Abstract
We used a sensitive real-time reverse transcription–polymerase chain reaction assay to quantify cyclin D1 mRNA levels in bone marrow samples collected at diagnosis from 74 newly diagnosed multiple myeloma (MM) patients who were randomized to undergo either single or double autologous peripheral blood stem cell transplantation as part of first-line therapy for their malignancy. In 46 cases, fluorescence in situ hybridization (FISH) analysis and/or conventional cytogenetics were performed to detect chromosome 11 abnormalities. Patients with the t(11;14) or trisomy 11 significantly overexpressed cyclin D1 (P < .0001) in comparison with patients without 11q abnormalities, who had cyclin D1 mRNA levels similar to healthy donors. Overall, 32 (43%) of 74 patients showed cyclin D1 overexpression. No difference was found between cyclin D1–positive (group A) and cyclin D1–negative (group B) patients with respect to presenting clinical and laboratory characteristics, including chromosome 13 abnormalities, as well as to response to therapy and overall survival, both of which were calculated on an intent-to-treat basis. Patients who overexpressed cyclin D1 had significantly longer duration of remission in comparison with patients who did not (41 vs 26 months, respectively; P = .02). As a result, median event-free survival (EFS) was longer in group A than in group B (33 vs 24 months, respectively; P = .055). We concluded that cyclin D1 overexpression is closely associated with 11q abnormalities and identifies a subset of MM patients who are more likely to have prolonged duration of remission and EFS following autologous transplantation.
Introduction
Although no specific genetic lesions are regularly associated with multiple myeloma (MM), recent studies have shown that reciprocal translocations involving the immunoglobulin heavy chain (IgH) gene locus are recurrent chromosomal abnormalities both in human myeloma cell lines and in patients with MM.1-3 More specifically, 3 nonrandom partner loci have been identified: 11q13 (cyclin D1),4-6 4p16 (FGFR3 and MMSET),7,8 and 16q23 (c-maf).9 The t(11;14)(q13;q32) is detected in approximately 5% of patients by conventional cytogenetic analysis10-12 and in as many as 15% to 20% by interphase fluorescence in situ hybridization (FISH).13-15 The t(11;14) results in the up-regulation of the oncogene encoding cyclin D1, since it is brought in close proximity to the powerful Eμ enhancer of the IgH locus. However, cyclin D1 overexpression may also be caused by mechanisms other than the t(11;14), including gene amplification.16 Cyclin D1 expression in MM has been so far investigated by means of immunohistochemistry,17,18 Northern blot,19,20 and semiquantitative and quantitative reverse transcription–polymerase chain reaction (RT-PCR).21,22 Conflicting data about its exact frequency in de novo MM have been reported, with values ranging from 17% to 50%. Moreover, the prognostic significance of the t(11;14) and/or of cyclin D1 up-regulation is still controversial. Some studies have suggested that MM patients with either the t(11;14) or other 11q abnormalities, as detected by standard karyotype, and/or showing cyclin D1 gene amplification had a worsened prognosis.12,16,23 On the other hand, Fonseca et al14 recently reported that conventionally treated patients harboring the t(11;14), as detected by FISH, had a slightly longer overall survival (OS) in comparison with patients without this abnormality. These conclusions were subsequently extended and confirmed at a statistically significant level by Moreau et al15 in a series of patients treated with high-dose chemotherapy.
To shed further light on the frequency and prognostic significance of cyclin D1 overexpression in MM, we applied a sensitive and straightforward real-time RT-PCR assay to quantify cyclin D1 mRNA levels in a consecutive series of previously untreated patients who were randomly assigned to undergo either single or double autologous peripheral blood stem cell (PBSC) transplantation. We found that more than one third of patients overexpressed cyclin D1 and had a better outcome in terms of extended duration of remission and event-free survival (EFS) in comparison with patients who did not overexpress cyclin D1.
Patients, materials, and methods
Patients
The study was performed on a total of 74 patients with newly diagnosed MM. These patients were part of a larger series of patients who were enrolled in the “Bologna 96” clinical trial.24 Criteria for entry into the molecular study required the availability of bone marrow (BM) samples collected at diagnosis and stored for subsequent RT-PCR quantification of cyclin D1 mRNA expression. The main features at diagnosis of the 74 patients are summarized in Table 1 and were not different from the demographics of the entire series of patients (data not shown).
Patient clinical and laboratory characteristics
Characteristic . | . |
|---|---|
| No. of patients | 74 |
| Median age, y (range) | 52.5 (36-60) |
| Males:females, n | 52:22 |
| Durie & Salmon stage, n | |
| I | 14 |
| II | 12 |
| III | 48 |
| M component isotype, n | |
| IgG | 43 |
| IgA | 18 |
| BJ | 13 |
| L chain, n | |
| κ | 48 |
| λ | 26 |
| Median M component concentration | |
| IgG, g/L (range) | 41.0 (10.0-111.7) |
| IgA, g/L (range) | 38.0 (22.0-78.0) |
| BJ, g/24 h (range) | 4.00 (1.90-18.60) |
| Median BMPC, % (range) | 45 (10-100) |
| Median β2-m level, mg/L (range) | 2.40 (1.10-16.00) |
| Median CRP level, mg/L (range) | 3.10 (1.0-70.20) |
| Median creatinine level, μM (range) | 88.4 (61.88-265.2) |
Characteristic . | . |
|---|---|
| No. of patients | 74 |
| Median age, y (range) | 52.5 (36-60) |
| Males:females, n | 52:22 |
| Durie & Salmon stage, n | |
| I | 14 |
| II | 12 |
| III | 48 |
| M component isotype, n | |
| IgG | 43 |
| IgA | 18 |
| BJ | 13 |
| L chain, n | |
| κ | 48 |
| λ | 26 |
| Median M component concentration | |
| IgG, g/L (range) | 41.0 (10.0-111.7) |
| IgA, g/L (range) | 38.0 (22.0-78.0) |
| BJ, g/24 h (range) | 4.00 (1.90-18.60) |
| Median BMPC, % (range) | 45 (10-100) |
| Median β2-m level, mg/L (range) | 2.40 (1.10-16.00) |
| Median CRP level, mg/L (range) | 3.10 (1.0-70.20) |
| Median creatinine level, μM (range) | 88.4 (61.88-265.2) |
BJ indicates Bence Jones; BMPC, bone marrow plasma cells; β2-m, β2-microglobulin; and CRP, C-reactive protein.
All 74 patients had symptomatic MM and were enrolled in the “Bologna 96” phase 3 clinical trial designed to compare single autologous PBSC transplantation (arm A) versus double autologous PBSC transplantation (arm B) as first-line therapy for MM.24 In both arms of the study, the treatment plan included conventional remission induction chemotherapy with vincristine, adriamycin, and dexamethasone (VAD), followed by collection of PBSCs with high-dose cyclophosphamide (7 g/m2) and a subsequent course of PBSC-supported high-dose melphalan (MEL; 200 mg/m2). In arm B, MEL was followed within 3 to 6 months by a second autologous transplantation prepared with the combination of melphalan (120 mg/m2) and busulphan (12 mg/kg). Maintenance therapy with α-interferon was offered to all patients who underwent a single or double autologous transplantation. All patients provided signed informed consent for participation in the study. The study was approved by the ethics committees of the participating institutions.
Bone marrow samples
BM samples were obtained at the time of diagnosis from all the patients who entered the study. Mononuclear cells were obtained by Ficoll-Hypaque density gradient centrifugation and then stored at –80°C in guanidium thiocyanate until use.
Healthy controls and cell lines
For standardization procedures, we used BM samples collected from 10 healthy donors as negative controls and the following myeloma cell lines: KMS12, incorporating the t(11;14); U266, overexpressing cyclin D1; and KMS26, carrying trisomy 11.
RNA extraction
Total cellular RNA was obtained by phenol/chloroform extraction, isopropanol precipitation, and 70% ethanol washing, as previously described.25 RNA was spectrophotometrically quantified at 260 nm, and its integrity was assessed by electrophoresis on 2% agarose gel.
Real-time RT-PCR
As previously described, 1 μg total cellular RNA was reverse transcribed to cDNA in a 20 μL final volume containing 25 mM random examers as primers.26 Real-time RT-PCR was performed on an ABI PRISM 7700 Sequence Detector (Perkin Elmer, Foster City, CA). The principles and procedure of quantification using the TaqMan probe (Applied Biosystems, Foster City, CA) have been described.27 To compensate for differences in the RNA quality or RT efficacy, parallel TaqMan assays were run on each sample for the ABL housekeeping gene, and the absolute levels of cyclin D1 mRNA were normalized with respect to ABL mRNA content.
TaqMan probes and primers
For cyclin D1, we designed a forward primer positioned on exon 4 and a reverse primer on exon 5 (5′ to 3′ sequences, ACC TGA GGA GCC CCA ACA A and TCT GCT CCT GGC AGG CC, respectively); a probe was positioned on exon 4 (5′ to 3′ sequence, TCC TAC TAC CGC CTC ACA CGC TTC CTC). For ABL, a forward primer was positioned on exon 1a (5′ to 3′ sequence, TCC TCC AGC TGT TAT CTG GAA GA), and a reverse primer and probe were both positioned on exon 2 (5′ to 3′ sequences, TGG GTC CAG CGA GAA GGT T and CCA GTA GCA TCT GAC TTT GAG CCT CAG GG, respectively). TaqMan probes were labeled at the 5′ end with the reporter dye molecule FAM (6-carboxy-fluorescein), and at the 3′ end with the quencher dye molecule TAMRA (6-carboxy-tetramethylrhodamine). Primers and probes were all selected using Primer Express software (Perkin Elmer), according to the manufacturer's guidelines.
Real-time amplification
Reaction mixture (25 μL) contained 12.5 μL TaqMan buffer A with the ROX dye as the passive reference, 5 mM MgCl2, 200 μM deoxy adenosine triphosphate (dATP), deoxycytosine triphosphosphate (dCTP), deoxyguanosine triphosphate (dGTP), 400 μM deoxyuridine triphosphate (dUTP), 1.25 U AmpliTaq Gold DNA polymerase, 0.5 U AmpErase uracil N-glycosylase (UNG), 300 nM forward and reverse primers, 200 nM specific TaqMan probe, and 6 μL cDNA (diluted 1:3). All the reagents were purchased from Perkin Elmer/Applied Biosystems. After 2 minutes of incubation at 50°C to allow destruction by UNG of potential contaminant RT-PCR products, and 10 minutes at 95°C to denature UNG and activate AmpliTaq Gold, amplification was performed by 50 cycles at 95°C for 15 seconds followed by 50 cycles at 65°C for 60 seconds. Triplicate experiments were done for each sample, and threshold cycle (Ct) values were averaged. In accordance with the comparative Ct method,28 for each sample the average Ct value for the endogenous reference (the ABL housekeeping gene) was subtracted from the average Ct value for cyclin D1, to yield the ΔCt. The ΔCt value obtained from the KMS12 cell line (chosen as the calibrator) was then subtracted from the ΔCt value for each patient to give the ΔΔCt.
Conventional cytogenetic and FISH analyses
Conventional chromosome studies were performed at diagnosis on BM cells after a 3- to 5-day culture with interleukin-3 and interleukin-6. G banding with Wright stain was done for all karyotypes. At least 30 metaphases were examined for each patient. Chromosome abnormalities were defined according to the International System for Human Cytogenetic Nomenclature. FISH analyses were performed on cells obtained from cytogenetic analysis and kept frozen in suspension in Carnoy fixative (methanol–acetic acid, 3:1 [vol/vol]). The slides were prepared immediately before hybridization. For the detection of the t(11;14), we used 2 sets of probes: LSI IgH, which hybridizes to chromosome 14q32.3 (IgH Spectrum Green; Vysis, Downers Grove, IL), and CCND1, which hybridizes to chromosome 11q13 (CCND1 Spectrum Orange; Vysis). The normal FISH pattern was 2 pairs of nonassociated signals (2 red/2 green). Translocation was observed as fusions of 2 signals. FISH was performed according to the manufacturer's guide with some modifications. At least 300 cells were analyzed for each patient. Chromosome 13 abnormalities (monosomy and/or deletions, Δ13) were also investigated by FISH analysis using a D13S319-specific probe (Vysis) and/or by conventional cytogenetics.
Statistical analysis
To characterize patients in the study we used descriptive statistics. For continuous variables, the Mann-Whitney U test was used to test for differences between patients with and without cyclin D1 overexpression. Fisher exact or chi-square tests were used to test for differences among levels of categoric variables between patients with and without cyclin D1 overexpression. Response to therapy was established according to previously reported criteria.29 Disease progression was defined as follows: reappearance of M protein in the case of complete remission (CR); 25% or higher increase in M protein concentration from minimal tumor mass in the case of partial remission; 25% or higher increase in M protein concentration from baseline values in the case of nonresponse. Curves for OS, EFS, and time to relapse/progression were plotted according to the method of Kaplan and Meier starting from the time of first VAD. An event included relapse/progression of MM and death from any cause. The log-rank test was used to test for differences in survival among groups. All analyses were performed using the SPSS software package (SPSS, Chicago, IL).
Results
Sensitivity and reliability of the real-time RT-PCR set-up for quantification of cyclin D1 expression
Optimal amounts of forward and reverse primers were determined by means of the primer-matrix experiment. Tested in triplicate were 9 combinations of 50, 300, and 900 nM for each primer: 50/50, 50/300, 50/900, 300/50, 300/300, 300/900, 900/50, 900/300, and 900/900. The 300/300 nM primer combination was chosen for all the experiments, as it yielded the lowest Ct.
A validation experiment was run to confirm that the efficiencies of the target (ie, cyclin D1 transcript) and endogenous control (ie, ABL) amplifications were approximately equal. For this purpose, total cellular RNA quantities of 1, 5, 10, 50, and 100 ng were tested in triplicate, and the ΔCt was calculated from the average Ct values obtained for each input amount. The plot of log input amount versus ΔCt had a slope of 0.012 (data not shown). These results indicated that the amplification efficacy was virtually superimposable for target and endogenous control, thereby validating the ΔΔCt approach.
The sensitivity of the real-time RT-PCR assay was tested on the cellular level by diluting the myeloma cell lines U266 (overexpressing cyclin D1) and KMS26 (carrying trisomy 11) in mononuclear cells from a healthy donor BM. Serial dilutions from pure U266 or KMS26 to a 1:103 ratio were processed and analyzed in triplicate. We found a linear decrease of the ΔΔCt value in proportion to the log of the starting copy number of MM cell dilutions, with correlation coefficients of 0.96 to 0.98 in various repetitions of the same experiment (data not shown).
Regarding the reliability of our assay, analysis of 20 identical samples of cyclin D1 in a single run (intra-assay comparison) revealed a coefficient of variation (CV) of 0.07, the respective Ct crossing points giving a CV of 0.04. Analysis of the same sample in 10 runs on 10 different days (interassay comparison, day-to-day variation with new mixtures of reagents) revealed a CV of 0.06, with the Ct crossing points giving a CV of 0.038.
Correlation between 11q abnormalities and cyclin D1 expression
In 46 cases where sufficient material was available, conventional cytogenetic analysis was performed in parallel with the real-time RT-PCR assay. Detected were 11q abnormalities in 11 patients, of whom 7 showed the t(11;14) and 4 an extra copy of chromosome 11 (Figure 1). Of 35 patients for whom conventional karyotype failed to show any chromosome 11 abnormality, 21 were subsequently analyzed by FISH; an abnormal pattern indicative of the t(11;14) was recognized in 2 of these patients, whereas 5 more patients were found to have trisomy 11 (Figure 1). In the remaining 14 patients for whom there was no sufficient material available for subsequent FISH analysis, the lack of 11q abnormalities was established only by conventional karyotype.
Results of FISH analysis and/or conventional cytogenetics in 46 patients with de novo MM who were investigated for 11q abnormalities. +11 indicates trisomy 11.
Results of FISH analysis and/or conventional cytogenetics in 46 patients with de novo MM who were investigated for 11q abnormalities. +11 indicates trisomy 11.
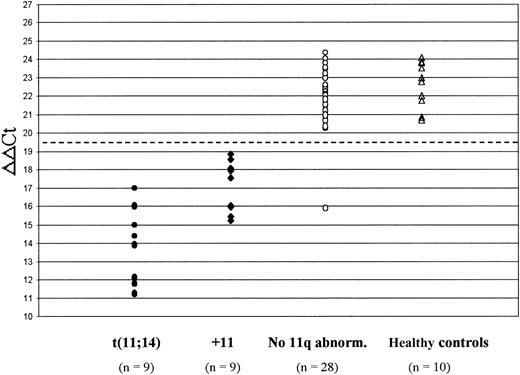
Figure 2 shows the expression of cyclin D1 in the 46 patients who were analyzed for the presence of 11q abnormalities and in the 10 healthy controls. Patients harboring the t(11;14) had significantly higher cyclin D1 mRNA levels (ie, lower ΔΔCt values) than those with trisomy 11 (median ΔΔCt, 13.92 vs 17.56, respectively; P = .001). Both of these subgroups significantly overexpressed cyclin D1 in comparison with patients who had no evidence of 11q abnormalities (median ΔΔCt, 15.71 vs 22.29, respectively; P < .0001) and to healthy controls (median ΔΔCt, 15.71 vs 22.88, respectively; P < .0001). In contrast, no difference in cyclin D1 expression was recognized between patients lacking 11q abnormalities and healthy controls (median ΔΔCt, 22.29 vs 22.88, respectively; P = .2). Therefore, the presence of the t(11;14) or trisomy 11 was found to be invariably associated with elevated cyclin D1 mRNA levels, except in a single patient who showed a very low ΔΔCt value (15.91) but no evidence of any chromosome 11 abnormality.
Cyclin D1 mRNA levels in 46 MM patients who were investigated for 11q abnormalities and 10 healthy controls. The dashed line at ΔΔCt = 19.5 indicates the cut-off level chosen to define cyclin D1 overexpression (ie, < 19.5). Patients with the t(11;14) had significantly lower ΔΔCt values (reflecting higher cyclin D1 levels) than patients with trisomy 11 (+11) (P = .001); both of these subgroups significantly overexpressed cyclin D1 in comparison with patients without 11q abnormalities (P < .0001) and with healthy controls (P < .0001).
Cyclin D1 mRNA levels in 46 MM patients who were investigated for 11q abnormalities and 10 healthy controls. The dashed line at ΔΔCt = 19.5 indicates the cut-off level chosen to define cyclin D1 overexpression (ie, < 19.5). Patients with the t(11;14) had significantly lower ΔΔCt values (reflecting higher cyclin D1 levels) than patients with trisomy 11 (+11) (P = .001); both of these subgroups significantly overexpressed cyclin D1 in comparison with patients without 11q abnormalities (P < .0001) and with healthy controls (P < .0001).
Correlation between cyclin D1 expression and 13q abnormalities
In 46 patients, FISH analysis and/or conventional cytogenetics were performed to detect 13q abnormalities. The frequency of Δ13 among 22 patients who overexpressed cyclin D1 was 41%; the corresponding value for 24 patients who did not overexpress cyclin D1 was 38% (P = .8).
Frequency of cyclin D1 overexpression in de novo MM
A cutoff value of ΔΔCt less than 19.5 was chosen to identify those patients who overexpressed cyclin D1 among the 28 patients for whom cytogenetic/FISH data on 11q abnormalities were unavailable. Overall, cyclin D1 overexpression was detected in 32 (43%) of 74 patients. Patients who overexpressed cyclin D1 (group A) had a median ΔΔCt value of 14.97 (range, 11.17-18.89), compared with a median value of 22.36 (range, 20.21-24.35) in the other patients (group B).
Clinical and prognostic relevance of cyclin D1 overexpression
Cyclin D1 overexpression was correlated with the presenting clinical features and disease outcome in the 74 patients who form the basis of the present study. As can be seen from Table 2, no difference was apparent in terms of age, sex, immunoglobulin isotype and concentration, stage at diagnosis, serum β2-microglobulin level, BM plasmacytosis, C-reactive protein level, and creatinine level between patients who overexpressed cyclin D1 (group A; n = 32) and patients who did not (group B; n = 42).
Correlation between cyclin D1 expression and patient clinical and laboratory characteristics
. | Cyclin D1 . | . | . | |
|---|---|---|---|---|
. | Overexpressed, group A . | Not overexpressed, group B . | P . | |
| No. of patients | 32 | 42 | ||
| Median age, y | 55.0 | 51.5 | .33 | |
| Male-to-female ratio, n | 21:11 | 31:11 | .12 | |
| Durie & Salmon stage, n | ||||
| I | 3 | 11 | ||
| II | 4 | 8 | .27 | |
| III | 25 | 23 | ||
| M component isotype, n | ||||
| IgG | 17 | 26 | .66 | |
| IgA | 8 | 10 | ||
| BJ | 7 | 6 | ||
| L chain, n | ||||
| κ | 20 | 28 | .92 | |
| λ | 12 | 14 | ||
| Median M component concentration | ||||
| IgG, g/L (range) | 39.0 (10.0-111.7) | 42.0 (25.5-84.0) | ||
| IgA, g/L (range) | 38.0 (22.0-42.0) | 36.5 (25.6-78.0) | .35 | |
| BJ, g/24 h (range) | 3.40 (1.90-6.40) | 4.25 (1.90-18.60) | ||
| Median BMPC, % (range) | 50 (10-100) | 40 (10-100) | .20 | |
| Median β2-m level, mg/L (range) | 2.80 (1.20-16.00) | 2.20 (1.10-6.90) | .15 | |
| Median CRP level, mg/L (range) | 3.60 (1-62.5) | 3.10 (1-70.20) | .80 | |
| Median creatinine level, μM (range) | 79.56 (61.88-123.76) | 97.24 (61.88-265.2) | .12 | |
. | Cyclin D1 . | . | . | |
|---|---|---|---|---|
. | Overexpressed, group A . | Not overexpressed, group B . | P . | |
| No. of patients | 32 | 42 | ||
| Median age, y | 55.0 | 51.5 | .33 | |
| Male-to-female ratio, n | 21:11 | 31:11 | .12 | |
| Durie & Salmon stage, n | ||||
| I | 3 | 11 | ||
| II | 4 | 8 | .27 | |
| III | 25 | 23 | ||
| M component isotype, n | ||||
| IgG | 17 | 26 | .66 | |
| IgA | 8 | 10 | ||
| BJ | 7 | 6 | ||
| L chain, n | ||||
| κ | 20 | 28 | .92 | |
| λ | 12 | 14 | ||
| Median M component concentration | ||||
| IgG, g/L (range) | 39.0 (10.0-111.7) | 42.0 (25.5-84.0) | ||
| IgA, g/L (range) | 38.0 (22.0-42.0) | 36.5 (25.6-78.0) | .35 | |
| BJ, g/24 h (range) | 3.40 (1.90-6.40) | 4.25 (1.90-18.60) | ||
| Median BMPC, % (range) | 50 (10-100) | 40 (10-100) | .20 | |
| Median β2-m level, mg/L (range) | 2.80 (1.20-16.00) | 2.20 (1.10-6.90) | .15 | |
| Median CRP level, mg/L (range) | 3.60 (1-62.5) | 3.10 (1-70.20) | .80 | |
| Median creatinine level, μM (range) | 79.56 (61.88-123.76) | 97.24 (61.88-265.2) | .12 | |
Abbreviations are explained in Table 1.
Regarding treatment assignments, the 2 groups were well balanced: in group A, 16 (50%) patients were randomized to undergo a single transplantation (Tx-1), and 16 (50%) to undergo double transplantation (Tx-2); in group B, 21 (50%) were assigned to Tx-1 and 21 (50%) to Tx-2. The probability of actually undergoing double transplantation for patients who were randomized to Tx-2 was 71% in group A and 68% in group B. An intention-to-treat analysis revealed that the probability of attaining CR was similar for patients in groups A and B (data not shown). With a median follow-up of 34 months (range, 9-64 months), the median OS was not reached after 48 months in group A and was 43 months in group B (P = .6). On the other hand, patients who overexpressed cyclin D1 had a significantly longer duration of remission (median, 41 months) compared with patients who did not (median, 26 months) (P = .02) (Figure 3).
Time to disease progression according to cyclin D1 expression. Median time to relapse/progression was 41 months for patients who overexpressed cyclin D1 (group A) versus 26 months for patients who did not (group B) (P = .02).
Time to disease progression according to cyclin D1 expression. Median time to relapse/progression was 41 months for patients who overexpressed cyclin D1 (group A) versus 26 months for patients who did not (group B) (P = .02).
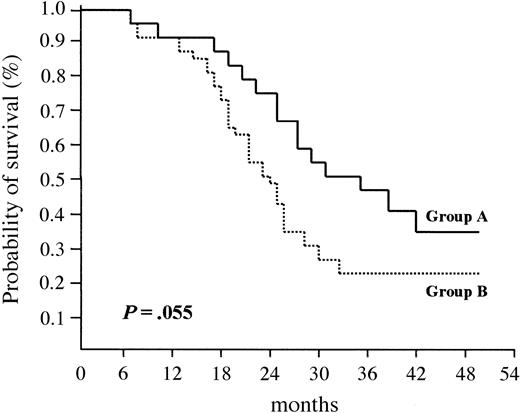
As a result, median EFS was longer in group A (33 months) than in group B (24 months); the difference between the 2 groups was of borderline statistical significance (P = .055) (Figure 4).
Event-free survival (EFS) according to cyclin D1 expression. Median EFS was 33 months for patients who overexpressed cyclin D1 (group A) versus 24 months for patients who did not (group B) (P = .055).
Event-free survival (EFS) according to cyclin D1 expression. Median EFS was 33 months for patients who overexpressed cyclin D1 (group A) versus 24 months for patients who did not (group B) (P = .055).
As reported above, we demonstrated that patients who overexpressed cyclin D1 and patients who did not were equally likely to have 13q abnormalities (41% vs 38%, respectively). This finding suggests that the different outcome we observed for cyclin D1–positive and cyclin D1–negative patients was unlikely to be influenced by the presence of Δ13. Analysis of EFS for a subgroup of patients with Δ13 revealed that those patients who overexpressed cyclin D1 (n = 9) fared better than those who did not (n = 9) (median, 41 vs 23 months, respectively). As expected, a similar outcome was observed when the end point analyzed was the duration of remission; the median value was 26 months for the Δ13-positive/cyclin D1–negative subgroup, as opposed to 41 months for the Δ13-positive/cyclin D1–positive subgroup. Because of the small sample size, detailed statistical analysis was not performed.
Discussion
In the present study we analyzed the frequency and the clinical and prognostic relevance of cyclin D1 expression in a large series of patients with de novo MM who were homogeneously treated with autologous PBSC-supported high-dose chemotherapy. For this purpose, we developed a novel and straightforward real-time RT-PCR assay. This method was shown to have a large dynamic range and to be highly reproducible, sensitive, and specific. In particular, the sensitivity of the assay was tested on the cellular level by checking cyclin D1 mRNA levels in serial dilutions of U266 and KMS26 myeloma cell lines (overexpressing cyclin D1 and carrying trisomy 11, respectively) in healthy donor BM mononuclear cells. Our method proved to be capable of detecting MM cells with dysfunction of cyclin D1 in all diagnostic BM samples containing more than 1% plasma cells, since we obtained ΔΔCt values less than 19.5 (ie, the cut-off chosen to define cyclin D1 overexpression) in all the dilutions up to 1:102 for both the cell lines. Such a high sensitivity was further demonstrated by the fact that we were able to discriminate patients who overexpressed cyclin D1 from those who did not, even in cases with minimal BM plasma cell infiltration (10%). Furthermore, the specificity of our assay was tested using BM samples from healthy donors as negative controls and the KMS12 MM cell line carrying the t(11;14) as positive control. Since normal B lymphocytes are known to express cyclin D2 and, albeit to a lesser extent, cyclin D3, but almost no cyclin D1,30 the up-regulation of cyclin D1 we found in BM samples from patients with MM can be reasonably attributed to the neoplastic clone, as suggested also in other studies addressing the same issue.17,21
Results of our analysis showed that cyclin D1 overexpression could be detected in 43% of patients with de novo MM, in agreement with previous data reported by others using similar methods of analysis21,22 or immunohistochemistry techniques.17,18,31 Such an incidence of cyclin D1 up-regulation is much higher than that expected on the basis of the frequency of the t(11;14), which is detectable in only 5% of patients by conventional karyotype,10-12 and in up to 15% to 20% by FISH analysis.13-15 A limited number of studies among those so far reported provided additional information on the presence of karyotypic abnormalities that could ultimately lead to cyclin D1 up-regulation in MM.17,31 In order to address this poorly investigated issue, in the present study we correlated the results of the real-time RT-PCR assay with those of conventional karyotype and FISH analyses for the detection of 11q abnormalities. We found that patients carrying either the t(11;14) or an extra copy of chromosome 11 had significantly higher cyclin D1 mRNA levels than those without apparent 11q abnormalities. Within this latter subgroup, cyclin D1 levels were not different from those found in healthy individuals. Thus, a close relationship between cyclin D1 overexpression, as detected by the real-time RT-PCR assay, and abnormalities involving 11q were demonstrated in 95% of patients analyzed. Similar results were previously reported by Pruneri et al,17 who found the deregulated expression of cyclin D1, as evaluated by immunohistochemistry, in 100% of patients with the t(11;14), but in only 15% of those with trisomy 11. In another study by Wilson et al,31 cyclin D1 overexpression was reported to correlate with the t(11;14) only in patients with low to intermediate labeling index, but not in patients with high proliferative activity of the myeloma clone. In our series, among patients who were analyzed for the correlation between cyclin D1 expression and 11q abnormalities, we found only a single case who had high levels of cyclin D1 mRNA and neither the t(11;14) nor trisomy 11. Alternative mechanisms, other than 11q abnormalities or gene amplification, may rarely cause the deregulated expression of cyclin D1. For instance, an insertion into the 11q13 region of excised Ig enhancers originating during the process of isotype switching may occur, as it has been previously demonstrated in the U266 cell line.32 Alternatively, the occurrence of a mutation causing the up-regulation of cyclin D1 mRNA transcript may be hypothesized. Unfortunately, the lack of sufficient DNA material from our patient hampered further investigations aimed to confirm these hypotheses.
Another objective of our study was to investigate whether cyclin D1 overexpression was correlated with particular clinical features and influenced the outcome of MM. The uniqueness of MM patients carrying the t(11;14), as detected by FISH analysis, has been recently reported,14 the most striking features being the higher prevalence of Bence-Jones isotype and lower serum M protein concentration, lower plasma cell labeling index, greater likelihood of being pseudodiploid or hypodiploid, and a higher frequency of lymphoplasmacytic morphology. In another study,16 the amplification of the cyclin D1 gene was found to correlate with bone marrow plasma cell infiltration and morphology, labeling index, and serum β2-microglobulin and C-reactive protein levels. The higher frequency of cyclin D1 overexpression in patients with higher bone marrow plasma cell infiltration was also reported by Pruneri et al.17 Unlike these observations, we were unable to demonstrate any significant relationship between cyclin D1 overexpression and any of the routinely available presenting features that were analyzed. Similar results were recently reported by Rasmussen et al,22 who investigated a series of patients for cyclin D1 overexpression by an RT-PCR method similar to that herein described.
Cyclin D1 protein plays a key role in the G1- to S-phase transition and is also involved in tumor development. These peculiar characteristics and the recognition that up-regulation of cyclin D1 may be found in up to 50% of MM patients prompted a series of studies aimed to investigate whether abnormalities of cyclin D1 and/or of chromosome 11 influenced the outcome of patients with MM. Results of these studies were conflicting, reflecting differences in patient characteristics, patients' disease phases (ie, at diagnosis or at relapse), treatment for MM (ie, standard- or high-dose chemotherapy), methods used to detect chromosomal abnormalities (ie, conventional karyotype or FISH), and/or cyclin D1 expression. In initial studies, the presence of the t(11;14) and/or other 11q abnormalities were investigated by means of conventional karyotype and were found to be associated with shortened survival,12,13,23 a finding confirmed also in a study on amplification of the cyclin D1 gene.16 Whether in these cases chromosomal abnormalities involving 11q were an independent adverse prognostic variable or the poor outcome was simply related to the higher proliferative activity of the myeloma clone that made the karyotype easily informative has been for a long time the matter of discussion. In order to circumvent the limitations of conventional cytogenetics and to obtain information in almost every patient analyzed, interphase FISH with probes specific for the t(11;14)(q13;q32) was systematically performed in 2 subsequent studies involving large series of patients.14,15 In one of them,14 the analysis was carried out in the context of a clinical trial of conventional chemotherapy and showed that patients carrying the translocation had a longer survival—both overall and progression-free—than the others, even though the difference was not statistically significant. In the other study,15 the t(11;14) was associated with a statistically significant longer OS following high-dose chemotherapy. Results of the present analysis are in agreement with these latter observations and provide demonstration that high cyclin D1 mRNA levels do not predict for worsened prognosis, rather identifying a subset of patients who are more likely to have prolonged duration of remission and EFS following autologous transplantation. The better outcome of patients overexpressing cyclin D1 in comparison with those who did not was not related to a more favorable response to the treatment program, nor to a close association between cyclin D1 overexpression and other variables favorably influencing the prognosis. In particular, we demonstrated that patients who overexpressed cyclin D1 versus patients who did not were equally likely to carry Δ13, as previously reported by others.14,15 Thus, in contrast to the t(4;14) that is closely associated with Δ13,15,33,34 coexistence of this latter chromosomal abnormality with cyclin D1 overexpression is likely unrelated. This finding suggests that the different outcome we observed for cyclin D1–positive and cyclin D1–negative patients was unlikely to be influenced by Δ13. Furthermore, analysis of a small subgroup of our patients seemed to suggest that cyclin D1 overexpression counterbalanced the adverse prognostic relevance of Δ13. Taken together, all of these findings give further support to the conclusions of recent studies reporting that cyclin D1 overexpression is not related to an increased proliferative activity of the myeloma clone31 and that the t(11;14) favorably influences the outcome of even the most aggressive plasma cell neoplasms, including plasma cell leukemia.35 Further analyses of prospective studies on large series of patients homogeneously treated and followed for adequate time periods are needed to confirm the favorable prognostic relevance of cyclin D1 overexpression or t(11;14) in de novo MM and to definitely assess the role of high-dose chemotherapy with autologous transplantation in the management of this peculiar subset of patients.
Appendix
The following is a list of investigators who participated in the “Bologna 96” trial and referred BM samples from their patients for the present study: R. Battista (Dolo); F. Di Raimondo (Catania); R. Fanin (Udine); P. P. Fattori (Rimini); V. Forcellini (S. Marino); P. Gentilini (Forlì); R. Giustolisi (Catania); L. Guardigni (Cesena); L. Gugliotta (Reggio Emilia); D. Mamone (Messina); L. Masini (Reggio Emilia); C. Musolino (Messina); A. L. Molinari (Ravenna); F. Patriarca (Udine); F. Ricciuti (Potenza); S. Ronconi (Forlì); D. Vertone (Potenza), E. Volpe (Avellino); and A. Zaccaria (Ravenna).
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2002-12-3789.
Supported by Ministero dell'Università e Ricerca Scientifica (MIUR); progetto FIRB, RBAU012E9A_001 (M.C.); Università di Bologna, Progetti di Ricerca ex-60% (M.C.); and Fondazione Carisbo.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
A list of investigators who referred BM samples from their patients for molecular studies appears in “Appendix.” We wish to thank Dr Takemi Otzuki (Okayama University, Japan) who kindly provided the KMS26 cell line. We also wish to thank Robin Cooke for his careful editing of the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal