Abstract
During lymphoid development, Notch1 plays a critical role in the T-cell/B-cell lineage decision, while Notch2 is essential for marginal zone B-cell (MZB) development. Notch pathway activation induces translocation of intracellular Notch (ICN) to the nucleus, where it interacts with the transcription factor CSL (CBF1/RBP-Jk, Suppressor of Hairless, Lag-1). In vitro, ICN binds Mastermind-like proteins, which act as potent Notch coactivators. Three MAML family members (MAML1-3) have been identified in mammals, but their importance in vivo is unknown. To investigate the function of MAMLs in hematopoietic development, we introduced a dominant negative (DN) mutant of MAML1, capable of inhibiting Notch1-4, in murine hematopoietic stem cells. DNMAML1 resulted in early inhibition of T-cell development and the appearance of intrathymic B cells, phenotypes consistent with Notch1 inhibition. The T-cell differentiation block was as profound as that produced by enforced expression of the Notch modulator Deltex1. In DNMAML1-transduced spleen cells, a dramatic decrease in MZB cells was present, consistent with Notch2 inhibition. In contrast, Deltex1 did not decrease MZB cell numbers. These results suggest a critical role for MAMLs during Notch-mediated cell fate decisions in vivo and indicate that DNMAML1, but not Deltex1, can be used to interfere with the function of multiple Notch family members. (Blood. 2004;104:1696-1702)
Introduction
Notch signaling plays a central role in multiple developmental processes. During blood and lymphoid development, Notch proteins influence several hematopoietic lineages. At the T-cell/B-cell branchpoint, Notch promotes T-cell at the expense of B-cell differentiation.1,2 More recent data implicate Notch1 in the early generation of the hematopoietic stem cell (HSC) pool during embryogenesis3 and Notch2 in marginal zone B-cell (MZB) development.4-6 Notch family members also have been postulated to play a role in HSC self-renewal and homeostasis.7,8 Other putative roles of Notch in the CD4/CD8 lineage choice, in αβ versus γδ T-cell differentiation and in the function of peripheral T cells are more controversial (reviewed in Radtke et al9 and Maillard et al10 ).
Part of the lingering uncertainty over the role of Notch signals in hematopoiesis is related to the existence of multiple receptors and ligands with complex patterns of expression. Notch proteins are transmembrane receptors interacting with ligands of the Jagged and Delta-like families. Mammals have 4 different Notch receptors (Notch1-4) with a similar overall structure (reviewed in Radtke et al9 and Maillard et al10 ). Notch-mediated transcriptional activation involves proteolytic cleavage and physical translocation of the Notch intracellular domain (ICN) into the nucleus. Once in the nucleus, ICN binds to CSL (CBF1/RBP-Jk, Suppressor of Hairless, Lag-1), a transcription factor that mediates most of the well-characterized Notch functions. ICN binding has 2 consequences. First, it displaces co-repressors from CSL, resulting in derepression of promoters with CSL binding sites.11 Secondly, ICN recruits Mastermind-like (MAML) proteins,12 which appear to function as a scaffold for the formation of a large multiprotein transcriptional activation complex.13 Three MAML family members (MAML1-3) have been identified in mammals.14,15 MAMLs are nuclear proteins with short basic N-terminal sequences that bind the ankyrin repeat domain of ICN, and C-termini that recruit p300 and other transcriptional activators. In vitro experiments suggest that MAMLs are critical for Notch-mediated stimulation of CSL-dependent transcription12,14-16 and have shown that truncated MAML mutants consisting of only the N-terminal ICN-binding domain have potent dominant negative effects, presumably due to their inability to recruit other components of the Notch transcriptional activation complex.16,17 The minimal region of MAML1 required for ICN binding, a basic region comprising amino acids 13 to 74, is sufficient to form a stable ternary complex with purified CSL and ICN (Nam et al18 ) and when overexpressed in transfection assays, generates a profound block in Notch1-mediated transcriptional activation.17 Furthermore, this minimal MAML113-74 region causes growth arrest and apoptosis of multiple T-ALL cell lines bearing constitutively active Notch1 alleles.17
Although these data suggest that MAML proteins may be critical components of the Notch signaling pathway, the importance of MAMLs in vivo is unknown. As MAMLs appear to form stable complexes with all 4 ICNs,14,15 dominant negative MAMLs would be predicted to act as pan-Notch inhibitors, enabling a broad screen for Notch/MAML function within particular developing tissues or lineages. To test this approach, we generated dominant negative mutants of MAML consisting of the N-terminal basic regions of MAML1-3 and characterized their ability to bind and down-regulate the activity of Notch1-4 in cultured cells. We then studied the effect of the most potent of these inhibitors, MAML1(13-74) fused to green fluorescence protein (GFP) (DNMAML1) in vivo through transduction of murine hematopoietic stem cells. We show that expression of DNMAML1 leads to marked inhibition of early T-cell differentiation and to the appearance of intrathymic B cells, as well as a marked decrease in the pool of marginal zone B cells in the spleen, phenotypes consistent with inhibition of Notch1 and Notch2, respectively. This contrasts with constitutive expression of Dtx1, which blocks T-cell development, but not MZB cell development. These observations suggest that MAML proteins play an important role in mediating the effects of several Notch family members in vivo. More generally, our results suggest that DNMAML will be useful in screening for CSL/ICN1-4/MAML1-3 complex function in vivo, thus providing an approach that complements strategies relying on the knockout of individual Notch signaling components.
Materials and methods
Mice
C57BL/6 mice (B6) were obtained from Charles River (Wilmington, MA) or Taconic Laboratories (Germantown, NY). Timed pregnant C57BL/6 females were obtained from the National Cancer Institute, Frederick, MD. Experiments were performed according to guidelines from the National Institutes of Health and with an approved protocol from the University of Pennsylvania Animal Care and Use Committee.
Constructs and retroviruses
The dominant negative DNMAML1-GFP fusion construct in EGFP-N3 has been described previously.17 Homologous regions of MAML2 (residues 73-128 AVPKHSTVVERL-RAKKSGAG) and MAML3 (residues 34-92 VTPRVHSAIVERL-GARKAGKH) were inserted into EGFP-N3 immediately 3′ of a consensus Kozak sequence. High-titer retroviral supernatant was produced as previously described using transient transfection of 293T cells.2
Reporter assays
To test dominant negative MAML1 peptides, U2OS cells in 24-well dishes were cotransfected in triplicate with expression plasmids encoding ICN1-4, various pEGFP-N3 plasmids encoding MAML peptides fused C-terminally to enhanced green fluorescence protein (EGFP), a CSL-sensitive luciferase reporter plasmid, and a Renilla luciferase internal control plasmid. The following amounts of plasmids were used: 10 ng ICN1/ICN2; 50 ng ICN3/ICN4; 25-50 ng mMAML1-3 with ICN1/2 and 50-100 ng with ICN3/4; 250 ng CSL reporter and 5 ng Renilla luciferase plasmid; and sufficient empty pcDNA3 to hold the total input DNA constant across all wells. Luciferase assays were performed with cell extracts prepared 40 to 44 hours after transfection, as described.19
Immunoprecipitation
293T cells in 6-well dishes were transfected with pcDNA3 plasmids encoding myc-tagged CSL,20 various pcDNA3 plasmids encoding the intracellular domain of Notch1, and pEGFP-N3 plasmids encoding the N-terminal basic regions of human MAML1-3 or EGFP, using Lipo-fectamine Plus (Invitrogen, Carlsbad, CA). The following amounts of plasmids were used: 1 μg CSL; 100 ng ICN1; 250 ng MAML1-GFP; 250 ng MAML2-GFP, and 250 ng MAML3-GFP. Cell lysates prepared 44 hours after transfection were immunoprecipitated with anti-myc 9E10 antibody, as described.20 Western blots were stained with anti-myc monoclonal 9E10, polyclonal rabbit anti-Notch1,20 or anti-GFP monoclonal JL8 (Clontech, Palo Alto, CA) using a chemiluminescent method (PicoSignal, Pierce, Rockford, IL).
Fetal liver cell transduction and fetal thymic organ culture
Transduction of E15.5 fetal liver cells with retroviral supernatant and reconstitution of irradiated fetal thymic lobes in hanging drop cultures were performed as described.21 Briefly, the fetal liver cells (FLCs) were purified on a Ficoll gradient, washed, and resuspended at 1 × 106/mL in Iscove modified Dulbecco medium supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U/mL penicillin G, 100 μg/mL streptomycin, 6 ng/mL interleukin-3 (IL-3), 1 ng/mL interferonγ (IFNγ), 4 ng/mL IL-1β, and 50 ng/mL stem cell factor (SCF). After overnight culture, the FLCs were washed, resuspended in retroviral supernatant in the presence of the same cytokines and polybrene (4 μg/mL), and spinoculated at 2500 rpm for 50 minutes at room temperature. The next day, FLCs were cocultured for 24 hours with irradiated thymic lobes (2700 rads) as 30 μL hanging drop cultures in Terasaki wells. The fetal lobes were then transferred to 5-μm polycarbonate membrane filters (Isopore, Millipore, Ireland) and cultured for 15 to 16 days before analysis.
Bone marrow transduction and transplantation
Transduction of B6 bone marrow cells with retroviral supernatant and transfer of these cells into lethally irradiated recipients were performed as described previously.2 Briefly, bone marrow (BM) cells were collected from 6- to 10-week-old mice 4 days after intravenous administration of 5-FU (5 mg), as described previously.22 The cells were cultured overnight in the presence of IL-3 (6 ng/mL), IL-6 (5-10 ng/mL), and SCF (100 ng/mL). On the next day, the cells were washed, resuspended in retroviral supernatant, the same cytokine cocktail plus polybrene (4 μg/mL), and centrifuged at 2500 rpm for 90 minutes. A second round of spinoculation was performed on the following day. After washing 3 times in phosphate-buffered saline (PBS), at least 0.5 × 106 cells were injected intravenously into lethally irradiated (900 rads) B6 recipients. Mice were maintained on antibiotics in drinking water for 2 weeks and analyzed at least 6 weeks after transplantation.
Antibodies
Phycoerythrin (PE)-anti-T-cell receptor (TCR)β, PE-anti-CD23, biotin-anti-CD8α, allophycocyanin (APC)-anti-CD4, biotin-anti-sIgM, and APC-anti-B220 were obtained from Pharmingen (San Diego, CA). APC-anti-CD21/CD35 and APC-anti-AA4.1 were conjugated in our laboratory.
Flow cytometry
After incubation with 2.4G2 hybridoma supernatant to block Fc receptors, cell suspensions were stained with primary antibodies and washed in PBS with 2% FCS and 0.01% NaN3. Biotinylated antibodies were revealed with a streptavidin-PerCP conjugate (Pharmingen). Data were acquired on a Becton Dickinson FACS Calibur (San Jose, CA) and analyzed using FlowJo software (Treestar, San Carlos, CA).
Results
Truncation mutants of Mastermind behave as pan-Notch inhibitors in vitro
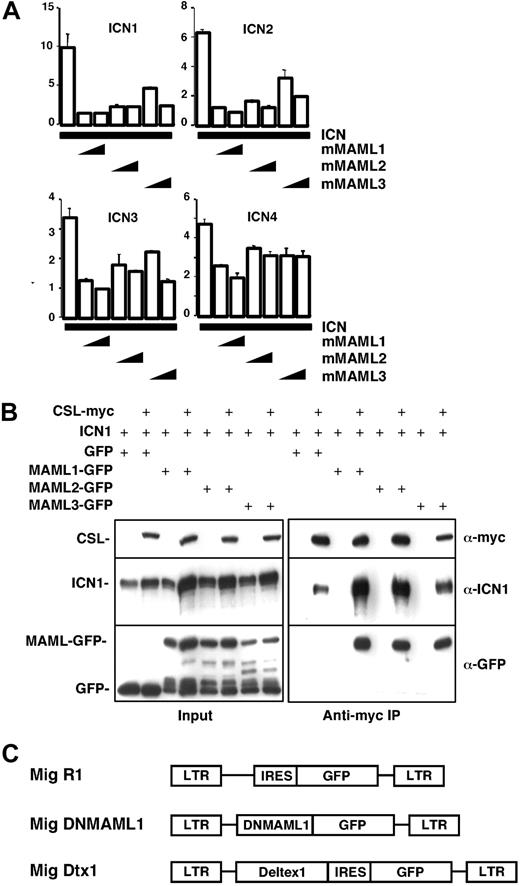
Previous studies show that Mastermind-like proteins are an important component of the transcriptional activation complex during Notch-mediated signaling, in association with the transcription factor CSL.12,14,16 We previously noted that a truncated MAML1 mutant (DNMAML1) preserving a short N-terminal Notch-binding domain, but lacking C-terminal transcriptional activation domains, efficiently blocked Notch1-mediated activation of CSL.17 In an effort to develop a maximally effective pan-Notch inhibitor that would be active against all Notch family members (Notch1-4), we constructed analogous MAML truncation mutants for MAML2 and MAML3, the 2 other MAML family members, and compared the ability of all 3 DNMAMLs to bind and inhibit Notch1-4 (Figure 1). U2OS cells were transiently transfected with combinations of plasmids encoding the constitutively active intracellular domain of Notch1-4 (ICN1-4), the MAML1-3 N-terminal region (DNMAML1-3), and a luciferase reporter driven by a CSL recognition element (Figure 1A). Cotransfection of ICN1 and DNMAML1 resulted in a profound inhibition of reporter activity. DNMAML1 also inhibited the activation of CSL by ICN2-4 (albeit slightly less potently for ICN4). While DNMAML2-3 inhibited ICN1-4 activity as well, they were somewhat less potent than DNMAML1.
Inhibition of Notch family members in vitro by MAML1-3 dominant negative mutants. Dominant negative mutants were generated from homologous regions in the N-terminal Notch-binding domain of MAML1, MAML2, and MAML3 (mMAML1-3). (A) Transient transfection of U2OS cells with a CSL reporter plasmid and combinations of plasmids encoding the constitutively active intracellular domain of Notch family members (ICN1-4) and mMAML1-3. Firefly luciferase activity was normalized to Renilla luciferase activity. The amounts of each plasmid used are indicated in “Materials and methods.” Relative activity is shown on the y-axis. Results are shown as the mean ± SD of triplicate samples. (B) 293 T cells were transfected with plasmids encoding ICN1 and DNMAML-GFP fusion constructs versus GFP alone, in the presence or absence of myc-tagged CSL. The amounts of each plasmid used are reported in “Materials and methods.” Immunoprecipitates prepared with anti-myc antibody were analyzed on Western blots stained with anti-myc, anti-Notch, or anti-GFP antibodies. The lower molecular weight bands revealed by anti-GFP antibodies in the DNMAML-GFP transfected samples are protein degradation products. (C) Schematic map of the MSCV-based retroviral constructs used in this study.
Inhibition of Notch family members in vitro by MAML1-3 dominant negative mutants. Dominant negative mutants were generated from homologous regions in the N-terminal Notch-binding domain of MAML1, MAML2, and MAML3 (mMAML1-3). (A) Transient transfection of U2OS cells with a CSL reporter plasmid and combinations of plasmids encoding the constitutively active intracellular domain of Notch family members (ICN1-4) and mMAML1-3. Firefly luciferase activity was normalized to Renilla luciferase activity. The amounts of each plasmid used are indicated in “Materials and methods.” Relative activity is shown on the y-axis. Results are shown as the mean ± SD of triplicate samples. (B) 293 T cells were transfected with plasmids encoding ICN1 and DNMAML-GFP fusion constructs versus GFP alone, in the presence or absence of myc-tagged CSL. The amounts of each plasmid used are reported in “Materials and methods.” Immunoprecipitates prepared with anti-myc antibody were analyzed on Western blots stained with anti-myc, anti-Notch, or anti-GFP antibodies. The lower molecular weight bands revealed by anti-GFP antibodies in the DNMAML-GFP transfected samples are protein degradation products. (C) Schematic map of the MSCV-based retroviral constructs used in this study.
Prior work showed that MAMLs do not bind CSL but associate indirectly through ICN,12,18 which binds CSL through its RBP-jk association motif (RAM) and ankyrin repeat domains. To confirm that this extends to DNMAML1-3, we investigated whether these polypeptides form trimolecular complexes with CSL and ICN in cells (Figure 1B). GFP-DNMAML1-3, but not GFP, coimmunoprecipitated with CSL and ICN1 with similar efficiency. Comparable results were observed in coimmunoprecipitation experiments with ICN2-4 (J.C.A., data not shown).
Based on these data, DNMAML1 was selected for use in further studies. DNMAML1 was subcloned as a GFP fusion construct into a murine stem cell virus (MSCV)-based retroviral vector lacking the internal ribosomal entry site-green fluorescence protein (IRES-GFP) sequence (Figure 1C). The resulting virus was compared to the retroviral backbone expressing only GFP (Mig R1), as well as to a retrovirus expressing GFP and Deltex1 (Mig Dtx1), a Notch modulator that has previously been shown to potently interfere with Notch1-mediated T-lineage commitment.21,23
Effect of DNMAML1 in fetal thymic organ cultures
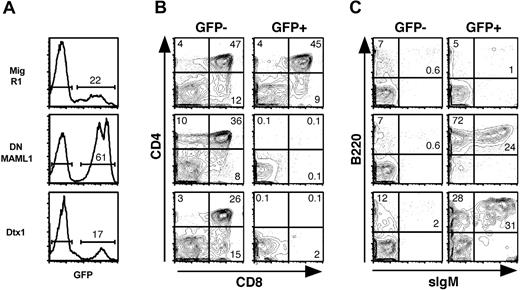
To evaluate the role of Mastermind during lymphoid development, we first studied Notch1-mediated T-lineage commitment in fetal thymic organ cultures (FTOCs). Fetal liver hematopoietic progenitors were transduced with MigR1, Mig DNMAML1, and Mig Dtx1 viruses. Irradiated fetal thymic lobes were reconstituted with transduced progenitors and analyzed by flow cytometry after 2 weeks of culture (Figure 2). Adequate levels of transduction (as assessed by GFP+ cells) were obtained with each virus (Figure 2A). Normal T-cell differentiation, with generation of CD4+CD8+ double-positive and CD4+ and CD8+ single-positive thymocytes, was observed in control MigR1 FTOCs and in the GFP- fraction of the other cultures (Figure 2B). In contrast, T-cell development was profoundly inhibited in the GFP+ fraction transduced with DNMAML1, with most cells falling within the CD4-CD8- double-negative (DN) gate. The inhibition was of comparable intensity to the block in differentiation induced by Dtx1 (Figure 2B). Analysis of the GFP+ DN cells showed that most of the DNMAML1-transduced cells were B cells, expressing B220 and CD19 (Figure 2C and data not shown). A fraction of the cells also expressed sIgM (Figure 2C). Generation of B cells with a similar phenotype was observed in Mig Dtx1 FTOCs, although it tended to be less efficient than for DNMAML1, with more cells remaining B220 negative as well as CD4/CD8 negative. Altogether, the profound block in T-cell differentiation and the generation of intrathymic B cells are consistent with efficient inhibition of Notch1 signals by DNMAML1, suggesting a critical role for Mastermind in the T/B lineage decision.
Inhibition of T-cell development and generation of intrathymic B cells in fetal thymic organ cultures reconstituted with DNMAML1 or Dtx1-transduced fetal liver cells. Day 15.5 fetal liver cells were transduced with the control retrovirus MigR1, Mig DNMAML1, or Mig Dtx1. Irradiated fetal lobes were reconstituted with the transduced fetal progenitors and cultured for 2 weeks before flow cytometric analysis. (A) GFP expression. (B) T-cell differentiation as assessed by anti-CD4 and anti-CD8 staining in the GFP- (internal control) and GFP+ populations. (C) Generation of intrathymic B cells as assessed with antibodies against B220 and sIgM. Results are representative of 3 independent experiments. The numbers in quadrants and above brackets indicate the percentage of cells.
Inhibition of T-cell development and generation of intrathymic B cells in fetal thymic organ cultures reconstituted with DNMAML1 or Dtx1-transduced fetal liver cells. Day 15.5 fetal liver cells were transduced with the control retrovirus MigR1, Mig DNMAML1, or Mig Dtx1. Irradiated fetal lobes were reconstituted with the transduced fetal progenitors and cultured for 2 weeks before flow cytometric analysis. (A) GFP expression. (B) T-cell differentiation as assessed by anti-CD4 and anti-CD8 staining in the GFP- (internal control) and GFP+ populations. (C) Generation of intrathymic B cells as assessed with antibodies against B220 and sIgM. Results are representative of 3 independent experiments. The numbers in quadrants and above brackets indicate the percentage of cells.
Effect of DNMAML1 in vivo
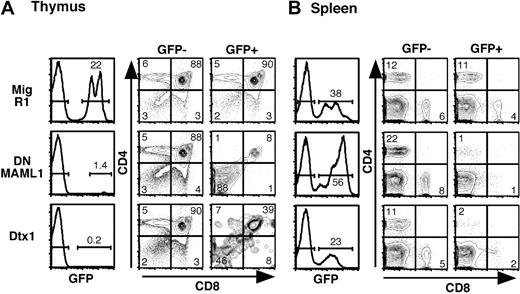
The role of Mastermind in the hematolymphoid system was evaluated further in vivo by reconstituting lethally irradiated mice with DNMAML1-transduced bone marrow (Figure 3). Flow cytometric analysis was performed at least 6-10 weeks after transplantation. The percentage of GFP+ cells was much lower in the thymus than in the peripheral lymphoid organs of DNMAML1 reconstituted mice, while being similar in control MigR1 mice (Figure 3A-B). As reported previously, the percentage of Dtx1-transduced thymocytes also was very low.21,23 Analysis of the GFP+ fraction revealed a profound block in thymocyte differentiation both for DNMAML1 and Dtx1 (Figure 3A). As in FTOCs, a large proportion of the GFP+ double-negative cells in DNMAML1 mice and in Dtx1 mice were intrathymic B cells (data not shown). The developmental block extended to peripheral lymphoid organs, as very few GFP+ T cells were observed in the spleen (Figure 3B). The inhibition of T-cell differentiation was as profound in the thymus as in FTOCs, although the percentage of GFP+ cells was strikingly lower in mice (compare Figure 2A and Figure 3A). This finding may result from several factors, including a greater propensity of fetal hematopoietic progenitors to differentiate to the B-cell lineage, a less permissive environment for B-cell differentiation in the adult thymus, and/or emigration of GFP+ B cells out of the thymus in vivo.
Impairment of Notch1-mediated T-cell development in vivo by DNMAML1 or Dtx1. C57BL/6 mice were lethally irradiated and reconstituted with BM cells transduced with MigR1, Mig DNMAML1, and Mig Dtx1. Analysis was performed at least 6 weeks after transplantation. (A) Flow cytometric analysis of the thymus showing GFP positivity and CD4/CD8 profile. (B) Flow cytometric analysis of the spleen showing GFP positivity in the same mice, as well as peripheral CD4+ and CD8+ cells in the GFP- and GFP+ fractions. Results are representative of 3 independent experiments. The numbers in quadrants and above brackets indicate the percentage of cells.
Impairment of Notch1-mediated T-cell development in vivo by DNMAML1 or Dtx1. C57BL/6 mice were lethally irradiated and reconstituted with BM cells transduced with MigR1, Mig DNMAML1, and Mig Dtx1. Analysis was performed at least 6 weeks after transplantation. (A) Flow cytometric analysis of the thymus showing GFP positivity and CD4/CD8 profile. (B) Flow cytometric analysis of the spleen showing GFP positivity in the same mice, as well as peripheral CD4+ and CD8+ cells in the GFP- and GFP+ fractions. Results are representative of 3 independent experiments. The numbers in quadrants and above brackets indicate the percentage of cells.
Role of Mastermind during marginal zone B-cell development
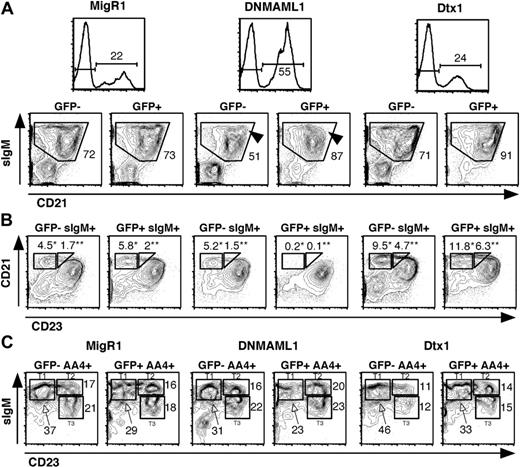
In addition to its effects on Notch1, we investigated whether DNMAML1 was able to efficiently inhibit other Notch family members in vivo (Figure 4). Recent reports have described a critical role for Notch2 in marginal zone B (MZB)-cell differentiation, although the details of this process are still poorly understood.4-6 MZB cells can be identified using flow cytometric criteria as being sIgMhi, CD21hi, and CD23low/neg. In spleens of mice transduced with DNMAML1, a selective loss of sIgMhi CD21hi cells was observed in the GFP+ fraction (arrowheads in Figure 4A), while the bulk of sIgMint CD21int splenic B cells, containing follicular B (FoB) cells, appeared normal. Further analysis of the sIgM+ cells using a combination of CD21 and CD23 showed a drastic reduction in the percentage of CD21hiCD23low cells, indicating a marked decrease in MZB cells (Figure 4B, indicated by “*”). Moreover, another population of sIgMhiCD21hiCD23int cells, different from MZB cells, also had virtually disappeared among DNMAML1 GFP+ splenocytes (Figure 4B, indicated by “**”). These cells were initially described by Loder and Carsetti as a population of spontaneously cycling cells that was thought to represent a subset of transitional cells, referred to as T2 or “cycling T2” B cells, and to give rise to mature FoB and MZB cells.25 However, the absence of cycling T2 B cells in the presence of a normal FoB-cell compartment in our DNMAML1-transduced mice strongly suggests that cycling T2 B cells are not a critical intermediate precursor to FoB cells.
Inhibition of Notch2-mediated marginal zone B-cell development by DNMAML1 but not Dtx1. C57BL/6 mice were lethally irradiated and reconstituted with BM cells transduced with MigR1, Mig DNMAML1, and Mig Dtx1. Analysis was performed at least 6 weeks after transplantation. (A) Expression of sIgM and CD21 in GFP- and GFP+ spleen cells. A selective loss of sIgMhi CD21hi B cells is observed in the GFP+ fraction of DNMAML1-transduced mice (arrowheads). (B) Analysis of CD21 and CD23 profiles among GFP- and GFP+ sIgM+ cells, showing the selective loss of 2 distinct populations of cells after transduction with DNMAML1. sIgM+CD21hiCD23low cells (*) are marginal zone B cells. sIgM+CD21hiCD23int cells(**) were originally thought to be a subset of transitional B cells referred to as T2 or cycling T2 B cells,25 but their exact nature is controversial. The decrease in GFP+ relative to GFP- MZB and cycling T2 B cells observed in DNMAML1 mice was statistically significant (P < .05). The slight increase observed in mice transduced with Mig Dtx1 was not statistically significant when compared to Mig R1 mice. (C) Analysis of AA4+ transitional B cell subsets as defined by a combination of sIgM and CD23 staining.26 Results are representative of 2 independent experiments with at least 3 mice. The numbers in the quadrants and above the brackets indicate the percentage of cells.
Inhibition of Notch2-mediated marginal zone B-cell development by DNMAML1 but not Dtx1. C57BL/6 mice were lethally irradiated and reconstituted with BM cells transduced with MigR1, Mig DNMAML1, and Mig Dtx1. Analysis was performed at least 6 weeks after transplantation. (A) Expression of sIgM and CD21 in GFP- and GFP+ spleen cells. A selective loss of sIgMhi CD21hi B cells is observed in the GFP+ fraction of DNMAML1-transduced mice (arrowheads). (B) Analysis of CD21 and CD23 profiles among GFP- and GFP+ sIgM+ cells, showing the selective loss of 2 distinct populations of cells after transduction with DNMAML1. sIgM+CD21hiCD23low cells (*) are marginal zone B cells. sIgM+CD21hiCD23int cells(**) were originally thought to be a subset of transitional B cells referred to as T2 or cycling T2 B cells,25 but their exact nature is controversial. The decrease in GFP+ relative to GFP- MZB and cycling T2 B cells observed in DNMAML1 mice was statistically significant (P < .05). The slight increase observed in mice transduced with Mig Dtx1 was not statistically significant when compared to Mig R1 mice. (C) Analysis of AA4+ transitional B cell subsets as defined by a combination of sIgM and CD23 staining.26 Results are representative of 2 independent experiments with at least 3 mice. The numbers in the quadrants and above the brackets indicate the percentage of cells.
To further investigate this question, we studied AA4+ transitional B-cell subsets in the spleen of DNMAML1-transduced mice (Figure 4C). Expression of the AA4 marker has been shown previously to reliably identify a pool of splenic B cells with typical characteristics of transitional B cells.26 Immunophenotypic analysis of the AA4+ B cells identified 3 populations of transitional B cells on the basis of differential expression of sIgM and CD23 (Figure 4C).26 The distribution of these populations was not significantly different between GFP- and GFP+ fractions of DNMAML1-transduced mice. In particular, the AA4+ T2 transitional B-cell population was not diminished, in contrast to the drastic reduction in cycling T2 cells. In summary, our data indicate that cycling T2 B cells and MZB cells are both Notch-dependent, while AA4+ transitional B-cell subsets are present in normal numbers in the absence of Notch signaling.
Deltex1 does not inhibit marginal zone B-cell development
In view of the similar effect of Mig DNMAML1 and Mig Dtx1 on early T-cell development (Figures 2, 3), we investigated whether overexpression of Dtx1 in vivo has the ability to interfere with Notch2-mediated MZB development. In contrast to DNMAML1, the splenic B-cell subsets of Mig Dtx1-transduced mice were identical in the GFP- and GFP+ fractions, with no significant change in the MZB compartment, cycling T2 B cells, or AA4+ transitional B-cell subsets (Figure 4B-C). These findings were observed in the same animals in which expression of Dtx1 resulted in a profound inhibition of T-cell development. These observations indicate that DNMAML1 and Dtx1 exert their activity through different mechanisms and that, unlike DNMAML1, Dtx1 cannot be used as a pan-Notch inhibitor in vivo.
Discussion
In vitro studies using both transfection assays and chromatin-reconstituted templates have assigned an important role to Mastermind-like proteins during Notch-mediated transcriptional activation. The data reported in this study indicate that MAML-mediated recruitment of co-activators to CSL target genes play a critical role in Notch1- and Notch2-mediated lymphoid cell fate decisions in vivo. This is the first report confirming a critical role for these MAML proteins in physiologically relevant and well-defined Notch-mediated cell fate decisions.
The ability of DNMAML1 to specifically affect 2 characteristic functions of Notch1 and Notch2 in vivo while having no other effects on myeloid or B-cell development suggests that the function of Notch1-4/MAML1-3/CSL complexes is restricted to only a subset of hematopoietic cell fate decisions. Furthermore, it indicates that expression of DNMAML1 specifically inhibits Notch-mediated transcriptional activation. While no definitive roles for Notch3 and Notch4 have been identified during hematopoiesis, the data presented here suggest that DNMAML1 may be a useful reagent to screen for the function of all 4 Notch receptors in a wide variety of in vivo contexts. In doing so, it will complement approaches employing the knockout of individual signaling components. For example, knockout of CSL removes not only an important component of the Notch signaling pathway, but also Notch-independent CSL transcriptional repressor and activator functions,11,27-29 leaving some uncertainty in the interpretation of phenotypes. Because DNMAMLs bind only CSL in the presence of ICN,18 they will not interfere with Notch-independent CSL activities. To date, phenotypes produced by the DNMAML and CSL knockout approaches are similar, however, it is possible that analysis of additional functions will reveal differences. A precedent for this approach was recently demonstrated in studies of Ikaros, where a point mutation in a critical zinc finger gave rise to an Ikaros mutant that lacked DNA binding activity but maintained the basic structure of the transcriptional complex, revealing novel phenotypes that were not evident in the Ikaros knockout mice.30
Our observations on the effect of DNMAML1 on splenic B-cell subsets have potentially important implications for the understanding of the cellular intermediates giving rise to marginal zone B cells. So far, the precise pathways of differentiation in the generation of mature MZB cells and follicular B cells in the spleen remain poorly characterized and controversial. Strikingly, expression of DNMAML1 preserves the follicular B-cell compartment while resulting in a severe loss of both cycling T2 and MZB cells. These observations indicate that follicular B cells can be generated normally in the absence of cycling T2 B cells. Thus, cycling T2 B cells are unlikely to be transitional B-cell precursors. In contrast, the concomitant reduction of cycling T2 and MZB cells by DNMAML1 suggests that these 2 subsets are developmentally related, with cycling T2 B cells serving as precursors to the MZB cells. Such a hypothesis is supported by the fact that MZB cells and cycling T2 B cells are immunophenotypically very similar, with the exception of surface CD23 levels. Our data are consistent with previously published observations of Saito et al in Notch2 conditional knockout mice and Witt et al in Notch2 heterozygous knockout mice, where the concomitant absence of MZB cells and cycling T2 B cells was reported.5,6 Furthermore, it is likely that cycling T2 B cells are identical to the sIgMhisIgDhiCD21hi B-cell subset described by Cariappa et al that was severely diminished in Aiolos knockout mice.31 In these mice also, both MZB cells and cycling T2 B cells are absent, while the follicular B-cell compartment is preserved. The exact interrelationship of MZB, follicular B cells, and cycling T2 B cells remains an important question to be addressed in future studies. Transduction with DNMAML1 may be a useful approach, as the nontransduced B-cell population preserves the normal microarchitecture of the marginal zone, unlike in Notch2 or CSL conditional knockout animals. This will permit clearer delineation of the cell-autonomous activity of Notch2, as well the interaction of the Notch pathway with B-cell receptor signaling, NF-κB, and other pathways implicated in the generation of MZB cells (reviewed in Martin and Kearney32 ).
The differential effects of Deltex1 between Notch1-mediated T-cell commitment and Notch2-mediated marginal zone B-cell development suggest that, in peripheral B cells, Notch2 is resistant to inhibition by Dtx1. This is in contrast to the potent inhibitory effect of this molecule in early thymocytes. Although it could be argued that these differential effects may be related to different levels of expression of DNMAML1 and Dtx1 after retroviral delivery, we think this is unlikely, as profound inhibition of T-cell development was observed in the same animals in which the MZB numbers were unaffected. Although it could be argued that more profound inhibition of Notch2 is necessary to block MZB development, this does not appear to be the case as decreased MZB cell numbers were observed in Notch2+/- mice, whereas Notch1+/- mice display normal T-cell development.1,5,33 The molecular mechanisms underlying the differential effect of DNMAML1 and Dtx1 are unknown. In addition, Dtx1 also has been well described as a direct Notch1 target.34 Saito et al have shown that Dtx1 mRNA levels are reduced in splenic B-cell subsets by inactivation of Notch2, leading them to suggest that Dtx1 may mediate the effects of Notch2 on MZB differentiation.5 Although our data do not exclude a role for Dtx1 in this process, they suggest that Dtx1 is not sufficient to drive MZB differentiation, as no significant expansion of the MZB cell pool was observed in Dtx1-transduced mice (Figure 4 and data not shown).
Notch signaling can be regulated by availability of ligands, expression of Notch receptors, expression of modulators, such as Deltex and Numb, and expression of critical components of the enhanceosome complex, such as MAML. In future studies, it will be important to determine in which circumstances these individual components are rate-limiting. The expression pattern of Notch family members in the hematolymphoid system has been reviewed recently.10 Briefly, both Notch1 and Notch2 are expressed in hematopoietic progenitors, while Notch1 and Notch3 are the predominant molecules expressed at high levels during early T-cell development and are down-regulated subsequently. In contrast, Notch2 but not other Notch family members is expressed in developing and in mature B cells.5,10 The expression pattern of Deltex1 approximates Notch signaling, where it is likely to be a direct Notch target.5,21,23,34 For example, Deltex1 is highly expressed during early thymocyte development and in MZB cells, both of which require Notch signaling. However, the physiological role of Deltex remains unknown. Finally, the respective role of each of the 3 Mastermind-like family members has not been determined. Since DNMAML1 interferes to a similar extent with all 3 family members, the data reported here indicate that they are collectively critical for Notch-mediated cell fate decisions in the hematolymphoid system in vivo. However, it remains to be determined if MAML1, MAML2, and MAML3 play specific roles in the regulation of the Notch pathway. While each MAML appears redundant in its ability to bind ICN1-4, our preliminary data shows that MAML1-3 are differentially expressed during lymphocyte development. For example, MAML1 and MAML2 seem to be the predominant MAMLs expressed during early T-cell development and in mature B cells, while all 3 MAMLs are expressed in mature T cells (data not shown). These findings raise the possibility that individual MAMLs may be critical at particular developmental junctures. Thus, specific loss of function approaches will be needed to determine the role of individual MAML family members in future experiments.
Prepublished online as Blood First Edition Paper, June 8, 2004; DOI 10.1182/blood-2004-02-0514.
Supported in part by a grant from the Swiss Society for Grants in Medicine and Biology, a grant from Ortho-Biotech (I.M.), and by grants from the National Institutes of Health (D.A., J.C.A., and W.S.P.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Avinash Bhandoola and Gary Koretzky for helpful comments and critical reading of the manuscript. W.S.P. is the recipient of Scholar and Score awards from the Leukemia and Lymphoma Society.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal