Abstract
Gene expression profiles of bone marrow (BM) CD34-derived megakaryocytic cells (MKs) were compared in patients with essential thrombocythemia (ET) and healthy subjects using oligonucleotide microarray analysis to identify differentially expressed genes and disease-specific transcripts. We found that proapoptotic genes such as BAX, BNIP3, and BNIP3L were down-regulated in ET MKs together with genes that are components of the mitochondrial permeability transition pore complex, a system with a pivotal role in apoptosis. Conversely, antiapoptotic genes such as IGF1-R and CFLAR were up-regulated in the malignant cells, as was the SDF1 gene, which favors cell survival. On the basis of the array results, we characterized apoptosis of normal and ET MKs by time-course evaluation of annexin-V and sub-G1 peak DNA stainings of immature and mature MKs after culture in serum-free medium with an optimal thrombopoietin concentration, and annexin-V–positive MKs only, with decreasing thrombopoietin concentrations. ET MKs were more resistant to apoptosis than their normal counterparts. We conclude that imbalance between proliferation and apoptosis seems to be an important step in malignant ET megakaryocytopoiesis.
Introduction
Essential thrombocythemia (ET) is a malignant megakaryocytic cell (MK) disorder principally characterized by abnormal proliferation of a malignant MK clone and persistent thrombocytosis.1-3 ET is usually associated with elevated numbers of bone marrow (BM) MKs and of morphologically and functionally abnormal circulating platelets.1,4,5 ET-affected BM displays enlarged and mature MKs with hyperploid nuclei.6 Even though ET is usually considered to be a clonal disease, it has recently been suggested that some patients do not have a clonal disorder7 ; this issue is further complicated by the recent observation of monoclonal hematopoiesis in healthy elderly women.8 The molecular events underlying the commitment and differentiation of normal and malignant megakaryocytes are poorly understood.9-13
The cause of ET is unknown. It is thought that molecular lesions in critical genes regulate the balance between proliferation/differentiation and apoptosis of MK progenitors. The few reported consistent cytogenetic abnormalities14 include trisomies of chromosomes 8 and 9 and deletions in 13q and 20q.15,16 Since little is known about specific molecular abnormalities, further molecular and transcriptional studies are required to better elucidate the development of the malignant MKs.
We used microarray technology to study the malignant megakaryocytopoiesis of ET. In particular, CD34+ hematopoietic progenitor cells taken from the BM of patients with ET and healthy subjects were induced to differentiate along the megakaryocytic lineage in liquid suspension cultures by continuous addition of 100 ng/mL thrombopoietin. The pattern of gene expression identified in normal CD34-derived MKs was compared with that of ET CD34-derived MKs to identify differentially expressed genes and disease-specific transcripts. We found that expression of genes involved in the apoptotic pathway is impaired in ET CD34-derived MKs. Moreover, CD34-derived MKs of patients with ET were more resistant to apoptosis than their normal counterparts.
Patients, materials, and methods
Patients
Routinely collected BM and peripheral blood (PB) samples from 15 patients (9 men, 6 women; median age, 43 years; range, 24-66 years) with ET diagnosed according to polyeythemia vera study group (PVSG) criteria17 were studied. The median platelet count was 1098 × 109/L (range, 810-1832 × 109/L). Karyotype analysis did not show any chromosome aberrations. At the time of the study, no patient showed hemorrhagic or thrombotic complications. The patients were either newly diagnosed (n = 8) or off cytotoxic treatment for at least 3 months (n = 7). BM samples from 15 healthy donors were used as controls. All subjects (patients/controls) provided written informed consent for the study, which was performed in accordance with the Helsinki declaration of 1975. In line with national and European Union guidelines, ethical approval was not required for this study.
Purification of CD34+ cells
Mononuclear cells were isolated from BM (20-30 mL) by Ficoll-Hypaque (D = 1.077 g/mL; Pharmacia, Uppsala, Sweden), rinsed, and adherence-depleted overnight. After recovery of suspended cells, CD34+ cells were isolated using a magnetic cell-sorting program (Mini-MACS; Miltenyi Biotec, Auburn, CA) and the CD34 isolation kit in accordance with the manufacturer's recommendations. The purity of CD34-selected cells was determined for each selection as previously described,18 and the percentage of CD34+ cells ranged from 89% to 98%.
Liquid suspension cultures and purification of CD34-derived MKs
CD34+ cells (from 12 patients and 12 controls) were differentiated in thrombopoietin-treated liquid suspension cultures as previously described.19 Briefly, CD34+ cells (80 000/mL) were resuspended in a serum-free medium in the presence of 100 ng/mL human recombinant thrombopoietin (TPO; Genzyme, Boston, MA). Every 3 days, viable cells were scored by trypan blue dye exclusion and cultures were amplified with fresh serum-free medium. Each well was then supplemented with 100 ng/mL TPO. After 14 to 16 days of liquid culture, CD34-derived MKs were purified by means of an anti-CD41a monoclonal antibody (MoAb; Dako, Milan, Italy) directed against the glycoproteic αIIb-β3 complex and immunobeads (MPC 450 Dynabeads; Dynal, Oslo, Norway), as previously described.19 The purity of the MKs was determined for each isolation by indirect immunofluorescence, using an anti-CD41b MoAb that reacts with a different epitope of the αIIb-β3 subunit (Immunotech, Westbrook, ME), followed by a goat anti–mouse immunoglobulin G (IgG), covalently linked to fluorescein (Becton Dickinson, San Jose, CA).
In a second set of experiments, CD34+ cells (from 3 patients and 3 healthy subjects) were also cultured in a serum-free medium with various concentrations of TPO (0.1, 1, 10, 100 ng/mL) for 14 days.
Microarray set-up and analysis
Total cellular RNA was isolated from 4 CD34-derived MK pools (from 2 groups of 6 patients and 2 groups of 6 healthy subjects) using the guanidinium-cesium chloride centrifugation technique,20 with slight modifications. RNA was assessed by formaldehyde agarose gel electrophoresis and quantified by ultraviolet (UV) absorbance. Biotin-labeled target synthesis reactions were performed using standard protocols supplied by the manufacturer (Affymetrix, Santa Clara, CA). Briefly, 5 μg of the RNA was converted into double-stranded cDNA by reverse transcription using a cDNA synthesis kit (SuperScript Double-Stranded cDNA Synthesis; Invitrogen, Paisley, United Kingdom), following the protocol supplied by the manufacturer, with a T7-(dT)24 primer (Affymetrix). After the second-strand synthesis, labeled cRNA was generated from the purified cDNA sample (Gene Chip Sample Cleanup Module; Affymetrix) by an in vitro transcription reaction (Enzo bio array HY RNA transcript labeling kit; Enzo Biochem, Farmingdale, NY) supplemented with biotin-11–cytidine triphosphate and biotin-16–uridine triphosphate, as described by the manufacturer. The labeled cRNA was purified using the provided Affymetrix spin columns. The concentration of biotin-labeled cRNA was determined by UV absorbance. In all cases, 15 μg of each biotinylated cRNA preparation was fragmented, assessed by gel electrophoresis, and put in the hybridization cocktail containing 4 biotinylated hybridization controls (BioB, BioC, BioD, and Cre), as recommended by the Affymetrix protocol. Samples were hybridized to an identical lot of Affymetrix HG-U133A Gene Chip arrays for 16 hours. Gene Chips were washed and stained following the instrument's standard Eukaryotic GE WS2v4 protocol and using antibody-mediated signal amplification as already described.21
The images from the scanned chips were processed by means of Affymetrix Microarray Analysis Suite 5.0 (MAS 5.0). The amount of a transcript mRNA (ie, the expression level [signal]) was determined with the MAS 5.0 absolute analysis algorithm,22 as well as the presence or the absence of a transcript. The MAS 5.0 comparison analysis algorithm was used to compare gene expression levels between 4 samples and to evaluate change in the expression level of a transcript: increase; decrease; or no change; and the fold change in transcript expression levels, expressed as the log2 ratio (signal log ratio; a log2 ratio of 1 is equal to a fold change of 2). All expression values for the genes in the absolute and comparison analysis were determined using the MAS 5.0 global scaling option. The MAS 5.0–generated absolute and comparison analysis data were then uploaded onto Affymetrix Micro DB 3.0 database and processed with Affymetrix Data Mining Tool (DMT) 3.0 software. DMT 3.0 was used to query, filter, and sort data from data sets. The gene lists created by DMT software were transferred to Microsoft Excel (Microsoft, Seattle, WA) and linked to the Internet genome databases (ie, NetAffx, GenBank, Swiss Prot, Online Mendelian Inheritance in Man, and GeneCards) and also uploaded onto Affymetrix Gene Ontology Mining Tool, which, in addition to providing the Gene Ontology (GO) terms for annotated genes, also provides graphic interactive views of the biologic process and molecular function, as well as cellular component gene ontology subgraphs. GO is widely accepted as the standard for vocabulary describing the biologic process, molecular function, and cellular component of genes.23
Quantitative RT-PCR
Based on the array results, we evaluated expression levels of selected genes by quantitative reverse-transcriptase–polymerase chain reaction (RT-PCR) analysis. It should be noted that quantitative RT-PCR was not possible for certain genes because the ABI Assays-on-Demand Gene Expression Assay (Applied Biosystems, Foster City, CA) was not available for Homo sapiens and the system is working only on the basis of Assays-on-Demand. cDNAs were reverse transcribed from total RNA samples (100 ng per sample) using High Capacity cDNA Archive Kit (Applied Biosystems) as described by the manufacturer. TaqMan PCR reactions were performed on cDNA samples using the TaqMan Universal PCR Master Mix (Applied Biosystems) according the manufacturer's instructions, in conjunction with custom 7900 microfluidic cards (Applied Biosystems) and ABI PRISM 7900 HT Sequence Detection Systems. The TaqMan strategies for each gene have been developed as Assay–on–Demand by Applied Biosystems. Gene expression profiling was achieved using the comparative cycle threshold (CT) method of relative quantification (the calibrator samples were MK pools from healthy subjects, with glyceraldehyde-3-phosphate dehydrogenase [GAPDH] used as the endogenous control). To easily compare the arrays' signal log ratios (SLR) with the ABI results, the relative quantity (RQ) was log transformed, with real-time SLR being the log2 RQ.
Identification of apoptotic MKs
Apoptosis of normal and malignant CD34-derived MKs (from 6 healthy subjects and 5 patients) was evaluated by both propidium iodide (PI) (DNA PrepStain, containing RNAse and PI; Beckman-Coulter, Miami, FL) and CD41 phycoerythrin (PE; BD Biosciences, San Jose, CA)/annexin-V–fluorescein isothiocyanate (FITC; Actiplate Valter Occhiena, Turin, Italy) staining following the manufacturers' instructions, in conjunction with flow cytometry analysis. The assays were performed after 7, 14, and 21 days of incubation in a serum-free medium in the presence of 100 ng/mL TPO. Briefly, MKs were recovered, counted, and washed. When apoptosis was determined by annexin-V–FITC (phosphatidylserine exposure), cells were stained with a MoAb directed against the glycoproteic αIIb-β3 complex (CD41a-PE; BD Biosciences) and then resuspended in binding buffer 1x and labeled with 5 μL annexin-V–FITC for 10 minutes. For PI staining (sub-G1 peak), MKs were stained with DNA Prepstain and analyzed with a dedicated program. Both analyses were performed with Cytomics FC500 (Beckman-Coulter) using the dedicated flow cytometry software. Statistical analysis among the groups was performed by Wilcoxon test. P values less than .05 were considered significant.
Protein analysis
Platelet-rich plasma (PRP) was obtained from PB (6 healthy subjects and 6 patients) anticoagulated with EDTA (ethylenediaminetetraacetic acid) by centrifugation at 200g for 10 minutes. To avoid leukocyte contamination, only the upper 80% of the PRP was used for filtration through a 5-μm no-wetting nylon filament filter (BioDesign, Carmel, NY) and harvested by centrifugation at 1000g for 10 minutes. Platelets were then washed twice with phosphate-buffered saline (PBS) containing 3.0 mM EDTA and 5% (wt/vol) bovine serum albumin (BSA; Sigma, St Louis, MO).
Expression of platelet proteins was investigated by the following antibodies: PE-conjugated anti-CD14 (BD Biosciences); anti-BAX (Bcl2-associated X protein; BD Biosciences); and anti-IGF1R (insulin-like growth factor 1 receptor) alpha subunit (BioSource International, Camarillo, CA) and evaluated by flow-cytometry. CD14 and IGF1R expression were investigated both at surface and intracellular levels. The intracellular expression pattern of BAX protein was evaluated. Platelets (2 × 106) were permeabilized by means of the Intraprep Permeabilization Reagent Kit (Immunotech, Marseilles, France) in accordance with the manufacturer's recommendations. The cells were incubated for 30 minutes at room temperature in the dark with the primary antibodies. After extensive washings, samples labeled with BAX (10 μL) and IGF1R (10 μL) were incubated for 30 minutes with 5 μL PE/FITC-conjugated goat anti–mouse immunoglobulins (BD Biosciences). Aspecific fluorescence was assessed using 5 μL decomplemented human AB serum (1:50 dilution with PBS) followed by PE/FITC-conjugated goat anti–mouse immunoglobulins (BD Biosciences). After additional washing, platelets were resuspended in 300 μL PBS containing 0.3 mM EDTA and 0.1% BSA prior to analysis with a FACScalibur flow cytometer (BD Biosciences). For each sample, up to 10 000 events were collected. Platelets were identified on the basis of their size and granularity, and an electronic gate was drawn around the platelet. A sample from a healthy donor was run with each patient sample.
Results
Characterization of the cultured MKs
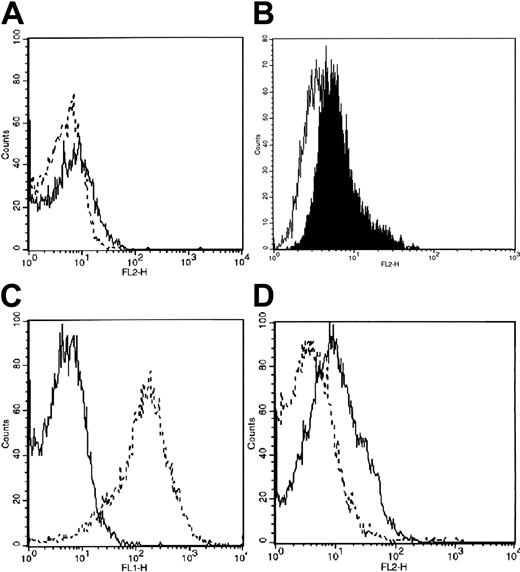
To characterize MK maturation, CD34+ BM cells from healthy or patients with ET were cultured in serum-free medium in the presence of TPO. MK differentiation was assessed by light microscopy evaluation after May-Grunwald Giemsa staining, and by analysis of αIIbβ3 expression with a CD41a MoAb and flow cytometry. At morphologic analysis after 14 to 16 days of culture, different degrees of maturation were observable with small mononucleated megakaryoblasts appearing alongside fully developed MKs (Figure 1A). The MK maturation was confirmed by the high frequency (92%-96%) of αIIbβ3+ cells in both patients and healthy subjects (Figure 1B). Morphologic analysis of purified MKs is shown in Figure 1C. Note the presence of small megakaryoblasts near mature MKs.
Analysis of morphology and αIIb-β3 expression of normal MKs. (A) Cytospins were observed at light microscopy after May-Grunwald staining (× 20 magnification, 0.50 aperture objective lenses [Carl Zeiss, Oberkochen, Germany]). Note the presence of cells belonging to various maturational stages of megakaryocytopoiesis. (B) αIIb-β3 expression (white area) of normal MK cells was examined with an anti-CD41a MoAb directly conjugated to phycoerythrin, and fluorescence was analyzed by flow cytometry. Cells treated with an isotype-matched irrelevant MoAb directly conjugated to phycoerythrin represent the negative control (black area). The x-axis indicates fluorescence intensity; the y-axis, relative number of cells. (C) Morphologic analysis of purified MKs. May-Grunwald staining (× 40 magnification, 1.0 aperture objective lenses [Carl Zeiss]). Note the presence of small megakaryoblasts near mature megakaryocytes. Morphology was evaluated with an Ultraphot universal photomicroscope (Carl Zeiss, Oberkochen, Germany). The camera was inserted within the microscope. No software was used to optimize the figure. Objective lenses 20 × and 40 ×.
Analysis of morphology and αIIb-β3 expression of normal MKs. (A) Cytospins were observed at light microscopy after May-Grunwald staining (× 20 magnification, 0.50 aperture objective lenses [Carl Zeiss, Oberkochen, Germany]). Note the presence of cells belonging to various maturational stages of megakaryocytopoiesis. (B) αIIb-β3 expression (white area) of normal MK cells was examined with an anti-CD41a MoAb directly conjugated to phycoerythrin, and fluorescence was analyzed by flow cytometry. Cells treated with an isotype-matched irrelevant MoAb directly conjugated to phycoerythrin represent the negative control (black area). The x-axis indicates fluorescence intensity; the y-axis, relative number of cells. (C) Morphologic analysis of purified MKs. May-Grunwald staining (× 40 magnification, 1.0 aperture objective lenses [Carl Zeiss]). Note the presence of small megakaryoblasts near mature megakaryocytes. Morphology was evaluated with an Ultraphot universal photomicroscope (Carl Zeiss, Oberkochen, Germany). The camera was inserted within the microscope. No software was used to optimize the figure. Objective lenses 20 × and 40 ×.
Microarray data
Gene expression analysis. We profiled gene expression of the 2 CD34-derived MK pools from healthy subjects and the 2 pools from patients with ET using Affymetrix HG-U133A Gene Chip array, which covers about 22 200 transcripts of known genes. All these data have been deposited in the Gene Expression Omnibus public database, at http://www.ncbi.nlm.nih.gov/geo (accession numbers are GSM8649 and GSM15648 for healthy subjects; GSM8650 and GSM15650 for patients with ET; and GSE567 and GSE997 for comparisons of ET MKs vs normal MKs).
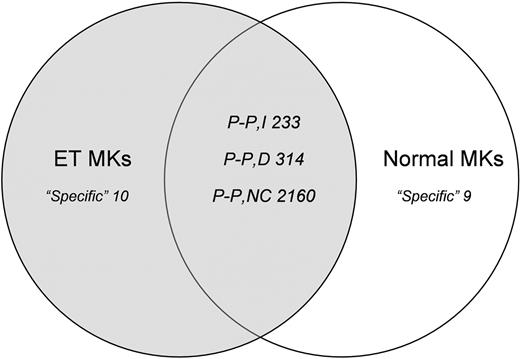
The complexity of the mRNAs was similar in the normal and malignant MKs, with about 10 000 genes (almost 50%) expressed in each setting. Excluding the low-abundance transcripts (signal < 100), the ET MK pools expressed 4637 genes and the normal pools expressed 4655. To identify genes that were differentially expressed in the 2 populations, we compared their expression profiles using the Affymetrix comparison analysis algorithm, which distinguished in the malignant MK pools those genes whose expression was “increased,” “not changed,” and “decreased.” We performed pair-wise comparisons of individual genes and of their expression changes in ET MKs relative to normal MKs. Figure 2 reports the numbers of genes that were specific to either the normal MKs (both pools) or the ET MKs (both pools), or that were expressed in the 2 pools of each population (increased, decreased, and not changed).
Gene expression analysis. Venn diagram summarizing pairwise comparisons of individual genes and of their expression changes in ET MKs relative to normal MKs (“low-abundance” genes and genes showing only borderline changes in expression were excluded from the analysis). “Specific” refers to genes expressed in only one cell population; “P-P” refers to genes detected in both populations, which are subdivided into those that were increased (I), decreased (D), or not changed (NC) in the ET MKs relative to their normal counterparts.
Gene expression analysis. Venn diagram summarizing pairwise comparisons of individual genes and of their expression changes in ET MKs relative to normal MKs (“low-abundance” genes and genes showing only borderline changes in expression were excluded from the analysis). “Specific” refers to genes expressed in only one cell population; “P-P” refers to genes detected in both populations, which are subdivided into those that were increased (I), decreased (D), or not changed (NC) in the ET MKs relative to their normal counterparts.
GO Mining Tool functional analysis of differentially expressed genes. We uploaded onto the NetAffx GO Mining Tool lists of those genes that were increased (n = 233) and decreased (n = 314) in ET MKs relative to their normal counterparts. As can be seen from Table 1, most of the increased genes are involved in the transcriptional process, and particularly in RNA metabolism and processing, apoptosis, and cell-cycle regulation; their molecular functions include nucleic acid binding and RNA binding. By contrast, most of the decreased genes are involved in macromolecule and protein biosynthesis, and are structural constituents of ribosomes. A significant number of decreased genes were also addressed to the mitochondrial membrane, some of which play key roles in the induction of apoptosis (Table 2). The latter include BAX, BNIP3, and BNIP3L (all Bcl2 associated), and members of the mitochondrial permeability transition (PT) pore complex, which plays a pivotal role in apoptosis.
Gene Ontology Mining Tool functional analysis
GO functional category . | Percentage of mapped transcripts, % . | No. of transcripts represented on the array that map to the GO category . |
|---|---|---|
| Increased genes in ET | ||
| Biologic process | ||
| RNA metabolism/RNA processing | 3.0 | 388 |
| Macromolecule/protein catabolism | 2.7 | 622 |
| Cell cycle/regulation of cell cycle | 2.1 | 518 |
| Programmed cell death/apoptosis | 1.9 | 519 |
| Cellular component | ||
| Nucleus | 1.4 | 3248 |
| Cytoplasm | 1.2 | 3562 |
| Integral to membrane | 0.5 | 3174 |
| Plasma membrane | 0.5 | 2180 |
| Molecular function | ||
| Nucleic/RNA binding | 2.8 | 711 |
| Hydrolase/peptidase activity | 1.9 | 601 |
| Decreased genes in ET | ||
| Biologic process | ||
| Macromolecule/protein biosynthesis | 7.5 | 572 |
| Carbohydrate metabolism | 5.4 | 367 |
| Cellular component | ||
| Ribosome | 16.3 | 220 |
| Eukaryotic 43S preinitiation complex | 14.4 | 69 |
| Cytosol | 10.3 | 386 |
| Mitochondrial membrane | 7.0 | 141 |
| Molecular function | ||
| Structural constituent of ribosome | 15.6 | 273 |
| RNA binding | 5.4 | 711 |
GO functional category . | Percentage of mapped transcripts, % . | No. of transcripts represented on the array that map to the GO category . |
|---|---|---|
| Increased genes in ET | ||
| Biologic process | ||
| RNA metabolism/RNA processing | 3.0 | 388 |
| Macromolecule/protein catabolism | 2.7 | 622 |
| Cell cycle/regulation of cell cycle | 2.1 | 518 |
| Programmed cell death/apoptosis | 1.9 | 519 |
| Cellular component | ||
| Nucleus | 1.4 | 3248 |
| Cytoplasm | 1.2 | 3562 |
| Integral to membrane | 0.5 | 3174 |
| Plasma membrane | 0.5 | 2180 |
| Molecular function | ||
| Nucleic/RNA binding | 2.8 | 711 |
| Hydrolase/peptidase activity | 1.9 | 601 |
| Decreased genes in ET | ||
| Biologic process | ||
| Macromolecule/protein biosynthesis | 7.5 | 572 |
| Carbohydrate metabolism | 5.4 | 367 |
| Cellular component | ||
| Ribosome | 16.3 | 220 |
| Eukaryotic 43S preinitiation complex | 14.4 | 69 |
| Cytosol | 10.3 | 386 |
| Mitochondrial membrane | 7.0 | 141 |
| Molecular function | ||
| Structural constituent of ribosome | 15.6 | 273 |
| RNA binding | 5.4 | 711 |
Genes decreased in ET MKs and involved in apoptosis
GenBank accession no. . | Gene description . | Normal MK signal* . | ET MK signal* . | SLR† . |
|---|---|---|---|---|
| NM_004324.1 | BAX (BCL2-associated X protein) | 305.7 | 162.6 | -1.0 |
| NM_004052.2 | BNIP3 (BCL2/adenovirus E1B 19-kDa interacting protein 3) | 436.8 | 125.2 | -1.8 |
| AL132665.1 | BCL2/adenovirus E1B 19-kDa interacting protein 3-like | 727.0 | 299.0 | -1.2 |
| NM_000714.2 | Benzodiazapine receptor (peripheral) | 623.1 | 413.8 | -0.5 |
| NM_021130.1 | Peptidylprolyl isomerase A (cyclophilin A) | 4932.4 | 4319.3 | -0.2 |
| BE962749 | Peptidylprolyl isomerase C (cyclophilin C) | 130.8 | 53.3 | -1.2 |
| NM_014765.1 | Translocase of outer mitochondrial membrane 20 (yeast) homolog | 541.7 | 276.8 | -1.0 |
| AL515918 | Voltage-dependent anion channel 1 | 469.8 | 270.9 | -0.8 |
| U90943.1 | Voltage-dependent anion channel 3 | 448.3 | 432.3 | -0.03 |
| NM_001152.1 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 1509.6 | 1149.9 | -0.3 |
| AA916851 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | 919.2 | 451.5 | -0.7 |
GenBank accession no. . | Gene description . | Normal MK signal* . | ET MK signal* . | SLR† . |
|---|---|---|---|---|
| NM_004324.1 | BAX (BCL2-associated X protein) | 305.7 | 162.6 | -1.0 |
| NM_004052.2 | BNIP3 (BCL2/adenovirus E1B 19-kDa interacting protein 3) | 436.8 | 125.2 | -1.8 |
| AL132665.1 | BCL2/adenovirus E1B 19-kDa interacting protein 3-like | 727.0 | 299.0 | -1.2 |
| NM_000714.2 | Benzodiazapine receptor (peripheral) | 623.1 | 413.8 | -0.5 |
| NM_021130.1 | Peptidylprolyl isomerase A (cyclophilin A) | 4932.4 | 4319.3 | -0.2 |
| BE962749 | Peptidylprolyl isomerase C (cyclophilin C) | 130.8 | 53.3 | -1.2 |
| NM_014765.1 | Translocase of outer mitochondrial membrane 20 (yeast) homolog | 541.7 | 276.8 | -1.0 |
| AL515918 | Voltage-dependent anion channel 1 | 469.8 | 270.9 | -0.8 |
| U90943.1 | Voltage-dependent anion channel 3 | 448.3 | 432.3 | -0.03 |
| NM_001152.1 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 1509.6 | 1149.9 | -0.3 |
| AA916851 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | 919.2 | 451.5 | -0.7 |
All present.
All decreased.
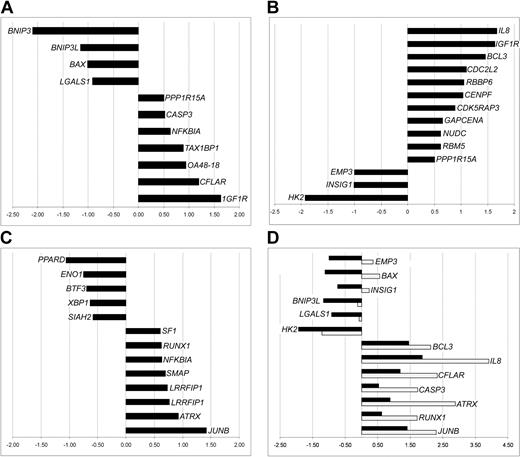
Functional categorization of differentially expressed genes. To shed further light on expression profile differences between ET and normal MKs, we divided the genes that were increased/decreased in ET MKs into functional categories. Expression levels of comprehensive sets of the differentially expressed genes involved in apoptosis, cell-cycle regulation, and transcription are shown in Figure 3A-C. None of the other categories thought to be likely candidates for association with the molecular pathology of ET—including growth factors, growth factors receptors, and tumor suppressors—contained increased/decreased genes (data not shown).
Functional classification of differentially expressed genes. Genes are broadly grouped into selected, known functional categories: apoptosis regulators (A), cell-cycle control (B), and transcription factors (C). The X-axis refers to the signal log ratio (SLR), representing the log2 of the average change in the expression level of a transcript between ET and donor MK pools. Positive/negative SLRs denote increases/decreases in expression in ET MKs, and an SLR of 1 corresponds to a 2-fold change. (D) Expression of selected genes was also monitored by quantitative RT-PCR (gray histograms) as well as by the array (black histograms). On the x-axis, RT-PCR values are expressed as log2 of the RQ (relative quantity) in order to allow comparison with the microarray SLRs.
Functional classification of differentially expressed genes. Genes are broadly grouped into selected, known functional categories: apoptosis regulators (A), cell-cycle control (B), and transcription factors (C). The X-axis refers to the signal log ratio (SLR), representing the log2 of the average change in the expression level of a transcript between ET and donor MK pools. Positive/negative SLRs denote increases/decreases in expression in ET MKs, and an SLR of 1 corresponds to a 2-fold change. (D) Expression of selected genes was also monitored by quantitative RT-PCR (gray histograms) as well as by the array (black histograms). On the x-axis, RT-PCR values are expressed as log2 of the RQ (relative quantity) in order to allow comparison with the microarray SLRs.
Where possible, gene expression was confirmed by quantitative RT-PCR. As can be seen from Figure 3D, the expression levels of most of the analyzed genes were broadly similar to those observed in the microarray assay. Among the major apoptosis-related genes, BNIP3L showed decreased expression both at quantitative RT-PCR and microarray analysis. However, the significant decrease of BAX at microarray analysis was not confirmed at quantitative RT-PCR, where a slight increase was recorded.
Functional classification of the genes “specific” to ET/normal MKs. Those genes specific to ET MKs (n = 10) and to normal MKs (n = 9) are listed according to their molecular function in Table 3 (functional clusters were not apparent for all the genes). Where possible, the expression patterns were also evaluated at quantitative RT-PCR. The results are consistent with array data (Table 3).
Functional classification of the specific genes
Description . | GenBank accession no. . | Quantitative RT-PCR SLR . |
|---|---|---|
| De novo-expressed genes in ET MKs | ||
| Enzymes | ||
| Phospholipase A2, group IVC (cytosolic, calcium-independent) | AF065214.1 | 2.9 |
| Receptors | ||
| CD14 antigen | NM_000591.1 | 4.1 |
| Gamma-aminobutyric acid (GABA) B receptor, 1 | NM_001470.1 | 2.7 |
| Tyrosine kinase with immunoglobulin and epidermal growth factor homology domains | NM_005424.1 | 2.2 |
| Transcription regulators | ||
| Chromodomain helicase DNA binding protein 3 | BE379542 | 1.8 |
| Others | ||
| Seven in absentia homolog 1 (Drosophila) | U70056 | NA |
| Splicing factor, arginine/serine-rich 2, interacting protein | AI984932 | NA |
| Tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | NM_000362.2 | NA |
| KIAA0285 gene product | R61539 | NA |
| KIAA0542 gene product | AB011114.2 | NA |
| Absent genes in ET MKs present in normal counterparts | ||
| Enzymes | ||
| Tryptase beta 1 | AF206665.1 | NA |
| Tryptase beta 2 | NM_024164.2 | NA |
| Transcription factors | ||
| HIF-1 responsive RTP801 | NM_019058.1 | -2.7 |
| Creatin transporters | ||
| Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | NM_005629.1 | -2.5 |
| Others | ||
| Rag D protein | AF272036.1 | NA |
| Tubulin beta-5 | BC002654.1 | NA |
| Centaurin, beta 1 | NM_014716.1 | NA |
| ESTs, Weakly similar to N-WASP (H sapiens) | BF968134 | NA |
| Consensus includes AF099143/DEF = Homo sapiens mast cell tryptase beta III gene, complete cds | AF099143 | NA |
Description . | GenBank accession no. . | Quantitative RT-PCR SLR . |
|---|---|---|
| De novo-expressed genes in ET MKs | ||
| Enzymes | ||
| Phospholipase A2, group IVC (cytosolic, calcium-independent) | AF065214.1 | 2.9 |
| Receptors | ||
| CD14 antigen | NM_000591.1 | 4.1 |
| Gamma-aminobutyric acid (GABA) B receptor, 1 | NM_001470.1 | 2.7 |
| Tyrosine kinase with immunoglobulin and epidermal growth factor homology domains | NM_005424.1 | 2.2 |
| Transcription regulators | ||
| Chromodomain helicase DNA binding protein 3 | BE379542 | 1.8 |
| Others | ||
| Seven in absentia homolog 1 (Drosophila) | U70056 | NA |
| Splicing factor, arginine/serine-rich 2, interacting protein | AI984932 | NA |
| Tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | NM_000362.2 | NA |
| KIAA0285 gene product | R61539 | NA |
| KIAA0542 gene product | AB011114.2 | NA |
| Absent genes in ET MKs present in normal counterparts | ||
| Enzymes | ||
| Tryptase beta 1 | AF206665.1 | NA |
| Tryptase beta 2 | NM_024164.2 | NA |
| Transcription factors | ||
| HIF-1 responsive RTP801 | NM_019058.1 | -2.7 |
| Creatin transporters | ||
| Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | NM_005629.1 | -2.5 |
| Others | ||
| Rag D protein | AF272036.1 | NA |
| Tubulin beta-5 | BC002654.1 | NA |
| Centaurin, beta 1 | NM_014716.1 | NA |
| ESTs, Weakly similar to N-WASP (H sapiens) | BF968134 | NA |
| Consensus includes AF099143/DEF = Homo sapiens mast cell tryptase beta III gene, complete cds | AF099143 | NA |
NA indicates not applicable.
Expression of genes involved in MK development. We also analyzed expression levels of genes known to play a role in MK development (Table 4). A wide range of signal intensity levels was recorded among the various genes in normal MKs. Notably, genes encoding for structural proteins like GPIB (glycoprotein Ib, alpha polypeptide), GPIIb (integrin alpha 2b), and CD9, which all play a pivotal role in MKs and platelet function,24-27 were highly expressed in normal MKs. As regards some essential regulators of distinct stages in MK differentiation,13 NF-E2 (nuclear factor-erythroid 2) showed high expression levels, whereas GATA 1, FOG-1 (friend of GATA 1), and Fli-1 (friend leukemia virus integration 1) did not. The high expression of JUND, Mcl1 (myeloid cell leukemia sequence 1), and TAL-1 (T-cell acute lymphocytic leukemia 1) in normal MKs has never been described before. Genes encoding for the chemokine PF4 (platelet factor 4), PBP (proplatelet basic protein), as well as the CXCR4 (chemokine receptor 4) were also highly expressed.
Expression levels of genes important for MK development
Description . | GenBank accession no. . | Normal MK signal . | ET MK signal . | SLR . | Quantitative RT-PCR SLR . |
|---|---|---|---|---|---|
| Selected proteins | |||||
| Glycoprotein Ib (platelet), alpha polypeptide | NM_000173.1 | 3028.7, P | 3630.25, P | 0.28, NC | 1.5 |
| Glycoprotein VI (platelet) | AB043821.1 | 314.9, P | 381.1, P | 0.22, NC | NA |
| Glycoprotein IX (platelet) | NM_000174.1 | 983.3, P | 619.7, P | -0.54, NC | NA |
| Integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41B) | NM_000419.2 | 3829.7, P | 4136.4, P | 0.33, NC | 1.1 |
| Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | AI189839 | 469.9, P | 288.8, P | -0.38, NC | 1.5 |
| CD9 antigen (p24) | NM_001769.1 | 2652.9, P | 2360, P | -0.17, NC | NA |
| Inhibitor | |||||
| Transforming growth factor, beta 1 (Camurati-Engelmann disease) | BC000125.1 | 2953.9, P | 3440.9, P | 0.2, NC | 1.1 |
| Growth factors | |||||
| Interleukin 1, beta | M15330 | 208.4, P | 154.6, P | -0.3, NC | NA |
| Interleukin 1, beta | NM_000576.1 | 329.4, P | 285, P | -0.27, NC | NA |
| Interleukin 6 (interferon, beta 2) | NM_000600.1 | —, A | 16.9, P | -0.11, NC | NA |
| Interleukin 11 | NM_000641.1 | —, A | —, A | —, NC | NA |
| Platelet-derived growth factor alpha polypeptide | NM_002607.1 | 223.9, P | 267.5, P | 0.4, I | 1.5 |
| Platelet-derived growth factor beta polypeptide | AK022920.1 | 21.7, P | 23.7, P | -0.26, NC | NA |
| Platelet-derived growth factor C | NM_016205.1 | 651.6, P | 708.8, P | 0.21, NC | 1.4 |
| Vascular endothelial growth factor | AF022375.1 | 568.6, P | 467.2, P | -0.32, NC | NA |
| Growth factors receptors | |||||
| Interleukin 1 receptor | NM_000877.1 | —, A | —, A | —, NC | NA |
| Interleukin 3 receptor | NM_002183.1 | —, A | —, A | —, NC | NA |
| Interleukin 6 receptor | NM_000565.1 | 67.1, P | 43.2, P | -0.69, NC | 0.1 |
| Interleukin 11 receptor | NM_004512.1 | —, A | —, A | —, NC | NA |
| Myeloproliferative leukemia virus oncogene | NM_005373.1 | 474.8, P | 554.7, P | -0.01, NC | NA |
| V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | NM_000222.1 | 223.7, P | 165.1, P | -0.9, D | NA |
| Chemokine (C-X-C motif) receptor 4 | AJ224869 | 1505.5, P | 957.7, P | -0.48, NC | 1.0 |
| Thromboxane A2 receptor | D38081 | 653.6, P | 690.8, P | 0.38, NC | 1.5 |
| Cell cycle regulators | |||||
| Cyclin B1 | BE407516 | 128.6, P | 159.5, P | 0.15, NC | NA |
| Cyclin D3 | NM_001760.1 | 3247.9, P | 3241.9, P | 0, NC | 0.7 |
| Cyclin E1 | AI671049 | 31.1, P | —, A | -0.52, NC | NA |
| Cyclin E2 | NM_004702.1 | 44.7, P | 34, P | -0.24, NC | NA |
| Cell division cycle 2, G1 to S and G2 to M | AL524035 | 287, P | 136.8, P | -0.65, D | NA |
| Cyclin-dependent kinase 2 | M68520.1 | 240.9, P | 172.4, P | -0.12, NC | NA |
| Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | BC001971.1 | 309, P | 232.5, P | -0.44, NC | NA |
| RAS p21 protein activator (GTPase activating protein) 1 | M23612.1 | 339.9, P | 271.2, P | -0.45, D | NA |
| Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4 | NM_000077.1 | —, A | —, A | —, NC | NA |
| RAS guanyl releasing protein 2 (calcium and DAG-regulated) | NM_005825.1 | 637.9, P | 868.2, P | 0.36, NC | 1.6 |
| Apoptosis regulators | |||||
| BCL2-antagonist of cell death | U66879 | 37.6, P | 23, A | -0.02, NC | NA |
| BCL2-antagonist/killer 1 | NM_001188.1 | 66.2, P | 82.1, P | -0.13, NC | NA |
| BCL2-like 2 | D87461.1 | 112, P | 174.2, P | 0.42, NC | NA |
| B-cell CLL/lymphoma 2 | NM_000633.1 | 16.6, P | 21.1, P | 0.3, NC | NA |
| Caspase-3, apoptosis-related cysteine protease | NM_004346.1 | 115.3, P | 155.4, P | 0.53, I | 1.7 |
| Caspase-9, apoptosis-related cysteine protease | U60521.1 | 171.8, P | 196.5, P | 0.07, NC | 0.9 |
| Myeloid cell leukemia sequence 1 (BCL2-related) | NM_021960.1 | 874.6, P | 1078.7, P | 0.3, NC | 1.5 |
| Transcriptional regulators | |||||
| Friend leukemia virus integration 1 | NM_002017.2 | 538.1, P | 322, P | -0.52, D | NA |
| GATA binding protein 1 (globin transcription factor 1) | M30601.1 | 606, P | 677.6, P | 0.18, NC | NA |
| GATA binding protein 2 | NM_002050.1 | 67.7, P | 83.2, P | -0.09, NC | NA |
| Jumonji homolog (mouse) | BG029530 | 1139.2, P | 911.1, P | 0.19, NC | NA |
| Jun D proto-oncogene | NM_005354.2 | 2139.5, P | 2371.8, P | 0.24, NC | 1.1 |
| V-jun sarcoma virus 17 oncogene homolog (avian) | BE327172 | 8.45, P | 27.6, P | 1.26, NC | NA |
| Kruppel-like factor 1 (erythroid) | U65404.1 | 96, P | 83, P | 0.05, NC | NA |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) | NM_002359.1 | 299.5, P | 286.9, P | 0.39, NC | NA |
| V-myb myeloblastosis viral oncogene homolog (avian) | NM_005375.1 | 85.2, P | 96.35, P | -0.16, NC | NA |
| V-myc myelocytomatosis viral oncogene homolog (avian) | NM_002467.1 | 317.6, P | 353.6, P | 0.42, NC | NA |
| V-fos FBJ murine osteosarcoma viral oncogene homolog | BC004490.1 | —, A | —, A | —, NC | 0.0 |
| Nuclear factor (erythroid-derived 2), 45 kDa | L13974.1 | 1386.8, P | 1419.3, P | 0.03, NC | 1.1 |
| T-cell acute lymphocytic leukemia 1 | NM_003189.1 | 1873.2, P | 1763.8, P | 0.03, NC | 1.1 |
| Ecotropic viral integration site 1 | S82592.1 | —, A | —, A | —, NC | NA |
| Core-binding factor, runt domain, alpha subunit 2; translocated to 3 | NM_005187.2 | 234.7, P | 377.1, P | 0.57, I | 1.6 |
| Core-binding factor, runt domain, alpha subunit 2; translocated to 2 | NM_005093.1 | 69.1, P | 80.3, P | 0.33, NC | NA |
| Chemokines | |||||
| GRO1 oncogene (melanoma growth stimulating activity, alpha) | NM_001511.1 | 20.65, P | 157.8, P | 2.33, I | 3.2 |
| Stromal cell-derived factor 1 | U19495.1 | 30.8, P | 62.9, P | 0.5, I | 2.6 |
| Interleukin 8 | NM_000584.1 | 435.8, P | 1601.3, P | 1.87, I | 3.9 |
| Proplatelet basic protein | R64130 | 4476.1, P | 4161.7, P | 0.06, NC | 0.7 |
| Small inducible cytokine A5 (Rantes) | M21121 | 302.1, P | 524.3, P | 0.44, NC | NA |
| Platelet factor 4 | NM_002619.1 | 6642.2, P | 7639.8, P | 0.12, NC | 0.7 |
| Signal transducers | |||||
| Signal transducer and activator of transcription | M97935 | 148.8, P | 132, P | 0.05, NC | NA |
| Signal transducer and activator of transcription 3 | BC000627.1 | 503.3, P | 375.2, P | -0.46, NC | NA |
| Signal transducer and activator of transcription 5A | NM_003152.1 | 229.9, P | 248.5, P | 0.11, NC | NA |
| Mitogen-activated protein kinase 14 | NM_001315.1 | 681, P | 535.2, P | -0.57, D | 0.9 |
| Janus kinase 2 (a protein tyrosine kinase) | NM_004972.2 | 45.5, P | 34.6, P | -0.08, NC | NA |
| Phosphoinositide-3-kinase, class 2, beta polypeptide | NM_002646.1 | —, A | —, A | —, NC | NA |
Description . | GenBank accession no. . | Normal MK signal . | ET MK signal . | SLR . | Quantitative RT-PCR SLR . |
|---|---|---|---|---|---|
| Selected proteins | |||||
| Glycoprotein Ib (platelet), alpha polypeptide | NM_000173.1 | 3028.7, P | 3630.25, P | 0.28, NC | 1.5 |
| Glycoprotein VI (platelet) | AB043821.1 | 314.9, P | 381.1, P | 0.22, NC | NA |
| Glycoprotein IX (platelet) | NM_000174.1 | 983.3, P | 619.7, P | -0.54, NC | NA |
| Integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41B) | NM_000419.2 | 3829.7, P | 4136.4, P | 0.33, NC | 1.1 |
| Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | AI189839 | 469.9, P | 288.8, P | -0.38, NC | 1.5 |
| CD9 antigen (p24) | NM_001769.1 | 2652.9, P | 2360, P | -0.17, NC | NA |
| Inhibitor | |||||
| Transforming growth factor, beta 1 (Camurati-Engelmann disease) | BC000125.1 | 2953.9, P | 3440.9, P | 0.2, NC | 1.1 |
| Growth factors | |||||
| Interleukin 1, beta | M15330 | 208.4, P | 154.6, P | -0.3, NC | NA |
| Interleukin 1, beta | NM_000576.1 | 329.4, P | 285, P | -0.27, NC | NA |
| Interleukin 6 (interferon, beta 2) | NM_000600.1 | —, A | 16.9, P | -0.11, NC | NA |
| Interleukin 11 | NM_000641.1 | —, A | —, A | —, NC | NA |
| Platelet-derived growth factor alpha polypeptide | NM_002607.1 | 223.9, P | 267.5, P | 0.4, I | 1.5 |
| Platelet-derived growth factor beta polypeptide | AK022920.1 | 21.7, P | 23.7, P | -0.26, NC | NA |
| Platelet-derived growth factor C | NM_016205.1 | 651.6, P | 708.8, P | 0.21, NC | 1.4 |
| Vascular endothelial growth factor | AF022375.1 | 568.6, P | 467.2, P | -0.32, NC | NA |
| Growth factors receptors | |||||
| Interleukin 1 receptor | NM_000877.1 | —, A | —, A | —, NC | NA |
| Interleukin 3 receptor | NM_002183.1 | —, A | —, A | —, NC | NA |
| Interleukin 6 receptor | NM_000565.1 | 67.1, P | 43.2, P | -0.69, NC | 0.1 |
| Interleukin 11 receptor | NM_004512.1 | —, A | —, A | —, NC | NA |
| Myeloproliferative leukemia virus oncogene | NM_005373.1 | 474.8, P | 554.7, P | -0.01, NC | NA |
| V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | NM_000222.1 | 223.7, P | 165.1, P | -0.9, D | NA |
| Chemokine (C-X-C motif) receptor 4 | AJ224869 | 1505.5, P | 957.7, P | -0.48, NC | 1.0 |
| Thromboxane A2 receptor | D38081 | 653.6, P | 690.8, P | 0.38, NC | 1.5 |
| Cell cycle regulators | |||||
| Cyclin B1 | BE407516 | 128.6, P | 159.5, P | 0.15, NC | NA |
| Cyclin D3 | NM_001760.1 | 3247.9, P | 3241.9, P | 0, NC | 0.7 |
| Cyclin E1 | AI671049 | 31.1, P | —, A | -0.52, NC | NA |
| Cyclin E2 | NM_004702.1 | 44.7, P | 34, P | -0.24, NC | NA |
| Cell division cycle 2, G1 to S and G2 to M | AL524035 | 287, P | 136.8, P | -0.65, D | NA |
| Cyclin-dependent kinase 2 | M68520.1 | 240.9, P | 172.4, P | -0.12, NC | NA |
| Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | BC001971.1 | 309, P | 232.5, P | -0.44, NC | NA |
| RAS p21 protein activator (GTPase activating protein) 1 | M23612.1 | 339.9, P | 271.2, P | -0.45, D | NA |
| Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4 | NM_000077.1 | —, A | —, A | —, NC | NA |
| RAS guanyl releasing protein 2 (calcium and DAG-regulated) | NM_005825.1 | 637.9, P | 868.2, P | 0.36, NC | 1.6 |
| Apoptosis regulators | |||||
| BCL2-antagonist of cell death | U66879 | 37.6, P | 23, A | -0.02, NC | NA |
| BCL2-antagonist/killer 1 | NM_001188.1 | 66.2, P | 82.1, P | -0.13, NC | NA |
| BCL2-like 2 | D87461.1 | 112, P | 174.2, P | 0.42, NC | NA |
| B-cell CLL/lymphoma 2 | NM_000633.1 | 16.6, P | 21.1, P | 0.3, NC | NA |
| Caspase-3, apoptosis-related cysteine protease | NM_004346.1 | 115.3, P | 155.4, P | 0.53, I | 1.7 |
| Caspase-9, apoptosis-related cysteine protease | U60521.1 | 171.8, P | 196.5, P | 0.07, NC | 0.9 |
| Myeloid cell leukemia sequence 1 (BCL2-related) | NM_021960.1 | 874.6, P | 1078.7, P | 0.3, NC | 1.5 |
| Transcriptional regulators | |||||
| Friend leukemia virus integration 1 | NM_002017.2 | 538.1, P | 322, P | -0.52, D | NA |
| GATA binding protein 1 (globin transcription factor 1) | M30601.1 | 606, P | 677.6, P | 0.18, NC | NA |
| GATA binding protein 2 | NM_002050.1 | 67.7, P | 83.2, P | -0.09, NC | NA |
| Jumonji homolog (mouse) | BG029530 | 1139.2, P | 911.1, P | 0.19, NC | NA |
| Jun D proto-oncogene | NM_005354.2 | 2139.5, P | 2371.8, P | 0.24, NC | 1.1 |
| V-jun sarcoma virus 17 oncogene homolog (avian) | BE327172 | 8.45, P | 27.6, P | 1.26, NC | NA |
| Kruppel-like factor 1 (erythroid) | U65404.1 | 96, P | 83, P | 0.05, NC | NA |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) | NM_002359.1 | 299.5, P | 286.9, P | 0.39, NC | NA |
| V-myb myeloblastosis viral oncogene homolog (avian) | NM_005375.1 | 85.2, P | 96.35, P | -0.16, NC | NA |
| V-myc myelocytomatosis viral oncogene homolog (avian) | NM_002467.1 | 317.6, P | 353.6, P | 0.42, NC | NA |
| V-fos FBJ murine osteosarcoma viral oncogene homolog | BC004490.1 | —, A | —, A | —, NC | 0.0 |
| Nuclear factor (erythroid-derived 2), 45 kDa | L13974.1 | 1386.8, P | 1419.3, P | 0.03, NC | 1.1 |
| T-cell acute lymphocytic leukemia 1 | NM_003189.1 | 1873.2, P | 1763.8, P | 0.03, NC | 1.1 |
| Ecotropic viral integration site 1 | S82592.1 | —, A | —, A | —, NC | NA |
| Core-binding factor, runt domain, alpha subunit 2; translocated to 3 | NM_005187.2 | 234.7, P | 377.1, P | 0.57, I | 1.6 |
| Core-binding factor, runt domain, alpha subunit 2; translocated to 2 | NM_005093.1 | 69.1, P | 80.3, P | 0.33, NC | NA |
| Chemokines | |||||
| GRO1 oncogene (melanoma growth stimulating activity, alpha) | NM_001511.1 | 20.65, P | 157.8, P | 2.33, I | 3.2 |
| Stromal cell-derived factor 1 | U19495.1 | 30.8, P | 62.9, P | 0.5, I | 2.6 |
| Interleukin 8 | NM_000584.1 | 435.8, P | 1601.3, P | 1.87, I | 3.9 |
| Proplatelet basic protein | R64130 | 4476.1, P | 4161.7, P | 0.06, NC | 0.7 |
| Small inducible cytokine A5 (Rantes) | M21121 | 302.1, P | 524.3, P | 0.44, NC | NA |
| Platelet factor 4 | NM_002619.1 | 6642.2, P | 7639.8, P | 0.12, NC | 0.7 |
| Signal transducers | |||||
| Signal transducer and activator of transcription | M97935 | 148.8, P | 132, P | 0.05, NC | NA |
| Signal transducer and activator of transcription 3 | BC000627.1 | 503.3, P | 375.2, P | -0.46, NC | NA |
| Signal transducer and activator of transcription 5A | NM_003152.1 | 229.9, P | 248.5, P | 0.11, NC | NA |
| Mitogen-activated protein kinase 14 | NM_001315.1 | 681, P | 535.2, P | -0.57, D | 0.9 |
| Janus kinase 2 (a protein tyrosine kinase) | NM_004972.2 | 45.5, P | 34.6, P | -0.08, NC | NA |
| Phosphoinositide-3-kinase, class 2, beta polypeptide | NM_002646.1 | —, A | —, A | —, NC | NA |
P indicates present; NC, not changed; NA, not applicable; A, absent; I, increased; D, decreased; and —, absent gene showing aspecific signal.
The SLRs indicate that the expression of the majority of the genes, listed in Table 4, was not changed in malignant MKs relative to their normal counterparts. Interestingly, only 2 genes showed increased expression in ET MKs, namely IL-8 (interleukin 8) and the GRO1 oncogene (bold in Table 4). Remarkably, expression of the TPO receptor gene (myeloproliferative leukemia virus oncogene [Mpl]), which is thought to be implicated in the pathogenesis of ET,1,3 remained unchanged in malignant MKs. Quantitative RT-PCR confirmed the array SLRs in the majority of the genes studied (Table 4). Of the 24 genes, 4, namely ITGB3 (integrin, beta3), IL6R (interleukin 6 receptor), CXCR4, and MAPK14 (mitogen/activated protein kinase 14), turned out to be increased in RT-PCR but not changed in array analysis, since TaqMan analysis can show a greater dynamic range in testing differences in gene expression.
Gene profiling of normal platelets has already been studied.28 We found that more than 90% of the reported top-50 platelet genes were also highly expressed both in normal and ET MKs, and showed similar levels of expression in the 2 cell populations (Table S1; see the Supplemental Table link at the top of the online article on the Blood website).
Protein analysis of selected genes
To further confirm the gene-profiling data and in order to investigate whether the expression of genes in MKs mirrors that found in platelets, we used flow cytometry to analyze expression of selected proteins in platelets from healthy subjects and patients with ET. Protein analysis broadly confirmed the gene expression results. As regards de novo–expressed genes, the CD14 protein was not expressed on the extracellular platelet membrane of either normal or ET platelets; however, low intracellular levels of CD14 were found in ET, but not normal, platelets (Figure 4A). To confirm this finding, we analyzed CD14 expression in a megakaryoblastic cell line (MK-1)29 : this cell line was markedly positive for CD14 with an intracellular distribution (Figure 4B). Among the differentially expressed genes, we analyzed protein levels of BAX and IGF1R because the former induces a proapoptotic signal,30 while the latter is associated with an increased cell-survival signal in hematopoietic cells.31 BAX protein expression was markedly reduced in ET platelets compared with their normal counterparts (Figure 4C), whereas IGF1R protein expression was increased (Figure 4D).
Protein analysis of CD14, BAX, and IGF1R in normal and ET platelets. (A) Intracellular CD14 expression was slightly increased in ET platelets (solid line) relative to normal platelets (dotted line), as shown by shifting of the curves to the right; staining was performed with anti-CD14 MoAb directly conjugated to phycoerythrin. (B) Intracellular CD14 expression was also slightly raised in the MK1 megakaryoblastic cell line (black area) relative to the isotype-matched MoAb-treated negative control (white area). (C) Intracellular BAX expression was markedly reduced in ET platelets (solid line) relative to their normal counterparts (dotted line). The cells were stained with BAX antibody followed by a fluorescein-conjugated goat anti–mouse immunoglobulin. (D) Membrane IGF1R expression was increased in ET platelets (solid line) relative to their normal counterparts (dotted line). Cells were labeled with IGF1R antibody followed by a phycoerythrin-conjugated goat anti–mouse immunoglobulin. The x-axis represents the relative fluorescence intensity; the y-axis, the relative number of cells. Results of 1 representative experiment (from 5 separate experiments) are shown for each protein.
Protein analysis of CD14, BAX, and IGF1R in normal and ET platelets. (A) Intracellular CD14 expression was slightly increased in ET platelets (solid line) relative to normal platelets (dotted line), as shown by shifting of the curves to the right; staining was performed with anti-CD14 MoAb directly conjugated to phycoerythrin. (B) Intracellular CD14 expression was also slightly raised in the MK1 megakaryoblastic cell line (black area) relative to the isotype-matched MoAb-treated negative control (white area). (C) Intracellular BAX expression was markedly reduced in ET platelets (solid line) relative to their normal counterparts (dotted line). The cells were stained with BAX antibody followed by a fluorescein-conjugated goat anti–mouse immunoglobulin. (D) Membrane IGF1R expression was increased in ET platelets (solid line) relative to their normal counterparts (dotted line). Cells were labeled with IGF1R antibody followed by a phycoerythrin-conjugated goat anti–mouse immunoglobulin. The x-axis represents the relative fluorescence intensity; the y-axis, the relative number of cells. Results of 1 representative experiment (from 5 separate experiments) are shown for each protein.
Characterization of apoptosis in normal and ET MKs
Since microarray analysis revealed impaired expression of genes involved in the regulation of the apoptotic pathway in ET MKs, we also performed functional assays to evaluate the biologic consequences.
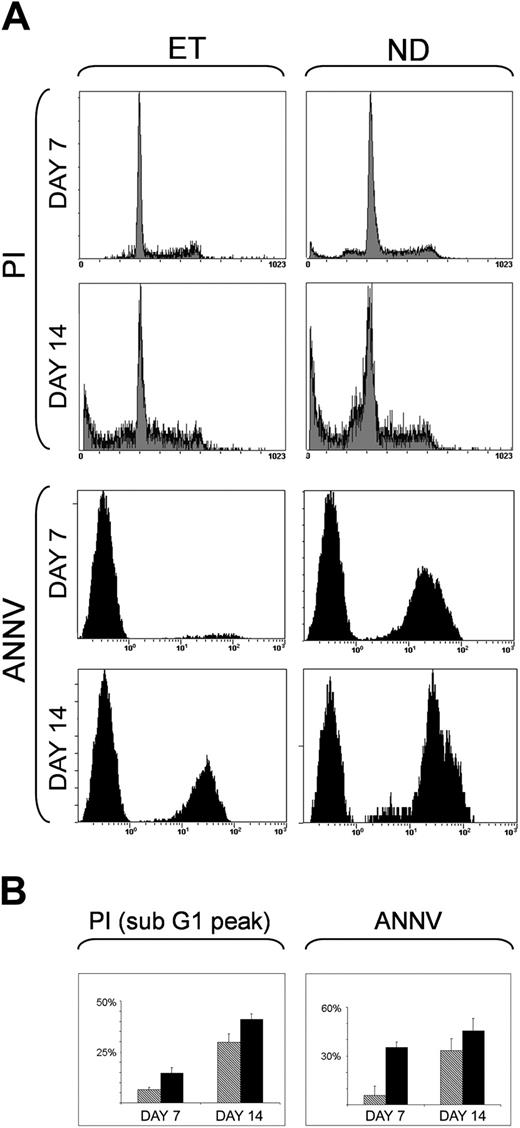
We first characterized apoptosis of normal and ET MKs in serum-free culture at 7, 14, and 21 days after stimulation with 100 ng/mL TPO by means of both PI and annexin-V staining, as evaluated by flow cytometry (Figure 5). The mean percentages of sub-G1 peak apoptosis after PI staining were significantly decreased in ET MKs relative to normal cells, both after 7 days (6.5 ± 1.2% vs 14.61 ± 2.6%; P < .04) and 14 days (29.72% ± 4.2% vs 40.9% ± 2.7%; P < .03). At annexin-V staining, significant reductions in apoptosis were also found in ET MKs after 7 days (5.8 ± 5.6% vs 35.33 ± 3.43%; P < .02) and 14 days (33.36 ± 4.5 vs 45.46 ± 4.8; P < .05). At 21 days, apoptosis was similar in the 2 populations at PI/annexin-V staining (data not shown).
Apoptosis of normal and ET MK cells after 7 and 14 days of culture with 100 ng/mL TPO. Apoptosis in normal donors (ND) and ET MKs was quantified by PI and annexin-V–FITC stainings after 7 and 14 days of culture. (A) The results of 1 representative experiment, the x-axis representing the relative fluorescence intensity and the y-axis, the relative number of cells. (B) Histograms showing mean ± SD percentages of positive ET ( ; 5 cases) and normal (▪; 6 cases) MKs for PI (sub-G1 peak DNA evaluation) and annexin-V–FITC.
; 5 cases) and normal (▪; 6 cases) MKs for PI (sub-G1 peak DNA evaluation) and annexin-V–FITC.
Apoptosis of normal and ET MK cells after 7 and 14 days of culture with 100 ng/mL TPO. Apoptosis in normal donors (ND) and ET MKs was quantified by PI and annexin-V–FITC stainings after 7 and 14 days of culture. (A) The results of 1 representative experiment, the x-axis representing the relative fluorescence intensity and the y-axis, the relative number of cells. (B) Histograms showing mean ± SD percentages of positive ET ( ; 5 cases) and normal (▪; 6 cases) MKs for PI (sub-G1 peak DNA evaluation) and annexin-V–FITC.
; 5 cases) and normal (▪; 6 cases) MKs for PI (sub-G1 peak DNA evaluation) and annexin-V–FITC.
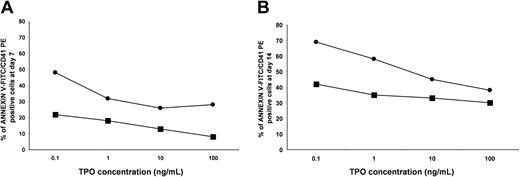
To confirm these results, in parallel experiments, we assayed the effects of various concentrations of TPO (0.1, 1, 10, 100 ng/mL) on the apoptosis (annexin-V staining) of normal and ET MKs after 7 and 14 days of liquid culture. As shown in Figure 6, ET MKs showed markedly increased resistance to apoptosis relative to their normal counterparts; at all concentrations, the percentage of apoptotic MKs was greater in normal MKs. This pattern was documented both in immature MKs (day 7; Figure 6A) and in more mature cells (day 14; Figure 6B).
Apoptosis of normal and ET MKs relative to different TPO concentrations. CD34+ cells from 3 healthy subjects (•) and 3 patients with ET (▪) were cultured with various concentrations of TPO in serum-free conditions. After 7 (A) and 14 (B) days of liquid culture, cells were dual-labeled with annexin-V–FITC/CD41-PE. The percentages of apoptotic MKs from patients with ET were compared with those of healthy subjects after incubation with 0.1, 1, 10, and 100 ng/mL of TPO. Data are expressed as means (standard deviations were always within 6% of the means).
Apoptosis of normal and ET MKs relative to different TPO concentrations. CD34+ cells from 3 healthy subjects (•) and 3 patients with ET (▪) were cultured with various concentrations of TPO in serum-free conditions. After 7 (A) and 14 (B) days of liquid culture, cells were dual-labeled with annexin-V–FITC/CD41-PE. The percentages of apoptotic MKs from patients with ET were compared with those of healthy subjects after incubation with 0.1, 1, 10, and 100 ng/mL of TPO. Data are expressed as means (standard deviations were always within 6% of the means).
Discussion
Malignant megakaryocytopoiesis is the hallmark of ET. Numerous studies have characterized the in vitro growth of the malignant MK progenitors,32 with spontaneous MK growth observed in the majority of patients. This property does not appear to be due either to paracrine stimulation, as can be observed in limit dilutions,33 or to autocrine stimulation by TPO.34 Nevertheless, circulating MK progenitor cells from patients with ET are hypersensitive in vitro to TPO.34 Dysregulation of megakaryocytopoiesis in ET has been found to involve defective binding of TPO by platelets and MKs, due to reduced or abnormal MPL expression; this results in increased levels of plasma TPO and increased sensitivity of MKs to TPO, in turn leading to MK hyperproliferation.2,12,35,36 However, no mutations of the TPO gene have yet been identified in families with hereditary ET,37,38 and no mutations in the TPO receptor39 have been found in patients with acquired ET. New approaches are needed to better elucidate the mechanisms of malignant megakaryocytopoiesis in ET. To our knowledge, the present study provides the first microarray-based comparison of the molecular phenotypes of CD34-derived MKs taken from malignant BM of patients with ET and from healthy subjects.
Analysis of our microarray data reveals that the expression of certain genes involved in the apoptotic pathway is impaired in ET MKs (Tables 1-2). Activation of the apoptotic cell machinery in normal MKs is known to play a key role in MK development. Zauli et al40 showed that cultured MKs undergo apoptosis in vitro and that the terminal phase of the MK life span is characterized by onset of apoptosis. Subsequent reports document close relationships between apoptosis, MK maturation, and platelet release.41,42 However, no data have previously been reported on the mechanism of apoptosis in ET MKs.
In the present study, down-regulation was apparent in genes with proapoptotic activity, such as BAX, BNIP3, BNIP3L, and members of the mitochondrial PT pore complex—a composite channel that is involved in the regulation of mitochondrial membrane permeability during apoptosis.43-45 We also observed increased expression of the CFLAR (CASP8 and FADD-like apoptosis regulator), which exerts known antiapoptotic activity,46 along with reduced expression of LGALS1 (galectin 1), another proapoptotic factor47 (Figure 3). Moreover, IGF1R overexpression, which was also apparent in ET MKs, could play a role by deregulating the apoptotic process. IGF1R is overexpressed in most malignant tissues, where it functions as an antiapoptotic agent by enhancing cell survival48 (Figure 3). Interestingly, we also found increased expression of SDF1 (stromal cell-derived factor 1) in the malignant MKs (Table 4). Recently, Hodohara et al49 demonstrated that SDF1α exerts direct growth-promoting effect on MK progenitors, and Guerriero et al50 found that it increases ploidy of MKs. Since MK proliferation in ET seems to be partially autonomous,51 increased expression of SDF1 might provide a proliferative advantage to the malignant MKs.
To evaluate the biologic consequences of the impaired expression of genes involved in the regulation of the apoptotic pathway in ET MKs, we also performed functional assays such as phosphatidylserine exposure, and sub-G1 peak DNA and limiting dilutions assays of TPO in normal and ET MKs. Despite the limited magnitude of the up-/down-regulation of the genes involved in apoptosis (as recorded at microarray analysis and/or quantitative RT-PCR), the functional assays provided strong confirmation that ET MKs are indeed more resistant to apoptosis than their normal counterparts. Whether this decreased propensity to apoptosis plays a causative role in the genesis of ET MKs or is the consequence of the malignancy remains a matter of speculation.
Extension of these findings to in vivo megakaryocytopoiesis must be considered with particular caution. In fact, cytokines other than TPO and complex cell-cell and cell–BM-matrix interactions are known to play a primary role in the development of MKs in vivo. However, Zauli et al40 found that primary megakaryocytic cells undergo apoptotic cell death in culture more rapidly than CD34-derived MKs. Furthermore, there is increasing evidence that chronic myeloproliferative diseases are associated with perturbed apoptotic programs and increased cell-survival signals. In polycythemia vera, dysregulated expression of BCL-xL has been implicated in erythropoietin-independent survival of erythroid-lineage cells and could therefore play a role in the pathogenesis of the disease.52 Similarly, the FKBP51 gene is overexpressed in idiopathic myelofibrosis, where it could be partially responsible for the accumulation of MKs by down-regulation of their apoptotic program.53
We also performed a subanalysis of genes that are known to be involved in megakayocytopoiesis.54 As expected, the majority of these genes were expressed at high levels in both normal and malignant MKs. Recent advances have highlighted the particular importance of 4 transcriptional regulators—GATA-1, FOG-1, NF-E2, and Fli-1 (Friend leukemia virus integration 1)—in distinct stages of MK commitment and differentiation.13 However, our gene profiling analysis did not reveal significant differences in expression between normal and ET MKs for any of these 4 genes (Table 4). By contrast, we found overexpression of the JunB proto-oncogene and RUNX1 (runt-related transcription factor 1) in ET MKs. Up-regulation of JunB in ET MKs could be particularly interesting since evidence exists that the related genes, c-Jun and c-Fos, are strongly expressed by developing MKs,55,56 while in chickens v-Jun and v-ErbB cooperate in induction of transformation of normal BM progenitors into leukemic megakaryoblasts.57 As regards RUNX1, recent data suggest that this myeloid transcription factor is expressed in MKs, where it cooperates with GATA1 in MK commitment and differentiation,58 and that it seems to play a role in malignant megakaryocytopoiesis.59,60 Interestingly, we found that the IL-8 and GRO-1 genes were also overexpressed in ET MKs. The possible role of these genes in the malignant megakaryocytopoiesis of ET is unclear. Their production has recently been described in normal MKs,61 and, although the IL-8 cytokine is known to down-regulate proliferation of human myeloid and megakaryocytic progenitor cells,62 it may also function as an autocrine growth factor and apoptosis resistance factor.63
In conclusion, our results suggest that the expression of several genes, interacting to regulate the balance between proliferation and apoptosis, are altered in ET MKs compared with normal MKs. As a consequence, ET MKs show resistance to apoptosis. This functional finding could represent a relevant step in ET tumorigenesis.
Prepublished online as Blood First Edition Paper, July 22, 2004; DOI 10.1182/blood-2003-07-2597.
Supported by Fondi ex 60% 2002 (L.C.), Cofin 2003 (L.C.), by Fondazione del Monte di Bologna e Ravenna grants, and by AIL (Italian association against Leukemia: Bologna and Modena, Italy).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Robin M. T. Cooke for scientific editing. We gratefully acknowledge the generous support of the “Cassa di Risparmio Foundation of Modena” for the acquisition of the “Affymetrix system.”

![Figure 1. Analysis of morphology and αIIb-β3 expression of normal MKs. (A) Cytospins were observed at light microscopy after May-Grunwald staining (× 20 magnification, 0.50 aperture objective lenses [Carl Zeiss, Oberkochen, Germany]). Note the presence of cells belonging to various maturational stages of megakaryocytopoiesis. (B) αIIb-β3 expression (white area) of normal MK cells was examined with an anti-CD41a MoAb directly conjugated to phycoerythrin, and fluorescence was analyzed by flow cytometry. Cells treated with an isotype-matched irrelevant MoAb directly conjugated to phycoerythrin represent the negative control (black area). The x-axis indicates fluorescence intensity; the y-axis, relative number of cells. (C) Morphologic analysis of purified MKs. May-Grunwald staining (× 40 magnification, 1.0 aperture objective lenses [Carl Zeiss]). Note the presence of small megakaryoblasts near mature megakaryocytes. Morphology was evaluated with an Ultraphot universal photomicroscope (Carl Zeiss, Oberkochen, Germany). The camera was inserted within the microscope. No software was used to optimize the figure. Objective lenses 20 × and 40 ×.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/10/10.1182_blood-2003-07-2597/6/m_zh80220469300001.jpeg?Expires=1769101162&Signature=jDCHCNFnEmEVxKMOw4uTcNOSvWmujSWyGndUzZsITPPZIOAaqB28NxBpyIxHbiyxSHvkMPbQB5dRCCJ-tpR3kiaK0YCZEAQpKmJXxD49Ue5zAJ~TcpHcHTL3fLkIeRibu-GVzPYcswZjFd56wxHbswrDe~-GDaqnQyL6U5gLjnlVH8ILQfWthk1ncPjUj2S4dI423AdIU7Ap1s~MJhuOMRHMe5ccw7VpgoRjgUI0xdLGJ0ptU3kPttBupj2jsWPd~1oN7c2~6VxJNgygwHFJMKoqknxcR-e2BJwpmFTm2KGJoOVsTZSL8Qu53jfpJNfWrpytEv8S-0W78vwn7beokg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal