Abstract
Substantial research has been dedicated to the study of the relationship between genetic mechanisms regulating cell functions in tumors and how those tumors respond to various treatment regimens. Because these mechanisms are still not well understood, we have chosen to study the genetic makeup of 57 tumor samples from patients with follicular lymphoma (FL). Our goal was to develop a prognostic tool, which can be used as an aid in determining FL patients with tumors genetically predisposed to a successful treatment with the CHOP (cyclophosphamide, vincristine, doxorubicin, prednisone) regimen. To select relevant genes, high-density oligonucleotide arrays were used. There were 14 genes highly expressed in FL patients that responded well to CHOP chemotherapy, and 11 of these were involved in G2/M transition of the cell cycle, in mitosis, or in DNA modulation. A high expression of CCNB1 (cyclin B1), CDC2, CDKN3A, CKS1B, ANP32E, and KIAA0101, but not of the proliferation-related antigen Ki-67, was associated with better survival rate in a univariate analysis. CCNB1 expression had an independent prognostic value when included in a multivariate analysis together with the 5 parameters of the follicular lymphoma international prognostic index.
Introduction
Follicular lymphomas (FLs) are the second largest group of non-Hodgkin lymphomas (NHLs) in the Western world. The clinical course of FL is often indolent, with a median survival of 10 years.1 Some patients can remain untreated with no signs of progression for several years, while others may suffer a rapid progression of the disease.2 The heterogeneity of the clinical course has led to various options of treatment including watchful waiting, oral alkylating agents, purine nucleoside analogues, combination chemotherapy, interferon, and monoclonal antibodies.3 However, it is still unclear which patients will benefit from aggressive treatment. The International Prognostic Index, originally developed for aggressive NHL,4 was not very useful in predicting outcome for FL NHL patients since only few of the FL patients were categorized in the high-risk group.5 In the recently published Follicular Lymphoma International Prognostic Index (FLIPI),5 8 parameters had independent prognostic value: age; Ann Arbor stage; number of nodal sites; hemoglobin level; lactate-dehydrogenase (LDH); sex; bone marrow involvement; and peripheral blood lymphocyte count. Age was the strongest predictor of survival and was selected together with stage, number of nodal sites, hemoglobin level, and LDH to build the FLIPI.
Prediction of treatment outcome is important to avoid overtreatment of some patients and to identify others in whom less intensive treatment is likely to fail. Initial response to chemotherapy has been shown to be associated with prolonged survival in patients with FL.6 Also, several studies tried to define which of the so-called tumor-related factors have impact on survival; patients with FL grade 3 with a significant diffuse component had a shorter survival compared with those with FL grades 1 and 2.7 Overexpression of the BCL-XL or BCL-2 mRNA, or mdm2 protein has been reported to correlate with an inferior survival, while patients with tumors demonstrating high expression of bcl-6 had a favorable prognosis.8-11 Molecular and cytogenetic studies have indicated that a partial deletion of chromosome 6, or deletion of the short arm of chromosome 17, or trisomy 12 have been associated with adverse clinical outcome.12,13
The potential link between a good response to chemotherapy and a good clinical outcome prompted us to apply the response to cyclophosphamide, vincristine, doxorubicin, prednisone (CHOP)14 as criteria to define the groups of FL patients in whom the differential gene expression in tumor samples taken at diagnosis was investigated. A group of differentially expressed genes has been defined, and the impact of these genes' expression on patients' outcome was further investigated in a cohort of 57 patients with FL.
Patients, materials, and methods
Patient characteristics
Lymph node tumor samples taken at diagnosis from 57 FL patients were included in this study. The 44 samples that were diagnosed at Karolinska Hospital, Stockholm, Sweden, were a part of a consecutive series of 66 diagnostic specimens frozen from 1994 to 1999. Of 66 samples, 22 were excluded due to poor RNA quality (n = 14), insufficient material (n = 5), and insufficient microarray data quality due to low number of genes with a present call (16.2%, 22.3%, 22.4%) (n = 3). To increase the number of investigated tumors, we have also included 13 samples obtained from patients diagnosed at Lund University Hospital, Sweden (1996-2001). Tumor characteristics and clinical data of included patients are given in Table 1. There were 31 women and 26 men. The median age at diagnosis was 60 years (range, 28-85 years). Clinical staging was performed according to the Ann Arbor classification15 ; 21 patients had clinical stages I to II and 36 patients, stages III to IV. According to the FLIPI,5 28 patients belonged to the low-risk group, 16 patients to the intermediate-risk group, and 13 patients to the high-risk group. The distribution among risk groups did not differ from that of the patients with FL that were used to design the FLIPI. At last follow-up, 35 patients were alive and 22 patients had died. Median follow-up time for patients still alive was 65 months (range, 20-117 months). The histopathologic slides of all cases were revised and classified according to WHO classification as FL grade 1 (n = 12), grade 2 (n = 32), and grade 3 (n = 13).1 Approval for this study was obtained from the ethics committee at Karolinska Institutet; informed consent was not required since the analyses were performed on retrospective patient material.
Characteristics of 57 follicular lymphoma patients studied
Patient . | Cyclin B1* . | Age, y . | Sex . | Stage . | Hemoglobin level, g/L . | FLIPI . | WHO grade . | Ki-67† . | Cause of death . | Follow-up, mo . | Treatment in temporal order‡ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 11.4 | 81 | F | IIA | 102 | 1 | 3A | 3 | Colon cancer | 15 | KNOSPE |
| 2 | 11.8 | 68 | F | IIIA | 123 | 3 | 2 | 1 | Lymphoma | 56 | KNOSPE, CHOPx3, MIME |
| 3 | 17.7 | 42 | M | IA | 160 | 0 | 2 | 1 | Alive | 82 + | RT, rituximab |
| 4 | 26.7 | 49 | F | IIIA | 124 | 2 | 2 | 2 | Alive | 65 + | KNOSPE |
| 5 | 26.8 | 76 | M | IA | 172 | 1 | 2 | 1 | Alive | 87 + | No treatment |
| 6 | 28.1 | 67 | M | IA | 159 | 1 | 2 | 1 | Alive | 84 + | RT |
| 7 | 30.6 | 61 | F | IIA | 146 | 1 | 1 | 0 | Lymphoma | 51 | KNOSPE, RT, CNOPx5 + MTX, LD-ACOP-B |
| 8 | 32.7 | 83 | M | IVB | 127 | 2 | 2 | 1 | Lymphoma | 8 | CHOPx6, RT |
| 9 | 34.2 | 28 | F | IIIA | 143 | 2 | 2 | 1 | Lymphoma | 56 | KNOSPE, RT, rituximab, fludarabine, CHOPx5, RT |
| 10 | 34.3 | 42 | M | IVA | 125 | 2 | 2 | 0 | Lymphoma | 89 | Flu/Cycl, CHOPx3, MIME, RT, gemcitabine |
| 11 | 34.5 | 60 | M | IVA | 115 | 4 | 1 | 0 | Alive | 53 + | Rituximab, CHV(O)Px6, RT, KNOSPE, MIME, Flu/Cycl |
| 12 | 35.9 | 53 | F | IIIA | 133 | 1 | 1 | 1 | Lymphoma | 80 | KNOSPE, CHOPx8, Flu/Cycl, MIME |
| 13 | 36.0 | 75 | M | IIIB | 153 | 3 | 2 | 1 | Lymphoma | 44 | CHOPx8, KNOSPE, ENAD |
| 14 | 36.7 | 77 | M | IVA | 111 | 5 | 3B | 3 | Lymphoma | 7 | CHOPx6 |
| 15 | 40.4 | 59 | F | IA | 124 | 0 | 2 | 1 | Lymphoma | 55 | RT, CHOPx8, ENAD, MIME |
| 16 | 40.6 | 57 | F | IVA | 138 | 1 | 1 | 1 | Lymphoma | 81 | KNOSPE, CHOPx6, rituximab, RT, Flu/Cycl |
| 17 | 43.0 | 75 | M | IIIA | 142 | 4 | 2 | 3 | Lymphoma | 41 | KNOSPE |
| 18 | 43.0 | 82 | F | IA | 118 | 2 | 2 | 1 | Lymphoma | 64 | KNOSPE |
| 19 | 46.6 | 66 | M | IVA | 135 | 2 | 1 | 0 | Lymphoma | 10 | Prednimustine, CHOPx2, RT-CEP |
| 20 | 47.0 | 79 | M | IIIB | 109 | 4 | 2 | 1 | Lymphoma | 8 | KNOSPE |
| 21 | 47.9 | 73 | M | IIIA | 172 | 4 | 1 | 1 | Lymphoma | 95 | KNOSPE |
| 22 | 47.9 | 60 | F | IA | 133 | 0 | 2 | 1 | Alive | 63 + | No treatment |
| 23 | 48.1 | 43 | F | IIIA | 118 | 2 | 2 | 1 | Alive | 20 + | No treatment |
| 24 | 50.7 | 45 | M | IIIA | 154 | 1 | 2 | 1 | Alive | 50 + | No treatment |
| 25 | 51.1 | 68 | F | IVB | 145 | 4 | 1 | 0 | Alive | 67 + | CHOP×8, KNOSPE, MIME |
| 26 | 51.8 | 47 | F | IVA | 136 | 1 | 2 | 2 | Alive | 72 + | KNOSPE |
| 27 | 52.6 | 56 | F | IVB | 139 | 1 | 1 | 0 | Lymphoma | 82 | CdA, KNOSPE |
| 28 | 54.1 | 61 | M | IA | 159 | 1 | 3A | 2 | Alive | 59 + | RT |
| 29 | 59.0 | 57 | F | IVA | 135 | 1 | 2 | 1 | Alive | 87 + | CHOPx1, RT |
| 30 | 63.1 | 28 | M | IVB | 114 | 3 | 3B | 3 | Alive | 65 + | CHOPx7 + SCT |
| 31 | 64.9 | 56 | M | IA | 147 | 0 | 1 | 1 | Alive | 56 + | No treatment |
| 32 | 68.3 | 43 | F | IVA | 142 | 3 | 2 | 1 | Alive | 112 + | CdA |
| 33 | 68.7 | 61 | F | IVA | 140 | 2 | 3B | 2 | Lymphoma | 54 | CHOPx2, CHVPx6 |
| 34 | 74.4 | 60 | F | IVA | 134 | 2 | 2 | 1 | Alive | 72 + | Chlorambucil, rituximab |
| 35 | 77.4 | 85 | F | IIA | 128 | 2 | 3B | 3 | Lymphoma | 38 | CHOPx5, gemcitabine |
| 36 | 80.2 | 56 | F | IVA | 140 | 1 | 1 | 1 | Alive | 29 + | No treatment |
| 37 | 81.5 | 59 | M | IIIA | 158 | 1 | 1 | 0 | Alive | 58 + | No treatment |
| 38 | 84.4 | 78 | F | IIIA | 147 | 3 | 2 | 1 | Alive | 117 + | CHOPx4 |
| 39 | 85.0 | 80 | M | IVB | 84 | 4 | 1 | 1 | Lymphoma | 1 | No treatment |
| 40 | 86.3 | 71 | F | IA | 144 | 1 | 2 | 1 | Alive | 56 + | RT |
| 41 | 88.7 | 44 | F | IVA | 140 | 2 | 2 | 1 | Alive | 54 + | No treatment |
| 42 | 88.8 | 70 | F | IA | 127 | 1 | 2 | 1 | Alive | 70 + | RT |
| 43 | 90.6 | 72 | M | IVA | 133 | 3 | 2 | 1 | Lymphoma | 43 | KNOSPE, CHOPx6 |
| 44 | 90.7 | 46 | M | IVB | 145 | 1 | 2 | 2 | Alive | 63 + | Rituximab + interferon alfa |
| 45 | 96.0 | 37 | F | IIB | 135 | 0 | 2 | 1 | Alive | 57 + | CHOPx3 |
| 46 | 104.6 | 56 | F | IIIA | 156 | 1 | 2 | 1 | Alive | 28 + | Chlorambucil, rituximab |
| 47 | 105.1 | 34 | M | IA | 160 | 0 | 2 | 1 | Alive | 65 + | RT |
| 48 | 105.6 | 54 | F | IVA | 137 | 1 | 2 | 1 | Alive | 53 + | Rituximab, chlorambucil, CHOPx6 |
| 49 | 119.8 | 54 | F | IIIA | 135 | 1 | 2 | 1 | Alive | 63 + | CHOPx6, MIME, rituximab |
| 50 | 130.8 | 61 | F | IIA | 136 | 2 | 3B | 3 | Alive | 68 + | CHOPx6 |
| 51 | 153.0 | 47 | M | IIIA | 97 | 2 | 3B | 3 | Alive | 82 + | EPOCHx8 |
| 52 | 153.5 | 67 | F | IA | 137 | 2 | 3A | 3 | Alive | 58 + | RT |
| 53 | 173.2 | 54 | M | IIB | 140 | 0 | 2 | 1 | Alive | 81 + | Prednimustin, chlorambucil, prednisone, fludarabine |
| 54 | 175.4 | 56 | M | IIA | 127 | 0 | 3B | 3 | Alive | 31 + | CHOPx5 |
| 55 | 181.8 | 64 | M | IIIB | 126 | 2 | 3B | 3 | Alive | 70 + | CHOPx7 |
| 56 | 200.6 | 69 | F | IA | 132 | 2 | 3B | 3 | Alive | 81 + | RT, CHOPx8 |
| 57 | 246.8 | 77 | M | IA | 151 | 1 | 3B | 3 | Cardiac | 48 | RT |
Patient . | Cyclin B1* . | Age, y . | Sex . | Stage . | Hemoglobin level, g/L . | FLIPI . | WHO grade . | Ki-67† . | Cause of death . | Follow-up, mo . | Treatment in temporal order‡ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 11.4 | 81 | F | IIA | 102 | 1 | 3A | 3 | Colon cancer | 15 | KNOSPE |
| 2 | 11.8 | 68 | F | IIIA | 123 | 3 | 2 | 1 | Lymphoma | 56 | KNOSPE, CHOPx3, MIME |
| 3 | 17.7 | 42 | M | IA | 160 | 0 | 2 | 1 | Alive | 82 + | RT, rituximab |
| 4 | 26.7 | 49 | F | IIIA | 124 | 2 | 2 | 2 | Alive | 65 + | KNOSPE |
| 5 | 26.8 | 76 | M | IA | 172 | 1 | 2 | 1 | Alive | 87 + | No treatment |
| 6 | 28.1 | 67 | M | IA | 159 | 1 | 2 | 1 | Alive | 84 + | RT |
| 7 | 30.6 | 61 | F | IIA | 146 | 1 | 1 | 0 | Lymphoma | 51 | KNOSPE, RT, CNOPx5 + MTX, LD-ACOP-B |
| 8 | 32.7 | 83 | M | IVB | 127 | 2 | 2 | 1 | Lymphoma | 8 | CHOPx6, RT |
| 9 | 34.2 | 28 | F | IIIA | 143 | 2 | 2 | 1 | Lymphoma | 56 | KNOSPE, RT, rituximab, fludarabine, CHOPx5, RT |
| 10 | 34.3 | 42 | M | IVA | 125 | 2 | 2 | 0 | Lymphoma | 89 | Flu/Cycl, CHOPx3, MIME, RT, gemcitabine |
| 11 | 34.5 | 60 | M | IVA | 115 | 4 | 1 | 0 | Alive | 53 + | Rituximab, CHV(O)Px6, RT, KNOSPE, MIME, Flu/Cycl |
| 12 | 35.9 | 53 | F | IIIA | 133 | 1 | 1 | 1 | Lymphoma | 80 | KNOSPE, CHOPx8, Flu/Cycl, MIME |
| 13 | 36.0 | 75 | M | IIIB | 153 | 3 | 2 | 1 | Lymphoma | 44 | CHOPx8, KNOSPE, ENAD |
| 14 | 36.7 | 77 | M | IVA | 111 | 5 | 3B | 3 | Lymphoma | 7 | CHOPx6 |
| 15 | 40.4 | 59 | F | IA | 124 | 0 | 2 | 1 | Lymphoma | 55 | RT, CHOPx8, ENAD, MIME |
| 16 | 40.6 | 57 | F | IVA | 138 | 1 | 1 | 1 | Lymphoma | 81 | KNOSPE, CHOPx6, rituximab, RT, Flu/Cycl |
| 17 | 43.0 | 75 | M | IIIA | 142 | 4 | 2 | 3 | Lymphoma | 41 | KNOSPE |
| 18 | 43.0 | 82 | F | IA | 118 | 2 | 2 | 1 | Lymphoma | 64 | KNOSPE |
| 19 | 46.6 | 66 | M | IVA | 135 | 2 | 1 | 0 | Lymphoma | 10 | Prednimustine, CHOPx2, RT-CEP |
| 20 | 47.0 | 79 | M | IIIB | 109 | 4 | 2 | 1 | Lymphoma | 8 | KNOSPE |
| 21 | 47.9 | 73 | M | IIIA | 172 | 4 | 1 | 1 | Lymphoma | 95 | KNOSPE |
| 22 | 47.9 | 60 | F | IA | 133 | 0 | 2 | 1 | Alive | 63 + | No treatment |
| 23 | 48.1 | 43 | F | IIIA | 118 | 2 | 2 | 1 | Alive | 20 + | No treatment |
| 24 | 50.7 | 45 | M | IIIA | 154 | 1 | 2 | 1 | Alive | 50 + | No treatment |
| 25 | 51.1 | 68 | F | IVB | 145 | 4 | 1 | 0 | Alive | 67 + | CHOP×8, KNOSPE, MIME |
| 26 | 51.8 | 47 | F | IVA | 136 | 1 | 2 | 2 | Alive | 72 + | KNOSPE |
| 27 | 52.6 | 56 | F | IVB | 139 | 1 | 1 | 0 | Lymphoma | 82 | CdA, KNOSPE |
| 28 | 54.1 | 61 | M | IA | 159 | 1 | 3A | 2 | Alive | 59 + | RT |
| 29 | 59.0 | 57 | F | IVA | 135 | 1 | 2 | 1 | Alive | 87 + | CHOPx1, RT |
| 30 | 63.1 | 28 | M | IVB | 114 | 3 | 3B | 3 | Alive | 65 + | CHOPx7 + SCT |
| 31 | 64.9 | 56 | M | IA | 147 | 0 | 1 | 1 | Alive | 56 + | No treatment |
| 32 | 68.3 | 43 | F | IVA | 142 | 3 | 2 | 1 | Alive | 112 + | CdA |
| 33 | 68.7 | 61 | F | IVA | 140 | 2 | 3B | 2 | Lymphoma | 54 | CHOPx2, CHVPx6 |
| 34 | 74.4 | 60 | F | IVA | 134 | 2 | 2 | 1 | Alive | 72 + | Chlorambucil, rituximab |
| 35 | 77.4 | 85 | F | IIA | 128 | 2 | 3B | 3 | Lymphoma | 38 | CHOPx5, gemcitabine |
| 36 | 80.2 | 56 | F | IVA | 140 | 1 | 1 | 1 | Alive | 29 + | No treatment |
| 37 | 81.5 | 59 | M | IIIA | 158 | 1 | 1 | 0 | Alive | 58 + | No treatment |
| 38 | 84.4 | 78 | F | IIIA | 147 | 3 | 2 | 1 | Alive | 117 + | CHOPx4 |
| 39 | 85.0 | 80 | M | IVB | 84 | 4 | 1 | 1 | Lymphoma | 1 | No treatment |
| 40 | 86.3 | 71 | F | IA | 144 | 1 | 2 | 1 | Alive | 56 + | RT |
| 41 | 88.7 | 44 | F | IVA | 140 | 2 | 2 | 1 | Alive | 54 + | No treatment |
| 42 | 88.8 | 70 | F | IA | 127 | 1 | 2 | 1 | Alive | 70 + | RT |
| 43 | 90.6 | 72 | M | IVA | 133 | 3 | 2 | 1 | Lymphoma | 43 | KNOSPE, CHOPx6 |
| 44 | 90.7 | 46 | M | IVB | 145 | 1 | 2 | 2 | Alive | 63 + | Rituximab + interferon alfa |
| 45 | 96.0 | 37 | F | IIB | 135 | 0 | 2 | 1 | Alive | 57 + | CHOPx3 |
| 46 | 104.6 | 56 | F | IIIA | 156 | 1 | 2 | 1 | Alive | 28 + | Chlorambucil, rituximab |
| 47 | 105.1 | 34 | M | IA | 160 | 0 | 2 | 1 | Alive | 65 + | RT |
| 48 | 105.6 | 54 | F | IVA | 137 | 1 | 2 | 1 | Alive | 53 + | Rituximab, chlorambucil, CHOPx6 |
| 49 | 119.8 | 54 | F | IIIA | 135 | 1 | 2 | 1 | Alive | 63 + | CHOPx6, MIME, rituximab |
| 50 | 130.8 | 61 | F | IIA | 136 | 2 | 3B | 3 | Alive | 68 + | CHOPx6 |
| 51 | 153.0 | 47 | M | IIIA | 97 | 2 | 3B | 3 | Alive | 82 + | EPOCHx8 |
| 52 | 153.5 | 67 | F | IA | 137 | 2 | 3A | 3 | Alive | 58 + | RT |
| 53 | 173.2 | 54 | M | IIB | 140 | 0 | 2 | 1 | Alive | 81 + | Prednimustin, chlorambucil, prednisone, fludarabine |
| 54 | 175.4 | 56 | M | IIA | 127 | 0 | 3B | 3 | Alive | 31 + | CHOPx5 |
| 55 | 181.8 | 64 | M | IIIB | 126 | 2 | 3B | 3 | Alive | 70 + | CHOPx7 |
| 56 | 200.6 | 69 | F | IA | 132 | 2 | 3B | 3 | Alive | 81 + | RT, CHOPx8 |
| 57 | 246.8 | 77 | M | IA | 151 | 1 | 3B | 3 | Cardiac | 48 | RT |
Relative expression of RNA of probe set 34736_at
Immunohistochemistry Ki-67: 0 indicates less than 10%; 1, 10% to 30%; 2, 30% to 50%; and 3, more than 50% cells positive
KNOSPE indicates chlorambucil, prednisone; CHOP, cyclophosphamide, doxorubicin, vincristine, prednisone; MIME, mitoguazone, ifosfamide, methotrexate, etoposide; RT, radiotherapy; CNOP, cyclophosphamide, mitoxantrone, vincristine, prednisone; MTX, methotrexate; LD-ACOP-B, low-dose doxorubicin, cyclophosphamide, vincristine, prednisone, bleomycin; Flu/Cycl, fludarabine, cyclophosphamide; CHVP, cyclophosphamide, doxorubicin, teniposide, prednisone; ENAD, etoposide, mitoxantrone, cytarabine, prednisone; RT-CEP, lomustine, etoposide, chlorambucil, prednisone; CdA, cladribine; EPOCH, etoposide, cyclophosphamide, doxorubicin, vincristine, prednisone; and SCT, stem cell transplantation
RNA extraction and microarray hybridization
Frozen tissue (30 to 50 mg; -70°C) from tumor samples was homogenized with a Polytron tissue tearer (Biospec Products, Bartlesville, OK) in 1 mL Trizol (Invitrogen, Carlsbad, CA). Prior to RNA isolation, 5 samples (patient nos. 23, 36, 46, 53, 54) were purified to 85% to 95% tumor content with T-cell depletion as previously described.16 RNA was extracted with Trizol according to the manufacturer's instructions, followed by purification with RNeasy mini kit (Qiagen, Hilden, Germany). To evaluate the RNA purity, the A260/280 ratio (1.9-2.1) was determined by spectrophotometry, followed by denaturating gel electrophoresis where the RNA quality was visually estimated by the presence of 2 sharp RNA bands. Furthermore, the RNA quality was confirmed by analysis in an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA). All preparation of mRNA, in vitro transcription and hybridization of labeled cRNA, and scanning of the U95Av2 oligonucleotide array (Affymetrix, Santa Clara, CA) were performed as previously described.17
Microarray analysis
The expression level for each probe set was given as a signal value and a detection call (present, absent, or marginal) by the Microarray Suite 5.0 software (Affymetrix). To enable interarray comparison, the signal values were scaled in Microarray Suite, against a target value of 100. The scaling factor was 0.68 to 3.43, and 46 of 57 samples had results within a factor of 3 (0.79-2.25). The range of probe sets with a present call was 34.3% to 54.9%. Microarray Suite comparison analysis (batch analysis) was used to compare the arrays. The created comparison files were imported into Genespring 6 (Silicon Genetics, Redwood City, CA) for gene selection and further analysis.
Selection of patients according to response to CHOP chemotherapy
The WHO criteria for evaluation of the response to chemotherapy were applied to define groups of patients with complete remission (CR), partial response (PR), and progressive disease (PD).18 The algorithms used to define probe sets that were differentially expressed in patients who responded to CHOP therapy with CR or PD are given in Table 2, and details of treatment are shown in Table 1. To define the probe sets with differential expression between CR and PD patients, 2 main comparisons were made. Tumors selected for the first comparison fulfilled all of the following strict criteria: (1) CHOP was given as the first-line treatment; (2) time to treatment was less than 6 months from diagnosis; (3) 4 or more courses of CHOP were given; and (4) treatment response was either CR or PD.
Algorithm used to find probe sets with differential expression in PD and CR patients
. | 1st comparison . | 2nd comparison . | Cross-comparison . |
|---|---|---|---|
| Compared groups of patients | 3 PD* vs 10 CR† | 5 PD‡ vs 3 CR§ | 3 PD* vs 3 CR§ + 10 PD† vs 3 CR‡ |
| Comparison files | 3 × 10 = 30 | 5 × 3 = 15 | 10 × 5 + 3 × 3 = 59 |
| Comparison files with probe sets altered (%) | 21/30 (70) | 10/15 (67) | 41/59 (69) |
| Change algorithm: probe sets decreased | 51 | 338 | 62 |
| Signal log ratio algorithm: probe sets of – 1 or less | 217 | 617 | 109 |
| Probe sets selected by both algorithms | 28 | 207 | 35 |
| Probe sets selected by 1st and 2nd comparison | 16 | 16 | 10∥ |
. | 1st comparison . | 2nd comparison . | Cross-comparison . |
|---|---|---|---|
| Compared groups of patients | 3 PD* vs 10 CR† | 5 PD‡ vs 3 CR§ | 3 PD* vs 3 CR§ + 10 PD† vs 3 CR‡ |
| Comparison files | 3 × 10 = 30 | 5 × 3 = 15 | 10 × 5 + 3 × 3 = 59 |
| Comparison files with probe sets altered (%) | 21/30 (70) | 10/15 (67) | 41/59 (69) |
| Change algorithm: probe sets decreased | 51 | 338 | 62 |
| Signal log ratio algorithm: probe sets of – 1 or less | 217 | 617 | 109 |
| Probe sets selected by both algorithms | 28 | 207 | 35 |
| Probe sets selected by 1st and 2nd comparison | 16 | 16 | 10∥ |
Patient numbers 8, 11, 14
Patient numbers 13, 25, 30, 33, 35, 38, 50, 51, 54, 55
Patient numbers 2, 7, 9, 10, 19
Patient numbers 45, 48, 56
Probe sets common in 1st, 2nd, and cross-comparison
In this comparison, 3 tumors from patients with PD (nos. 8, 11, 14) were compared with 10 tumors from patients who obtained CR (nos. 13, 25, 30, 33, 35, 38, 50, 51, 54, 55). The results of this comparison were used as a main selection of probe sets. To further select relevant probe sets and to narrow the number of probe sets, a second comparison was made using the following criteria: (1) CHOP was given either as the second-line treatment (minimum 2 courses) or as the first-line treatment with 3 courses given (patient no. 45); and (2) treatment response was either CR or PD.
In this second comparison, 5 tumors from patients with PD (nos. 2, 7, 9, 10, 19) were compared with 3 tumors from patients with CR (nos. 45, 48, 56). The average time between the biopsy and CHOP treatment was 1.7 months (range, 0-5 months) in the first comparison and 33 months (range, 1-84 months) in the second comparison. Since they did not fulfill the selection criteria, 6 tumors from patients who received CHOP were not included in the first or second comparison. These patients either responded with PR or received only one course of CHOP.
To compare samples from patients responding with PD with samples from CR patients, comparison files were created with Microarray Suite 5.0. The comparison analysis was based on 2 algorithms: change algorithm and signal log ratio algorithm. Briefly, the change algorithm indicates increase, decrease, or no change, and the signal log ratio algorithm calculates the log ratio (base 2), between the compared arrays. A detailed description of the algorithms has been published.19
To consider the expression of a probe set significantly lower in the PD group compared with CR group, it had to be down-regulated by both algorithms in both comparisons. To consider the expression of a probe set significantly higher in the PD group compared with the CR group, it had to be up-regulated by both algorithms in both comparisons. A cross-comparison was also made where the tumors from the CR patients from the first comparison were compared with tumors from PD patients of the second comparison and vice versa (Table 2).
Tumor samples used to identify genes coexpressed with KIAA0101
Since KIAA0101 was one of the genes selected by the previous comparisons and the function of this gene is unknown, we investigated the expression pattern of this gene and its correlation with the global gene expression pattern in other samples. To avoid bias from the 21 FLs of the previous comparisons, this correlation study was done using the results from the remaining 36 FL together with 21 mantle cell lymphomas and 11 samples representing 5 different B-cell populations from reactive tonsils. The mantle cell lymphomas and B-cell populations have previously been described in detail.16,17
Immunohistochemistry
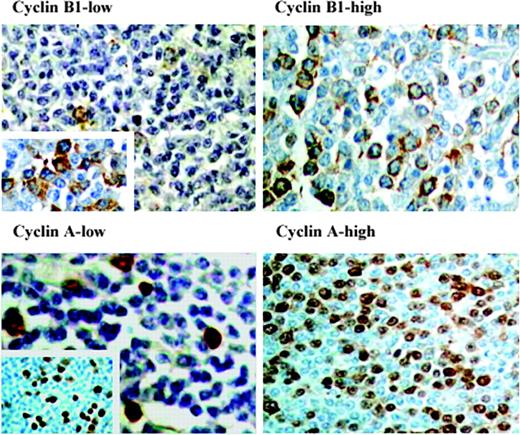
The protein expression of Ki-67, cyclin B1, and cyclin A was evaluated by immunohistochemistry (IHC). Briefly, 5-μm paraffin sections were deparaffinized, rehydrated, and cooked in a microwave oven (in citrate buffer, pH 6: for cyclin A, at 750 W until boiling then at 500 W for 10 minutes and for cyclin B1, 10 minutes at 750 W and 10 minutes at 450 W; in citrate buffer, pH 7.3: for Ki-67, at 300 W for 3 × 10 minutes). Incubation was carried out for 1 hour at room temperature with primary antibodies for cyclin B1 (Clone 7A9, dilution 1:40; Novocastra Laboratories, Newcastle upon Tyne, United Kingdom) and cyclin A2 (Clone 6E6, dilution 1:100; Novocastra). Cyclin B1 and cyclin A visualization was carried out using the immunoperoxidase method, with Strept ABComplex/HRP Duet Mouse/Rabbit and diaminobenzidine (DAB) used as substrate (DakoCytomation, Glostrup, Denmark). For Ki-67 (Clone M7240, dilution 1:50; DakoCytomation), the slides were stained using a NexES IHC staining module (Ventana Medical Systems, Tucson, AZ) with enhanced DAB paraffin procedure. Distinct nuclear staining pattern was obtained in all tumors with cyclin A and Ki-67 antibodies. For cyclin B1, only 51 of the 57 tumors were successfully stained, despite repeated staining attempts in the remaining 6 cases. Cyclin B1 showed preponderant cytoplasmic staining (Figure 1).
Immunohistochemistry of cyclin B1 and cyclin A. Examples of follicular lymphomas with high and low expression of cyclin A and cyclin B1. (Left) Tumor from patient no. 9 showing low fractions of cyclin B1-positive cells (magnification, × 200) and cyclin A-positive cells (magnification, × 600). Inserts show regions with higher expression, so called hot spots (magnifications, × 600 and × 100). (Right) Tumor from patient no. 55 with high percentages of cyclin B1-positive cells (magnification, × 400) and cyclin A-positive cells (magnification, × 200). Original magnifications, × 200 (top left panel and inset; bottom right panel); × 400 (top right panel; bottom left panel and inset). A Nikon Optishot microscope equipped with 20 ×/0.40 and 40 ×/0.65 objective lenses (Nikon, Tokyo, Japan) was used to visualize images. Pictures were taken with a Nikon CoolPix 990 camera, and Adobe Photoshop 6.0 software was used to zoom images to their present magnitude (Adobe, San Jose, CA).
Immunohistochemistry of cyclin B1 and cyclin A. Examples of follicular lymphomas with high and low expression of cyclin A and cyclin B1. (Left) Tumor from patient no. 9 showing low fractions of cyclin B1-positive cells (magnification, × 200) and cyclin A-positive cells (magnification, × 600). Inserts show regions with higher expression, so called hot spots (magnifications, × 600 and × 100). (Right) Tumor from patient no. 55 with high percentages of cyclin B1-positive cells (magnification, × 400) and cyclin A-positive cells (magnification, × 200). Original magnifications, × 200 (top left panel and inset; bottom right panel); × 400 (top right panel; bottom left panel and inset). A Nikon Optishot microscope equipped with 20 ×/0.40 and 40 ×/0.65 objective lenses (Nikon, Tokyo, Japan) was used to visualize images. Pictures were taken with a Nikon CoolPix 990 camera, and Adobe Photoshop 6.0 software was used to zoom images to their present magnitude (Adobe, San Jose, CA).
To evaluate cyclin B1 and cyclin A expression, 2 different counting methods were used. In the first method, 4 representative high-power fields (HPFs) were evaluated and the numbers of positive cells per field were calculated. We also applied a “hot-spot” (HS) approach where the percentages of positive cells were counted out of a total of 500 cells from the 3 to 4 most positive areas of the slide. All counting of cells was facilitated with a Weibul grid. Evaluation of the proliferative fraction with Ki-67 was performed as previously described.20
FISH analysis of topoisomerase II alpha
Fluorescent in situ hybridization (FISH) was performed with the Vysis LSI topoisomerase II alpha (TOPO2A) probe according to the manufacturer's instructions (Vysis, Downers Grove, IL) on tumor tissue imprints from 18 tumors (patient nos. 2, 8, 9, 10, 11, 13, 14, 18, 25, 29, 30, 33, 35, 39, 45, 50, 55, 56). Signals from 200 interphase nuclei of each imprint were analyzed with Smart Capture 2 (Digital Scientific, Cambridge, United Kingdom) software.
Statistical analysis
Survival curves were calculated with the Kaplan-Meier method and compared with log-rank test.21 In the univariate analysis, the cohort of 57 patients was dichotomized by the median expression of each studied probe set. Cox proportional hazard regression was used for multivariate analysis.22 Survival analysis was calculated both for cause-specific survival (CSS) and overall survival (OS). CSS was defined as the time from diagnosis to death of lymphoma or death judged to be related to treatment of the disease. OS was defined as the time from diagnosis to death, irrespective of cause. A chi-2 test and a Mann-Whitney test were used to compare the expression of Ki-67 antigen in tumors from patients with CR and patients with PD and between cases with high and low expression of cyclin B1, as well as to evaluate the distribution of patients with regard to FLIPI groups. To evaluate the correlation between expression of RNA (microarray analysis) and protein (IHC), the Pearson correlation coefficient was calculated. The statistical analysis was performed with Minitab 14.1 (Minitab, State College, PA) and Statistica 6.1 (StatSoft, Tulsa, OK). P values less than .05 were considered statistically significant.
Results
Differential gene expression in tumors with diverse response to chemotherapy
There were 16 probe sets representing 14 genes that showed high expression in the CR group and low expression in the PD group in both comparisons (Tables 2, 3). The cross-comparison analysis identified 10 probe sets in common with the selected 16 probe sets (Table 3). Of the 14 selected genes, 11 were involved in cell cycle, mitosis, or DNA modulation, while 3 had other or unknown functions. No probe set fulfilled the criteria of high expression in the PD group and low expression in the CR group with all algorithms in both the first and the second comparison.
Probe sets up-regulated in tumors from patients with a good response to CHOP
Function/probe set . | Gene symbol . | Gene name . | P, log-rank test* . |
|---|---|---|---|
| Cell-cycle related | |||
| 1945_at | CCNB1 | Cyclin B1† | .020 |
| 34736_at | CCNB1 | Cyclin B1† | .010 |
| 1803_at | CDC2 | Cell division cycle 2, G1 to S and G2 to M | .004 |
| 37347_at | CKS1B | CDC28 protein kinase regulatory subunit 1B | .005 |
| 1599_at | CDKN3 | Cyclin-dependent kinase inhibitor 3†‡ | .005 |
| 40348_s_at | ANP32E | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E† | .005 |
| 40347_at | ANP32E | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E† | NS |
| 32263_at | CCNB2 | Cyclin B2 | NS |
| Mitosis | |||
| 40041_at | KNTC2 | Kinetochore associated 2 (highly expressed in cancer, HEC)† | NS |
| 35699_at | BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast)† | NS |
| 36863_at | HMMR | Hyaluronan-mediated motility receptor (RHAMM) | NS |
| DNA modulation | |||
| 40145_at | TOP2A | Topoisomerase (DNA) II alpha 170 kDa† | NS |
| 38065_at | HMGB2 | High-mobility group box 2† | NS |
| Other/unknown | |||
| 38116_at | KIAA0101 | KIAA0101 gene product† | .004 |
| 35071_s_at | GMDS | GDP-mannose 4,6-dehydratase | NS |
| 39056_at | PAICS | Phosphoribosylaminoimidazole carboxylase§ | NS |
Function/probe set . | Gene symbol . | Gene name . | P, log-rank test* . |
|---|---|---|---|
| Cell-cycle related | |||
| 1945_at | CCNB1 | Cyclin B1† | .020 |
| 34736_at | CCNB1 | Cyclin B1† | .010 |
| 1803_at | CDC2 | Cell division cycle 2, G1 to S and G2 to M | .004 |
| 37347_at | CKS1B | CDC28 protein kinase regulatory subunit 1B | .005 |
| 1599_at | CDKN3 | Cyclin-dependent kinase inhibitor 3†‡ | .005 |
| 40348_s_at | ANP32E | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E† | .005 |
| 40347_at | ANP32E | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E† | NS |
| 32263_at | CCNB2 | Cyclin B2 | NS |
| Mitosis | |||
| 40041_at | KNTC2 | Kinetochore associated 2 (highly expressed in cancer, HEC)† | NS |
| 35699_at | BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast)† | NS |
| 36863_at | HMMR | Hyaluronan-mediated motility receptor (RHAMM) | NS |
| DNA modulation | |||
| 40145_at | TOP2A | Topoisomerase (DNA) II alpha 170 kDa† | NS |
| 38065_at | HMGB2 | High-mobility group box 2† | NS |
| Other/unknown | |||
| 38116_at | KIAA0101 | KIAA0101 gene product† | .004 |
| 35071_s_at | GMDS | GDP-mannose 4,6-dehydratase | NS |
| 39056_at | PAICS | Phosphoribosylaminoimidazole carboxylase§ | NS |
NS indicates not significant.
Log-rank test for cause-specific survival
Probe sets that were selected with cross-comparison; see Table 2 for details
Other name: CDK2-associated dual specificity phosphatase
Other name: phosphoribosylaminoimidazole succinocarboxamide synthetase
Prognostic significance of the identified gene expression
Levels of expression of 6 of the 14 selected genes showed in univariate analysis a statistically significant impact on both CSS and OS in the whole cohort of 57 FL patients (Table 3). Since the CCNB1 gene is a key regulator of the G2/M transition of the cell cycle, and the other genes that have significant impact on survival have a direct or indirect interaction with CCNB1,23-25 we have chosen this gene expression for further analysis. The Kaplan-Meier curves comparing the survival of patients with CCNB1 high and low tumors (Figure 2) show that a high expression of CCNB1 is associated with a better survival rate (P = .010). When CCNB1 expression and the FLIPI factors were tested in a multivariate analysis, a high expression of CCNB1 was independently associated with favorable prognosis (Table 4). Age, stage, hemoglobin, and CCNB1 still retained their independent prognostic values when the WHO histologic grade was entered in the multivariate analysis. To avoid a potential bias from data that might skew the results, 4 additional survival analyses were made: (1) to see if the importance of CCNB1 expression was independent of the response to first-line chemotherapy, we excluded the 13 patients of the first comparison; (2) to investigate if the prognostic significance of CCNB1 was valid in low-grade FL, we excluded the 13 patients with FL grade 3; (3) we excluded the 5 purified samples that might have given a false high value of CCNB1; and (4) to determine whether or not the scaling factor influenced the survival analysis, we excluded the 11 samples that were not within the recommended scaling factor of 3.19
Prognostic significance of CCNB1 expression in follicular lymphomas. Cause-specific survival depending on expression of probe set 34736_at. Patients were divided in groups (cyclin B-high, solid line; cyclin B-low, dashed line) by the median expression of CCNB1. Difference in survival was calculated with log-rank test.
Prognostic significance of CCNB1 expression in follicular lymphomas. Cause-specific survival depending on expression of probe set 34736_at. Patients were divided in groups (cyclin B-high, solid line; cyclin B-low, dashed line) by the median expression of CCNB1. Difference in survival was calculated with log-rank test.
Multivariate analysis of prognostic impact of cyclin B1 and FLIPI parameters on overall survival
. | Adverse factor . | Relative risk . | Range . | P . |
|---|---|---|---|---|
| Age | ≥ 60 | 5.6 | 1.7-18.7 | .005 |
| Ann Arbor stage | III-IV | 4.4 | 1.3-15.2 | .020 |
| Nodal sites | > 4 | 0.3 | 0.09-1.02 | .054 |
| Hemoglobin | < 12 g/dL | 3.0 | 1.0-8.9 | .043 |
| Serum LDH level | > ULN | 1.0 | 0.33-3 | .99 |
| Cyclin B1* | ≤ 59 | 3.0 | 1.1-8.3 | .036 |
. | Adverse factor . | Relative risk . | Range . | P . |
|---|---|---|---|---|
| Age | ≥ 60 | 5.6 | 1.7-18.7 | .005 |
| Ann Arbor stage | III-IV | 4.4 | 1.3-15.2 | .020 |
| Nodal sites | > 4 | 0.3 | 0.09-1.02 | .054 |
| Hemoglobin | < 12 g/dL | 3.0 | 1.0-8.9 | .043 |
| Serum LDH level | > ULN | 1.0 | 0.33-3 | .99 |
| Cyclin B1* | ≤ 59 | 3.0 | 1.1-8.3 | .036 |
ULN indicates upper limit of normal.
Relative expression of RNA of probe set 34736_at
In each of these 4 analyses, the significant prognostic impact of CCNB1 expression was retained.
High expression of each of the genes CDC2, CDKN3, CKS1B, ANP32E, and KIAA0101 was also associated with a good outcome in a multivariate analysis independent of the factors of FLIPI (relative risk: CDC2, 3.6; CDKN3, 3.0; CKS1B, 5.0; ANP32E, 3.7; and KIAA0101, 3.0). Since the expression levels of all these genes were significantly correlated with CCNB1, no further multivariate analyses were made including the factors of FLIPI. The results for ANP32E should be interpreted with caution since only one probe set (40348_s_at) had prognostic impact on survival.
Since both cyclin B1 (CCNB1) and cyclin A (CCNA2) are essential for G2/M transition in the cell cycle, the impact of CCNA2 expression on survival was analyzed. There were 2 probe sets for CCNA2 present on the u95Av2 array (1943_at and 40697_at). High expression of both was statistically significant for CSS, but only high expression of the 1943_at probe set was significant for OS (data not shown). Cyclin A did not have independent prognostic impact when included in a multivariate analysis combined with the parameters of the FLIPI. No significant difference in survival could be seen when the mRNA expression levels of other cyclins (C, D1, D3, E, F, G2, T1, T2) and of the proliferation marker Ki-67 were analyzed.
Protein expression of cyclin B, cyclin A, and Ki-67
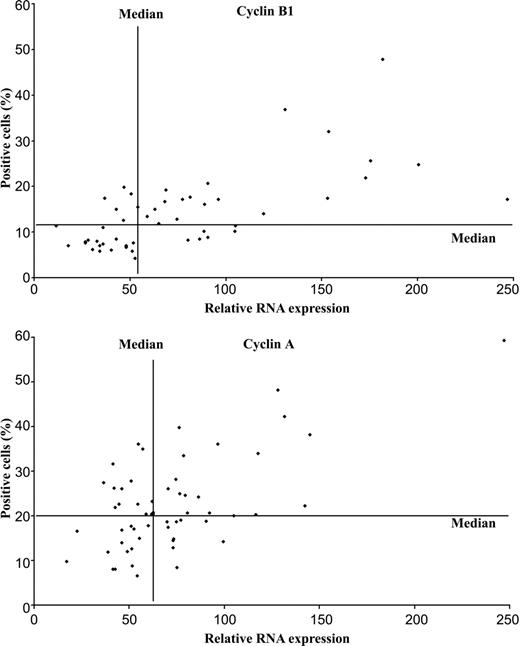
The FL samples showed a large variation in the number of cells showing protein expression of cyclin B1: the HPF method showed a range of 13 to 204 positive cells (median, 61 cells) and the HS method, 4.2% to 47.8% (median, 11.8%) positive cells. The HS method gave the best correlation when comparing RNA and protein expression, with a correlation coefficient of 0.67 (P = 6 × 10-5) (Figure 3). However, we could not define a cut-off level for cyclin B1 protein expression, which could divide the patients into 2 groups with statistically significant differences in survival. This was due to discrepant results found in 11 patients; 6 patients (nos. 36, 40, 41 43, 46, 47) had RNA expression above the median but low fractions of cells expressing protein, and 6 patients (nos. 14, 18, 19, 20, 24, 28) had RNA below the median but high fractions of cells expressing protein (Figure 3). Cyclin A expression with the HPF method varied between 31 and 520 positive cells, with a median of 91 cells. The results obtained with the HS method ranged between 6.6% and 59.2%, with a median of 20.4% positive cells. The HS method gave the best correlation when comparing RNA and protein expression, with the correlation coefficient of 0.63 (P = 2 × 10-7) (Figure 3). For cyclin A, a cut-off of 18% positive cells gave a statistically significant difference on the CSS (P = .033) but not on the OS. When this cut-off value was used in a multivariate analysis together with the prognostic factor of the FLIPI, cyclin A did not have an independent prognostic value.
Correlation between RNA and protein expression of cyclin B1 and cyclin A in follicular lymphomas. RNA expression for CCNB1 was measured by probe set 34736_at and for CCNA2 by probe set 1943_at. The percentages of cyclin B1-positive cells (top) and cyclin A-positive cells (bottom) with immunohistochemistry were calculated with the hot-spot method (see “Patients, materials, and methods”). Correlation coefficient between RNA and protein for cyclin B1 was 0.67 (P value, 1 × 10-7) and for cyclin A, 0.63 (P value, 2 × 10-7).
Correlation between RNA and protein expression of cyclin B1 and cyclin A in follicular lymphomas. RNA expression for CCNB1 was measured by probe set 34736_at and for CCNA2 by probe set 1943_at. The percentages of cyclin B1-positive cells (top) and cyclin A-positive cells (bottom) with immunohistochemistry were calculated with the hot-spot method (see “Patients, materials, and methods”). Correlation coefficient between RNA and protein for cyclin B1 was 0.67 (P value, 1 × 10-7) and for cyclin A, 0.63 (P value, 2 × 10-7).
The fractions of cells expressing Ki-67 were compared in groups of patients with different responses to CHOP chemotherapy and also in tumors with high and low expression of cyclin B1. A significantly higher expression of Ki-67 was seen both on RNA and protein level in the tumors from CR patients compared with the PD group (P < .05). Also, tumors with high expression of the CCNB1 gene had a significantly higher incidence of high Ki67 expression on RNA levels (P < .05) but not on protein levels.
FISH analysis of TOP2A
Since the TOP2A gene was overexpressed in tumors from patients with favorable response to CHOP, we performed FISH analysis to assess if the overexpression was due to genomic amplification. A normal pattern with 2 signals for chromosome 17 centromere and 2 signals from TOP2A was found in 85% to 95% of the cells of the investigated tumors.
Genes showing coexpression with KIAA0101
Since the function of KIAA0101 is unknown, we have attempted to identify the probe sets with expression pattern similar to KIAA0101 in order to elucidate the role of this gene. In the 68 samples that were used for this analysis, 21 genes had a correlation coefficient above 0.8 with KIAA0101. Of these genes, 11 were involved in mitosis (CENPA, TPX2, KIF11, KNTC2, BUB1R, KPNA2, ZWINT, KIF2C, DLG7, AURKB, PTCC1), 5 genes were cell-cycle related (CCNB2, UBE2C, CDC20, CDKN3, EZH2, CDC2), 2 had DNA modulating function (HMGB2, TOP2A), and 3 had other functions (TYMS, KIF14, EZH2). The strongest correlation was obtained with CCNB2, with a correlation coefficient of 0.89 (P = 1.3 × 10-22). This pattern of expression suggests that KIAA0101 is involved in mitosis or cell-cycle regulation.
Discussion
In this study we compared gene expression in diagnostic samples from FL patients with diverse response to CHOP chemotherapy and investigated the prognostic impact of such expression pattern.
There were 14 genes that showed a high expression level in the group with a good response to CHOP and low levels in the group with poor response. Of the 14 selected genes, 11 were involved in cell-cycle regulation (CKS1B, CDKN3, ANP32E, CCNB1, CCNB2, CDC2), mitosis (HMMR, BUB1B, KNTC2), or DNA modulation (TOP2A, HMGB1). This may indicate that the tumors responding well to CHOP chemotherapy had more cells in an active proliferating phase than the tumors from nonresponders. In agreement with this observation was the fact that the protein expression of the proliferation marker Ki-67 was higher in the group with good response to chemotherapy compared with the group of nonresponders. The components of the CHOP regimen interact in different modes with the proliferating cells. The active metabolite of cyclophosphamide binds to DNA and inhibits replication and cell division, and vincristine prevents the spindle formation of mitotic cells and causes cell-cycle arrest. Both of these drugs are most likely lethal when the target cells are in the replicative phase. In this context, it is interesting to note that TOP2A was among the selected genes. TOP2A alters the topologic state of DNA during transcription, and it is also the main target of doxorubicin.26 It is likely that a high expression of TOPO2A facilitated the response to doxorubicin in the studied CR patients. A high expression of TOPO2A has been found in Wilms tumor samples from patients with good response to doxorubicin.27 Järvinen et al showed that genomic amplification and overexpression of TOPO2A was correlated with higher sensitivity to doxorubicin in breast cancer cell lines.28 In our FL patients, the overexpression of TOPO2A was most likely posttranscriptionally regulated since no amplification could be seen at a genomic level of investigated tumors. Our observation indicating that more active proliferating cells can be found in FL sensitive to chemotherapy is in agreement with previous studies.20,29
Most of the 14 selected genes have their main function during the G2/M phase of the cell cycle. CCNB1 and CDC2 form a complex, which is essential for the G2/M transition of the cell cycle. This complex has a peak during the G2/M phase and must be degraded before the completion of the cell division. Other selected genes interacting with CDC2 were CKS1B, CDKN3, CCNB2, and TOPO2A.23,25,30 A link between ANP32E and CDC2 has also been established, since ANP32E inhibits the CDC2 inhibitor PP2A.24 BUB1B and KNTC2 are both attached to the kinetochore and involved in the spindle checkpoint.31,32 They are involved in 2 different inhibition pathways of the APC/C-CDC20 complex. An activated APC/C-CDC20 complex is believed to contribute to the degradation of the CCNB1/CDC2 complex,33,34 and thus high expression of BUB1B and KNTC3 is indirectly related to high expression of CCNB1/CDC2. Another gene involved with mitosis is RHAMM, which has a role in spindle pole stability. RHAMM has interestingly also been linked to the function of CCNB1.35,36 A link to CCNB1/CDC2 or to the mitotic spindle was not found for GMDS and PAICS. The function of KIAA0101 is not known, but interestingly its expression was strongly correlated to that of cyclin B2 and of other genes involved in mitosis or cell-cycle regulation. It is therefore reasonable to speculate that KIAA0101 might have a similar function.
We also investigated the impact of the mRNA expression levels for the 14 selected genes on prognosis. A high expression of 6 of these genes (CCNB1, CDC2, CDKN3, CKS1B, KIAA0101, and ANP32E) was associated with better prognosis in the univariate analysis (Table 1; Figure 2). Since all of these have their main function during the G2/M transition of the cell cycle and CCNB1 is a key regulator of this transition, CCNB1 was selected for further analysis. Multivariate Cox regression analysis of CCNB1 and the clinical parameters of the FLIPI revealed that age, stage, hemoglobin, and CCNB1 all had independent prognostic value (Table 4).
We could not show any significant impact on survival for levels of the proliferation marker Ki-67. The prognostic impact of proliferation in lymphomas measured by Ki-67 protein expression remains controversial.20,37-39 However, we could show that a high expression of cyclin A was correlated with a longer survival in the univariate analysis but not in the multivariate analysis. There are only a few clinical studies comparing expression of cyclin A and cyclin B1 in lymphoma. Low expression of cyclin A measured by Western blot has been shown to correlate to a longer survival in NHL.40 Cyclin B1 evaluated by IHC could not be related to survival in the study by Jin and Park.41
Our results showed a good correlation between the results of microarray and IHC for both cyclin B1 and cyclin A. Despite this good correlation, it was difficult to determine the prognostically significant cut-off for percentage of positive cells. A possible reason for this could be the uneven distribution of cyclin B1-positive and cyclin A-positive cells in the tissue. The strongest correlation between RNA levels and immunohistochemistry findings was obtained when the analysis was restricted to cells only from the most positive areas of the slide. This may suggest that the groups of cells with high expression may be of biologic importance for progression of the disease. It is therefore plausible to conclude that the possibilities to reproducibly quantify immunohistochemistry are limited, and that it would be valuable to study the impact on survival of cyclin A and cyclin B1 with other methods that would secure a more accurate measurement of the protein expression. Flow cytometry analysis of CCNB142 could be an attractive alternative method in a prospective study of follicular lymphomas. An alternative approach in the diagnostic setting is to measure RNA levels of these genes with quantitative polymerase chain reaction (PCR).43
In conclusion, we have shown that a high expression of genes associated with the G2/M transition of the cell cycle, mitosis, and DNA modulation indicates the possibility of a favorable response to CHOP chemotherapy in FL patients. High RNA expression of cyclin B1 was significantly correlated with longer survival. The prognostic value of cyclin B1 was independent when tested in a multivariate analysis including the established prognostic parameters of the FLIPI.
Prepublished online as Blood First Edition Paper, December 2, 2004; DOI 10.1182/blood-2004-07-2721.
Supported by grants from the Swedish Cancer Society, the Swedish Society of Medicine, the Stockholm County Council, the Swedish Society for Medical Research, the King Gustave V Jubilee Foundation, and BioInvent International AB (S.E., C.A.K.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Margareta Waern, Stefanie Bug, and Anna-Barbara Witte for skillful technical assistance; Ingrid Svensson for advice on statistical concerns; Marie Bohlin for help retrieving information on treatment of the patients; and Göran Björck and Lewis Edgel for linguistic consultation.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal