Abstract
Transcription factor GATA-1 is required for erythropoiesis, yet its full actions are unknown. We performed transcriptome analysis of G1E-ER4 cells, a GATA-1–null erythroblast line that undergoes synchronous erythroid maturation when GATA-1 activity is restored. We interrogated more than 9000 transcripts at 6 time points representing the transition from late burst forming unit–erythroid (BFU-E) to basophilic erythroblast stages. Our findings illuminate several new aspects of GATA-1 function. First, the large number of genes responding quickly to restoration of GATA-1 extends the repertoire of its potential targets. Second, many transcripts were rapidly down-regulated, highlighting the importance of GATA-1 in gene repression. Third, up-regulation of some known GATA-1 targets was delayed, suggesting that auxiliary factors are required. For example, induction of the direct GATA-1 target gene β major globin was late and, surprisingly, required new protein synthesis. In contrast, the gene encoding Fog1, which cooperates with GATA-1 in β globin transcription, was rapidly induced independently of protein synthesis. Guided by bioinformatic analysis, we demonstrated that selected regions of the Fog1 gene exhibit enhancer activity and in vivo occupancy by GATA-1. These findings define a regulatory loop for β globin expression and, more generally, demonstrate how transcriptome analysis can be used to generate testable hypotheses regarding transcriptional networks.
Introduction
Studies of red blood cell development provide an important basis for understanding general mechanisms of gene regulation and tissue formation. Erythrocyte maturation is largely dedicated to producing hemoglobin and supporting its major function of oxygen delivery. Acquisition of this specialized phenotype is controlled by a concerted differentiation program that serves both generalized and cell type–specific requirements. Production of hemoglobin is coordinated to avoid the toxic accumulation of globins or heme synthetic intermediates. Special adaptations are required for iron import and defense against reactive oxygen species. The cell membrane must undergo remodeling to present developmentally appropriate cytokine receptors and ion transporters. In addition, erythropoiesis requires active mechanisms to prevent apoptosis and coordinate cell-cycle arrest with terminal maturation.
Erythroid differentiation is highly regulated at the level of mRNA transcription, and factors controlling the expression of numerous erythroid genes, particularly globins, have been extensively characterized. Recent studies of primary hematopoietic precursors1-6 and immortalized cell lines7,8 are beginning to define global gene expression patterns during hematopoietic maturation, although a comprehensive view of erythropoiesis is lacking. This information is essential for new gene discovery and to better understand the regulatory networks that specify the formation and function of red blood cells. One general approach to understanding gene regulation during development is to identify lineage-determining transcription factors and then define the genes that they control. In this regard, studies of the nuclear protein GATA-1 have provided numerous insights into erythropoiesis.9-11
GATA-1 is a transcription factor essential for erythrocyte, megakaryocyte, mast cell, and eosinophil differentiation.12-17 Ablation of the GATA-1 gene in mice causes embryonic death from severe anemia due to maturation arrest and apoptosis of committed erythroid precursors.13,18,19 In humans, missense mutations in GATA-1 cause dyserythropoietic anemia and/or thalassemia.20,21 While virtually all erythroid genes examined are positively regulated directly by GATA-1, the full extent of its transcriptional actions is currently unknown. Defining the genes regulated by GATA-1 more comprehensively should better elucidate the molecular pathways that control erythropoiesis.
To examine the role of GATA-1 in erythroid development, we created G1E cells (for GATA-1–erythroid), an immortalized GATA-1–null line derived from gene-targeted embryonic stem cells.22 G1E cells proliferate in culture as immature erythroblasts and undergo terminal erythroid maturation when GATA-1 function is restored. Thus, complementation of a defined loss-of-function mutation generates a synchronized cohort of differentiating erythroid precursors, an ideal system in which to delineate gene expression at specific stages of erythropoiesis and to investigate the transcriptional effects of GATA-1 in a physiologic context. Previously, we used subtractive hybridization to analyze transcripts up-regulated by GATA-1 in G1E cells.23,24 These studies identified known GATA-1 targets and also new genes important for erythroid maturation, but examined a limited number of transcripts at only one point in differentiation.
Here we report transcriptome analysis using Affymetrix (Santa Clara, CA) oligonucleotide microarrays to simultaneously interrogate 12 451 murine transcripts corresponding to 9266 unique known genes and expressed sequence tages (ESTs). RNA from G1E cells was examined at baseline and at 5 time points after synchronous terminal maturation was induced by an estradiol-activated form of GATA-1. This model system allowed us to define both immediate and delayed actions of GATA-1 at specific developmental stages of erythropoiesis.
Numerous erythroid markers and known GATA-1 target genes were induced, which, together with prior studies,22,24-29 validate the use of G1E cells as a model system for erythropoiesis. Our work illustrates several new aspects of GATA-1 function and erythroid biology. A surprising number of ESTs and transcripts encoding known proteins were altered at early time points, identifying genes likely to be regulated directly by GATA-1 and providing new insights into the control of erythropoiesis. Moreover, GATA-1 rapidly inhibited as many transcripts as it induced, highlighting an underappreciated role in gene repression. In addition, up-regulation of a subset of established GATA-1 targets was relatively delayed, suggesting that their activation requires induction of accessory factors. One important example was β major globin, whose mRNA accumulated at relatively late time points. β globin induction required new protein synthesis, whereas the gene encoding the GATA-1 cofactor, Fog-1 (Fog1, Zfpm1), was induced rapidly and independent of protein synthesis. Here we show that GATA-1 also binds the Fog1 locus in vivo at a site within an enhancer, suggesting a direct mechanism for gene activation. The latter tests were guided by bioinformatic predictions based on genomic sequence alignments and knowledge of factor-binding sites, illustrating the power of transcriptome analysis coupled with comparative genomics techniques. Together, these studies identify a regulatory hierarchy in which GATA-1 induces its transcriptional cofactor to cooperate in activation of β major globin gene expression. Hence, detailed kinetics of GATA-1–regulated gene expression provided by the current studies forms a basis to generate experimentally testable hypotheses regarding the transcriptional cascades that coordinate terminal erythrocyte maturation.
Materials and methods
Cell culture
Western blotting
GATA-1 was detected in Western blots with antibody N-6 from Santa Cruz Biotechnology (Santa Cruz, CA) at 1:10 000 dilution.
Microarray experiments
In 3 independent experiments, G1E-ER4 cells growing in log phase were induced for 0, 3, 7, 14, 21, or 30 hours with 10–7 M β-estradiol. RNA from 5 × 107 G1E-ER4 cells was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) and processed for hybridization to Affymetrix MG-U74Av2 GeneChips.30
Analysis was performed with Affymetrix MAS 5.0 software using the statistical algorithm.31,32 Chip-wide signal was scaled to a value of 150 using all probe sets, without additional normalization, using default parameters. The signal for the 3 replicates of each probe set at each time point was averaged. Average signal log2 ratios (SLRs) were computed for comparative analyses by averaging the linearized SLR values for each probe set triplicate, followed by reconversion to a base-2 logarithm. Annotations were extracted from the Unigene and GeneOntology (GO)33 consortium databases using GenBank accession numbers assigned by Affymetrix. Results and annotations were merged into a Microsoft Access database (Microsoft, Seattle, WA).
For comparative analyses between 2 time points, we adopted a stringent numeric filtering strategy based on the article by Cammenga et al4 : all P values for each comparison in a triplicate had to satisfy the condition that P was less than .005 or greater than .995, and at least 1 of the 2 average signals being compared (baseline or experimental) had to be above a threshold value of 100, with an SLR more than 1.5.
Hierarchic clustering of unfiltered data was performed using dCHIP.34 After normalization to the median intensity array, “PM only” model-based expression was calculated with default settings. Values obtained from triplicate samples were pooled for each time point, and genes were filtered using the following criteria: variation across samples after pooling: 0.6 less than standard deviation/mean less than 10; percent present calls in the arrays used 15% or more; replication variation such that 0 less than median less than 0.5 and signal level of 100 or more in 15% or more of samples. These criteria were selected to obtain a visual representation of about 100 genes that changed the most over the 30-hour time course.
Gene expression analysis
Northern blotting was performed as previously described.23 To detect β major globin (first described by Konkel et al35 ) and Fog1 (first described by Tsang et al26 ) RNAs, total RNA was converted to cDNA using reverse transcriptase according to standard methods. Real-time polymerase chain reaction (PCR) was performed in 25-μL reactions (1 × SYBR Green Master Mix [QIAGEN, Hilden, Germany], 10 ng cDNA, 0.6 μM primer) using an ABI prism 7000 (Applied Biosystems, Foster City, CA) with standard amplification conditions. mRNA levels were normalized to those of glyceraldehyde phosphate dehydrogenase (Gapdh). The primers were as follows: β major globin: forward, 5′TCCCGTCAACTTCAAGCTCCT3′, reverse, 5′GGAATTTGTCCAGAGAGGCATG3′; Fog1: forward, 5′ATCCCCTGAGAGAGAAGAACCG3′, reverse, 5′GGCGTCATCCTTCCTGTAGATC3′; and Gapdh: forward, 5′GATGCCCCCATGTTTGTGAT3′, reverse, 5′GGTCATGAGCCCTTCCACAAT3′.
Predicting cis-regulatory modules in Fog1 by comparative genomics
All bioinformatic predictions used whole-genome sequence alignments of mouse and human generated by blastZ36 or mouse, rat, and human generated by multiZ.37 For a measure of selection that indicates biologic function, we used L-scores, which give a log-likelihood estimate of the probability that an alignment would not be generated at the locally adjusted neutral rate.37 The conservation score data were plotted in the University of California Santa Cruz (UCSC) Browser display.39 The regulatory potential score (RP score) is a log-likelihood measure that estimates a probability that the function under selection is a cis-acting gene regulatory element.40 In this method, alignments are scored for how well they match patterns that distinguish alignments in regulatory regions from those in neutral DNA. The database of Genome Alignments and Annotations (GALA)41 was searched to find DNA intervals in the Fog1/Zfpm1 locus whose alignments had an RP score of at least 2.3, an empirically derived threshold based on identification of known regulatory elements in the Hbb locus.42 GALA was also queried for all mouse sequences in the Fog1 locus that match weight matrices for GATA-1 binding sites in TRANSFAC43 and for blocks in the mouse-rat-human 3-way alignments containing conserved matches to GATA-1 weight matrices (computed by the tffind comparative sequencing program).44 The DNA intervals exceeding the RP score threshold that also contain a conserved, predicted GATA-1 binding site were identified by intersection operations in GALA. Data from GALA were displayed as custom tracks in the UCSC Genome Browser to provide a consolidated view.
Generation of G1E-ER4 cells overexpressing bcl-xL
The coding region of mouse bcl-xL was cloned into the retroviral vector pGD and G1E-ER4 cells were transduced as described.22 A polyclonal population of cells expressing ectopic bcl-xL was generated by culturing the transduced cells for 24 hours in the absence of erythropoietin and kit ligand. Under these conditions virtually 100% of G1E-ER4 cells transduced with vector alone died, while about 50% of the bcl-xL–transduced cells survived. After this selection step, all of the bcl-xL–transduced cells survived 24-hour cytokine deprivation and underwent erythroid differentiation in response to estradiol-induced activation of GATA-1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described.45,46 PCR products were quantified using SYBR green dye on an ABI 7000 real-time PCR machine. PCR product signals were referenced to a dilution series of the relevant input. Primer pairs used for PCR were as follows: β globin HS3: forward, 5′ATGGGACCTCTGATAGACACATCTT3′, reverse, 5′CTAGGGACTGAGAGAGGCTGCTT3′; Fog1 upstream region: forward, 5′GGCAGATGTTCACTGTGGCA3′, reverse, 5′GGGAGGAGCCAGAGGTCAG3′; Fog1 preCRM1: forward, 5′TGCAAGTCCCATCCTGATAAGA3′, reverse, 5′GCACGCCAGATAAGATCACAATT3′; and Fog1 preCRM4: forward, 5′GCGATAACGGGCACTAGAGC3′, reverse, 5′AAGCGAGCGGAGCCG3′.
Enhancer assays by transient transfection
The ability of predicted cis-regulatory module 1 (preCRM1 or R1) to enhance the level of expression from an erythroid promoter was tested by measuring its effect on expression of a gamma-globin-promoter-luciferase reporter gene plasmid, gammaLuc,47 after transient transfection of uninduced K562 cells. The DNA segment from mouse Chromosome 8, positions 121 975 271 to 121 975 514 (February 2003 assembly, mm338 ) contains preCRM1, located in the first intron of Fog1. It was added upstream of the promoter in the plasmid gammaLuc to make R1gammaLuc. The parental and test plasmids were transfected in triplicate into K562 cells using Tfx50 (Promega, Madison, WI), along with a control plasmid with a Renilla luciferase reporter gene. The firefly luciferase activity encoded by gammaLuc plasmids and the Renilla luciferase activity from the control were measured using the Promega Dual-Luciferase Reporter (DLR) Assay kit, and the ratio of these activities (FF/Ren) was calculated for each replicate. The experiment was performed twice. In order to present the results on a similar scale, the FF/Ren ratios were normalized by dividing by the mean FF/Ren value for the parental gammaLuc samples in each experiment.
Microscopy
The image in Figure 1A was obtained using an Axioskop 2 microscope equipped with an AxioCam camera and a 100 ×/1.3 Plan-Neofluor oil immersion objective lens (Carl Zeiss, Munchen-Hallbergmoos, Germany). Images were processed with Axiovision 3.1 software (Carl Zeiss).
GATA-1–mediated erythroid maturation of G1E-ER4 cells. (A) Morphologic maturation and accumulation of hemoglobin after activation of GATA-1. GATA-1 was activated by estradiol at time zero and cells were examined by cytocentrifugation and histologic staining at the indicated time points. MGG indicates May–Grunwald Giemsa stain; BZ, benzidine stain for hemoglobin. Original magnification, × 400. (B) GATA-1–ER expression in G1E-ER4 cells. Western blots of whole cell lysates at various time points after activation of GATA-1–ER by estradiol. MEL cell lysates at 0 and 96 hours after induction of differentiation with 2% dimethyl sulfoxide (DMSO) were used for comparison. There is 10 μg protein per lane. MW indicates molecular weight in kilodaltons.
GATA-1–mediated erythroid maturation of G1E-ER4 cells. (A) Morphologic maturation and accumulation of hemoglobin after activation of GATA-1. GATA-1 was activated by estradiol at time zero and cells were examined by cytocentrifugation and histologic staining at the indicated time points. MGG indicates May–Grunwald Giemsa stain; BZ, benzidine stain for hemoglobin. Original magnification, × 400. (B) GATA-1–ER expression in G1E-ER4 cells. Western blots of whole cell lysates at various time points after activation of GATA-1–ER by estradiol. MEL cell lysates at 0 and 96 hours after induction of differentiation with 2% dimethyl sulfoxide (DMSO) were used for comparison. There is 10 μg protein per lane. MW indicates molecular weight in kilodaltons.
Results and discussion
The complete microarray dataset was deposited at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO; accession no. GSE628). From this point forward, “supplemental data” refers to analysis, raw data, and the G1EDb database containing results and annotations found at http://stokes.chop.edu/web/weiss/G1Eindex.html and its subdirectories.49
We used G1E-ER4 cells, a G1E subclone that constitutively expresses a conditional, estradiol-activated form of GATA-1 (GATA-1–ER, GATA-1 fused to the estrogen receptor ligand binding domain).25,26,48 Upon addition of estradiol, G1E-ER4 cells undergo synchronous and homogeneous erythroid maturation (Figure 1A). G1E-ER4 cells express GATA-1–ER at a level similar to that of endogenous GATA-1 in murine erythroleukemia (MEL) cells (Figure 1B) and estradiol has no apparent effects on parental G1E cells that do not express GATA-1–ER (Rylski et al25 ; Gregory et al48 ; and not shown). These control experiments indicate that erythroid maturation of G1E-ER4 cells is caused by physiologic restoration of GATA-1 and is not due to its overexpression or irrelevant estradiol effects.
We used Affymetrix GeneChips (MG-U74Av2) to profile mRNA expression in G1E-ER4 cells at 0, 3, 7, 14, 21, and 30 hours after activation of GATA-1. These time points were selected to examine both early and late effects of GATA-1 and to encompass several key aspects of erythroid maturation, including loss of proliferative capacity, extensive hemoglobin synthesis, and characteristic morphologic changes25,48 (Figure 1A).
For each time point, 3 biologic repeats were performed to minimize random variations in gene expression. There was strong agreement in datasets from identical time points of separate experiments (supplemental data49 ). In filtering genes that changed versus time zero, we favored specificity over sensitivity, using stringent criteria requiring large fold induction and highly significant P values; for a full listing of genes considered to be induced/repressed at each time point, see supplemental data.49
Among the 12 451 murine probe sets printed on the microarray, 3419 (27.4%) were designated present (in triplicate) in at least one time point. In general, activation of GATA-1 triggered numerous rapid changes in gene expression: at 3 hours, 1.3% of the probe sets represented on the microarray were increased and 0.8% were decreased versus time zero. By 30 hours, 5.6% were increased, and 10.5% were decreased, consistent with a more restricted pattern of gene expression during cellular maturation.
Data were compiled into a custom database, G1EDb, available for public download (supplemental data49 ). Expression data and corresponding annotations can be retrieved gene by gene in a visually intuitive manner and filtered according to present/absent calls, numeric criteria, and functional categories. The G1EDb also includes hypertext links to public and Affymetrix databases. Our microarray data have also been deposited at the NCBI Gene Expression Omnibus (GEO)49 (www.ncbi.nlm.nih.gov/geol) (accession no. GSE628).
Validating the experimental system
We addressed 2 important validation concerns: does the experimental system recapitulate GATA-1–dependent erythropoiesis and does the microarray accurately reflect transcript levels? Regarding the first issue, numerous erythroid-expressed genes, many of which are known GATA-1 targets, were up-regulated after GATA-1 activation (Figure 2). In addition, our dataset is in agreement with an earlier study that examined expression of 600 genes during erythroid maturation of primary fetal liver cells3 (Figure 3). Although erythroid and megakaryocytic lineages arise from a common precursor and are both GATA-1 dependent, platelet-expressed transcripts such as platelet factor 4, glycoprotein 1b, coagulation factors 8 and 10, and integrin β3 were not detected in G1E-ER4 cells (supplemental data49 ). Microarray transcript profiles were in agreement with the kinetics of RNAexpression demonstrated by Northern blotting, as described previously,23,25,48 and in Figure 4.Inall cases examined, the microarray data accurately reflected trends in gene expression. For both α and β globins, the microarrays tended to underestimate the magnitude of RNA induction, possibly due to saturation of the detection system by these extremely abundant transcripts.
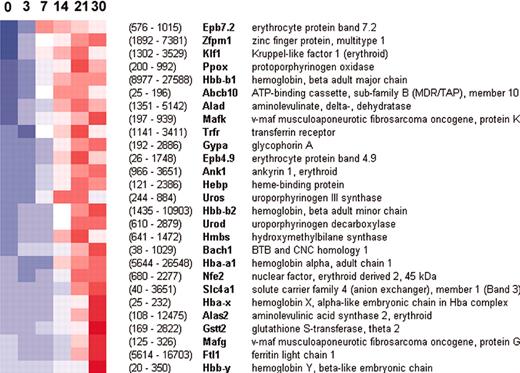
Activation of erythroid genes by GATA-1 in G1E-ER4 cells. Each column represents a time point from 0 to 30 hours after GATA-1 activation, and each row is assigned to a transcript. The color scale ranges from blue to red, corresponding to values ranging from –2.5 to +2.5 standard deviations about each transcript's mean, with white representing the mean value. Hence, a shift from blue to red indicates induction. Signal values (average of 3 triplicates) at 0 and 30 hours are shown in parentheses; these correspond roughly to transcript abundance. Transcripts shown here underwent at least a 2-fold change from time zero and had an average signal value of at least 100 in at least one time point.
Activation of erythroid genes by GATA-1 in G1E-ER4 cells. Each column represents a time point from 0 to 30 hours after GATA-1 activation, and each row is assigned to a transcript. The color scale ranges from blue to red, corresponding to values ranging from –2.5 to +2.5 standard deviations about each transcript's mean, with white representing the mean value. Hence, a shift from blue to red indicates induction. Signal values (average of 3 triplicates) at 0 and 30 hours are shown in parentheses; these correspond roughly to transcript abundance. Transcripts shown here underwent at least a 2-fold change from time zero and had an average signal value of at least 100 in at least one time point.
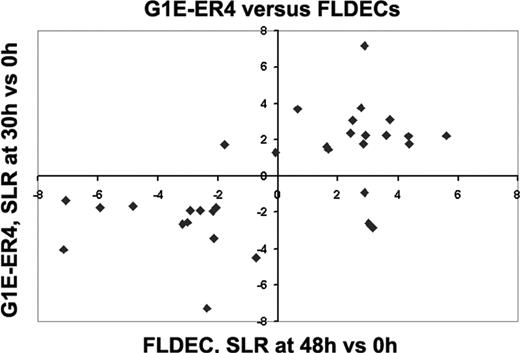
Concordance of gene expression during terminal maturation of G1E-ER4 cells and primary fetal liver–derived erythroid cells (FLDECs). Signal log ratio (SLR) of transcript levels in differentiated versus undifferentiated G1E-ER4 cells (Y-axis, 0 versus 30 hours) and FLDECs3 induced to undergo erythroid maturation (X-axis, 0 versus 48 hours). Each symbol represents one gene that met nonbiased filtering criteria (SLR > 2, P < .005, threshold 100 in both systems). Top right and bottom left quadrants show transcripts with concordant expression trends in both experimental systems: bottom left quadrant (down-regulated in both): Brca2, Cdc20, Hsp60, Mybl2, Gstp2, Msh2, Akt (Pkb), Myb, Myc, Kit, Pcna, Mre11a, Nme2; top right quadrant (up-regulated in both): Ccng2, Pim1, Ube2b (Hr6b), Zfp36, Gadd45a, Prkcd, Bcl2l (Bcl-XL), Ddit3 (Chop10), Mafk, U2af1-rs1, Cdkn1b, Mad, Csf2rb2. Transcripts not expressed concordantly in both systems: bottom right quadrant: Slc2a1, Pold1, Il4ra; top left quadrant: Myln, Ccnd3. An expanded description of these genes is available in supplemental data.49
Concordance of gene expression during terminal maturation of G1E-ER4 cells and primary fetal liver–derived erythroid cells (FLDECs). Signal log ratio (SLR) of transcript levels in differentiated versus undifferentiated G1E-ER4 cells (Y-axis, 0 versus 30 hours) and FLDECs3 induced to undergo erythroid maturation (X-axis, 0 versus 48 hours). Each symbol represents one gene that met nonbiased filtering criteria (SLR > 2, P < .005, threshold 100 in both systems). Top right and bottom left quadrants show transcripts with concordant expression trends in both experimental systems: bottom left quadrant (down-regulated in both): Brca2, Cdc20, Hsp60, Mybl2, Gstp2, Msh2, Akt (Pkb), Myb, Myc, Kit, Pcna, Mre11a, Nme2; top right quadrant (up-regulated in both): Ccng2, Pim1, Ube2b (Hr6b), Zfp36, Gadd45a, Prkcd, Bcl2l (Bcl-XL), Ddit3 (Chop10), Mafk, U2af1-rs1, Cdkn1b, Mad, Csf2rb2. Transcripts not expressed concordantly in both systems: bottom right quadrant: Slc2a1, Pold1, Il4ra; top left quadrant: Myln, Ccnd3. An expanded description of these genes is available in supplemental data.49
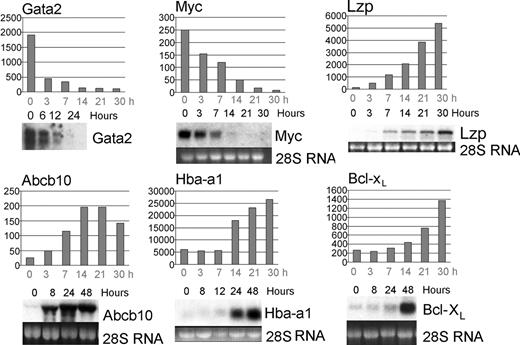
The microarray studies reflect gene expression kinetics accurately. Genes previously shown to exhibit altered expression during G1E-ER4 maturation include Gata-2,48 Myc,25 Abcb10 (Abc-me),23 and Bcl-xL.48 Northern blots are compared with transcript levels predicted by microarrays. The Y-axes of the graphs show absolute signal values from the microarrays. Hours after estradiol treatment are indicated.
The microarray studies reflect gene expression kinetics accurately. Genes previously shown to exhibit altered expression during G1E-ER4 maturation include Gata-2,48 Myc,25 Abcb10 (Abc-me),23 and Bcl-xL.48 Northern blots are compared with transcript levels predicted by microarrays. The Y-axes of the graphs show absolute signal values from the microarrays. Hours after estradiol treatment are indicated.
Developmental stages of G1E cell maturation defined by gene profiling
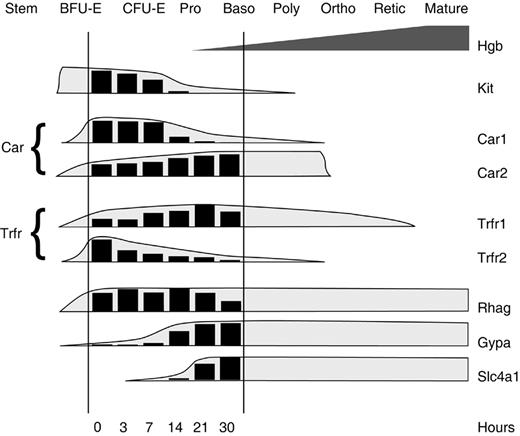
Erythroid precursors develop through distinct stages that are defined by proliferative capacity, cytokine requirements, cell-surface markers, morphology, and patterns of gene expression.50 G1E cells without GATA-1 most closely resemble the late burst forming unit–erythroid (BFU-E) stage.5,27,48 They exhibit an immature morphology, are negative for the proerythroblast marker Ter 119,51 and require kit ligand (KL, stem cell factor) for proliferation and survival. By 24 hours after GATA-1 activation, G1E cells become division arrested, down-regulate c-Kit, the receptor for KL, and require erythropoietin to prevent apoptosis. By 30 hours, most of the cells stain for hemoglobin and resemble basophilic erythroblasts. The maturation of G1E cells in relation to designated developmental stages of erythropoiesis is further defined by the current gene expression studies (Figure 5). For example, expression kinetics of erythrocyte membrane protein Rh50 (Rhag), glycophorin A (Gypa), and band III (Slc4a1) mRNAs during G1E cell maturation resembled those observed in primary erythroid cultures, with Rh50 appearing first and band III appearing last on basophilic erythroblasts.52 Several developmental switches in protein isoforms that occur during normal erythroid maturation, including the shifts from carbonic anhydrase 1 (Car1) to carbonic anhydrase 2 (Car2)53 and transferrin receptor 2 (Trfr2) to transferrin receptor 1 (Trfr1),54 were recapitulated during GATA-1–induced maturation of G1E cells (Figure 5).
Approximate developmental stages recapitulated during G1E-ER4 cell maturation. The histograms represent the average signal level of selected erythroid transcripts at indicated time points, as predicted by the current microarray study. The shaded envelopes drawn around the histograms are based on previously reported expression kinetics determined in other models of erythropoiesis.52-54 The expression pattern observed during G1E-ER4 cell maturation is consistent with the transition from late BFU-E to basophilic erythroblast stages. Pro indicates proerythroblast; Baso, basophilic erythroblast; Poly, polychromatophilic erythroblast; Ortho, orthochromic erythroblast; and Retic, reticulocyte.
Approximate developmental stages recapitulated during G1E-ER4 cell maturation. The histograms represent the average signal level of selected erythroid transcripts at indicated time points, as predicted by the current microarray study. The shaded envelopes drawn around the histograms are based on previously reported expression kinetics determined in other models of erythropoiesis.52-54 The expression pattern observed during G1E-ER4 cell maturation is consistent with the transition from late BFU-E to basophilic erythroblast stages. Pro indicates proerythroblast; Baso, basophilic erythroblast; Poly, polychromatophilic erythroblast; Ortho, orthochromic erythroblast; and Retic, reticulocyte.
Global GATA-1–induced changes in gene expression
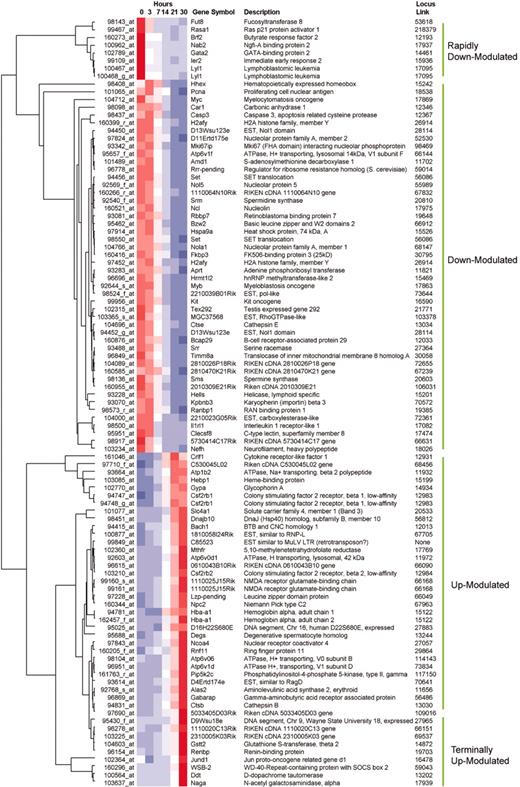
Analysis of transcripts according to their expression kinetics provides a basis to define the regulatory hierarchy through which GATA-1 controls erythroid development. A broad overview of transcripts that change during GATA-1–induced maturation of G1E cells is illustrated by hierarchic clustering (Figure 6). This method selects the genes that change the most over the entire 30-hour time course and graphically sorts them according to the similarity of their transcriptional profiles. Since each transcript is scaled relative to its own median value for the experiment, all transcripts can be viewed at once, despite a range of absolute signal intensities. As a complementary approach, we identified the most significant changes in gene expression at each time point relative to the baseline level at time zero. This pairwise comparison was particularly useful to identify rapidly modulated genes (Tables 1 and 2; supplemental data49 ). Together, these studies define groups of genes that are regulated by GATA-1 at early and late time points. These genes represent various functional classes including transcription factors, signaling molecules, enzymes, and structural proteins. Insights into GATA-1 actions and erythroid development deriving from these analyses are discussed below.
Overview of gene expression by hierarchic clustering. The color scale is identical to that in Figure 2. Filtering parameters were selected to illustrate approximately 100 genes whose expression changed most markedly over the entire time course (see “Materials and methods”). Full listings of all transcripts are provided in the supplemental data.49
Overview of gene expression by hierarchic clustering. The color scale is identical to that in Figure 2. Filtering parameters were selected to illustrate approximately 100 genes whose expression changed most markedly over the entire time course (see “Materials and methods”). Full listings of all transcripts are provided in the supplemental data.49
Genes induced or repressed by GATA-1: 3 hours
Probe . | Gene . | Description . | Function . | 0 h → 3 h . | SLR . |
|---|---|---|---|---|---|
| 94747_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 32 → 514 | 4.3 |
| 99922_at | Slc9a1 | Solute carrier family 9, member 1 | Membrane transporter | 55 → 453 | 2.9 |
| 94415_at | Bach1 | BTB and CNC homology 1 | Transcription factor | 38 → 171 | 2.5 |
| 94748_g_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 111 → 402 | 1.9 |
| 103210_at | Csf2rb2 | Colony-stimulating factor 2 receptor, beta 2 | Signaling | 62 → 175 | 1.8 |
| 103773_at | 1110020K19Rik | RIKEN cDNA 1110020K19 gene | EST | 192 → 773 | 1.7 |
| 104283_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | EST | 129 → 427 | 1.7 |
| 97228_at | Lzf | Leucine zipper domain protein | Transcription factor? | 125 → 505 | 1.6 |
| 160225_at | Gtf2b | General transcription factor IIB | Transcription factor | 409 → 1432 | 1.6 |
| 98415_at | Crlf3 | Cytokine receptor—like factor 3 (crème-9) | Signaling | 384 → 1051 | 1.5 |
| 102787_at | Gpr56 | G protein—coupled receptor 56 | Signaling | 809 → 251 | -1.6 |
| 93836_at | Bnip3 | BCL2/adenovirus E1B 19 kDa—interacting protein 1, NIP3 | Apoptosis pathway | 274 → 78 | -1.7 |
| 161067_at | Ifld2 | Induced in fatty liver dystrophy 2 | Apoptosis pathway | 236 → 51 | -1.7 |
| 102789_at | Gata2 | GATA-binding protein 2 | Transcription factor | 1909 → 454 | -1.9 |
| 104032_at | NO_GENE | Mus musculus, clone MGC:37981 IMAGE 5137303 | EST | 117 → 29 | -1.9 |
| 102397_at | Cbfa2t3h | Eto-2 | Transcription factor | 350 → 57 | -2.1 |
| 100468_g_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 3505 → 793 | -2.1 |
| 100467_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 1638 → 289 | -2.3 |
| 99603_g_at | Tieg | TGFB inducible early growth response 1 | Transcription factor | 108 → 21 | -2.4 |
| 92300_at | Mnt | Max binding protein | Transcription factor | 113 → 16 | -2.5 |
| 102021_at | Il4ra | Interleukin 4 receptor, alpha | Signaling | 142 → 22 | -2.5 |
| 100962_at | Nab2 | Ngfi-A binding protein 2 | Transcription factor | 680 → 72 | -3.1 |
Probe . | Gene . | Description . | Function . | 0 h → 3 h . | SLR . |
|---|---|---|---|---|---|
| 94747_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 32 → 514 | 4.3 |
| 99922_at | Slc9a1 | Solute carrier family 9, member 1 | Membrane transporter | 55 → 453 | 2.9 |
| 94415_at | Bach1 | BTB and CNC homology 1 | Transcription factor | 38 → 171 | 2.5 |
| 94748_g_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 111 → 402 | 1.9 |
| 103210_at | Csf2rb2 | Colony-stimulating factor 2 receptor, beta 2 | Signaling | 62 → 175 | 1.8 |
| 103773_at | 1110020K19Rik | RIKEN cDNA 1110020K19 gene | EST | 192 → 773 | 1.7 |
| 104283_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | EST | 129 → 427 | 1.7 |
| 97228_at | Lzf | Leucine zipper domain protein | Transcription factor? | 125 → 505 | 1.6 |
| 160225_at | Gtf2b | General transcription factor IIB | Transcription factor | 409 → 1432 | 1.6 |
| 98415_at | Crlf3 | Cytokine receptor—like factor 3 (crème-9) | Signaling | 384 → 1051 | 1.5 |
| 102787_at | Gpr56 | G protein—coupled receptor 56 | Signaling | 809 → 251 | -1.6 |
| 93836_at | Bnip3 | BCL2/adenovirus E1B 19 kDa—interacting protein 1, NIP3 | Apoptosis pathway | 274 → 78 | -1.7 |
| 161067_at | Ifld2 | Induced in fatty liver dystrophy 2 | Apoptosis pathway | 236 → 51 | -1.7 |
| 102789_at | Gata2 | GATA-binding protein 2 | Transcription factor | 1909 → 454 | -1.9 |
| 104032_at | NO_GENE | Mus musculus, clone MGC:37981 IMAGE 5137303 | EST | 117 → 29 | -1.9 |
| 102397_at | Cbfa2t3h | Eto-2 | Transcription factor | 350 → 57 | -2.1 |
| 100468_g_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 3505 → 793 | -2.1 |
| 100467_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 1638 → 289 | -2.3 |
| 99603_g_at | Tieg | TGFB inducible early growth response 1 | Transcription factor | 108 → 21 | -2.4 |
| 92300_at | Mnt | Max binding protein | Transcription factor | 113 → 16 | -2.5 |
| 102021_at | Il4ra | Interleukin 4 receptor, alpha | Signaling | 142 → 22 | -2.5 |
| 100962_at | Nab2 | Ngfi-A binding protein 2 | Transcription factor | 680 → 72 | -3.1 |
Microarray results were filtered at an SLR of 1.5, P < .005, and threshold of 100 for the 3-hour time points versus time 0. The penultimate column indicates signal values for the indicated time points (average for triplicate experiments). 3 versus 0; SLR = 1.5; threshold = 100; P < .005.
Genes induced or repressed by GATA-1: 7 hours
Probe . | Gene . | Description . | Function . | 0 h → 7 h . | SLR . |
|---|---|---|---|---|---|
| 94747_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 32 → 1466 | 5.8 |
| 99922_at | Slc9a1 | Solute carrier family 9, member 1 | Membrane transporter | 55 → 724 | 3.7 |
| 94748_g_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 111 → 1150 | 3.5 |
| 104650_at | Ache | Acetylcholinesterase | Membrane glycoprotein | 27 → 272 | 3.4 |
| 103600_at | Epb4.9 | Erythrocyte protein band 4.9 | Cytoskeletal | 26 → 260 | 3.3 |
| 103210_at | Csf2rb2 | Colony-stimulating factor 2 receptor, beta 2 | Signaling | 62 → 568 | 3.3 |
| 94415_at | Bach1 | BTB and CNC homology 1 | Transcription factor | 38 → 200 | 3.2 |
| 104394_at | Abcb10 | ATP-binding cassette, sub-family B, member 10 | Heme metabolism | 25 → 115 | 3.0 |
| 97228_at | Lzf | Leucine zipper domain protein | Transcription factor? | 125 → 1198 | 3.0 |
| 160853_at | Ptdss2 | Phosphatidylserine synthase 2 | Membrane | 90 → 919 | 2.6 |
| 103085_at | Hebp | Heme-binding protein | Heme metabolism | 121 → 677 | 2.5 |
| 94438_at | Pfkm | Phosphofructokinase, muscle | Energy | 189 → 1161 | 2.1 |
| 97710_f_at | 1110004B06Rik | RIKEN cDNA 1110004B06 gene | ? | 278 → 992 | 2.0 |
| 92286_g_at | Il4 | Interleukin 4 | Signaling | 205 → 796 | 2.0 |
| 99849_at | NO_GENE | VL30-like retrotransposon | Retrotransposon | 537 → 2039 | 1.9 |
| 92768_s_at | Alas2 | Aminolevulinic acid synthase 2, erythroid | Heme metabolism | 108 → 453 | 1.9 |
| 93664_at | Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | Membrane transporter | 61 → 201 | 1.9 |
| 98596_s_at | Siat9 | Sialyltransferase 9 | Glycoprotein synthesis | 275 → 1086 | 1.9 |
| 103773_at | 1110020K19Rik | RIKEN cDNA 1110020K19 gene | ? | 192 → 774 | 1.8 |
| 104283_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | ? | 129 → 521 | 1.7 |
| 103471_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | ? | 33 → 106 | 1.7 |
| 94753_at | Gna15 | Guanine nucleotide binding protein, alpha 15 | Signaling | 100 → 338 | 1.6 |
| 101583_at | Btg2 | B-cell translocation gene 2, antiproliferative | Transcription regulator | 291 → 1253 | 1.6 |
| 95052_at | 1110035L05Rik | RIKEN cDNA 1110035L05 gene | ? | 231 → 638 | 1.6 |
| 103833_at | Hipk2 | Homeodomain interacting protein kinase 2 | Transcription regulator | 85 → 207 | 1.6 |
| 96353_at | 1110021D01Rik | RIKEN cDNA 1110021D01 gene | ? | 1553 → 4876 | 1.6 |
| 101947_at | Nakap95-pending | Neighbor of A-kinase anchoring protein 95 | ? | 186 → 510 | 1.5 |
| 102383_at | 5730593F17Rik | RIKEN cDNA 5730593F17 gene | ? | 348 → 596 | 1.5 |
| 99535_at | NO_GENE | Intracisternal A—particle retrotransposon | Retrotransposon | 355 → 905 | 1.5 |
| 160225_at | Gtf2b | General transcription factor IIB | Transcription factor | 409 → 1403 | 1.5 |
| 101973_at | Cited2 | Cbp/p300-interacting transactivator, Glu/Asp-rich C-term., 2 | Transcription factor | 343 → 101 | -1.5 |
| 93680_at | Stk10 | Serine/threonine kinase 10 | Signaling? | 215 → 71 | -1.6 |
| 99109_at | Ier2 | Immediate early response 2 | Transcription factor | 490 → 139 | -1.6 |
| 93793_at | Lasp1 | LIM and SH3 protein 1 | ? | 259 → 72 | -1.7 |
| 98143_at | Fut8 | Fucosyltransferase 8 | Glycoprotein synthesis | 305 → 75 | -1.7 |
| 99603_g_at | Tieg | TGFB inducible early growth response | Transcription factor | 108 → 25 | -1.9 |
| 102318_at | Siat8d | Sialyltransferase 8 (alpha-2, 8-sialytransferase) D | Glycoprotein synthesis | 121 → 30 | -2.0 |
| 161067_at | Ifld2 | Induced in fatty liver dystrophy 2 | Apoptosis pathway | 236 → 59 | -2.1 |
| 102021_at | Il4ra | Interleukin 4 receptor, alpha | Signaling | 142 → 30 | -2.1 |
| 102789_at | Gata2 | GATA-binding protein 2 | Transcription factor | 1909 → 338 | -2.2 |
| 100467_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 1638 → 259 | -2.3 |
| 100468_g_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 3505 → 764 | -2.3 |
| 98500_at | Il1rl1 | Interleukin 1-receptor—like 1 | Signaling | 222 → 38 | -2.4 |
| 100962_at | Nab2 | Ngfi-A binding protein 2 | Transcription factor | 680 → 114 | -2.5 |
| 94419_at | Slc19a1 | Solute carrier family 19, member 1 | Membrane transporter | 290 → 45 | -2.6 |
| 97473_at | Tm4sf7 | Transmembrane 4 superfamily member 7 | Membrane glycoprotein | 295 → 25 | -2.8 |
Probe . | Gene . | Description . | Function . | 0 h → 7 h . | SLR . |
|---|---|---|---|---|---|
| 94747_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 32 → 1466 | 5.8 |
| 99922_at | Slc9a1 | Solute carrier family 9, member 1 | Membrane transporter | 55 → 724 | 3.7 |
| 94748_g_at | Csf2rb1 | Colony-stimulating factor 2 receptor, beta 1 | Signaling | 111 → 1150 | 3.5 |
| 104650_at | Ache | Acetylcholinesterase | Membrane glycoprotein | 27 → 272 | 3.4 |
| 103600_at | Epb4.9 | Erythrocyte protein band 4.9 | Cytoskeletal | 26 → 260 | 3.3 |
| 103210_at | Csf2rb2 | Colony-stimulating factor 2 receptor, beta 2 | Signaling | 62 → 568 | 3.3 |
| 94415_at | Bach1 | BTB and CNC homology 1 | Transcription factor | 38 → 200 | 3.2 |
| 104394_at | Abcb10 | ATP-binding cassette, sub-family B, member 10 | Heme metabolism | 25 → 115 | 3.0 |
| 97228_at | Lzf | Leucine zipper domain protein | Transcription factor? | 125 → 1198 | 3.0 |
| 160853_at | Ptdss2 | Phosphatidylserine synthase 2 | Membrane | 90 → 919 | 2.6 |
| 103085_at | Hebp | Heme-binding protein | Heme metabolism | 121 → 677 | 2.5 |
| 94438_at | Pfkm | Phosphofructokinase, muscle | Energy | 189 → 1161 | 2.1 |
| 97710_f_at | 1110004B06Rik | RIKEN cDNA 1110004B06 gene | ? | 278 → 992 | 2.0 |
| 92286_g_at | Il4 | Interleukin 4 | Signaling | 205 → 796 | 2.0 |
| 99849_at | NO_GENE | VL30-like retrotransposon | Retrotransposon | 537 → 2039 | 1.9 |
| 92768_s_at | Alas2 | Aminolevulinic acid synthase 2, erythroid | Heme metabolism | 108 → 453 | 1.9 |
| 93664_at | Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | Membrane transporter | 61 → 201 | 1.9 |
| 98596_s_at | Siat9 | Sialyltransferase 9 | Glycoprotein synthesis | 275 → 1086 | 1.9 |
| 103773_at | 1110020K19Rik | RIKEN cDNA 1110020K19 gene | ? | 192 → 774 | 1.8 |
| 104283_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | ? | 129 → 521 | 1.7 |
| 103471_at | 4432405K22Rik | RIKEN cDNA 4432405K22 gene | ? | 33 → 106 | 1.7 |
| 94753_at | Gna15 | Guanine nucleotide binding protein, alpha 15 | Signaling | 100 → 338 | 1.6 |
| 101583_at | Btg2 | B-cell translocation gene 2, antiproliferative | Transcription regulator | 291 → 1253 | 1.6 |
| 95052_at | 1110035L05Rik | RIKEN cDNA 1110035L05 gene | ? | 231 → 638 | 1.6 |
| 103833_at | Hipk2 | Homeodomain interacting protein kinase 2 | Transcription regulator | 85 → 207 | 1.6 |
| 96353_at | 1110021D01Rik | RIKEN cDNA 1110021D01 gene | ? | 1553 → 4876 | 1.6 |
| 101947_at | Nakap95-pending | Neighbor of A-kinase anchoring protein 95 | ? | 186 → 510 | 1.5 |
| 102383_at | 5730593F17Rik | RIKEN cDNA 5730593F17 gene | ? | 348 → 596 | 1.5 |
| 99535_at | NO_GENE | Intracisternal A—particle retrotransposon | Retrotransposon | 355 → 905 | 1.5 |
| 160225_at | Gtf2b | General transcription factor IIB | Transcription factor | 409 → 1403 | 1.5 |
| 101973_at | Cited2 | Cbp/p300-interacting transactivator, Glu/Asp-rich C-term., 2 | Transcription factor | 343 → 101 | -1.5 |
| 93680_at | Stk10 | Serine/threonine kinase 10 | Signaling? | 215 → 71 | -1.6 |
| 99109_at | Ier2 | Immediate early response 2 | Transcription factor | 490 → 139 | -1.6 |
| 93793_at | Lasp1 | LIM and SH3 protein 1 | ? | 259 → 72 | -1.7 |
| 98143_at | Fut8 | Fucosyltransferase 8 | Glycoprotein synthesis | 305 → 75 | -1.7 |
| 99603_g_at | Tieg | TGFB inducible early growth response | Transcription factor | 108 → 25 | -1.9 |
| 102318_at | Siat8d | Sialyltransferase 8 (alpha-2, 8-sialytransferase) D | Glycoprotein synthesis | 121 → 30 | -2.0 |
| 161067_at | Ifld2 | Induced in fatty liver dystrophy 2 | Apoptosis pathway | 236 → 59 | -2.1 |
| 102021_at | Il4ra | Interleukin 4 receptor, alpha | Signaling | 142 → 30 | -2.1 |
| 102789_at | Gata2 | GATA-binding protein 2 | Transcription factor | 1909 → 338 | -2.2 |
| 100467_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 1638 → 259 | -2.3 |
| 100468_g_at | Lyl1 | Lymphoblastomic leukemia | Transcription factor | 3505 → 764 | -2.3 |
| 98500_at | Il1rl1 | Interleukin 1-receptor—like 1 | Signaling | 222 → 38 | -2.4 |
| 100962_at | Nab2 | Ngfi-A binding protein 2 | Transcription factor | 680 → 114 | -2.5 |
| 94419_at | Slc19a1 | Solute carrier family 19, member 1 | Membrane transporter | 290 → 45 | -2.6 |
| 97473_at | Tm4sf7 | Transmembrane 4 superfamily member 7 | Membrane glycoprotein | 295 → 25 | -2.8 |
Microarray results were filtered at an SLR of 1.5, P < .005, and threshold of 100 for the 7-hour time point versus time 0. The penultimate column indicates signal values for the indicated time points (average for triplicate experiments). 7 versus 0; SLR = 1.5; threshold = 100; P < .005.
? indicates unknown function.
GATA-1 as a repressor of transcription. One of the most striking findings is that numerous transcripts were rapidly down-regulated, highlighting a potential role for GATA-1 in gene repression. Traditionally, GATA-1 is viewed as an activator of erythroid-specific transcripts; potential roles in gene repression have been implicated in prior studies, but are poorly understood.25,55-58 The current findings indicate that gene repression by GATA-1 is more prevalent than previously recognized and likely to play a critical role in hematopoiesis. We recently demonstrated that repression of Gata259 and Myc25 in erythroid cells is associated with GATA-1 binding to the respective genes, indicating potentially direct mechanisms for transcriptional inhibition. GATA-1 may repress gene expression through its interaction partner Fog-1 (Zfpm1, “Friend of GATA-1”), which binds corepressors and inhibits GATA-1–mediated transcription in specific promoter and cellular contexts.46,58,60
Transcription factors were prominent among the most rapidly repressed transcripts, for example: Eto-2 (Cbfa2t3h), Gata2, Lyl1, Nab2, and Tieg (Table 1). It is likely that many pleiotropic effects of GATA-1 are transmitted through modulation of some or all of these nuclear proteins, although their specific roles in erythropoiesis require further study. GATA-2 is of particular interest; this GATA-1 relative is expressed in hematopoietic stem cells and/or early progenitors61 and is derepressed in GATA-1– erythroblasts, which may account for their baseline transcription of GATA-dependent target genes.13,62 These findings suggest functional overlap between GATA-1 and GATA-2 in vivo. However, distinct roles for these proteins are illustrated by the ability of GATA-1, but not GATA-2, to repress transcription through a defined cis-element in the GATA-2 gene.46,59,60 Hence, it is possible that competition between GATA-1 and GATA-2 regulates the transcriptional output of some GATA-regulated genes and reflects a requirement for GATA-2 repression during normal erythropoiesis.
Also of note, several genes associated with oncogenic transformation were down-regulated after GATA-1 activation. These include Myc, Myb, Kit, and Nab2. GATA-1 binding to the Myc gene is associated with its transcriptional repression during erythroid maturation.25 GATA-1 may also repress Myb through cognate elements in the proximal promoter.63 Hence, GATA-1 may promote hematopoietic maturation by repressing mitogenic genes. This hypothesis is consistent with recent findings that link GATA-1 mutations to a subset of megakaryocytic leukemias.64
Genes that are rapidly induced by GATA-1. Rapidly induced genes are shown in Tables 1 and 2. Some of the earliest up-regulated genes might encode proteins that participate in the initial cascade of GATA-1–induced maturation. For example, transcripts encoding EKLF (Klf1) and Fog-1 (Zfpm1), nuclear proteins that augment GATA-1 activities, were induced at early time points. Another rapidly up-regulated gene was Bach1, which encodes a CNC-type transcription factor that interacts with control elements in the β globin gene locus to coordinate transcription with heme availability.65,66 Surprisingly, cytokine receptor genes Csf2rb1 and Csf2rb2 were strongly up-regulated within the first 3 hours (Table 1). Csf2rb2 encodes a shared β subunit (βc), which pairs with unique α chains to form interleukin-3 (IL-3), IL-5, or granulocyte-macrophage colony-stimulating factor (GM-CSF) receptors, while Csf2rb1 encodes βIL-3, which pairs only with the IL-3α subunit.67 As IL-3 is known to stimulate erythropoiesis in vitro at the BFU-E stage,68 it is unexpected to find that its receptor subunits are up-regulated at later stages of erythropoiesis in G1E-ER4 cells. Nonetheless, similar findings were observed during maturation of fetal liver primary erythroid precursors.3 This suggests potential functions for Csf2rb1 and Csf2rb2 in erythropoiesis. Supporting this, Csf2rb2 was observed to potentiate erythropoietin signaling through direct physical interaction with the erythropoietin receptor,69 although no defects in steady-state erythropoiesis or erythropoietin signaling were demonstrated in mice with targeted mutations in Csf2rb1 and Csf2rb2.70 In addition to these examples, numerous other signaling molecules and transcription factors whose role in erythroid development is currently unexplored were rapidly up-regulated (Table 1).
Facilitating new insights into erythroid function and physiology. Many of the genes up-regulated by GATA-1 at early and late time points participate in specialized aspects of terminal erythropoiesis (Figure 2). Examination of additional induced and repressed genes not previously implicated in erythroid development or function should provide further insight into these processes (Tables 1 and 2; Figure 6; and supplemental data49 ). To facilitate the discovery of genes that participate in erythrocyte biology, we annotated transcripts in the G1EDb according to functional classes as predicted by prior studies or the presence of signature protein motifs. These include transcription factors, signaling molecules, antioxidants, and others. Data retrieval according to gene function and expression patterns provides a broad overview of specific aspects of erythropoiesis and illustrates potential new pathways that can be tested experimentally in future studies. Analysis of GATA-1–regulated gene expression according to functional categories is discussed in supplemental data.49 In addition, several poorly characterized ESTs were up-regulated by GATA-1 in G1E-ER4 cells. As indicated by our prior studies,23,24 characterization of these ESTs should identify novel proteins that participate in erythroid development or function.
Regulatory hierarchies defined by the kinetics of GATA-1–regulated gene expression
Numerous established erythroid GATA target genes including those encoding alpha globin (Hba-a1), band 3 (Slc4a1), and β major globin (Hbb-b1) exhibited relatively delayed up-regulation(Figures 4, 6-7A). Therefore, it is possible that some GATA-1 targets require modulation of additional transcription factors and/or intracellular signaling pathways for full activation. We examined this possibility experimentally, focusing on β major globin.
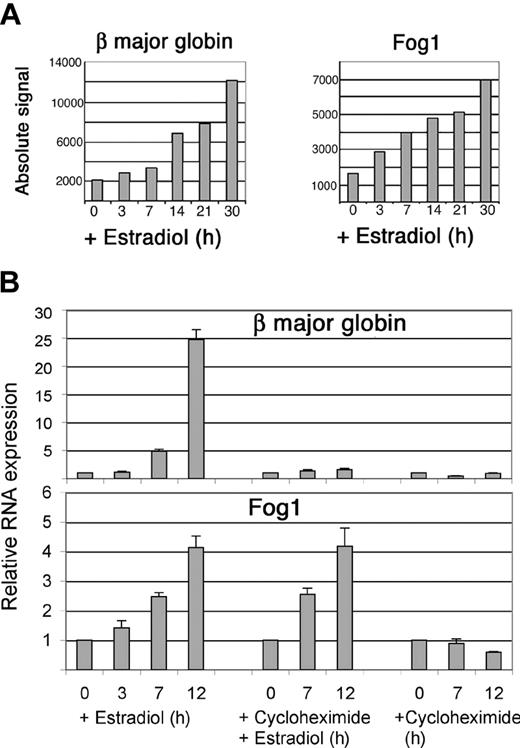
Induction of β major globin and Fog1 by GATA-1. (A) Messenger RNA levels in G1E-ER4 cells predicted by microarray experiments. (B) Messenger RNA levels quantitated by real-time polymerase chain reaction (PCR) in G1E-ER4 cells overexpressing bcl-xL to overcome cycloheximide-induced death. Each data point represents 6 separate PCR reactions in 2 independent experiments. Cycloheximide (0.5 mM) was added as indicated. Note that the magnitude of β globin RNA induction appears to be underestimated by the microarrays, as was also observed for α globin (Figure 4). This is likely due to saturation of the microarray system by these abundant transcripts. Error bars indicate standard deviations for triplicate experiments.
Induction of β major globin and Fog1 by GATA-1. (A) Messenger RNA levels in G1E-ER4 cells predicted by microarray experiments. (B) Messenger RNA levels quantitated by real-time polymerase chain reaction (PCR) in G1E-ER4 cells overexpressing bcl-xL to overcome cycloheximide-induced death. Each data point represents 6 separate PCR reactions in 2 independent experiments. Cycloheximide (0.5 mM) was added as indicated. Note that the magnitude of β globin RNA induction appears to be underestimated by the microarrays, as was also observed for α globin (Figure 4). This is likely due to saturation of the microarray system by these abundant transcripts. Error bars indicate standard deviations for triplicate experiments.
Delayed kinetics of β major globin induction predicted by the microarrays is shown in Figure 7A, left panel. In contrast, RNA encoding Fog-1, a cofactor that participates with GATA-1 in β major globin transcription,26,46,71 was activated more rapidly (Figure 7A, right panel). These data suggest that GATA-1 could facilitate β major globin production by first stimulating transcription of Fog1. To explore this further, we investigated whether either of these genes is induced by GATA-1 independently of new protein synthesis. Initially, it was not possible to examine gene expression in G1E-ER4 cells treated with the protein synthesis inhibitors cycloheximide or anisomycin, as these drugs induced rapid apoptosis (data not shown). To circumvent this problem, we transduced G1E-ER4 cells with retrovirus encoding the antiapoptotic protein bcl-xL. In response to GATA-1 activation, G1E–ER4–bcl-xL cells underwent erythroid maturation similar to the G1E-ER4 parental line, but survived for at least 12 hours in the presence of cycloheximide (not shown). We used real-time PCR to determine whether cycloheximide inhibits activation of Fog1 or β major globin by GATA-1 in G1E–ER4–bcl-xL cells (Figure 7B). After treatment with estradiol alone, both mRNAs were strongly induced. Cycloheximide treatment, which blocked more than 95% of protein synthesis (not shown), completely inhibited estradiol-induced β major globin RNA up-regulation, indicating that additional downstream GATA-1 targets are required. In contrast, Fog1 mRNA was fully induced after activation of GATA-1 in the presence of cycloheximide (Figure 7B). These findings raise the possibility that GATA-1 activates Fog1 transcription directly and that this might be necessary for full β major globin gene induction.
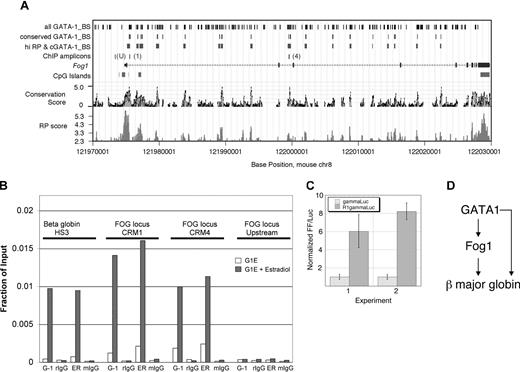
To test whether Fog1 is a direct GATA-1 target gene, we first applied bioinformatic predictions to find candidate GATA-1 binding sites in Fog1 that are likely to be involved in transcriptional regulation (Figure 8A). Whole-genome sequence alignments between mouse and human were analyzed to find sequences likely to be functional (conservation score) and sequences more likely to be involved in regulation of gene expression (regulatory potential40 ). Also, matches to weight matrices for GATA-1 binding sites that are conserved in human, mouse, and rat were identified. As shown in Figure 8A, 17 intronic segments have prominent peaks for both regulatory potential and conservation and all of these contain one or more conserved GATA-binding sites. The presence of potential GATA sites in regulatory regions of erythroid-expressed genes is not unexpected. However, the Fog1 locus is remarkable in the large number of predicted regulatory regions that all contain conserved GATA motifs. This suggests that GATA-1 may be a critical protein in regulating Fog1 and that the multiple binding sites could mediate concentration-dependent effects.
GATA-1 regulates Fog1 directly. (A) Cis-regulatory modules in Fog1 as predicted by comparative genomics. The mouse Fog1 gene (Zfpm1) is plotted in the middle, using base positions on chromosome 8 from the February 2003 (mm3) assembly. Coding exons are taller boxes, untranslated regions are shorter boxes, and introns are shown as lines with arrowheads pointing in the direction of transcription. Positions of cytosine-phosphate-guanosine (CpG) islands are on the line underneath the gene map. The Conservation Score38 estimates a log-likelihood that an alignment is in functional sequences. The RP score is a log-likelihood measurement that estimates the probability that a sequence is involved in regulating expression.40 Matches to weight matrices for GATA-1 binding sites were identified in the mouse sequence (track labeled “all GATA-1_BS”), and these were filtered to find those that are conserved in mouse, rat, and human alignments (track labeled “conserved GATA-1_BS”). The DNA segments whose alignments exceed a calibrated threshold for RP score and also contain at least one conserved, predicted GATA-1 binding site are shown in the track labeled “hi RP & cGATA-1_BS”; these are the predicted cis-regulatory modules for the Fog1 locus. There were 2 tested for occupancy by GATA-1 in vivo (B); these are shown as ChIP amplicons R1 (preCRM1 in intron 1) and R2 (preCRM4 in intron 2) along with a negative control (“U”) located upstream of the Fog1 start site for transcription. The information in the first 3 tracks was obtained by queries to GALA41 and the data were visualized in the UCSC Genome Browser.39 (B) Chromatin immunoprecipitation (ChIP) experiments showing GATA-1 occupancy at the Fog1 locus in G1E-ER4 cells at 24 hours after GATA-1 activation. The regions of the Fog1 gene examined are indicated as “ChIP amplicons” in line 4 of panel A.Antibodies used are indicated below the figure: G-1 indicates anti–GATA-1; ER, anti–estrogen receptor (used to detect the GATA-1/ER fusion protein). Control antibodies: rIgG indicates rat IgG; mIgG, mouse IgG. Each data point represents the average value obtained from 2 independent ChIP experiments. (C) Predicted cis-regulatory module 1 enhances expression from an erythroid promoter. The plasmid gammaLuc contains a firefly (FF) luciferase gene driven by the γ globin gene promoter, and preCRM1 (R1) was added upstream of the promoter to make the plasmid R1 gammaLuc. The normalized levels of expression are plotted for 2 experiments, with each sample tested in triplicate in both; the error bars indicate standard deviations. The activity from R1 gammaLuc is significantly higher than that from gammaLuc, with a P value no more than .005 for either by Student t test. (D) GATA1 and Fog1 act in a feed-forward loop to activate the β major globin gene.
GATA-1 regulates Fog1 directly. (A) Cis-regulatory modules in Fog1 as predicted by comparative genomics. The mouse Fog1 gene (Zfpm1) is plotted in the middle, using base positions on chromosome 8 from the February 2003 (mm3) assembly. Coding exons are taller boxes, untranslated regions are shorter boxes, and introns are shown as lines with arrowheads pointing in the direction of transcription. Positions of cytosine-phosphate-guanosine (CpG) islands are on the line underneath the gene map. The Conservation Score38 estimates a log-likelihood that an alignment is in functional sequences. The RP score is a log-likelihood measurement that estimates the probability that a sequence is involved in regulating expression.40 Matches to weight matrices for GATA-1 binding sites were identified in the mouse sequence (track labeled “all GATA-1_BS”), and these were filtered to find those that are conserved in mouse, rat, and human alignments (track labeled “conserved GATA-1_BS”). The DNA segments whose alignments exceed a calibrated threshold for RP score and also contain at least one conserved, predicted GATA-1 binding site are shown in the track labeled “hi RP & cGATA-1_BS”; these are the predicted cis-regulatory modules for the Fog1 locus. There were 2 tested for occupancy by GATA-1 in vivo (B); these are shown as ChIP amplicons R1 (preCRM1 in intron 1) and R2 (preCRM4 in intron 2) along with a negative control (“U”) located upstream of the Fog1 start site for transcription. The information in the first 3 tracks was obtained by queries to GALA41 and the data were visualized in the UCSC Genome Browser.39 (B) Chromatin immunoprecipitation (ChIP) experiments showing GATA-1 occupancy at the Fog1 locus in G1E-ER4 cells at 24 hours after GATA-1 activation. The regions of the Fog1 gene examined are indicated as “ChIP amplicons” in line 4 of panel A.Antibodies used are indicated below the figure: G-1 indicates anti–GATA-1; ER, anti–estrogen receptor (used to detect the GATA-1/ER fusion protein). Control antibodies: rIgG indicates rat IgG; mIgG, mouse IgG. Each data point represents the average value obtained from 2 independent ChIP experiments. (C) Predicted cis-regulatory module 1 enhances expression from an erythroid promoter. The plasmid gammaLuc contains a firefly (FF) luciferase gene driven by the γ globin gene promoter, and preCRM1 (R1) was added upstream of the promoter to make the plasmid R1 gammaLuc. The normalized levels of expression are plotted for 2 experiments, with each sample tested in triplicate in both; the error bars indicate standard deviations. The activity from R1 gammaLuc is significantly higher than that from gammaLuc, with a P value no more than .005 for either by Student t test. (D) GATA1 and Fog1 act in a feed-forward loop to activate the β major globin gene.
We examined 2 regions in the Fog1 locus, designated preCRM1 and preCRM4 (for predicted cis-regulatory module), for in vivo GATA-1 occupancy by chromatin immunoprecipitation (ChIP) (Figure 8B). An upstream region of the Fog1 promoter without any conserved GATA sites was selected as a negative control (labeled as “U” in Figure 8A and as “Upstream” in Figure 8B). ChIP was performed in G1E-ER4 cells before and 24 hours after estradiol treatment. Occupancy of the GATA-1–ER fusion protein was measured using 2 different antibodies, 1 directed against GATA-1 and 1 against the ER moiety. Both CRM1 and CRM4 regions exhibited significant GATA-1 occupancy after estradiol treatment, as detected with both antibodies, roughly 50- to 100-fold greater than isotype control or irrelevant antibodies and comparable with GATA-1 occupancy at hypersensitive site 3 (HS3) of the β major globin locus.45 No GATA-1 occupancy was detected at the negative control site upstream of Fog1 exon 1.
Next, we tested preCRM1 for its ability to augment gene expression in erythroid cells. We inserted this region upstream of a minimal γ globin gene promoter linked to a luciferase reporter gene and measured the level of expression after transient transfection into erythroid cells. As illustrated in Figure 7D, preCRM1 augmented expression of the linked reporter gene an average of about 7-fold in erythroid cells in 2 independent experiments. Together these results demonstrate that GATA-1 occupies functionally important regions of the Fog-1 locus and is likely to play a direct role in activating its transcription. More generally, the results verify the utility of our microarray database in combination with bioinformatics to predict genomic sites where GATA-1 acts to facilitate the erythroid program of transcription.
Considering that Fog-1 interacts with GATA-1 both functionally and physically and that both nuclear proteins are required for optimal β globin synthesis,26,46,60,72 our data are consistent with a feed-forward loop73 in which GATA-1 induces Fog1 to activate β major globin transcription (Figure 8D). This is in accord with prior studies demonstrating that Fog1 is induced by GATA-1 independent of its interaction with the Fog-1 protein.74 The model predicts that GATA-1 binds the Fog1 locus at early time points prior to β major globin induction during induced maturation of G1E-ER4 cells; detailed kinetic studies are under way to test this. The feed-forward loop is a common mode of gene regulation described in prokaryotes and in the development of muscle, liver, and pancreas.73,75,76 The current example illustrates how a tissue-specific transcription factor (GATA-1) can positively regulate the production of its own cofactor (Fog-1), which functions in both gene activation and repression. Of note, low levels of β globin mRNA are expressed without GATA-1 in G1E-ER4 cells and in primary GATA-1– erythroblasts.13,62 Also, Fog1 mRNA is expressed basally in G1E-ER4 cells prior to activation of GATA-1. Therefore, it seems likely that GATA-1 and Fog-1 act in a dose-dependent fashion and that relatively high threshold levels are required for optimal β major globin gene activation. Specific dosage dependency thresholds also probably exist for genes that are repressed by GATA-1 in a Fog-1–dependent manner.46,60 Finally, our data do not exclude the possibility that additional GATA-1–induced signaling pathways or transcription factors are required for β major globin transcription. If so, these additional genes should be represented among the cohort of rapidly induced transcripts in G1E-ER4 cells. One such candidate is EKLF, which is activated by GATA-1 in G1E-ER4 cells (Crispino et al;74 Figure 2; and Table 1), is known to be a direct GATA-1 target,46,77-79 and cooperates with GATA-1 to activate β major globin transcription.80-84
Defining erythroid development through GATA-1 gene complementation
Use of microarrays for mRNA profiling is a powerful tool for examining gene expression during hematopoiesis.4,5,85-87 The current study validates the utility of G1E cells as a biologically relevant model system and provides a comprehensive transcriptome analysis of the transition through several well-defined stages of erythropoiesis, illustrating new aspects of its metabolic and developmental control. In addition, our experimental system provides a unique view into erythroid maturation from the perspective of GATA-1, further defining its regulatory hierarchy and mechanisms of action. In this regard, the kinetics of gene expression in G1E cells, which are arrested by loss of GATA-1, may differ somewhat from that in maturing primary erythroid cells where GATA-1 is induced in a more gradual and physiologic manner. Therefore, it will be interesting and important to contrast the current findings to comprehensive transcriptome studies performed using other model systems in which the GATA-1 gene is intact and under endogenous control.1-3 Such comparisons should better define the developmental block conferred by loss of GATA-1. Ultimately, it is likely that these complementary experimental approaches will intersect to provide a greater understanding of the transcriptional program in erythropoiesis. In the future, it will also be important to analyze G1E-ER4 cells and other model systems using proteomic approaches, as many aspects of erythroid gene expression are regulated at the level of protein translation, modification, or degradation.88-92
Prepublished online as Blood First Edition Paper, August 5, 2004; DOI 10.1182/blood-2004-04-1603.
Supported by National Institutes of Health (NIH) grant R33 CA94393 (L.A.C.), NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R01 DK065806 (R.C.H. and M.J.W.), Abramson Cancer Center of the University of Pennsylvania Pilot Project Grant, and Johnson and Johnson Focused Giving Award (M.J.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ernst Müller and Jeffrey Miller for sharing their cDNA and subtraction hybridization data. We are also grateful to Mortimer Poncz for reviewing this manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal