Abstract
Recurrent chromosomal rearrangements are associated with the development of acute myeloid leukemia (AML). The frequent inversion of chromosome 16 creates the CBFB-MYH11 fusion gene that encodes the fusion protein CBFβ-SMMHC. This fusion protein inhibits the core-binding factor (CBF), resulting in a block of hematopoietic differentiation, and induces leukemia upon the acquisition of additional mutations. A recent genetic screen identified Plag1 and Plagl2 as CBFβ-SMMHC candidate cooperating proteins. In this study, we demonstrate that Plag1 and Plagl2 independently cooperate with CBFβ-SMMHC in vivo to efficiently trigger leukemia with short latency in the mouse. In addition, Plag1 and Plagl2 increased proliferation by inducing G1 to S transition that resulted in the expansion of hematopoietic progenitors and increased cell renewal in vitro. Finally, PLAG1 and PLAGL2 expression was increased in 20% of human AML samples. Interestingly, PLAGL2 was preferentially increased in samples with chromosome 16 inversion, suggesting that PLAG1 and PLAGL2 may also contribute to human AML. Overall, this study shows that Plag1 and Plagl2 are novel leukemia oncogenes that act by expanding hematopoietic progenitors expressing CbFβ-SMMHC.
Introduction
The core-binding factor (CBF) is the most common target of chromosomal translocations in acute myeloid leukemia (AML). Frequent rearrangements affecting CBF include the chromosome 16 inversion inv(16)(p13;q22) [hereafter inv(16)] and the translocation t(8;21)(q22;q22).1 CBF is a heterodimeric transcription factor composed of 1 of 3 DNA-binding α subunits (encoded by RUNX1, RUNX2, and RUNX3) and a non-DNA binding β subunit (encoded by CBFB). In hematopoiesis, the RUNX1:CBFβ complex is a key regulator of lymphoid and myeloid differentiation. For example, studies using knock-out mice demonstrated that embryonic definitive hematopoiesis is defective in the absence of Cbfβ or Runx1.2-5 In adult hematopoiesis, Runx1 knockouts show deficient differentiation of T cells, B cells, and myeloid cells.6
The inv(16) breaks and joins CBFB with the smooth muscle myosin heavy chain (MYH11) gene, creating the CBFB-MYH11 fusion gene, which encodes the CBFβ-SMMHC fusion protein.7 Interestingly, Cbfb+/MYH11 knock-in embryos expressing this fusion protein lack definitive hematopoiesis, as shown in Cbfb and Runx1 knock-out embryos, thus indicating that Cbfβ-SMMHC is a dominant inhibitor of CBF function.8 Normal lymphoid and myeloid differentiation was also impaired in hematopoietic stem cells from Cbfb+/MYH11 knock-in chimeras expressing Cbfβ-SMMHC. In addition, these mice developed AML after induction of additional mutations.9 However, little is known about the genes and pathways that cooperate with CBFβ-SMMHC in leukemogenesis. We have recently performed a genetic screen, by means of retroviral insertional mutagenesis (RIM), in Cbfb+/MYH11 knock-in chimeras to identify genes that could synergize with Cbfβ-SMMHC in leukemia development.10 The study identified retroviral insertions at the Plag1 locus in as many as 40% of the samples and at the Plagl2 locus in 10% of the samples. Interestingly, the proviral insertions mapped upstream of the translation-start site, thus suggesting that up-regulation of these genes may contribute to AML.
The PLAG family is composed of 3 members (PLAG1, PLAGL1/LOT1/ZAC1, and PLAGL2) with highly conserved structure and function.11 These proteins are transcription factors that include a C-terminal trans-activation domain preceded by 7 C2H2 zinc fingers with DNA binding function. Genetic studies have associated PLAG1 with benign tumors harboring 8q12 translocations, including pleomorphic adenomas of the salivary gland, lipoblastomas, and hepatoblastomas.12-14 PLAGL2 has not been previously associated with human cancer, but it has been reported to have similar DNA-binding affinity to the Plag1 consensus sequence and to induce proliferation in NIH-3T3 cells.15 Conversely, PLAGL1 seems to function as a tumor suppressor protein that is found mutated in breast and pituitary tumors16,17 and that regulates apoptosis and G cell cycle arrest.18
In the present study we tested whether Plag1 and PLAGL2 are oncogenes in AML using a combination of in vivo and in vitro assays. We found that these transcription factors are specifically up-regulated in RIM-induced leukemic samples with retroviral insertions within these loci. To functionally validate their role in leukemogenesis we used a bone marrow transduction assay followed by transplantation (tBMT). Coexpression of either Plag1 or PLAGL2 and Cbfb-MYH11 efficiently induced AML in 100% of recipient mice with a latency of 3 to 12 weeks. Furthermore, Plag1 or PLAGL2 induced expansion of hematopoietic progenitors and increased proliferation by inducing G1 to S transition in vitro. Importantly, PLAG1 and PLAGL2 are overexpressed in 20% of human AML. Among these, PLAGL2 is preferentially overexpressed in samples carrying inv(16). These results are in concordance with our in vivo studies in the mouse and highlight PLAG1 and PLAGL2 oncogenic function in human leukemia.
Materials and methods
Reverse transcriptase and quantitative PCR (qPCR) analyses
RT and qPCR of murine samples. RNA was extracted with guanidium thiocyanate followed by centrifugation in cesium chloride solution or with Trizol (Invitrogen, Carlsbad CA) according to the manufacturer's protocol. First-stand cDNA was generated by using 2 μg RNA, 1 U Superscript-2 reverse transcriptase (RT; Invitrogen), and 0.1 μg oligo dT primer in a 20-μL reaction. Each polymerase chain reaction (PCR) included 1 μL cDNA, 1 μL 10 mM primers, 1 μL 10 mM deoxyribonucleoside triphosphates (dNTPs), 5 μL10 × buffer, 1 U Taq Polymerase (Fisher Scientific, Pittsburgh PA). PCR conditions included a 2-minute step at 95°C followed by a 30-cycle amplification (each with a 30-second denaturing step at 95°C, a 30-second annealing step at 56°C, and a 30-second extension step at 72°C). SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) was used for qPCR according to the manufacturer's instructions. Plag1 primers were mP1 × 1 (GGTTCACTCCTTCTCTCACACG) and mP1 × 2 (TGAGTAGCCATGTGCCTTTGTA). Plagl2 primers were mPL2 × 1 (TATAGGCACATGGCCACCCACT) and mPl2 × 2 (TGACGACGGTATCCCAGCTTT). The long terminal repeat (LTR) primer was LTR1 (CTCTTGCTGTTGCATCGGACT). Igf2 primers were mIgf2-1 (AGTCCGAGAGGGACGTGTCTAC) and mIgf2-2 (CGTCCCGCGGACTGTCTC). Actb (β-actin) primers were b-actinF1(CGAGGCCCAGAGCAAGAGAG) and b-actinR1 (CGGTTGGCCTTAGGGTTCAG). QPCR was performed in an ABI PRISM 7000 sequence detection system (Applied Biosystems). Samples were normalized to beta-actin expression levels, and relative values were determined by the standard curve method.
qPCR of human samples. Bone marrow samples from 162 patients with AML at diagnosis (classified according to the French-American-British nomenclature; Table 1) and 6 healthy volunteers were obtained after informed consent. Approval was obtained from the Erasmus Medical Center institutional review board for these studies. Blasts from patients with AML and mononucleated fractions from normal bone marrow specimens were isolated by Ficoll-Hypaque (Nygaard, Oslo, Norway) centrifugation and then cryopreserved. After thawing, cells were washed with Hanks Balanced Salt Solution (HBSS) and further processed for RNA isolation. AML samples treated according to this procedure usually contain more than 90% blasts after thawing. Total RNA was extracted with guanidium thiocyanate followed by centrifugation in cesium chloride solution. RNA (1 μg) was transcribed into cDNA by using Superscript (Life Technologies, Merelbeke, Belgium) and random hexamers in a 40-μL reaction, under standard conditions. The qPCR amplification was performed in an ABI PRISM 7900 HT Sequence Detector, using 12.5 μL SYBR Green PCR Master Mix (PE Biosystems, Nieuwekerk a/d IJssel, The Netherlands), 2 μL (1/20th aliquot) cDNA, 2.5 pmol primer mix (Life Technologies), and 10 μL water. The PCR conditions included 2 minutes at 50°C and 10 minutes at 95°C followed by 45 cycles of amplification (each with a 15-second denaturation step at 95°C and a 1-minute annealing/extension step at 60°C). The PLAG1 primers were hP1Taq1 (ACACAGGAGAGAGGCCCTACA) and hP1Taq2 (ATGAGTAGCCATGTGCCTTTGT); the PLAGL2 primers were hPL2Taq1 (CACTGTGGCAAGGCTTTTGC) and hPL2Taq2 (GATGGTCCTTGCGGTGAAACAT). qPCR for normalization control was carried as described previously.19
Clinical characteristics of the AML samples
Characteristics . | Number . |
|---|---|
| Gender | |
| Male | 77 |
| Female | 85 |
| Age, median (range), y | 42.3 (15.2-60.8) |
| Age groups | |
| Younger than 35 | 51 |
| 35-50 | 62 |
| 50 and older | 49 |
| FAB | |
| M0 | 4 |
| M1 | 39 |
| M2 | 33 |
| M3 | 13 |
| M4 | 32 |
| M5 | 36 |
| M6 | 2 |
| Unclassified | 3 |
| Cytogenetic risk group | |
| Favorable | 34 |
| inv(16) | 12 |
| t(8;21) | 11 |
| t(15;17) | 11 |
| Intermediate | 111 |
| Unfavorable | 17 |
| WBC count, median (range), 109/L | 29.8 (0.3-263) |
| Blast count, median (range), % | 66 (0-98) |
Characteristics . | Number . |
|---|---|
| Gender | |
| Male | 77 |
| Female | 85 |
| Age, median (range), y | 42.3 (15.2-60.8) |
| Age groups | |
| Younger than 35 | 51 |
| 35-50 | 62 |
| 50 and older | 49 |
| FAB | |
| M0 | 4 |
| M1 | 39 |
| M2 | 33 |
| M3 | 13 |
| M4 | 32 |
| M5 | 36 |
| M6 | 2 |
| Unclassified | 3 |
| Cytogenetic risk group | |
| Favorable | 34 |
| inv(16) | 12 |
| t(8;21) | 11 |
| t(15;17) | 11 |
| Intermediate | 111 |
| Unfavorable | 17 |
| WBC count, median (range), 109/L | 29.8 (0.3-263) |
| Blast count, median (range), % | 66 (0-98) |
FAB indicates French-American-British; WBC, white blood count.
To determine PLAG1 and PLAGL2 relative expression levels, the average Ct (threshold cycle) values from duplicate readings were normalized for endogenous reference (dCt = Ct target - Ct PBGD) and compared with a calibrator using the “delta-delta Ct method” (δδCt = δCt Sample - δCt Calibrator). As calibrator we used the average Ct value of PLAG1 or PLAGL2 in the 6 CD34+ healthy volunteer samples. Using the δδCt value, relative expression was calculated (2-δδCt).
Retroviral production
The Plag1 cDNA was cloned from testis RNA by using PFU polymerase (Stratagene, La Jolla, CA) and primers mP1FlF (GATAATCTCGAGACCATGGATTACAAGGATGACGACGACGATAAGGCCACTGTCATTCCTGGTGAT) and mP1F1R (GAATGAGAAGCGGCCGCACTACTGAAAAGCTTGATGGA). The PLAGL2 cDNA was cloned by using primers hPL2FF (GATAATCTCGAGACCATGGATTACAAGGATGACGACGACGATAAGACCACATTTTTCACCAGCGT) and hPL2FR (GAATGAGAAGCGGCCGCACTACTGGAATGCTTGGTGGA). The PCR amplicons were cut with Xho1 and Not1 and cloned into pMSCVire-sEGFP(MIG).20 Phoenix packaging cells (generously provided by Gary Nolan, Stanford University, CA) were cotransfected with 2 μg retroviral constructs, 2 μg Ψ-Eco packaging plasmid with Effectene reagent (Quiagen, Valencia CA) according to manufacturer's protocol. Retrovirus supernatants were taken at 24, 48, and 72 hours and titered in 3T3 cells by fluorescence-activated cell sorting (FACS) analysis for green fluorescent protein (GFP).
Bone marrow transduction and transplantation (tBMT)
Cbfb-MYH11 expression was induced in a Cbfb56M/+ conditional knock-in mouse model recently generated in our laboratory with the use of the Cre-Lox system. Its characterization will be published elsewhere. Cbfb-MYH11 was induced by using the Mx1Cre transgenic mouse.21 Briefly, Cbfb56M/+/Mx1Cre double transgenics or wild-type control mice were injected with 3 doses of polyinosinic-polycytidylic acid (pI-pC; Sigma, St Louis, MO) at 3 weeks of age, and cells were harvested 6 days after treatment with 150 mg/kg 5-fluorouracil. Double transgenic bone marrow cells consistently rendered more than 90% of Cre-mediated Cbfb-MYH11 induction. Cells were spin-infected twice with retrovirus supernatant. Following infection, 5 × 105 to 1 × 106 bone marrow cells were transplanted into 4- to 6-week-old sublethally irradiated (650 rads [6.5 Gy]) 129SvEv wild-type mice intravenously. Mice were under daily observation for early signs of leukemia. These signs included limited motility, pale paws, and dehydration. At first signs of illness, peripheral blood was analyzed for cell number and morphology for the presence of immature cells. FACS analysis of peripheral blood was performed by using antibodies to cell-surface markers Gr-1, CD11b, B220, CD3, Ter119, and c-kit (Pharmingen, San Diego CA). Leukemic cells were harvested from the bone marrow or spleen of affected mice in RPMI 1640 (Invitrogen, Carlsbad, CA) and 20% fetal bovine serum (FBS; Invitrogen), and single-cell suspension aliquots of 1 × 106 cells were transplanted intravenously in sublethally irradiated 4- to 6-week-old 129Sv/Ev recipients.
Southern blot analysis
Southern blot analysis was performed following standard protocols. Briefly, spleen DNA from leukemic and control mice was cut with HindIII (to identify viral insertions in sample) separated in a 0.8% agarose gel and transferred onto Hybond-XL nylon membrane (Amersham-Pharmacia Biotech, Buckinghamshire, England). The 800-base pair (bp) GFP probe was an EcoRI/NcoI fragment from MIG. Hybridizations were performed overnight at 45°C in formamide solution (Hybrisol-I; Serologicals, Norcross, GA).
Methylcellulose colony-forming assay and cell-cycle analysis
129SvEv bone marrow cells expressing Cbfb-SMMHC were harvested 6 days after treatment with 150 mg/kg 5-fluorouracil, spin-infected with 2 rounds of retrovirus supernatant (either MIG, MIG-Plag1, or MIG-PLAGL2), and sorted for GFP expression. GFP-positive cells (1 × 104) were plated (in triplicate) in methylcellulose supplemented with interleukin 3 (IL3), IL6, and stem cell factor (SCF; Methocult 3534). Colonies were counted 7 days after plating and analyzed by FACS and cytospin. Remaining cells were recovered and 1 × 104 cells were serially replated for 3 rounds. For cell-cycle analysis, sorted cells were fixed in 70% ice-cold ethanol, stained with 50 μg/mL propidium iodide (Sigma), and analyzed by FACS.
Cytology and histology analysis
Morphology analysis of peripheral blood cells was carried out on modified Wright-Giemsa-stained smears and cytospins. Histology analysis was conducted on sections from paraffin-embedded blocks and stained with Hematoxylin and Eosin (Histoserv, Gaithersburg, MD). Images were taken using a Zeiss Axioskop 40 microscope (Carl Zeiss, Jena, Germany) with 40 × or 100 × (under Zeiss Immersol 518N oil) lenses, or a Zeiss Stemi 2000-C stereoscope at 6.5 × magnification. The images were acquired with a Zeiss AxioCam MRc camera and MR-Green acquisition software.
Results
Retroviral insertions induce Plag1 and Plagl2 overexpression in leukemic samples expressing Cbfb-MYH11
Our recent RIM study identified 8 retroviral insertions at the Plag1 locus and 2 at the Plagl2 locus in Cbfb-MYH11-associated leukemia samples. These results suggested that the transcription factors may play a critical role in inv(16) AML.10 To determine the precise location of retroviral insertions in the Plag1 locus, we analyzed the Plag1 gene structure in the mouse genome. The sequence of the 5′ Plag1 cDNA was cloned from embryo RNA (embryonic day 14.5) by using a combination of RT-PCR and 5′-rapid amplification of cDNA ends (RACE; GenBank accession no. AY574219). Analysis of this sequence using the public mouse genome database (http://genome.ucsc.edu) confirmed that the 8 Plag1-associated RIM retroviral insertions were located in its introns 1 and 2, upstream of the translation start codon in exon 4, and in the transcriptional orientation10 (Figure 1A). Notably, these introns are also involved in the breakpoints of 8q21-associated translocations in salivary gland adenomas.22 Considering that the retroviral insertions mapped upstream of Plag1 and Plagl2 translational start site and in the same transcriptional orientation, we reasoned that these insertions may up-regulate Plag1 and Plagl2 transcription. Therefore, we analyzed Plag1 and Plagl2 expression in normal and RIM-AML samples. The Plag1 transcript was not detected in mouse hematopoietic tissues by RT-PCR (Figure 1B). Furthermore, expression was not detected in a panel of 10 RIM-AML samples with retroviral insertions in other loci (Figure 1B, sample V65; data not shown). Conversely, Plag1 was expressed in all samples with retroviral insertions in the Plag1 locus. Finally, since these AML samples were induced in Cbfb+/MYH11 chimeras, Cbfb-MYH11 expression was confirmed in leukemia samples by RT-PCR (Figure 1B, middle panel).
Plag1 and PLAGL2 are overexpressed in RIM-AML samples. (A) Representation of Plag1-RIM insertions (V62, Vc6, V84, V85, VC4, V61, VC2, V89). Thin arrows indicate location of retroviral insertions and direction of LTR-transcription. Triangles indicate regions of chromosomal breakpoints in pleomorphic adenomas.22 Boxes indicate exons, including untranslated (▦) and translated (□) regions, and alternatively spliced exon 2b (▤). (B) RT-PCR analysis of Plag1 (top panel), Cbfb-MYH11 (middle panel), andActb (β-actin) (bottom panel), including whole embryo day 14.5 (E14.5), testis (te), bone marrow (bm), hematopoietic progenitor-enriched bone marrow (bmp; bone marrow 6 days after 5-fluorouracil treatment), spleen (sp), peripheral blood white blood cells (pb), Plag1-associated RIM-AML samples (V62, Vc6, V85, Vc4, Vc2, V89, and V84), Vc6 transplant (Vc6T), Plagl2-associated AML sample (V65), and 0.1 ng Plag1 plasmid control (+c). (C) Representation of Plagl2-RIM insertions (Vc1 and V65). Arrows indicate location of retroviral insertions and direction of LTR-transcription. Boxes indicate exons, including untranslated (▦) and translated (□) regions. (D) RT-PCR analysis of Plagl2 (top panel), Plagl2 transcribed from viral LTR (middle panel), and Actb (bottom panel); including hematopoietic tissues, Plag1-associated RIM-AML samples (V62, Vc6, V85, Vc4, Vc2, V89, and V84), Plagl2 RIM-AML sample V65, and a representative RIM-AML sample with no viral insertion near Plag1 or Plagl2 (V68). (E) Quantitative PCR analysis of Plagl2, using Plagl2 specific primers, in normal and leukemic samples. Expression levels were normalized to Actb and shown relative to sample E14.5 (E14.5 = 1). Error bars indicate standard errors from duplicate experiments.
Plag1 and PLAGL2 are overexpressed in RIM-AML samples. (A) Representation of Plag1-RIM insertions (V62, Vc6, V84, V85, VC4, V61, VC2, V89). Thin arrows indicate location of retroviral insertions and direction of LTR-transcription. Triangles indicate regions of chromosomal breakpoints in pleomorphic adenomas.22 Boxes indicate exons, including untranslated (▦) and translated (□) regions, and alternatively spliced exon 2b (▤). (B) RT-PCR analysis of Plag1 (top panel), Cbfb-MYH11 (middle panel), andActb (β-actin) (bottom panel), including whole embryo day 14.5 (E14.5), testis (te), bone marrow (bm), hematopoietic progenitor-enriched bone marrow (bmp; bone marrow 6 days after 5-fluorouracil treatment), spleen (sp), peripheral blood white blood cells (pb), Plag1-associated RIM-AML samples (V62, Vc6, V85, Vc4, Vc2, V89, and V84), Vc6 transplant (Vc6T), Plagl2-associated AML sample (V65), and 0.1 ng Plag1 plasmid control (+c). (C) Representation of Plagl2-RIM insertions (Vc1 and V65). Arrows indicate location of retroviral insertions and direction of LTR-transcription. Boxes indicate exons, including untranslated (▦) and translated (□) regions. (D) RT-PCR analysis of Plagl2 (top panel), Plagl2 transcribed from viral LTR (middle panel), and Actb (bottom panel); including hematopoietic tissues, Plag1-associated RIM-AML samples (V62, Vc6, V85, Vc4, Vc2, V89, and V84), Plagl2 RIM-AML sample V65, and a representative RIM-AML sample with no viral insertion near Plag1 or Plagl2 (V68). (E) Quantitative PCR analysis of Plagl2, using Plagl2 specific primers, in normal and leukemic samples. Expression levels were normalized to Actb and shown relative to sample E14.5 (E14.5 = 1). Error bars indicate standard errors from duplicate experiments.
In humans, PLAGL2 is expressed during embryogenesis and not detected in adult tissues.11 In the mouse, however, Plagl2 is detected in several tissues, showing higher expression in lung, spleen, and testes.23 The RIM-AML samples Vc1 and V65 included retroviral insertions in Plagl2 intron 1, in the same transcription orientation and upstream of its translation start codon (Figure 1C). Plagl2 expression was detected in all normal tissues analyzed (Figure 1D, top panel, lanes 1-6). The RIM-AML samples analyzed also expressed Plagl2 (Figure 1D, top panel, lanes 7-4), with higher levels in the Plagl2-associated sample V65 (RNA from sample Vc1 was not available). Similar analyses that used LTR-forward and Plagl2-reverse primers identified a strong band only in sample V65 (Figure 1D, middle panel), confirming that the increase in Plagl2 expression was transcribed from the retroviral LTR. Furthermore, increased Plagl2 relative expression levels (RELs) were confirmed by qPCR using Plagl2-specific primers (Figure 1E). Therefore, upstream retroviral insertions up-regulated Plag1 and Plagl2 transcription in Plag-associated leukemic samples.
Plag1 and Plagl2 cooperate with Cbfb-MYH11 to induce leukemia
The Plag1 and Plagl2 overexpression in mouse RIM-AML samples suggests that these zinc finger transcription factors may participate in leukemogenesis. Therefore, we tested whether Cbfb-MYH11 and Plag1 or Plagl2 coexpression is sufficient to trigger AML in the mouse by using a tBMT assay (described in “Materials and methods”). All mice that received transplants with bone marrow cells expressing Cbfb-MYH11/MIG-Plag1 (n = 22) or Cbfb-MYH11/MIG-PLAGL2 (n = 19) readily developed leukemia 3 to 12 weeks after transplantation (Figure 2A). Control groups remained healthy up to 5 months (experimental end point), including mice that received transplants with cells expressing Cbfb-MYH11/MIG (n = 24), MIG-Plag1 (n = 17), or MIG-PLAGL2 (n = 17).
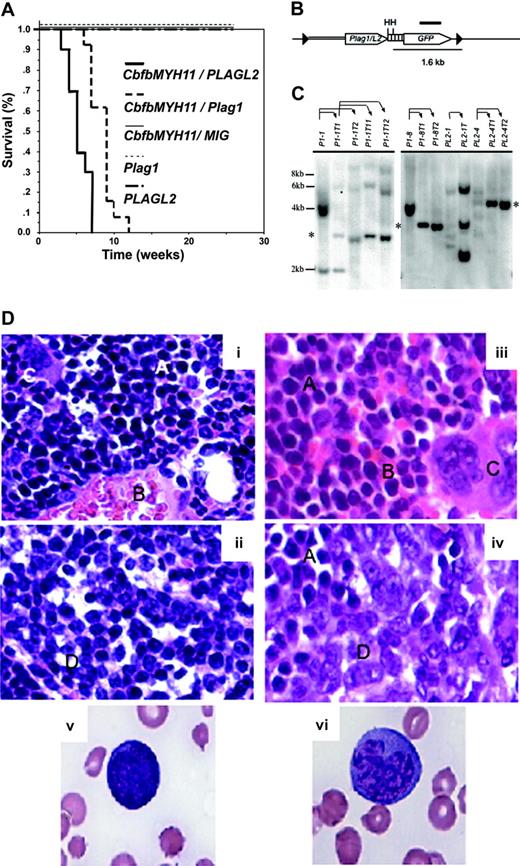
Plag1 and PLAGL2 cooperate with Cbfb-MYH11 to induce AML in mice. (A) Kaplan-Meier survival curve of mice that received transplants with bone marrow cells expressing: Cbfb-MYH11 and MIG-Plag1 (thick dashed line), Cbfb-MYH11 and MIG-PLAGL2 (thick solid line), Cbfb-MYH11 and MIG (thin solid line), MIG-Plag1 (thin dashed line), or MIG-PLAGL2 (dotted-dashed line). (B) Representation of the MIG-Plag1/L2 provirus. The LTRs (▸), coding sequence for Plag1/L2 (sequence encoding Plag1 or PLAGL2) and GFP (open arrows), internal ribosome entry site (IRES; ▥), probe (thick line), HindIII restriction sites (H), genomic (thin line) and viral (double thin line) sequences, are detailed. (C) Representative Southern blot analysis of transplantation of leukemic cells. DNA from PLAG-associated primary leukemia (lanes 1, 6, 9, 11), secondary transplanted leukemias (lanes 2, 3, 7, 8, 10, 12, 13), and tertiary transplanted leukemias (lanes 4 and 5) were probed with the retroviral GFP sequence. Thin arrows indicate which primary leukemic cells were used for secondary and tertiary transplants. Asterisk shows a band that represents the clonal expansion of a population of leukemic cells that became predominant in mice that receive transplants. The expected molecular weight of expected fragments (in kb) is indicated at the left. (D) Pathologic characteristics of leukemic cells. Wright-Giemsa staining of (× 10 magnification) normal (panel i) and leukemic (panel iii) bone marrow, normal (panel ii) and leukemic spleen (panel iv), and representative blastlike (panel v) and monocytic-like (panel vi) cells (× 100 magnification). Note area with lymphoid (A), red (B), megakaryocytes (C), and leukemic (D) cells.
Plag1 and PLAGL2 cooperate with Cbfb-MYH11 to induce AML in mice. (A) Kaplan-Meier survival curve of mice that received transplants with bone marrow cells expressing: Cbfb-MYH11 and MIG-Plag1 (thick dashed line), Cbfb-MYH11 and MIG-PLAGL2 (thick solid line), Cbfb-MYH11 and MIG (thin solid line), MIG-Plag1 (thin dashed line), or MIG-PLAGL2 (dotted-dashed line). (B) Representation of the MIG-Plag1/L2 provirus. The LTRs (▸), coding sequence for Plag1/L2 (sequence encoding Plag1 or PLAGL2) and GFP (open arrows), internal ribosome entry site (IRES; ▥), probe (thick line), HindIII restriction sites (H), genomic (thin line) and viral (double thin line) sequences, are detailed. (C) Representative Southern blot analysis of transplantation of leukemic cells. DNA from PLAG-associated primary leukemia (lanes 1, 6, 9, 11), secondary transplanted leukemias (lanes 2, 3, 7, 8, 10, 12, 13), and tertiary transplanted leukemias (lanes 4 and 5) were probed with the retroviral GFP sequence. Thin arrows indicate which primary leukemic cells were used for secondary and tertiary transplants. Asterisk shows a band that represents the clonal expansion of a population of leukemic cells that became predominant in mice that receive transplants. The expected molecular weight of expected fragments (in kb) is indicated at the left. (D) Pathologic characteristics of leukemic cells. Wright-Giemsa staining of (× 10 magnification) normal (panel i) and leukemic (panel iii) bone marrow, normal (panel ii) and leukemic spleen (panel iv), and representative blastlike (panel v) and monocytic-like (panel vi) cells (× 100 magnification). Note area with lymphoid (A), red (B), megakaryocytes (C), and leukemic (D) cells.
To confirm that these cells were leukemic, bone marrow cells from affected mice (Plag1, n = 10; PLAGL2, n = 9) were transplanted into sublethally irradiated secondary recipients (2 transplants per donor cell population). In all cases, the leukemic cells were readily transplantable with an average disease onset of 4 weeks.
As Plag1 and PLAGL2 are expressed from the MIG-LTR (Figure 2B), we were able to examine the clonality of disease by Southern blot analysis using primary, secondary, and tertiary leukemic samples. Multiple bands were detected in most primary leukemic samples (Figure 2C, lanes 1, 6, 9, and 11), while secondary (Figure 2C, lanes 2, 3, 7, 8, 10, 12, and 13) and tertiary (Figure 2C, lanes 3-5) transplants frequently displayed enrichment of subclones underrepresented in the primary transplant (note asterisks). These data reveal that Plag1 and PLAGL2 efficiently cooperate with Cbfb-MYH11 to induce AML in mice. In addition, the short latency and oligoclonality of disease is consistent with the hypothesis that leukemia may rise from few genetic alterations.
Leukemic mice exhibited an increase of immature cells and a decrease of red blood cells and megakaryocytes in bone marrow (Figure 2D, panels i and iii). Likewise, spleens were consistently enlarged (with spleen weight of 601.5 ± 234.7 mg for Plag1, and 197 ± 106.8 mg for PLAGL2) with an altered architecture and evident invasion of leukemic cells (Figure 2D, panels ii and iv). Leukemic cells were progressively predominant in peripheral blood (Plag1 = 23.5 × 106, range 7.3 × 106 to 74.4 × 106 cells/mL; PLAGL2 = 12.5 × 106, range 0.3 × 106 to 61.7 × 106 cells/mL), including blastlike and monocytic-like cells (Figure 2D, panels v-vi). Occasionally, enlargement of liver resulting from focal infiltrations of leukemic cells was also observed. However, enlargement of the thymus and lymph nodes was not observed (data not shown), indicating a nonlymphoid disease.
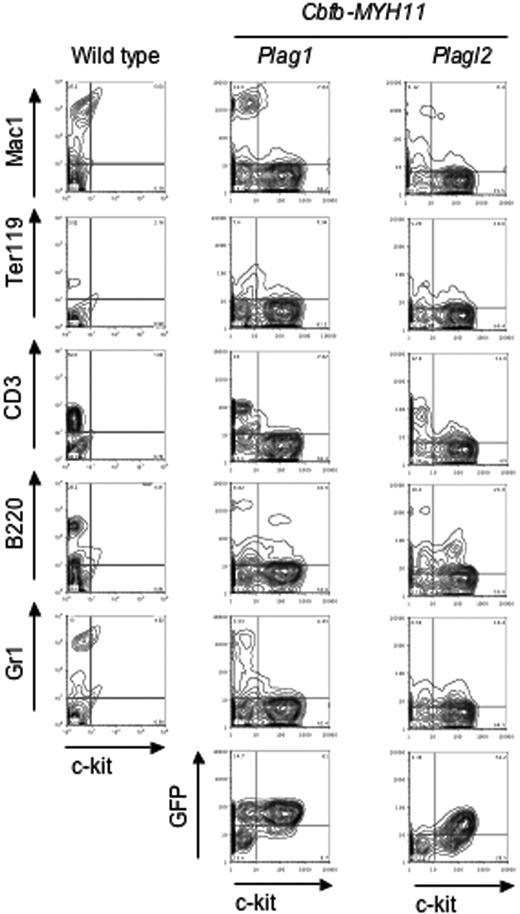
We characterized the leukemic cells by FACS analysis. The predominant cells in peripheral blood were c-kit+ and Lin- (Lin: CD3, Gr1, B220, Ter119, Mac1; Figure 3). Furthermore, the leukemic cells were consistently GFP+, confirming the presence of the provirus (Figure 3). Finally, Cbfb-MYH11 and either Plag1 or PLAGL2 expression in leukemic cells was confirmed by RT-PCR (data not shown).
Peripheral blood leukemic cells are Lin-/c-kit+ and GFP+. Representative examples of peripheral blood leukemic cell FACS analysis. FACS analysis of lineage markers (Gr1, B220, CD3, Ter119, and Mac1) and GFP, versus hematopoietic progenitor marker (c-kit) for wild-type (left column) and leukemic cells expressing either Cbfb-MYH11/Plag1 (middle column) or Cbfb-MYH11/PLAGL2 (right column).
Peripheral blood leukemic cells are Lin-/c-kit+ and GFP+. Representative examples of peripheral blood leukemic cell FACS analysis. FACS analysis of lineage markers (Gr1, B220, CD3, Ter119, and Mac1) and GFP, versus hematopoietic progenitor marker (c-kit) for wild-type (left column) and leukemic cells expressing either Cbfb-MYH11/Plag1 (middle column) or Cbfb-MYH11/PLAGL2 (right column).
Taken together, the morphology and FACS analysis data indicate that these leukemic cells are predominantly blastlike and monocytic-like cells and confirm that they express Plag1 and Plagl2. Finally, the leukemic cells resemble Cbfb-MYH11-associated leukemic cells previously described,9,10 supporting the idea that Cbfβ-SMMHC directs the phenotypic characteristics of leukemic cells.
Plag1 and PLAGL2 expand hematopoietic progenitors in Cbfb-MYH11-expressing cells by increasing G1 to S phase transition of cell cycle
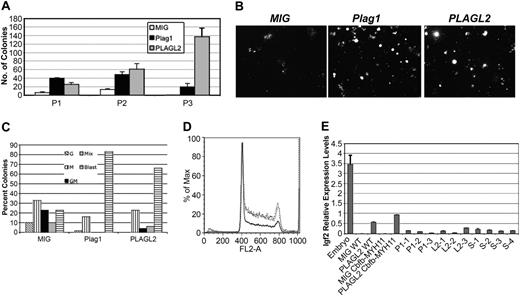
To further understand the role of Plag1 and Plagl2 in AML development, in vitro differentiation assays were performed in GFP-sorted bone marrow progenitors expressing Cbfb-MYH11 and either MIG-Plag1, MIG-PLAGL2, or MIG. At day 7, the number of Plag1- and PLAGL2-expressing colonies increased significantly (Figure 4A, P1). In addition, Plag1 and PLAGL2 colonies were predominantly compact and large when compared with diffuse colonies in controls, suggesting an increased progenitor proliferation (Figure 4B). Furthermore, cytology analysis revealed that colonies expressing Plag1 or PLAGL2 included predominantly immature blasts when compared with MIG control colonies (Figure 4C). This enrichment for immature forms suggests that Plag1 and PLAGL2 selectively expanded Cbfb-MYH11 differentiation-blocked progenitors.
Plag1 and PLAGL2 increase proliferation of hematopoietic progenitor in vitro by inducing entry into S phase. (A) Serial replating (P1 to P3) of 104Cbfb-MYH11-expressing bone marrow cells infected with MIG (□), MIG-Plag1 (▪), or MIG-PLAGL2 (▦) and cultured in methylcellulose culture for 7 days. (B) Representative images of colony morphology at day 7. (C) Lineage distribution colony-forming units (CFUs) from P1 analyzed by cytospin of individual colonies. Colony types are indicated as granulocytic (G; ▧), monocytic (M; ▥), granulocytic and monocytic (GM; ▪), mix (▦), and blast (▤). (D) Cell-cycle analysis of sorted bone marrow cells expressing Cbfb-MYH11 and MIG-Plag1 (gray line), MIG-PLAGL2 (dotted black line), or MIG (solid black line) and stained with propidium iodine. (E) The Igf2 RELs determined by quantitative PCR in E14.5 total embryo (lane 1), MIG wild-type bone marrow (lane 2), MIG-PLAGL2 wild-type bone marrow (lane 3), MIG Cbfb-MYH11 bone marrow cells (lane 4), MIG-PLAGL2 Cbfb-MYH11 bone marrow (lane 5), Plag1/Cbfb-MYH11 leukemias (lanes 6-8), PLAGL2/Cbfb-MYH11 leukemias (lanes 9-11), and leukemias not overexpressing Plag1 or PLAGL2.12-15 Values were normalized to Actb expression levels. Error bars indicate standard error from triplicate experiments.
Plag1 and PLAGL2 increase proliferation of hematopoietic progenitor in vitro by inducing entry into S phase. (A) Serial replating (P1 to P3) of 104Cbfb-MYH11-expressing bone marrow cells infected with MIG (□), MIG-Plag1 (▪), or MIG-PLAGL2 (▦) and cultured in methylcellulose culture for 7 days. (B) Representative images of colony morphology at day 7. (C) Lineage distribution colony-forming units (CFUs) from P1 analyzed by cytospin of individual colonies. Colony types are indicated as granulocytic (G; ▧), monocytic (M; ▥), granulocytic and monocytic (GM; ▪), mix (▦), and blast (▤). (D) Cell-cycle analysis of sorted bone marrow cells expressing Cbfb-MYH11 and MIG-Plag1 (gray line), MIG-PLAGL2 (dotted black line), or MIG (solid black line) and stained with propidium iodine. (E) The Igf2 RELs determined by quantitative PCR in E14.5 total embryo (lane 1), MIG wild-type bone marrow (lane 2), MIG-PLAGL2 wild-type bone marrow (lane 3), MIG Cbfb-MYH11 bone marrow cells (lane 4), MIG-PLAGL2 Cbfb-MYH11 bone marrow (lane 5), Plag1/Cbfb-MYH11 leukemias (lanes 6-8), PLAGL2/Cbfb-MYH11 leukemias (lanes 9-11), and leukemias not overexpressing Plag1 or PLAGL2.12-15 Values were normalized to Actb expression levels. Error bars indicate standard error from triplicate experiments.
To assess the effect of Plag1 and PLAGL2 on the renewal capacity of progenitors, 104 cells from day 7 pooled colonies were serially replated. In contrast to MIG controls, MIG-Plag1 and MIG-PLAGL2 transduced cells exhibited a marked increase in replating potential (Figure 4A, P2-P3). As Plag1 and PLAGL2 seemed to expand progenitors, we next assessed their effect on cell-cycle progression. Bone marrow cells expressing Cbfb-MYH11 were sorted after infection with MIG-Plag1, MIG-PLAGL2, or MIG and analyzed with the use of propidium iodine staining by FACS. Notably, Plag1 and PLAGL2 induced entry into S phase and reduced the number of cells in G0/G1 phase (Figure 4D). The increase of cells in S phase was also confirmed by bromodeoxyuridine (BRDU) incorporation (data not shown). Overall, these data indicate that Plag1 and Plagl2 stimulate proliferation and renewal of hematopoietic progenitors expressing CbFb-MYHII.
PLAG1 and PLAGL2 regulate gene expression by binding to the same DNA consensus site.24 The insulin growth factor 2 gene (IGF2) harbors 8 PLAG binding sites in its promoter 3 and is up-regulated in cell lines ectopically expressing PLAG1 as well as in salivary gland tumors with PLAG1-associated translocations.15,24,25 In addition, AML samples frequently show loss of imprinting at the IGF2 locus associated with IGF2 overexpression.26 Therefore, it is possible that PLAG1 and PLAGL2 could expand hematopoietic precursors via the IGF2 pathway to induce AML in cooperation with Cbfβ-SMMHC. To test this hypothesis, we assessed Igf2 levels in GFP-sorted bone marrow cells and leukemic samples expressing Plag1, PLAGL2, or without expression of these genes by qPCR. Igf2 RELs were increased by Plag1 and PLAGL2 in bone marrow (Figure 4E; data not shown). However, RELs were not increased in leukemia samples when compared with non-Plag1- or -PLAGL2-induced AMLs. These results indicate that Igf2 transcripts are up-regulated by Plag1 and PLAGL2 in hematopoietic progenitors, but that Plag1 and PLAGL2 do not maintain increased levels of Igf2 in leukemic progression.
PLAG1 and PLAGL2 are overexpressed in human AML
AML is a heterogeneous disorder with regard to both morphology and chromosomal aberrations detected in the leukemic cells. Currently, AML is classified according to pretreatment karyotype and treatment outcome. For example, cytogenetic groups with inv(16)/t(16;16), t(15;17), and t(8;21) define an AML subset with increased survival (favorable outcome).
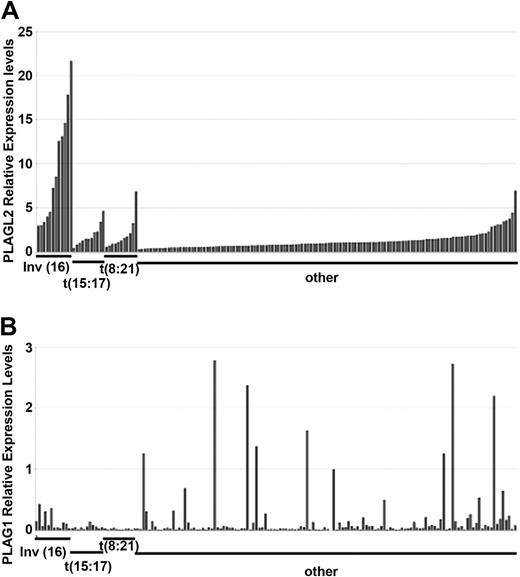
To investigate whether PLAG1 or PLAGL2 could also play a role in human AML, we asked whether PLAG1 and PLAGL2 are up-regulated in human AML samples. The RELs of PLAG1 and PLAGL2 were analyzed in a panel of 162 human primary AML samples by qPCR. The REL values were normalized to PBGD expression and compared with expression levels in normal CD34+ bone marrow cells. The panel was divided into 4 cytogenetic groups: 12 samples with inv(16)(p13q22)/t(16;16) expressing CBFB-MYH11, 11 samples with t(15:17), 11 samples with t(8:21), and 128 samples without these translocations. The characteristics of the AML samples are detailed in Table 1.
Overall, PLAGL2 was overexpressed in 15% of the samples. Strikingly, higher RELs were associated with inv(16) than with the other cytogenetic subgroups (Figure 5A). The association between PLAGL2 and CBFb-MYH11 was highlighted, as no other cytogenetic change was common in inv(16) samples (Table 2). In addition, PLAGL2 expression was significantly higher in t(8:21) and t(15:17) subgroups (with P < .01 and P < .05, respectively; 2-tailed paired t test) when compared with samples without these 3 chromosomal rearrangements or inv(16).
Quantitative PCR analysis of PLAG1 and PLAGL2 in a panel of 162 human AML samples.PLAGL2 (A) and PLAG1 (B) RELs. Samples are clustered in 4 cytogenetic groups: samples showing inv(16)(p13;q22), t(15;17), t(8;21), and samples without these rearrangements (other). Within each group, samples are ordered by increased PLAGL2 RELs. RELs represent the average of duplicate values and are compared with the value of 6 healthy CD34+ bone marrow samples.
Quantitative PCR analysis of PLAG1 and PLAGL2 in a panel of 162 human AML samples.PLAGL2 (A) and PLAG1 (B) RELs. Samples are clustered in 4 cytogenetic groups: samples showing inv(16)(p13;q22), t(15;17), t(8;21), and samples without these rearrangements (other). Within each group, samples are ordered by increased PLAGL2 RELs. RELs represent the average of duplicate values and are compared with the value of 6 healthy CD34+ bone marrow samples.
Cytogenetics of inv(16) AML samples
ID . | Cytogenetics . |
|---|---|
| 1 | 46, XY, inv(16)(p13;q22) |
| 2 | 46, XY, inv(16)(p13;q22) |
| 3 | 46, XY, inv(16)(p13;q22)[37]/47,id, + 8[2]/47,id, + 21[4] |
| 4 | 46, XY, inv(16)(p13;q22) |
| 5 | 46, XY, t(16;16)(p13;q22) |
| 6 | 46, XX, inv(16)(p13;q22) |
| 7 | 46, XY, inv(16)(p13;q22) |
| 8 | 46, XX, inv(16)(15%)/47,XX,inv(16), + 22 (85%) |
| 9 | 46, XX, inv(16)(p13;q22) |
| 10 | 46, XX, inv(16)(p13;q22) |
| 11 | 46, XX, inv(16)(p13;q22) |
| 12 | 46, XX, inv(16)(p13;q22)[8]/47,idem, + 8[12] |
ID . | Cytogenetics . |
|---|---|
| 1 | 46, XY, inv(16)(p13;q22) |
| 2 | 46, XY, inv(16)(p13;q22) |
| 3 | 46, XY, inv(16)(p13;q22)[37]/47,id, + 8[2]/47,id, + 21[4] |
| 4 | 46, XY, inv(16)(p13;q22) |
| 5 | 46, XY, t(16;16)(p13;q22) |
| 6 | 46, XX, inv(16)(p13;q22) |
| 7 | 46, XY, inv(16)(p13;q22) |
| 8 | 46, XX, inv(16)(15%)/47,XX,inv(16), + 22 (85%) |
| 9 | 46, XX, inv(16)(p13;q22) |
| 10 | 46, XX, inv(16)(p13;q22) |
| 11 | 46, XX, inv(16)(p13;q22) |
| 12 | 46, XX, inv(16)(p13;q22)[8]/47,idem, + 8[12] |
The RELs of PLAG1 transcript were also significantly increased 1.4- to 2.4-fold (P < .0001; 2-tailed paired t test) in 5% (8 of 162) of the samples analyzed (Figure 5B). The overexpression did not correlate with a distinct cytogenetic subgroup, suggesting that PLAG1 may cooperate with other mutations associated with AML. PLAG1 overexpression has been associated in tumors with chromosome 8 polyploidy,14,29 and trisomy of chromosome 8 is a frequent rearrangement in human AML. However, we found no correlation between trisomy 8 and increased PLAG1 expression in this AML panel (trisomy 8 REL = 0.14 ± 0.13, n = 18; disomy 8 REL = 0.24 ± 0.44, n = 144). These data reveal that PLAG1 and PLAGL2 expression is deregulated in human AML. Furthermore, PLAGL2 overexpression strongly correlates with inv(16) AML.
Discussion
AML arises from the uncontrolled clonal expansion of hematopoietic progenitor cells that have acquired (at least) 2 types of genetic alterations. These cooperating mutations have been classified as “class-I” mutations, conferring hematopoietic progenitor expansion (such as increase in proliferation, cell survival, or self-renewal potential), and “class-II” mutations, inducing impairment of hematopoietic differentiation.30 Previously, we demonstrated that expression of the inv(16)-associated Cbfb-MYH11 fusion gene (a class-II mutation) impairs hematopoietic differentiation in the mouse, and that this effect is required for leukemogenesis.9 Our genetic study identified Plag1 and Plagl2 as class-I candidate genes for Cbfb-MYH11-associated leukemogenesis in the mouse.9 The current study demonstrates that up-regulation of Plag1 and PLAGL2 is a class-I mutation that induces AML in cooperation with Cbfβ-SMMHC.
Plag1 and Plagl2 were specifically overexpressed in RIM-AML samples with retroviral insertions at these loci, confirming the genetic data previously published and further implicating these genes in mouse leukemia. Using a tBMT assay, we demonstrate that both transcription factors can independently cooperate with Cbfβ-SMMHC to induce AML in 3 to 12 weeks with 100% penetrance. Mice consistently display expansion of immature cells and infiltration into peripheral blood, spleen, and liver, whereas lymph nodes and thymus remain unaffected. The leukemic cells in peripheral blood morphologically appear as blastlike and monocytic-like cells and are predominantly Lin-/c-kit+. Interestingly, although the leukemic cells have a myeloid morphology, their immunophenotype suggests that this population contains blastlike cells stalled at an early stage of differentiation. This phenotype is similar to the AML observed in Cbfb+/MYH11 knock-in mice treated with chemical or retroviral mutagens. Taken together, these studies suggest that Cbfb-MYH11, and not the cooperating gene(s), determines the phenotype of the disease.
Consistent with the oligoclonal and aggressive course of disease, coexpression of Cbfb-MYH11 and Plag1 or Plagl2 may be sufficient to trigger AML in the mouse. Alternatively, as the tBMT assays require insertion of the retroviral vector in the hematopoietic progenitor's genome, it is possible that a third hit could be required for AML development. We show that Plag1 and Plagl2 oncogenic function is associated with expansion of hematopoietic progenitors expressing Cbfβ-SMMHC. This is evident in the blastlike colony morphology, the increase of blast cells in these colonies, the increase of cells entering into S and G2/M phases, and the expanded replating potential. Interestingly, these results are similar to the mitogenic effect that Plag1 or Plagl2 overexpression provides NIH-3T3 cells.15
The effect of Plag1 and Plagl2 in leukemogenesis is similar to that of other class-I genes mutated in human inv(16) AML, including activating (oncogenic) mutations in KRAS and FLT3.31,32 For example, expression of activated Flt3 or Kras alone cannot induce AML in the mouse,33-35 but Flt3 efficiently induces AML in cooperation with promyelocytic leukemia-retinoic acid receptor α (PML-RARα).36 Conversely, activated Kras or Flt3 alone induces a myeloproliferative-like disease, while Plag1- and Plagl2-expressing mice remain healthy. Also, the in vitro expansion of hematopoietic precursors observed on Plag1 or PLAGL2 expression was not observed in oncogenic Kras cultures.34 These differences may indicate that distinct pathways are involved.
Previous studies have shown that PLAG1 and PLAGL2 up-regulate IGF2.15,24 In addition, AML samples frequently show loss of imprinting at the IGF2 locus associated with IGF2 overexpression.26 Surprisingly, our study shows that Plag1 and PLAGL2 induce moderate Igf2 transcript levels in bone marrow cells, but that this increase is not maintained in leukemic samples. These results suggest that the Plag transcription factors may affect an Igf2-independent pathway in leukemogenesis or play a role in initiation but not maintenance of leukemia.
We find PLAGL2 expression preferentially induced in human AML samples with inv(16), supporting our findings using the mouse model. Interestingly, PLAGL2 was also significantly induced in t(8;21)-and t(15;17)-associated AML samples, suggesting that PLAGL2 may also induce AML in cooperation with other fusion genes. This is the first study implicating PLAGL2 in human cancer and demonstrating that both Plag1 and PLAGL2 function as oncogenes in vivo. It will be, therefore, interesting to examine PLAGL2 function in other cancers. Considering the in vivo effect of Plag1 and Plagl2 in Cbfb-MYH11-dependent AML in mice and increased PLAG1 and PLAGL2 levels in human AML without inv(16) cytogenetics, it will be important to test whether these transcription factors can also induce AML in cooperation with other class-II fusion genes in vivo. Interesting examples could include transgenic mice models for AML1-ETO and PML-RARα.37-39 Finally, the finding that PLAG1 and PLAGL2 zinc finger proteins participate in AML development implicates novel pathways in leukemogenesis and may provide additional targets in the design of improved therapies.
Prepublished online as Blood First Edition Paper, December 7, 2004; DOI 10.1182/blood-2004-09-3630.
Supported by a grant from the National Institutes of Health (RO1 CA096983-01) (L.H.C.) and by a Special Fellow Award (Leukemia and Lymphoma Society) (L.H.C.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Michelle Kelliher for critical reading of the manuscript, and Weibo Li and C. J. Clark for technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal