoietic system in response to interferon or interferon-inducing agents such as pIpC.49 Runx1F/F and Runx1F/F—Tg(Mx1-Cre) littermates on the C57BL/6 background were induced for excision at 8 to 12 weeks of age by intraperitoneal injection of pIpC, and PB counts were monitored for 22 weeks. We compared 2 pIpC dosing regimens, 3 doses of 250 μg every other day and 7 doses of 600 μg every other day. The latter dosing regimen induced a more than 90% excision of the Runx1F allele throughout the observation period, as demonstrated by Southern blot analysis, and was subsequently used for all further studies (Figure 1B; data not shown). Although Runx1 is essential for the establishment of definitive hematopoiesis during development, we observed, as did Ichikawa et al,56 that adult hematopoietic development was maintained after the excision of Runx1F (Figure 1C). There were, however, several distinctive hematopoietic abnormalities affecting all lineages. Within this cohort of mice, these findings included an average approximately 80% decrease in platelets, an approximately 39% decrease in total white blood cell (WBC) counts, an approximately 22% decrease in the percentage of lymphocytes, and an approximately 28% increase in the percentage of neutrophils during the course of the observation period. We made similar observations after Runx1 excision in Runx1F/rd—Tg(Mx1-Cre) mice on a C57BL/6 × BALB/c F1 (C×B6F1) background (data not shown). PB counts of Runx1 heterozygous mice (Runx1F/rd and Runx1F/+—Tg(Mx1-Cre) were not significantly different from Runx1F/+ mice (data not shown).
Effects of Runx1 excision on the hematopoietic stem cell compartment
Because Runx1 is essential for definitive hematopoiesis during development, we first assayed for the presence of HSCs in Runx1-deficient animals by enumerating the IL-7Rα–Lin–Sca-1hic-Kithi population in total BM. The percentage of phenotypically defined HSCs was increased approximately 3-fold (P = .011) after Runx1F excision by Mx1-Cre (Table 1). This increase in HSCs was observed at 17, 27, and 39 weeks after pIpC, indicating that Runx1 is not required to maintain a phenotypic HSC compartment in adult mice. PCR analysis showed complete excision in the HSC compartment (Figure 2A).
Stem and progenitor cell populations in RunxIF/F—Tg(Mx1-Cre) BM
. | . | Genotype . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Runx1F/F . | . | Runx1F/F—Tg(Mx1-Cre) . | . | . | . | |||||
| Cell type . | Marker content . | BM, %, mean ± SD* . | No. mice . | BM, %, mean ± SD* . | No. mice . | Fold change . | P† . | |||||
| HSC | IL-7Rα-Lin-Sca-1hi c-Kithi | 0.34 ± 0.1 | 4 | 0.99 ± 0.5 | 6 | 2.9 | .011 | |||||
| CLP | IL-7Rα+Lin-Sca-1lo c-Kitlo | 0.07 ± 0.01 | 4 | <0.01 ± <0.01 | 6 | >-7.0 | .019 | |||||
| CMP | IL-7Rα-Lin-Sca-1- c-Kit+ CD34+ FcγRII/III+ | 0.37 ± 0.1 | 3 | 0.30 ± 0.2 | 4 | -0.8 | .377‡ | |||||
| GMP | IL-7Rα-Lin-Sca-1- c-Kit+ CD34+ FcγRII/IIIhi | 0.67 ± 0.1 | 3 | 1.98 ± 0.8 | 4 | 3.0 | .032 | |||||
. | . | Genotype . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Runx1F/F . | . | Runx1F/F—Tg(Mx1-Cre) . | . | . | . | |||||
| Cell type . | Marker content . | BM, %, mean ± SD* . | No. mice . | BM, %, mean ± SD* . | No. mice . | Fold change . | P† . | |||||
| HSC | IL-7Rα-Lin-Sca-1hi c-Kithi | 0.34 ± 0.1 | 4 | 0.99 ± 0.5 | 6 | 2.9 | .011 | |||||
| CLP | IL-7Rα+Lin-Sca-1lo c-Kitlo | 0.07 ± 0.01 | 4 | <0.01 ± <0.01 | 6 | >-7.0 | .019 | |||||
| CMP | IL-7Rα-Lin-Sca-1- c-Kit+ CD34+ FcγRII/III+ | 0.37 ± 0.1 | 3 | 0.30 ± 0.2 | 4 | -0.8 | .377‡ | |||||
| GMP | IL-7Rα-Lin-Sca-1- c-Kit+ CD34+ FcγRII/IIIhi | 0.67 ± 0.1 | 3 | 1.98 ± 0.8 | 4 | 3.0 | .032 | |||||
Percentage of total BM. Detection limit is 0.01%. Data are results from 17, 27, and 39 weeks after pIpC.
Mann-Whitney U test.
Not statistically different.
We next evaluated stem cell function by testing the ability of Mx1-Cre–excised Runx1 BM to competitively repopulate the adult hematopoietic compartment in lethally irradiated syngeneic mice. BM from a pIpC-treated Runx1F/F—Tg(Mx1-Cre) (CD45.2+) mouse or a control pIpC-treated Runx1F/F (CD45.2+) mouse was transplanted into lethally irradiated congenic B6.SJL (CD45.1+) mice with wild-type competitor B6.SJL (CD45.1+) BM (4:1 ratio of test cells to competitor cells). Mice were monitored for contributions of CD45.2+ donor cells to PB at 4, 8, and 19 weeks after transplantation (Figure 2B).57 Control Runx1F/F donor BM reconstituted B6.SJL recipients in all animals tested (n = 6), with contributions from the CD45.2+ donor cells increasing with time after transplantation (Figure 2C). The average contribution of Runx1F/F BM to total PB WBCs at 19 weeks after transplantation was 54.5%. BM from Mx1-Cre excised Runx1F/F mice also contributed to hematopoietic reconstitution at all time points, but reconstitution decreased with time. Two of 8 animals failed to show reconstitution at any time point. At 19 weeks after transplantation, Mx1-Cre–excised CD45.2+ donor cells (n = 6) accounted for an average of 10% of total PB WBCs. Three of 8 mice demonstrated significant contributions (3%, 17%, and 39%) to PB (not shown) at 19 weeks after transplantation. PCR of Runx1 loci from BM confirmed these findings (Figure 2D), indicating that Runx1-excised marrow has reduced competitive repopulating ability in assays for long-term repopulating activity.
We next tested whether Runx1-excised BM could repopulate syngeneic B6.SJL (CD45.1+) mice in the absence of competitor BM. Lethally irradiated B6.SJL (CD45.1+) mice underwent transplantation with 1 × 106 BM cells from a pIpC-treated Runx1F/F—Tg(Mx1-Cre) (CD45.2+) mouse or a control pIpC-treated Runx1F/F (CD45.2+) mouse. Mice receiving Runx1F/F—Tg(Mx1-Cre) marrow (n = 4) were viable at 14 weeks after transplantation, indicating that Runx1-excised cells are competent for reconstitution (Figure 2E). The alterations in PB counts observed in primary animals (Figure 1C), including an 80% decrease in platelet counts, 49% decrease in WBC counts, and relative changes in the differential, were observed in mice that underwent transplantation with excised Runx1F/F—Tg(Mx1-Cre) BM. (Table 2). In nonexcised recipients, approximately 1.3% of the PB cells were derived from the host (CD45.1+), whereas approximately 12.1% of the PB cells in excised recipients were derived from the host (Table 2). Thus, even under these stringent selective conditions for HSC repopulation, Runx1-excised HSCs are less competitive under repopulating conditions than wild-type HSCs.
Hematopoietic effects of Runx1 excision are transplantable
. | Donor cell genotype* . | . | |
|---|---|---|---|
| PB counts† . | Runx1F/F, mean ± SD . | Runx1F/F-Tg(Mx1-Cre), mean ± SD . | |
| WBCs, ×103/μL | 6.94 ± 2.3 | 3.55 ± 0.4 | |
| RBCs, ×106/μL | 7.53 ± 1.7 | 9.46 ± 0.5 | |
| Platelets, ×103/μL | 1808 ± 200 | 364 ± 140 | |
| Neutrophils, % | 32.9 ± 4.6 | 40.6 ± 2.6 | |
| Lymphocytes, % | 60.0 ± 4.6 | 49.0 ± 3.1 | |
| Lineage and origin of PB cells‡ | |||
| WBC donor derived, CD45.2+, %§ | 98.7 ± 0.5 | 87.9 ± 3.4 | |
| PB T cells | |||
| CD3+ %∥ | 3.6 ± 1.8 | 9.0 ± 1.7 | |
| CD3+ donor derived, %¶ | 70.9 ± 22.0 | 7.0 ± 4.5 | |
| PB B cells | |||
| B220+, %∥ | 35.4 ± 7.3 | 0.6 ± 0.3 | |
| B220+ donor derived, %¶ | 100.0 ± 0.0 | 97.1 ± 4.5 | |
| PB myeloid cells | |||
| Mac1+, %∥ | 6.4 ± 2.1 | 17.1 ± 3.7 | |
| Mac1+ donor derived, %¶ | 99.6 ± 0.2 | 99.1 ± 0.4 | |
| Gr1+/Mac1+, %∥ | 40.5 ± 71.2 | 67.8 ± 32.9 | |
| Gr1+/Mac1+ donor derived, %¶ | 99.8 ± 0.3 | 98.1 ± 0.8 | |
| Gr1+, %∥ | 1.0 ± 0.4 | 9.8 ± 1.3 | |
| Gr1+ donor derived, %¶ | 45.5 ± 34.7 | 4.9 ± 1.7 | |
. | Donor cell genotype* . | . | |
|---|---|---|---|
| PB counts† . | Runx1F/F, mean ± SD . | Runx1F/F-Tg(Mx1-Cre), mean ± SD . | |
| WBCs, ×103/μL | 6.94 ± 2.3 | 3.55 ± 0.4 | |
| RBCs, ×106/μL | 7.53 ± 1.7 | 9.46 ± 0.5 | |
| Platelets, ×103/μL | 1808 ± 200 | 364 ± 140 | |
| Neutrophils, % | 32.9 ± 4.6 | 40.6 ± 2.6 | |
| Lymphocytes, % | 60.0 ± 4.6 | 49.0 ± 3.1 | |
| Lineage and origin of PB cells‡ | |||
| WBC donor derived, CD45.2+, %§ | 98.7 ± 0.5 | 87.9 ± 3.4 | |
| PB T cells | |||
| CD3+ %∥ | 3.6 ± 1.8 | 9.0 ± 1.7 | |
| CD3+ donor derived, %¶ | 70.9 ± 22.0 | 7.0 ± 4.5 | |
| PB B cells | |||
| B220+, %∥ | 35.4 ± 7.3 | 0.6 ± 0.3 | |
| B220+ donor derived, %¶ | 100.0 ± 0.0 | 97.1 ± 4.5 | |
| PB myeloid cells | |||
| Mac1+, %∥ | 6.4 ± 2.1 | 17.1 ± 3.7 | |
| Mac1+ donor derived, %¶ | 99.6 ± 0.2 | 99.1 ± 0.4 | |
| Gr1+/Mac1+, %∥ | 40.5 ± 71.2 | 67.8 ± 32.9 | |
| Gr1+/Mac1+ donor derived, %¶ | 99.8 ± 0.3 | 98.1 ± 0.8 | |
| Gr1+, %∥ | 1.0 ± 0.4 | 9.8 ± 1.3 | |
| Gr1+ donor derived, %¶ | 45.5 ± 34.7 | 4.9 ± 1.7 | |
Results are for 2 Runx1F/F mice (2 others were lost to fighting) and 4 Runx1F/F-Tg(Mx1-Cre) mice.
Genotypes of donor cells (CD45.2+) transplanted into lethally irradiated B6.SJL (CD45.1+) recipient mice. Mice were killed 14 weeks after transplantation.
Results from automated blood counts.
Cells gated by scatter, CD45.1, and CD45.2.
Indicated is the percentage of total WBCs (pan-CD45+) that were donor derived (CD45.1- and CD45.2+).
Indicated is the percentage of total WBCs (pan-CD45+) that were CD3, B220, Mac-1, or Gr-1 positive.
Indicated is the percentage of total CD3-, B220-, Mac-1-, Mac-1/Gr-1-, or Gr-1-positive WBCs (pan-CD45+) that were donor derived (CD45.2+).
Stem cells from pIpC-treated Runx1F/F—Tg(Mx1-Cre) mice are reduced in competitive repopulation ability. (A) Ethidium bromide (EtBr)–stained 3% agarose gel of 3-primer PCR of Runx1 loci from sorted HSCs derived from fresh BM 17 weeks after pIpC (Table 1) and unfractionated BM and PB. 1 = Runx1F/F; 2 = RunxF/F—Tg(Mx1-Cre). Mr = 1-kb ladder, sizes indicated (bp). Control samples include tail DNA from Runx1+/+, Runx1F/+, and Runx1F/F mice and from Runx1Δ/Δ, Runx1F/Δ, and Runx1+/Δ embryos. Runx1Δ alleles were generated by mating Runx1F/F mice to Tg(EIIa-Cre) mice and intercrossing Runx1F/Δ mice. (B) Composite showing gating strategy and CD45.1/CD45.2 staining of PB from representative mice that underwent transplantation with Runx1F/F or Runx1F/F—Tg(Mx1-Cre) and competitor marrow 8 weeks after transplantation. Green indicates lymphocytic (R2), and red indicates granulocytic (R3) fractions based on scatter gating (SSC, side scatter; FSC, forward scatter). Gate assignments were confirmed by back-gating of control stainings with Gr1+–, Mac1+–, CD3+–, T-cell receptor β (TCRβ+)–, CD19+–, or B220+–stained PB (not shown). The percentage contribution to the granulocytic and lymphocytic lineages of PB from each marrow isotype (CD45.1/CD45.2) was determined. (C) Plotted is the mean (±SD) percentage contribution of donor-derived CD45.2+ cells in PB from mice that underwent transplantation with Runx1F/F—Tg(Mx1-Cre) (red symbols; n = 6) or Runx1F/F (black symbols; n = 6) and competitor bone marrow to total WBCs, granulocytes, and lymphocytes of recipient mice at 4, 8, and 19 weeks after transplantation. Two of 8 mice receiving Runx1F/F—Tg(Mx1-Cre) marrow that failed to show contribution of CD45.2+ marrow to PB of recipient mice at any time point are excluded. (D) Nineteen weeks after transplantation, EtBr-stained 3% agarose gel of 3-primer PCR of Runx1 loci of BM from mice that underwent competitive transplantation. (E) EtBr-stained 3% agarose gel of 3-primer PCR of Runx1 loci from BM of mice that underwent transplantation without competitor marrow.
Stem cells from pIpC-treated Runx1F/F—Tg(Mx1-Cre) mice are reduced in competitive repopulation ability. (A) Ethidium bromide (EtBr)–stained 3% agarose gel of 3-primer PCR of Runx1 loci from sorted HSCs derived from fresh BM 17 weeks after pIpC (Table 1) and unfractionated BM and PB. 1 = Runx1F/F; 2 = RunxF/F—Tg(Mx1-Cre). Mr = 1-kb ladder, sizes indicated (bp). Control samples include tail DNA from Runx1+/+, Runx1F/+, and Runx1F/F mice and from Runx1Δ/Δ, Runx1F/Δ, and Runx1+/Δ embryos. Runx1Δ alleles were generated by mating Runx1F/F mice to Tg(EIIa-Cre) mice and intercrossing Runx1F/Δ mice. (B) Composite showing gating strategy and CD45.1/CD45.2 staining of PB from representative mice that underwent transplantation with Runx1F/F or Runx1F/F—Tg(Mx1-Cre) and competitor marrow 8 weeks after transplantation. Green indicates lymphocytic (R2), and red indicates granulocytic (R3) fractions based on scatter gating (SSC, side scatter; FSC, forward scatter). Gate assignments were confirmed by back-gating of control stainings with Gr1+–, Mac1+–, CD3+–, T-cell receptor β (TCRβ+)–, CD19+–, or B220+–stained PB (not shown). The percentage contribution to the granulocytic and lymphocytic lineages of PB from each marrow isotype (CD45.1/CD45.2) was determined. (C) Plotted is the mean (±SD) percentage contribution of donor-derived CD45.2+ cells in PB from mice that underwent transplantation with Runx1F/F—Tg(Mx1-Cre) (red symbols; n = 6) or Runx1F/F (black symbols; n = 6) and competitor bone marrow to total WBCs, granulocytes, and lymphocytes of recipient mice at 4, 8, and 19 weeks after transplantation. Two of 8 mice receiving Runx1F/F—Tg(Mx1-Cre) marrow that failed to show contribution of CD45.2+ marrow to PB of recipient mice at any time point are excluded. (D) Nineteen weeks after transplantation, EtBr-stained 3% agarose gel of 3-primer PCR of Runx1 loci of BM from mice that underwent competitive transplantation. (E) EtBr-stained 3% agarose gel of 3-primer PCR of Runx1 loci from BM of mice that underwent transplantation without competitor marrow.
Effects of Runx1 excision on the lymphoid lineage
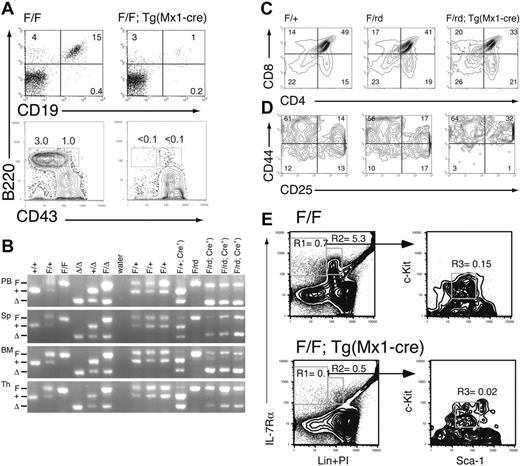
Mature B and T cells were present in pIpC-treated Runx1F/F—Tg(Mx1-Cre) mice, but the lymphocyte count in PB was reduced by an average of approximately 50% (Figure 1C). There was an average 19.7-fold (P = .007) reduction in the percentage of mature B cells (CD19+B220+) in the BM of excised mice (Figure 3A). Pre-B (IgM–NK1.1–Lin–CD43–B220+) and pro-B (IgM– NK1.1–Lin–CD43+B220+) cell precursors were nearly undetectable (less than 0.1%) in excised mice (Figure 3A), consistent with an early block in B-cell maturation.
There were also marked defects in T-lineage development in Mx1-Cre–excised Runx1 animals. The total cellularity of the thymus was reduced approximately 10-fold compared with littermate controls (not shown). Although there was 90% or greater excision of Runx1F allele in BM, the percentage of excised alleles was 50% or less in thymocytes from either Runx1F/F—Tg(Mx1-Cre) (not shown) or Runx1F/rd—Tg(Mx1-Cre) mice (Figure 3B). This suggests that Mx1-Cre expression in the thymus is lower than in BM. To distinguish between the effects of homozygous and heterozygous excision, we evaluated thymic T-cell maturation in Runx1F/rd—Tg(Mx1-Cre) (CXB6F1) and littermate mice. There was a 37% decrease (P = .033) in the relative percentage of CD4+CD8+ (double-positive [DP]) thymocytes in Mx1-Cre excised Runx1F/rd mice compared with Runx1F/+ mice (Figure 3C). We did not observe a significant expansion of the CD4–CD8– (DN) population. However, this was likely obscured by the approximately 40% increase (P = .024) in CD4+CD8– (SP4) cells compared with Runx1F/+ mice. This increase in SP4 cells likely resulted from the requirement for Runx1 repression of CD4 expression during thymocyte maturation from the DN to the DP stage.6,7 There was an approximate 3-fold decrease in the percentages of DN3 (CD44–CD25+)(P = .033) and DN4 (CD44–CD25–) (P = .011) cells relative to Runx1F/+ thymocytes (Figure 3D). Relative to Runx1F/rd and Runx1F/+—Tg(Mx1-Cre) mice, the percentage of DN3 and DN4 cells were reduced 4.3-fold (P = .018) and 2.3-fold (P = .018) in Runx1F/rd—Tg(Mx1-Cre) mice, respectively.
Lymphoid development is inhibited in Runx1-excised mice. (A) Representative B220/CD19 and B220/CD43 staining of BM from Runx1F/F and Runx1F/F—Tg(Mx1-Cre) mice. CD43 staining is shown for cells gated as IgM–NKK1.1–Lin–. Numbers indicate percentage of cells gated in each respective quadrant. (B) EtBr-stained 3% agarose gels with 3-primer PCR products of Runx1 loci in PB, spleen (Sp), BM, and thymocytes (Th) of mice 154 days after pIpC. Genotypes are as indicated. Control samples are as in Figure 2. The primers do not amplify a product from the Runx1rd allele. (C) Shown are CD4 and CD8 staining from representative animals of genotypes, Runx1F/+, Runx1F/rd, and Runx1F/rd—Tg(Mx1-Cre). Numbers indicate percentage of cells gated in each respective quadrant. (D) Shown are CD25 and CD44 staining of CD45.2+CD4–CD8– gated thymocytes from representative animals, as in panel C. (E) Composite of high-speed flow cytometric analysis of BM from representative Runx1F/F and Runx1F/F—Tg(Mx1-Cre) mice. Numbers indicate percentages of total BM for indicated populations. Gated populations indicated include IL-7Rα+Lin– (R1), IL-7Rα+Lin+ (R2) (which includes immature B cells), and CLP (R3), as indicated in Table 1. PI indicates propidium iodide.
Lymphoid development is inhibited in Runx1-excised mice. (A) Representative B220/CD19 and B220/CD43 staining of BM from Runx1F/F and Runx1F/F—Tg(Mx1-Cre) mice. CD43 staining is shown for cells gated as IgM–NKK1.1–Lin–. Numbers indicate percentage of cells gated in each respective quadrant. (B) EtBr-stained 3% agarose gels with 3-primer PCR products of Runx1 loci in PB, spleen (Sp), BM, and thymocytes (Th) of mice 154 days after pIpC. Genotypes are as indicated. Control samples are as in Figure 2. The primers do not amplify a product from the Runx1rd allele. (C) Shown are CD4 and CD8 staining from representative animals of genotypes, Runx1F/+, Runx1F/rd, and Runx1F/rd—Tg(Mx1-Cre). Numbers indicate percentage of cells gated in each respective quadrant. (D) Shown are CD25 and CD44 staining of CD45.2+CD4–CD8– gated thymocytes from representative animals, as in panel C. (E) Composite of high-speed flow cytometric analysis of BM from representative Runx1F/F and Runx1F/F—Tg(Mx1-Cre) mice. Numbers indicate percentages of total BM for indicated populations. Gated populations indicated include IL-7Rα+Lin– (R1), IL-7Rα+Lin+ (R2) (which includes immature B cells), and CLP (R3), as indicated in Table 1. PI indicates propidium iodide.
To determine the earliest step in lymphocyte development requiring Runx1, we enumerated the number of BM-derived CLPs (IL-7Rα+Lin–Sca-1loc-Kitlo) in Runx1F/F—Tg(Mx1-Cre)–excised mice. This revealed a greater than 7-fold decrease (P = .019) in the percentage of CLPs in the BM of Runx1-excised mice (Figure 3E; Table 1). We next evaluated lymphocyte commitment in mice after transplantation. In the competitive reconstitution assay, there was a complete lack of contribution of excised Runx1F/F—Tg(Mx1-Cre) donor cells to the lymphocyte lineages (Figure 2C). In noncompetitive repopulation assays, Runx1F/F—Tg(Mx1-Cre) contributed to T- and B-cell lineages to a limited extent (Table 2). The percentage of CD3+ T cells in Runx1-excised recipients was increased 2.5-fold compared with nonexcised recipients. However, 93% of CD3+ T cells were derived from the host BM (CD45.1+). Strikingly, peripheral blood B220+ B cells were decreased 58-fold in excised recipients compared with nonexcised recipients. In contrast to the T-cell lineage, 97% of PB B cells were donor derived. Together, these findings suggest that Runx1 is not required for lymphocyte maturation but that the loss of Runx1 inhibits lymphocyte maturation at early and late stages of development.
Effects of Runx1 excision on the myeloid lineage
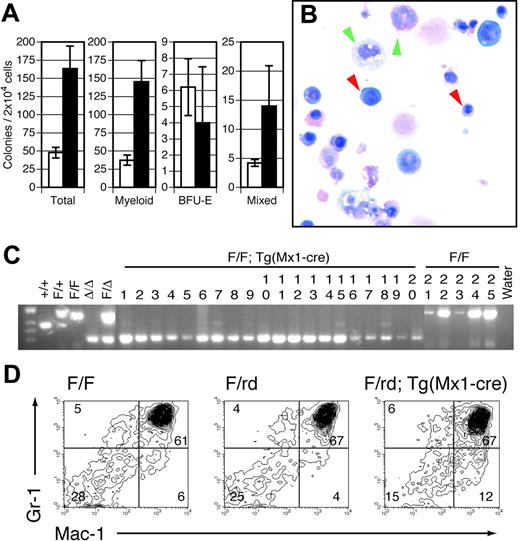
The marked defects in lymphocyte maturation observed in Runx1-excised mice are sharply contrasted with the effects of Runx1 excision on the myeloid lineage. All myeloid lineages, including granulocytes, monocytes, erythrocytes, and platelets, were represented in Mx1-Cre–excised Runx1F/F (Figure 1C) and Runx1F/rd (data not shown) animals. No significant abnormalities were observed in the erythroid lineage. However, we observed significant defects in the megakaryocytic lineage. Although there was no clinically evident bleeding diathesis, within 14 days of pIpC injection there was an approximately 80% reduction in average PB platelet counts in Runx1F/F—Tg(Mx1-Cre) (Figure 1C) and Runx1F/rd—Tg(Mx1-Cre) (not shown) mice compared with littermate controls. Histologic examination of BM from pIpC-treated Runx1F/F—Tg (Mx1-Cre) (Figure 4A-B) and Runx1F/rd—Tg(Mx1-Cre) (not shown) mice showed a marked decrease in the number of mature megakaryocytes compared with control mice, suggesting that the decrement in platelet count was attributed to decreased megakaryocyte maturation. This defect was observed in animals that underwent transplantation, indicating that the megakaryocyte maturation defect is cell autonomous (Figure 4C). Propidium iodide staining showed reduced polyploidization of CD41+ BM cells (Figure 4D). However, we paradoxically observed that in vitro megakaryocyte colony-plating efficiency was increased by an average of 2.9 (± 0.6)–fold (Figure 4E). Furthermore, megakaryocytic colonies displayed an increased number of acetylcholinesterase-positive cells per colony (Figure 4F-G).
Megakaryocyte maturation is inefficient in pIpC-treated Runx1-excised mice. BM from representative (A) Runx1F/F and (B) Runx1F/F—Tg(Mx1-Cre) mice 143 days after pIpC injection shows an absence of normal megakaryocytes in Runx1-excised mice (hematoxylin and eosin [H&E] staining; original magnification, × 600). (C) BM from representative Runx1F/F—Tg(Mx1-Cre) recipient mice 98 days after transplantation (H&E; original magnification, × 600). (A-C) Yellow arrows indicate representative megakaryocytes. Red and green arrowheads indicate representative erythroid and myeloid elements, respectively. Sections demonstrate a notable absence of megakaryocytes and an increased ratio of maturing myeloid-to-erythroid forms in Runx1-excised versus nonexcised marrows. (D) Histograms depicting number of cultured Runx1F/F and Runx1F/F—Tg (Mx1-Cre) bone marrow cells stained with propidium iodide to show ploidy. Colors indicate CD41 gates: green, CD41+; red, CD41–. (E, left) Plotted are mean ± SD numbers of acetylcholinesterase-positive colonies per 5 × 104 BM cells plated from pIpC-treated Runx1F/F (□) and Runx1F/F—Tg(Mx1-Cre) (▪) mice. Results shown are for 3 experiments performed in quadruplicate. 1 and 3, fresh marrow; 2, previously frozen marrow. (Right) Plotted is the average fold increase for Runx1F/F—Tg(Mx1-Cre) (▪) relative to Runx1F/F (□) in the 3 experiments (P ≤ .034). Representative megakaryocyte colonies from (F) Runx1F/F and (G) Runx1F/F—Tg(Mx1-Cre) mice show acetylcholinesterase staining (brown) of megakaryocytic cells (original magnification, × 100).
Megakaryocyte maturation is inefficient in pIpC-treated Runx1-excised mice. BM from representative (A) Runx1F/F and (B) Runx1F/F—Tg(Mx1-Cre) mice 143 days after pIpC injection shows an absence of normal megakaryocytes in Runx1-excised mice (hematoxylin and eosin [H&E] staining; original magnification, × 600). (C) BM from representative Runx1F/F—Tg(Mx1-Cre) recipient mice 98 days after transplantation (H&E; original magnification, × 600). (A-C) Yellow arrows indicate representative megakaryocytes. Red and green arrowheads indicate representative erythroid and myeloid elements, respectively. Sections demonstrate a notable absence of megakaryocytes and an increased ratio of maturing myeloid-to-erythroid forms in Runx1-excised versus nonexcised marrows. (D) Histograms depicting number of cultured Runx1F/F and Runx1F/F—Tg (Mx1-Cre) bone marrow cells stained with propidium iodide to show ploidy. Colors indicate CD41 gates: green, CD41+; red, CD41–. (E, left) Plotted are mean ± SD numbers of acetylcholinesterase-positive colonies per 5 × 104 BM cells plated from pIpC-treated Runx1F/F (□) and Runx1F/F—Tg(Mx1-Cre) (▪) mice. Results shown are for 3 experiments performed in quadruplicate. 1 and 3, fresh marrow; 2, previously frozen marrow. (Right) Plotted is the average fold increase for Runx1F/F—Tg(Mx1-Cre) (▪) relative to Runx1F/F (□) in the 3 experiments (P ≤ .034). Representative megakaryocyte colonies from (F) Runx1F/F and (G) Runx1F/F—Tg(Mx1-Cre) mice show acetylcholinesterase staining (brown) of megakaryocytic cells (original magnification, × 100).
Immunophenotypic analysis of myeloid progenitors derived from BM showed that the CMP population (IL-7Rα–Lin–Sca-1–c-Kit+CD34+FcγRII/III+) was not significantly altered in Runx1-excised mice. However, the GMP population (IL-7Rα–Lin–Sca-+–c-Kit+CD34+FcγRII/IIIhi) was expanded approximately 3-fold (P = .032) in Runx1-excised mice (Table 1). These observations are consistent with the increased production of myeloid colonies in vitro (Figure 5A). Unfractionated BM derived from pIpC-treated Runx1F/F—Tg(Mx1-Cre) mice had an approximately 3.4-fold increase in total myeloid colonies relative to pIpC-treated Runx1F/F mice. Microscopic evaluation of Wright-Giemsa–stained cytospins of granulocyte-macrophage (GM) and mixed-lineage colonies indicated that Runx1-excised colonies gave rise to the full range of myeloid cell types observed in control colonies (Figure 5B). GM and mixed-lineage colonies were increased 3.9- and 3.2-fold compared with controls, respectively. In contrast, there was no significant change in erythroid blast-forming unit (BFU-E) colony number. Evaluation of individual colonies by PCR indicated complete Runx1 excision in 92% of evaluable colonies from pIpC-treated Runx1F/F—Tg(Mx1-Cre) mice (Figure 5C) compared with Runx1F/F controls.
Perhaps the most unexpected finding in adult Runx1-excised mice was a mild myeloid expansion in hematopoietic tissues. Histopathologic examination of Runx1-excised BM showed subtle myeloid hyperplasia with a consistent increase in the ratio of maturing myeloid to erythroid forms compared with controls (Figure 4A-B). Runx1-excised marrows were hypercellular, yielding twice as many cells as control marrows. Similar findings were observed in mice after noncompetitive transplantation (Figure 4C). The histopathologic findings were supported by a 2.4-fold expansion of Mac1+ cells in excised BM of Runx1F/F—Tg(Mx1-Cre) (P = .021) (data not shown) and Runx1F/rd—Tg(Mx1-Cre) (P = .021) (Figure 5D) compared with controls.
Mature and progenitor myeloid cell populations are expanded in pIpC-treated Runx1-excised mice. (A) Plotted are the mean ± SD of total (P < .05), myeloid (P < .05), BFU-E (P = .5), and mixed-lineage colonies (P = <.05) per 2 × 104 BM cells plated in vitro from pIpC-treated Runx1F/F (□) and Runx1F/F—Tg(Mx1-Cre) (▪) mice. Results are the average of 3 experiments performed in duplicate. (B) Representative cytospin from mixed-lineage colony derived from Runx1F/F—Tg(Mx1-Cre) BM (Wright-Giemsa staining; original magnification, × 600). Red arrowheads indicate representative erythroid elements. Green arrowheads denote myeloid and monocytic forms. (C) Representative EtBr-stained 3% agarose gel of Runx1 PCR products from single GM or mixed-lineage colonies derived from Runx1F/F—Tg(Mx1-Cre) (lanes 1-20) and Runx1F/F (lanes 21-25) mice. Of 70 Runx1F/F—Tg(Mx1-Cre) colonies evaluated, 9 (12.8%) failed to show any amplification, 5 of 61 (8.2%) indicated partial excision, and 56 of 61 (91.8%) were completely excised. Tail and embryo genomic DNA controls are as indicated. (D) Gr1 and Mac1 staining of BM from representative CXB6F1 mice 154 days after pIpC induction. Numbers indicate percentages of cells gated in each respective quadrant.
Mature and progenitor myeloid cell populations are expanded in pIpC-treated Runx1-excised mice. (A) Plotted are the mean ± SD of total (P < .05), myeloid (P < .05), BFU-E (P = .5), and mixed-lineage colonies (P = <.05) per 2 × 104 BM cells plated in vitro from pIpC-treated Runx1F/F (□) and Runx1F/F—Tg(Mx1-Cre) (▪) mice. Results are the average of 3 experiments performed in duplicate. (B) Representative cytospin from mixed-lineage colony derived from Runx1F/F—Tg(Mx1-Cre) BM (Wright-Giemsa staining; original magnification, × 600). Red arrowheads indicate representative erythroid elements. Green arrowheads denote myeloid and monocytic forms. (C) Representative EtBr-stained 3% agarose gel of Runx1 PCR products from single GM or mixed-lineage colonies derived from Runx1F/F—Tg(Mx1-Cre) (lanes 1-20) and Runx1F/F (lanes 21-25) mice. Of 70 Runx1F/F—Tg(Mx1-Cre) colonies evaluated, 9 (12.8%) failed to show any amplification, 5 of 61 (8.2%) indicated partial excision, and 56 of 61 (91.8%) were completely excised. Tail and embryo genomic DNA controls are as indicated. (D) Gr1 and Mac1 staining of BM from representative CXB6F1 mice 154 days after pIpC induction. Numbers indicate percentages of cells gated in each respective quadrant.
Gross and histopathologic findings consistent with a myeloproliferative phenotype included splenomegaly attributable to extramedullary hematopoiesis in the spleen, with an increase in spleen weight of 1.4- to 2.1-fold in Runx1-excised mice compared with controls (Table 3). Histopathologic examination of spleen sections showed effacement of splenic architecture with a mild expansion of the red pulp resulting from extramedullary hematopoiesis composed predominantly of maturing myeloid forms and admixed erythroid elements, with only scant to rare megakaryocytes (Figure 6A-B). Liver sections from Runx1-excised mice also showed extramedullary hematopoiesis composed of maturing myeloid elements in a perivascular distribution (Figure 6C-D). Spleens of mice that underwent transplantation with excised Runx1F/F—Tg(Mx1-Cre) marrow in the absence of wild-type competitor cells also demonstrated extramedullary hematopoiesis (Figure 6E-F). Flow cytometric analysis of single cell suspensions derived from spleens of primary Runx1-excised animals and animals that underwent transplantation with Runx1-excised BM confirmed the increase of myeloid lineage cells in the spleen (Figure 6G; Table 4). In Runx1F/F—Tg(Mx1-Cre) mice and mice after transplantation, but not in Runx1F/rd—Tg(Mx1-Cre) mice, there was a significant expansion of CD45–Ter119+ cells in the spleen (Figure 6G; Table 4). Taken together, these data indicate that Runx1 is not required for neutrophil development, but its loss of function contributes to a myeloproliferative phenotype.
Spleen weights of pIpC-treated mice
Genotype . | No. mice . | Days after pIpC . | Spleen weight, mg, mean ± SD . | Fold increase . |
|---|---|---|---|---|
| Runx1F/F | 7 | 60-273 | 153 ± 48 | — |
| Runx1F/F—Tg(Mx1-Cre) | 9 | 91-273 | 318‡ ± 195 | 2.08 |
| Runx1F/+ | 4 | 154 | 122 ± 26 | — |
| Runx1F/rd and Runx1F/+—Tg(Mx1-Cre) | 5 | 154 | 129 ± 14 | — |
| Runx1F/rd—Tg(Mx1-Cre) | 6 | 154 | 185§ ± 42 | 1.52∥/1.43¶ |
| Runx1F/F recipients* | 2 | 98† | 84 ± 21 | — |
| Runx1F/F—Tg(Mx1-Cre) recipients* | 4 | 98† | 128 ± 10 | 1.53 |
Genotype . | No. mice . | Days after pIpC . | Spleen weight, mg, mean ± SD . | Fold increase . |
|---|---|---|---|---|
| Runx1F/F | 7 | 60-273 | 153 ± 48 | — |
| Runx1F/F—Tg(Mx1-Cre) | 9 | 91-273 | 318‡ ± 195 | 2.08 |
| Runx1F/+ | 4 | 154 | 122 ± 26 | — |
| Runx1F/rd and Runx1F/+—Tg(Mx1-Cre) | 5 | 154 | 129 ± 14 | — |
| Runx1F/rd—Tg(Mx1-Cre) | 6 | 154 | 185§ ± 42 | 1.52∥/1.43¶ |
| Runx1F/F recipients* | 2 | 98† | 84 ± 21 | — |
| Runx1F/F—Tg(Mx1-Cre) recipients* | 4 | 98† | 128 ± 10 | 1.53 |
Mice that underwent transplantation with Runx1F/F or Runx1F/FTg(Mx1-Cre) marrow in the absence of competitor marrow.
Days after transplantation.
Comparison with Runx1F/F (P < .01; Mann-Whitney U test).
Comparison with Runx1F/+ (P = .033). Comparison with Runx1F/rd and Runx1F/+—Tg(Mx1-Cre) (P = .068; Mann-Whitney U test).
Comparison with Runx1F/+.
Comparison with Runx1F/rd and Runx1F/+—Tg(Mx1-Cre).
Myeloid expansion in spleens of Runx1-excised mice
Genotype . | No. mice . | Time after pIpC, d . | Gr1+ splenocytes, mean ± SD, %*(P) . | Gr1+Mac1+ splenocytes, mean ± SD, %*(P) . | Mac1+ splenocytes, mean ± SD, %*(P) . | Ter119+/CD45- splenocytes, mean ± SD, %*(P)† . |
|---|---|---|---|---|---|---|
| Runx1F/F‡ | 4 | 186-190 | 2.9 ± 0.6 (.999∥¶) | 1.6 ± 0.4 (.008∥) | 6.1 ± 1.4 (.014∥) | 2.9 ± 1.6 (.023∥) |
| Runx1F/F—Tg(Mx1-Cre)‡ | 7 | 186-190 | 2.7 ± 1.9 | 10.2 ± 4.7 | 14.9 ± 8.8 | 20.6 ± 15.5 |
| Runx1F/+ | 4 | 154 | 5.7 ± 1.5 (.020#) | 2.5 ± 0.2 (.020#) | 1.8 ± 0.2 (.020#) | 2.0 ± 1.3 (.136¶#) |
| Runx1F/+—Tg(Mx1-Cre) and Runx1F/rd | 5 | 154 | 7.5 ± 1.2 (.465¶**) | 5.4 ± 2.8 (.029**) | 12.0 ± 0.6 (.006**) | 3.1 ± 2.1 (.715¶**) |
| Runx1F/rd—Tg(Mx1-Cre) | 6 | 154 | 8.3 ± 1.2 | 12.7 ± 5.3 | 4.0 ± 0.5 | 3.8 ± 2.0 |
| Runx1F/F (recipient)‡ | 2 | 96§ | 3.1 ± 1.2 (.098¶††) | 2.5 ± 0.1 (.040††) | 1.9 ± 0.5 (.061¶††) | 2.8 ± 1.7 (.003††) |
| Runx1F/F—Tg(Mx1-Cre) (recipient)‡ | 4 | 96§ | 5.8 ± 1.6 | 7.4 ± 2.2 | 3.9 ± 1.0 | 50.0 ± 9.3 |
Genotype . | No. mice . | Time after pIpC, d . | Gr1+ splenocytes, mean ± SD, %*(P) . | Gr1+Mac1+ splenocytes, mean ± SD, %*(P) . | Mac1+ splenocytes, mean ± SD, %*(P) . | Ter119+/CD45- splenocytes, mean ± SD, %*(P)† . |
|---|---|---|---|---|---|---|
| Runx1F/F‡ | 4 | 186-190 | 2.9 ± 0.6 (.999∥¶) | 1.6 ± 0.4 (.008∥) | 6.1 ± 1.4 (.014∥) | 2.9 ± 1.6 (.023∥) |
| Runx1F/F—Tg(Mx1-Cre)‡ | 7 | 186-190 | 2.7 ± 1.9 | 10.2 ± 4.7 | 14.9 ± 8.8 | 20.6 ± 15.5 |
| Runx1F/+ | 4 | 154 | 5.7 ± 1.5 (.020#) | 2.5 ± 0.2 (.020#) | 1.8 ± 0.2 (.020#) | 2.0 ± 1.3 (.136¶#) |
| Runx1F/+—Tg(Mx1-Cre) and Runx1F/rd | 5 | 154 | 7.5 ± 1.2 (.465¶**) | 5.4 ± 2.8 (.029**) | 12.0 ± 0.6 (.006**) | 3.1 ± 2.1 (.715¶**) |
| Runx1F/rd—Tg(Mx1-Cre) | 6 | 154 | 8.3 ± 1.2 | 12.7 ± 5.3 | 4.0 ± 0.5 | 3.8 ± 2.0 |
| Runx1F/F (recipient)‡ | 2 | 96§ | 3.1 ± 1.2 (.098¶††) | 2.5 ± 0.1 (.040††) | 1.9 ± 0.5 (.061¶††) | 2.8 ± 1.7 (.003††) |
| Runx1F/F—Tg(Mx1-Cre) (recipient)‡ | 4 | 96§ | 5.8 ± 1.6 | 7.4 ± 2.2 | 3.9 ± 1.0 | 50.0 ± 9.3 |
Numbers indicate mean percentage Mac1+, Gr1+, or Mac1+Gr1+ of total splenocytes. Primary pIpC-treated splenocytes were stained with CD45.2-FITC, Mac1-PE, 7AAD-PerCP-Cy5.5, and Gr1-APC and were gated by scatter and 7AAD negativity. Splenocytes from animals that underwent transplantation were stained with CD45.2-FITC, Mac1-PE, CD45.2-PerCP-Cy5.5, and Gr1-APC and were scatter gated.
Numbers indicate mean percentage CD45-Ter119+ of total splenocytes. Splenocytes were stained with CD45.2-FITC, c-Kit-PE, 7AAD-PerCP-Cy5.5, and Ter119-APC. Splenocytes were gated by scatter and 7AAD negativity.
Representative data in Figure 6.
Days after transplantation.
Mann-Whitney U test for comparison of primary Runx1F/F—Tg(Mx1-Cre) to primary Runx1F/F animals.
Not statistically significant.
Mann-Whitney U test for comparison of primary Runx1F/rd—Tg(Mx1-Cre) to primary Runx1F/+ animals.
Mann-Whitney U test for comparison of primary Runx1F/rd—Tg(Mx1-Cre) to primary Runx1F/+—Tg(Mx1-Cre) and Runx1F/rd animals.
Unpaired t test for comparison of mice that underwent transplantation with Runx1F/F—Tg(Mx1-Cre) to mice that underwent transplantation with Runx1F/F animals.
Discussion
Runx1 deficiency during development results in a failure of HSC emergence and lethal central nervous system (CNS) hemorrhage, but the data presented here indicate that Runx1 is not required in the adult murine hematopoietic compartment for HSC maintenance or survival of adult mice. However, there were a number of mild to severe hematopoietic abnormalities, including a block in lymphoid development, reduced platelet production, and development of a myeloproliferative phenotype.
Runx1-excised adult mice have an expanded, immunophenotypically defined HSC compartment. These mice survive with a follow-up of more than 5 months, indicating that there is long-term HSC activity in these animals. This finding is surprising in light of the hematopoietic defect observed in Runx1-deficient embryos. We note that homozygous loss of RUNX1 has been observed in humans with AML, though not in those with acute lymphoid leukemia.38 These observations suggest that AML cells with self-renewal ability and limited ability to differentiate also do not require RUNX1. Our findings that Runx1 is not required for HSC maintenance and survival in the adult are consistent with the persistence of RUNX1-deficient cells in human leukemias. BM transplantation studies indicate that Runx1-excised HSCs are competent for long-term repopulation. However, competitive repopulation studies indicate a significant reduction in Runx1-deficient HSC ability to compete with wild-type HSCs during repopulation.
Runx1-excised mice display myeloid expansion in spleen and liver. Spleen (A-B) and liver (C-D) sections from representative (A,C) Runx1F/F and (B,D) Runx1F/F—Tg(Mx1-Cre) mice 143 days after pIpC (H&E; original magnification, ×100). Insets' original magnification, × 600. (E-F) Spleen sections from representative mice 14 weeks after transplantation with (E) Runx1F/F and (F) Runx1F/F—Tg(Mx1-Cre) marrow (H&E; original magnification, × 100). Insets' original magnification, ×600. (A-F) Yellow arrows indicate representative lymphocytes. Red and green arrowheads indicate representative erythroid and myeloid elements, respectively. Sections demonstrate the presence of extramedullary hematopoiesis in the liver and splenic red pulp in Runx1-excised mice, absent in nonexcised control animals. (G) Gr1/Mac1 and CD45.2/Ter119 staining of splenocytes of indicated genotype from representative primary mice or mice that underwent transplantation. Numbers indicate percentage of cells gated in each respective quadrant.
Runx1-excised mice display myeloid expansion in spleen and liver. Spleen (A-B) and liver (C-D) sections from representative (A,C) Runx1F/F and (B,D) Runx1F/F—Tg(Mx1-Cre) mice 143 days after pIpC (H&E; original magnification, ×100). Insets' original magnification, × 600. (E-F) Spleen sections from representative mice 14 weeks after transplantation with (E) Runx1F/F and (F) Runx1F/F—Tg(Mx1-Cre) marrow (H&E; original magnification, × 100). Insets' original magnification, ×600. (A-F) Yellow arrows indicate representative lymphocytes. Red and green arrowheads indicate representative erythroid and myeloid elements, respectively. Sections demonstrate the presence of extramedullary hematopoiesis in the liver and splenic red pulp in Runx1-excised mice, absent in nonexcised control animals. (G) Gr1/Mac1 and CD45.2/Ter119 staining of splenocytes of indicated genotype from representative primary mice or mice that underwent transplantation. Numbers indicate percentage of cells gated in each respective quadrant.
Runx1 is required for efficient lymphoid maturation at multiple stages of differentiation. We observed a significant reduction in the number mature PB B cells and BM-derived B-cell precursors in primary Runx1-excised mice. This suggests a significant block in B-cell maturation, though it is unclear at which stage of B-cell maturation Runx1 functions. In the T-cell lineage, we report a specific block in T-cell maturation during the transition from the DN2 (CD44+CD25+) to the DN3 (CD44–CD25+) stage.56 This is consistent with reports describing a tightly regulated expression pattern for Runx1 and CBFβ in developing thymocytes.6,7,15,16,58 However, in contrast to the findings of Ichikawa et al,56 we observed a significant reduction in bone marrow CLP, suggesting that Runx1 is also important during the earliest stages of lymphoid lineage development. The difference in CLP production we observed compared with the observations of Ichikawa et al56 may be the result of higher pIpC dosing in our study, resulting in more efficient excision, although other explanations are possible. Noncompetitive reconstitution assays indicate that Runx1 is not required for commitment to and maturation of the lymphoid lineage. It is clear that the loss of Runx1 severely reduces the efficiency of CLP production and lymphocyte maturation. This conclusion is supported by the clinical observation that RUNX1 loss is not observed in patients with acute lymphoblastic leukemia (ALL).
In contrast to the pronounced inhibition of the lymphoid lineage, excision of Runx1 did not inhibit maturation of the myeloid lineage. Runx1 loss had no apparent effect on the erythroid lineage, but it did have a significant effect on megakaryocytic maturation. Runx1 is required for normal maturation of the megakaryocyte lineage but not for the establishment of this lineage. These differential requirements for Runx1 in erythroid and megakaryocyte maturation correlate with reports that the expression of Runx1 and its heterodimeric binding partner, CBFβ, are markedly decreased in erythroid lineages15,16,59 but are maintained in megakaryocytes.16,59,60 Humans with FPD/AML syndrome have heterozygous loss of RUNX1, with an associated quantitative and qualitative platelet defect, and a proclivity to develop AML.35 The platelet defect is reminiscent of the abnormalities observed in mice with homozygous loss of Runx1 and suggests that there may be compensation in mice for monoallelic loss of Runx1.
RUNX1 loss in humans is most often associated with M0 (undifferentiated) AML, suggesting that RUNX1 loss may have deleterious effects on hematopoietic differentiation.61,62 In contrast, Runx1-excised mice demonstrate expansion of the myeloid lineage by several phenotypic and functional criteria, with no evidence of a block in myeloid development. The specific expansion of the GMP, but not the CMP, population suggests that the myeloid expansion observed in Runx1-excised animals results from the loss of negative regulatory functions of Runx1 in the GMP population. The strain-specific differences in expansion of CD45–—Ter119+ cells in the spleens of Runx1-excised mice indicate that the myeloid expansion is not secondary to a compensatory drive for increased platelet production. Although these findings are surprising, there have been several other indications that Runx1 is not essential for adult hematopoiesis in mice. For example, expression of the leukemogenic fusion protein RUNX1-ETO, a dominant-negative inhibitor of CBF, during development results in an embryonic lethal phenotype with a lack of definitive hematopoiesis that is nearly identical to that observed in Runx1-deficient mice.44,45 However, expression of Runx1-ETO from a conditional allele in the adult hematopoietic compartment has only subtle effects on hematopoiesis, characterized by enhanced replating efficiency of myeloid progenitors in vitro and an increased susceptibility to develop leukemia after N-ethyl-N-nitrosurea (ENU) mutagenesis.63 Runx1-ETO or Runx1-deficient animals do not develop leukemia, suggesting that Runx1 loss of function must be complemented with additional leukemogenic genetic events for the development of a leukemia phenotype. The myeloproliferative phenotype associated with Runx1 excision suggests that Runx1 loss may constitute a preleukemic state, and it may explain the proclivity for the development of myeloid leukemias in the context of RUNX1 deficiency in humans. The ability of Runx1-deficient myeloid lineage cells to differentiate normally suggests that the block in differentiation observed in leukemias with loss of RUNX1 function cannot be attributed to the loss of RUNX1 alone and that it must be due to other genetic events.
Prepublished online as Blood First Edition Paper, March 22, 2005; DOI 10.1182/blood-2004-08-3280.
Supported in part by the American Cancer Society (postdoctoral fellowship grant PF-02-133-01-LIB) (J.D.G.), the Public Health Service (grant R01CA58343) (N.A.S.), the National Cancer Institute (grant CA66996), the National Institute of Diabetes and Digestive and Kidney Diseases (grant 50564), and the Leukemia and Lymphoma Society (D.G.G.). The transgenic mouse facility at Dartmouth is supported in part by a core grant from the Norris Cotton Cancer Center. D.G.G is an Investigator for the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Woo Jong Song, Terryl Stacy, Vivienne Rebel, and Hanno Hock for their insightful discussions and contributions to the completion of this work.

![Figure 1. Inducible Runx1 excision in adult mice induces thrombocytopenia. (A) Schematic representation of Runx1 gene-targeting strategy used to flank exon 4 with LoxP-targeting sites. B indicates BamHI; S, SspI; E4, Runt domain exon 4. (filled boxes) 5′ and 3′ targeting probes. (dashed arrows) BamHI-digested genomic DNA fragments for wild-type (7.3-kb), Floxed (2.8-kb), and excised (2.5-kb) DNA detected with excision (Δ) probe (open boxes) located 3′ to the distal LoxP site. (B) Southern blot analysis of BamHI-digested genomic DNA from bone marrow [B] and spleen [S] cells of representative mice killed at 14, 35, and 91 days after pIpC injection. The dose of pIpC is indicated. The blots are probed with Δ probe, which detects the Floxed (2.8-kb) and excised (2.5-kb) Runx1 alleles. Numbers indicate percentage excision. (C) Mean ± SD of total WBC counts, total red blood cell (RBC) counts, total platelet counts, percentage lymphocytes, and percentage neutrophils in PB of pIpC-treated Runx1F/F—Tg(Mx1-Cre) (red symbols) and Runx1F/F (black symbols) mice. Mice were bled 12 days before pIpC injection and 14, 21, 28, 35, 49, 91, 119, and 154 days after pIpC injection. The number of animals evaluated for each genotype at the respective time points is N (Runx1F/F—Tg(Mx1-Cre) = 14, 14, 12, 8, 10, 9, 8, 7 and 6; N (Runx1F/F) = 7, 5, 7, 7, 5, 7, 6, 6, and 6.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/2/10.1182_blood-2004-08-3280/6/m_zh80140581270001.jpeg?Expires=1769120969&Signature=AYYc7pXiS~DsS-6flvmOJO5~Ah~g4YOiNLaUMXMy7IA1~reV2T8fvYNlFVJR05qVodxT6INgfr86-s-bmkPGuCr0BWFKbyE5h0wpBmslMkS~xWHADZZGZFvs31JvW0YZ2xtV4TgR3DGzmKx-rHTOJnFeYpC~ADoTmlq-icU5s2djziG9tdY3w1hbSPpMj2z3xNhNVZo1~L0M8ae5W54gp4WfhscKrOekviLT9FAYqVolJDV4vbKV59uDKhRPicR7J1IbRow7MQkMbXTQzuJDek82Fzow~UcKl9iq6xoDZiH2batQcT1u6qlqnLl3RHJdBFccjZzEVO8rDWULOItI7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Megakaryocyte maturation is inefficient in pIpC-treated Runx1-excised mice. BM from representative (A) Runx1F/F and (B) Runx1F/F—Tg(Mx1-Cre) mice 143 days after pIpC injection shows an absence of normal megakaryocytes in Runx1-excised mice (hematoxylin and eosin [H&E] staining; original magnification, × 600). (C) BM from representative Runx1F/F—Tg(Mx1-Cre) recipient mice 98 days after transplantation (H&E; original magnification, × 600). (A-C) Yellow arrows indicate representative megakaryocytes. Red and green arrowheads indicate representative erythroid and myeloid elements, respectively. Sections demonstrate a notable absence of megakaryocytes and an increased ratio of maturing myeloid-to-erythroid forms in Runx1-excised versus nonexcised marrows. (D) Histograms depicting number of cultured Runx1F/F and Runx1F/F—Tg (Mx1-Cre) bone marrow cells stained with propidium iodide to show ploidy. Colors indicate CD41 gates: green, CD41+; red, CD41–. (E, left) Plotted are mean ± SD numbers of acetylcholinesterase-positive colonies per 5 × 104 BM cells plated from pIpC-treated Runx1F/F (□) and Runx1F/F—Tg(Mx1-Cre) (▪) mice. Results shown are for 3 experiments performed in quadruplicate. 1 and 3, fresh marrow; 2, previously frozen marrow. (Right) Plotted is the average fold increase for Runx1F/F—Tg(Mx1-Cre) (▪) relative to Runx1F/F (□) in the 3 experiments (P ≤ .034). Representative megakaryocyte colonies from (F) Runx1F/F and (G) Runx1F/F—Tg(Mx1-Cre) mice show acetylcholinesterase staining (brown) of megakaryocytic cells (original magnification, × 100).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/2/10.1182_blood-2004-08-3280/6/m_zh80140581270004.jpeg?Expires=1769120969&Signature=hw9sMsMX4l1VuW2JWx4KaTpNXBRj-5PursV62Aph8Bho3M8uICtzrHws8cKECPwNlx5o42xLLKXAMKLtmkQt1jSL05djbLUAPnLUTabOCtjpU9cbnj--ug6zqTOxFEEK-96O~G1mZgtVURHSjinX3J3spowJFA2RpyNk9lwoI-QdD8hdwvrHRrF0i5gkZAcP2BN2P~wB09epwLtSMOvA-eOz7pwC1AHq1agYdXA8S91dl7CyGTtZ4yO8cvxfcCyxSBfMQ3GXUSEd8i~MmEzeo6oDlBcUZ0dDnbXfob8zJyW4avjKTZJjWbGxB~aUHWLbzW58cz~aT60c-MgNwNIBfg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal