Abstract
The structural basis of the interaction between single-chain urokinase-type plasminogen activator (scuPA) and its receptor (uPAR) is incompletely defined. Several observations indicated the kringle facilitates the binding of uPA to uPAR. A scuPA variant lacking the kringle (ΔK-scuPA) bound to soluble uPAR (suPAR) with the similar “on-rate” but with a faster “off-rate” than wild-type (WT)-scuPA. Binding of ΔK-scuPA, but not WT-scuPA, to suPAR was comparably inhibited by its growth factor domain (GFD) and amino-terminal fragment (ATF). ATF and WT-scuPA, but not GFD, scuPA lacking the GFD (ΔGFD-scuPA), or ΔK-scuPA reconstituted the isolated domains of uPAR. ATF completely inhibited the enzymatic activity of WT-scuPA-suPAR unlike comparable concentrations of GFD. Variants containing mutations that alter the charge, length, or flexibility of linker sequence (residues 43-49) between the GFD and the kringle displayed a lower affinity for uPAR, were unable to reconstitute uPAR domains, and their binding to uPAR was inhibited by GFD in the same manner as ΔK-scuPA. A scuPA variant in which the charged amino acids in the heparin binding site (HBS) in the kringle domain were mutated to alanines behaved like ΔK-scuPA, indicating that that the structure of the kringle as well as its interaction with the GFD govern receptor binding. These data demonstrate an important role for the kringle in stabilizing the binding of scuPA to uPAR. (Blood. 2003;102:3600-3608)
Introduction
Urokinase-type plasminogen activator (uPA) has been implicated in diverse physiologic and pathophysiologic processes that involve cell adhesion, fibrinolysis, and signal transduction.1-3 Several of these processes are mediated by the catalytic activity of the molecule,4 while others involve the participation of the uPA receptor (uPAR)1,5-8 or other cell surface molecules.8-11
uPA is expressed as a single-chain molecule (scuPA) composed of an N-terminal fragment (ATF; amino acids 1-135) and a protease domain (amino acids 136-411), also known as low molecular weight uPA (LMW-uPA). The amino-terminal fragment (ATF) is itself composed of 2 independent domains, the amino-terminal growth factor-like domain (GFD; amino acids 1-43), which is known to bind to the uPA receptor, and a single kringle (K; amino acids 47-135), the function of which is uncertain.12-17
scuPA expresses plasminogen activator activity when converted to a 2-chain molecule (tcuPA) by plasmin18 or as a single-chain molecule when bound to its receptor.19-22 The mature urokinase receptor (uPAR) is a 283-amino acid glycolipid-anchored protein composed of 3 partial repeat domains.5,6,23 Cooperation among the 3 domains is required to optimize scuPA binding and activation.24-26 Binding of scuPA is inhibited by peptides that cross-link residues Arg53 and Leu66 in domain I and His251 in domain III of uPAR, respectively,27,28 or by mutations in a region of domain II, which has been implicated in interdomain cooperativity.26
There is compelling evidence to indicate that scuPA binds to uPAR through its GFD.28-34 However, the potential involvement of other sites in uPA in receptor binding has not been studied in detail. Evidence favoring the existence of such auxiliary sites comes from studies in which uPA-induced, uPAR-mediated adhesion to vitronectin (Vn) and other integrin ligands has been examined using purified proteins7,35,36 or whole cells.7,8,31,36,37 For example, we have reported that ATF enhances the binding of soluble uPAR (suPAR) to cellular Vn to a considerably greater extent than does GFD,35 and similar observations have been made by others.31
The induction of receptor-mediated adhesion and enzymatic activity by scuPA or its fragments likely involves conformational changes in either or both molecules that derive from multiple or more stable intermolecular contacts between the reactants.36 The observation that ATF induces greater uPAR-mediated adhesion than does GFD35 suggests that ATF promotes contacts with the receptor that induce the required conformation. Yet, there is little evidence to date that the interaction between GFD and uPAR is influenced by the other domains of uPA. Specifically, the function of the uPA kringle remains poorly understood, and its role in binding to uPAR, if any, is unknown.
Moreover, we have previously observed an unexpectedly prolonged retention of scuPA bound to uPAR on endothelial cell surface based on its off-rate.38 However, in the cellular environment, we could not distinguish the contribution of post-binding events to stabilization of the uPA-uPAR complex from events within the complex itself. Therefore, in this study we examined the contribution of the kringle to the binding of scuPA to purified recombinant soluble uPAR. The results of the present study indicate that the kringle helps to stabilize the binding of scuPA to uPAR, likely through an effect on the GFD.
Materials and methods
Chemicals and reagents
Chinese hamster ovary (CHO) cells and human embryonic kidney 293 cells were purchased from American Type Culture Collection (Rockville, MD). Tissue culture medium, bovine serum, and Lipofectin were from Life Technologies (Bethesda, MD). Phosphatidylinositol-specific phospholipase C (PI-PLC) was from Oxford GlycoSciences (Wakefield, MA). Chymotrypsin was from Sigma (St Louis, MO), Na125I from NEN Life Science Products (Boston, MA), and iodogen-precoated tubes from Pierce Chemical (Rockford, IL). Plasminogen, Spectrozyme PL, and monoclonal antihuman uPAR nonblocking antibody 3937 were kind gifts of American Diagnostica (Greenwich, CT). Recombinant scuPA and 2-chain uPA (tcuPA) expressed in mammalian cells and polyclonal anti-uPAR antibody were the kind gifts from Dr J. Henkin (Abbott Laboratories, Abbott Park, IL). Monoclonal antihuman LMWuPA and anti-uPA kringle antibodies immobilized to Sepharose were kindly provided by Dr S. Domogatsky (IMTEK, Moscow, Russia).
Production of recombinant proteins
Generation of wild-type uPA and uPA fragments. Wild-type scuPA, scuPA lacking the kringle (amino acids 47-135; designated ΔK-scuPA), and scuPA lacking the “connecting peptide” (amino acids 136-143, designated ΔCP-scuPA) were generated and subcloned into pMT/BiP/V5 (Invitrogen, Carlsbad, CA) as described previously.9,26 cDNA encoding ATF (amino acids 1-135), scuPA lacking the GFD (amino acids 1-46, designated ΔGFD-scuPA), or Flag-low molecular weight uPA (LMW-uPA) (amino acids 136-411) were generated in the same manner using full-length UK/pUN121 39 as the template. The polymerase chain reaction (PCR) products were digested with BamHI and Xho and subcloned into pMT/BiP/V5 at the BglII and Xho sites. The recombinant proteins were expressed using the Drosophila Expression System (Invitrogen) in Schneider S2 cells according to the manufacturer's recommendations. Wild-type scuPA, ΔK-scuPA, ΔCP-scuPA, ΔGFD-scuPA, and LMW-uPA were purified from S2 media by affinity chromatography using monoclonal anti-LMW-uPA immobilized to Sepharose. In some experiments wild-type scuPA was purified using cation exchange column (SP-Trisacryl M beads) followed by high-performance liquid chromatography (HPLC) separation on a reversed phase (RP)-C8 column, as described.9 ATF 1-135 was purified from S2 media by affinity chromatography using a monoclonal antikringle antibody immobilized on Sepharose.
Mutagenesis of scuPA. The active site mutant scuPA-Ser356Ala was generated using scuPA/pMT/BiP/V5 as the template with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The 6 residues within the kringle implicated in heparin binding (amino acids 105-110)40 were mutated to alanine in clusters in the same way using cDNA encoding wild-type scuPA/pMT/BiP/V5 as the template to generate scuPA-Lys105-110Ala, scuPA-Lys107-109Ala, scuPA-Lys108-110Ala, scuPA-Lys110Ala, and scuPA-Lys107Ala (Table 1). The same cloning strategy was used to mutate the “linker” region that connects GFD and kringle domain (amino acids 43-49) and to generate scuPA-Leu43-49Ala, ΔLeu45-48-scuPA, and 2 other point mutations in the linker region scuPA-Ser47Gly and scuPA-Lys48Pro (Table 1). All recombinant proteins were expressed in the Drosophila Expression System and purified as above.
scuPA kringle and linker mutants
Kringle mutants | |
| Wild-type-scuPA | 102-C R N P D N R R R P W C-112 |
| scuPA-Lys 105-110Ala | 102-C R N A A A A A A P W C-112 |
| scuPA-Lys 107-109Ala | 102-C R N P D A A A R P W C-112 |
| scuPA-Lys 108-110Ala | 102-C R N P D N A A A P W C-112 |
| scuPA-Lys 110Ala | 102-C R N P D N R R A P W C-112 |
| scuPA-Lys 107Ala | 102-C R N P D A R R R P W C-112 |
| Linker mutants | |
| Wild-type-scuPA | 42-C E I D K S K T C-50 |
| scuPA-Leu43-49Ala | 42-C A A A A A A A C-50 |
| ΔLeu45-48-scuPA | 42-C - - - - - - - - C-50 |
| scuPA-Lys48Pro | 42-C E I D K S P T C-50 |
| ScuPA-Ser47Gly | 42-C E I D K G K T C-50 |
Kringle mutants | |
| Wild-type-scuPA | 102-C R N P D N R R R P W C-112 |
| scuPA-Lys 105-110Ala | 102-C R N A A A A A A P W C-112 |
| scuPA-Lys 107-109Ala | 102-C R N P D A A A R P W C-112 |
| scuPA-Lys 108-110Ala | 102-C R N P D N A A A P W C-112 |
| scuPA-Lys 110Ala | 102-C R N P D N R R A P W C-112 |
| scuPA-Lys 107Ala | 102-C R N P D A R R R P W C-112 |
| Linker mutants | |
| Wild-type-scuPA | 42-C E I D K S K T C-50 |
| scuPA-Leu43-49Ala | 42-C A A A A A A A C-50 |
| ΔLeu45-48-scuPA | 42-C - - - - - - - - C-50 |
| scuPA-Lys48Pro | 42-C E I D K S P T C-50 |
| ScuPA-Ser47Gly | 42-C E I D K G K T C-50 |
Amino acids in the heparin binding site of uPA kringle (residues 105-110) and in the linker region (residues 43-49) between the growth factor domain and the kringle were either mutated to the indicated amino acids (bold) or deleted (-).
Generation of the growth factor-like domain of uPA (GFD). GFD was generated from scuPA as described.16 Briefly, scuPA was digested at 100:1 molar ratio with endoproteinase Glu-C (Staph V8) (Worthington Biochemical, Lakewood, NJ) in 0.1 M phosphate 0.6 M NaCl, pH 7.6, for 6 hours at 37°C. The mixture was concentrated and separated on a G50 column in the same buffer. As a final step, the GFD was further purified by HPLC on RP-C8.26 GFD migrated as a single band at Mr of about 4 to 5 kDa on reduced and nonreduced sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), confirming the integrity of the product (data not shown). All experiments were confirmed using a second source of GFD generated in the same manner and shown previously to migrate on mass spectroscopy as a single homogeneous protein with a mass of 4617 Da, consistent with the theoretical mass of uncleaved GFD amino acids 4-43.16
Production of wild-type (WT) soluble urokinase receptor (suPAR) and suPAR fragments. WT suPAR (amino acids 1-277) was expressed in Drosophila Schneider S2 cells, purified, and characterized as previously described.26 Full-length suPAR was digested with chymotrypsin to generate soluble domain I of suPAR (sDI) and sDII+DIII, as described.26 Briefly, chymotrypsin was incubated with suPAR (1 mg/mL in phosphate-buffered saline [PBS]) at a final molar ratio of suPAR/chymotrypsin of 1000:1 for 2 hours at room temperature (RT), and the digestion was quenched by adding Pefablock (100 μM final concentration; Sigma). sDI and sDII+DIII were separated by HPLC using RP-C8.
Radiolabeling of proteins. scuPA, suPAR, and sDI were radiolabeled with Na125I using iodogen-precoated tubes at a ratio of 100 μCi (3.7 MBq) of Na125I per 100 μg protein to avoid oxidative injury.26 Free 125I was removed by gel filtration using Sephadex G25 (PD-10; Amersham Pharmacia Biotech, Piscataway, NJ). The specific activity of 125I-scuPA was in the range of 3 × 104 to 8 × 104 cpm/pmol and 5 × 104 to 10 × 104 cpm/pmol for 125I-sD1.
Binding of soluble uPAR domain I to cells expressing uPAR domains II+III. Stable 293 cell lines expressing glycosyl phosphatidyl inositol (GPI)-linked uPAR DII-III were developed, as described.26 The expression of uPAR DII-DIII was confirmed by flow cytometry using affinity-purified rabbit polyclonal anti-uPAR immunoglobulin G (IgG) in the presence and absence of PI-PLC. The 293 cells expressing GPI-anchored uPAR domains II-III or mock-transfected cells were incubated with 125I-sDI (50 nM) in the absence or presence of scuPA, scuPA variants, or scuPA fragments (0 to 600 nM) for 1 hour at 4°C in PBS/1% bovine serum albumin (BSA). The cells were washed 3 times with ice-cold binding buffer, and the cell-associated radioactivity was measured.
Binding of scuPA to CHO cells. Specific binding of 125I-wild-type scuPA (10 nM) to CHO cells was measured in the presence of varying concentrations of unlabeled WT and variant scuPA or scuPA fragments (0.012 to 400 nM), as described.41
Radioligand binding experiments. Ninety-six-well plates were coated overnight with 0.5 μg/mL suPAR in PBS at 4°C. Unreacted sites were blocked with 3% BSA in PBS for 2 hours at 22°C and washed twice in binding buffer (1% BSA/PBS). To measure ligand binding, 125I-scuPA WT or 125I-ΔK-scuPA (2.5, 5, 10, and 20 nM) were added in binding buffer for the indicated time points at 22°C, wells were washed 4 times, and bound radioactivity was measured. Nonspecific binding was measured in the presence of 50-fold excess unlabeled ligand. Specific binding was measured as the difference between total and nonspecific binding. To measure dissociation, ligand binding proceeded in an identical manner. The wells were chilled to 4°C on ice, washed 4 times, and resuspended in ice-cold binding buffer. The plates were then shifted to 22°C, and the released radioactivity was measured at the indicated time points. Data were fitted to the following equations assuming a 1:1 reversible interaction. The dissociation rate constants were calculated by fitting the data to the equation (Bt = Bmax * e(-koff * t) + plateau, where Bt = amount bound at time “t,” Bmax is the maximal total binding at the start of dissociation, and “plateau” is the residual binding after dissociation) using GraphPad Prism 3.0 (GraphPad Software, San Diego, CA). The half-life (t1/2) = 0.69/koff. Association binding data were analyzed using GraphPad Prism 3.0 using the equation (Bt = Bmax * (1-e(-kobs*X)), where B = specific binding, kobs = the apparent association constant where [Kon = (kobs-koff)/ligand concentration]). The half-life to equilibrium (t1/2) = 0.69/kobs.
Binding of scuPA to suPAR measured as surface plasmon resonance. Surface plasmon resonance (SPR) experiments were performed using a BIACORE X optical Biosensor (Biacore, Uppsala, Sweden).42-44 This method detects binding interactions in real time by measuring changes in the refractive index at a biospecific surface, enabling association and dissociation rate constants to be calculated. All binding experiments were carried out at 25°C in PBS, pH 7.4, containing 0.005% Tween-20.
scuPA binding. suPAR (10 μg/mL) was coupled directly to a CM5-research grade sensor chip flow cell (Biacore) in 10 mM sodium acetate buffer, pH 5.0, via standard amine coupling procedures26,45 using N-hydroxysuccinimide/N-ethyl-N′-[3-(dimethylamino)propyl]carbodiimine hydrochloride (Pierce) to a level of 600 to 1000 response units (RUs). Unreacted groups were blocked with 1 M ethanolamine, pH 8.5. A second flow cell, similarly activated and blocked without protein immobilization, served as a control. Binding of scuPA was measured for 2 minutes at a flow rate of 50 μL/min, followed by dissociation for either 2 or 20 minutes. The bulk shift due to changes in refractive index measured on the blank surface was subtracted from the binding signal at each condition to correct for nonspecific binding. Surfaces were regenerated with one or more 18-second pulses of 1 M NaCl/HCl, pH 3.5, followed by an injection of running buffer for 1 minute to remove this high salt solution.
An antibody capture assay was performed as a second approach; 5100 RU of the mouse monoclonal antihuman uPAR antibody 3937 (American Diagnostica), which requires domains II + III of uPAR for recognition and does not block binding of uPA, was coupled to both flow cells of a CM5 chip as described in the preceding paragraph at pH 4.0. Unreacted groups were blocked with 1 M ethanolamine, pH 8.5. To study the binding of scuPA to suPAR, 20 nM suPAR (300 RU) was captured for 2 minutes at 5 μL/min to 1 of 2 flow cells. Five minutes later, binding of scuPA was measured on both flow cells for 2 minutes at a flow rate of 50 μL/min, followed by dissociation for 20 minutes. The bulk shift due to changes in refractive index measured on the monoclonal antibody surface (flow cell 1) was subtracted from the binding signal (flow cell 2) at each scuPA concentration to correct for nonspecific binding. Dissociation of suPAR from antibody 3937 was measured by injecting running buffer under the same conditions as scuPA and subtracting this value from each scuPA signal. Antibody 3937 surfaces were regenerated with one or more brief pulses of 1 M NaCl in 0.2 M sodium carbonate, pH 11, followed by an injection of running buffer for 1 minute to remove the high-salt solution.
Competition-inhibition. suPAR was immobilized as described in the preceding paragraph. To study the binding of WT-scuPA, ΔK-scuPA, and the heparin binding site (HBS) mutants of scuPA, buffer or 400 nM GFD or ATF (inhibitor) was injected at a flow rate of 30 μL/min for 3 minutes. This was followed immediately by injection of 2 nM WT or variant scuPA, either alone or mixed with GFD, for 3 minutes. Dissociation was measured over the next 3 minutes.
Chromogenic assay of scuPA variants bound to suPAR. The capacity of suPAR to promote scuPA-mediated plasminogen activation was measured using a chromogenic assay, as described.19 Briefly, 2 nM wild-type scuPA was incubated with 4 nM suPAR and the indicated concentrations of variants, scuPA fragments or variants, in the presence of 20 nM plasminogen and 500 μM chromogenic substrate (Spectrozyme PL). The optical density (405 nm) was measured over time in a Thermomax microplate reader (Molecular Devices, Sunnyvale, CA).
Results
The kringle of uPA contributes to the stability of the suPAR-scuPA complex
The purpose of this study was to explore the contribution of the kringle to the binding of scuPA to suPAR. To address this question, we compared the binding kinetics of radiolabeled WT-scuPA and a variant scuPA lacking the kringle (amino acids 47-135; ΔK-scuPA). The dissociation rate constant of WT-scuPA was lower than for ΔK-scuPA (0.23 × 10-4 ± 0.08 × 10-4 s-1 and 1.67 × 10-4 ± 0.50 × 10-4 s-1, respectively) (Figure 1). In contrast, the kon ofWT-scuPA and ΔK-scuPA calculated from the kobserved and the koff were indistinguishable (2.0 × 105 ± 0.66 × 105 and 2.33 × 105 ± 0.88 × 105 M-1s-1, respectively). These results show a clear difference in the stability of the complexes of suPAR with the 2 forms of scuPA. However, the use of radio ligand binding to calculate kinetic parameters is limited by the imprecision in measuring ligand binding during washing which, in turn, leads to imprecision in defining the onset of dissociation.
Dissociation of radiolabeled WT- and ΔK-scuPA from soluble uPAR. suPAR-coated wells were incubated with 125I-WT-scuPA (5 [▾], 10 [▴], and 20 nM [▪]) or 125I-ΔK-scuPA (5 [▿], 10 [▵], and 20 nM [□]) for 2 hours at 22°C. Wells were washed 4 times, and the released radioactive ligand was counted at the indicated times. The lines represent the best fit to the single exponential decay model as determined using GraphPad software.
Dissociation of radiolabeled WT- and ΔK-scuPA from soluble uPAR. suPAR-coated wells were incubated with 125I-WT-scuPA (5 [▾], 10 [▴], and 20 nM [▪]) or 125I-ΔK-scuPA (5 [▿], 10 [▵], and 20 nM [□]) for 2 hours at 22°C. Wells were washed 4 times, and the released radioactive ligand was counted at the indicated times. The lines represent the best fit to the single exponential decay model as determined using GraphPad software.
To circumvent this potential problem, ligand binding was also analyzed using real-time surface plasmon resonance, which enabled us to obtain a global fit analysis based on thousands of data points.46 Although the binding kinetics of WT- and ΔK-scuPA had appeared to be similar over the first 2 minutes of association and dissociation,9 the radioligand binding data led us to hypothesize that the contribution of the kringle to the stability of binding would become apparent if the dissociation phase were extended. Because soluble uPAR may oligomerize at high concentrations,47 binding of scuPA to immobilized suPAR was studied.
To generate an oriented, functional, and homogeneous suPAR reaction surface, we first covalently attached a nonblocking monoclonal anti-uPAR antibody to the sensor surface.48,49 suPAR was then captured on the surface, followed by either wild-type or mutant scuPA. After the dissociation phase of the experiment, the surface was regenerated such that both scuPA and suPAR were removed, leaving behind the antibody, permitting capture of fresh suPAR.
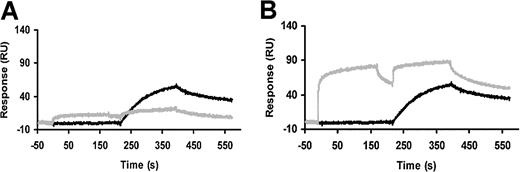
Consistent with the results of the radioligand binding experiments, the association rates of WT-scuPA and ΔK-scuPA determined by surface plasmon resonance were similar (Figure 2). In contrast, ΔK-scuPA dissociated from suPAR more rapidly than did WT-scuPA (Figure 2). As expected, the calculated kon's for WT and ΔK-scuPA were similar (4.8 × 105 and 11.0 × 105 M-1s-1, respectively) based on a global fit of the data, assuming a 1:1 Langmuir interaction model.9 This is consistent with the results obtained using labeled ligand, whereas the dissociation rate constant (koff) of ΔK-scuPA was about 10-fold faster than WT-scuPA (WT = 3.4 × 10-4 and ΔK = 2.8 × 10-3 s-1). The Kd of scuPA for suPAR calculated from these kinetic data were 7.1 × 10-10 M, a figure comparable to that reported previously by others. The calculated Kd for ATF (5.2 × 10-10 M) was almost identical to WT-scuPA, consistent with published data,28 whereas the Kd for ΔK-scuPA was approximately 5-fold lower (2.5 × 10-9 M).
Association and dissociation of WT- and ΔK-scuPA from suPAR measured as surface plasmon resonance. Antihuman-uPAR monoclonal nonblocking antibodies were coupled to a CM5-research grade sensor chip flow cell. suPAR was captured for 2 minutes. Five minutes later, binding of WT-scuPA and ΔK-scuPA was measured for 2 minutes, followed by dissociation for 20 minutes. The bulk shift due to changes in refractive index measured on the monoclonal antibody/buffer surface was subtracted from the binding signal at each scuPA concentration to correct for nonspecific binding. Dissociation of suPAR from anti-uPAR antibodies, determined under the same conditions as scuPA, was also subtracted. The dose-response of scuPA (0.63 to 5 nM) binding to suPAR is shown. The lines represent a 1:1 interaction model of the binding of WT-scuPA during the association and dissociation phases. (A) WT-scuPA concentrations are 5, 2.5, 1.3, and 0.6 nM. (B) ΔK-scuPA concentrations are 10, 5, 2.5, and 1.3 nM. The lines represent the best global fit to a 1:1 binding model.
Association and dissociation of WT- and ΔK-scuPA from suPAR measured as surface plasmon resonance. Antihuman-uPAR monoclonal nonblocking antibodies were coupled to a CM5-research grade sensor chip flow cell. suPAR was captured for 2 minutes. Five minutes later, binding of WT-scuPA and ΔK-scuPA was measured for 2 minutes, followed by dissociation for 20 minutes. The bulk shift due to changes in refractive index measured on the monoclonal antibody/buffer surface was subtracted from the binding signal at each scuPA concentration to correct for nonspecific binding. Dissociation of suPAR from anti-uPAR antibodies, determined under the same conditions as scuPA, was also subtracted. The dose-response of scuPA (0.63 to 5 nM) binding to suPAR is shown. The lines represent a 1:1 interaction model of the binding of WT-scuPA during the association and dissociation phases. (A) WT-scuPA concentrations are 5, 2.5, 1.3, and 0.6 nM. (B) ΔK-scuPA concentrations are 10, 5, 2.5, and 1.3 nM. The lines represent the best global fit to a 1:1 binding model.
Competition-inhibition experiments
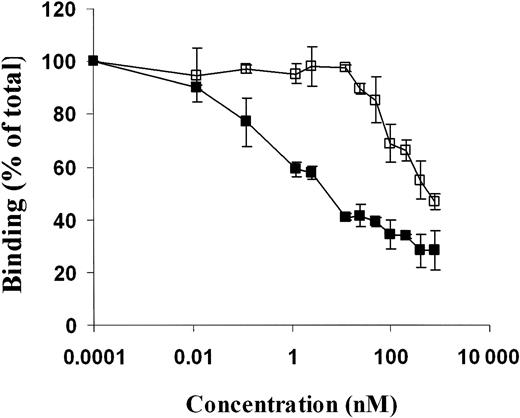
These binding experiments led us to conclude that the binding of scuPA to suPAR is mediated primarily by the GFD, as postulated by many groups, but that the kringle contributes to the stability of the resultant complex. Because the signal generated by GFD itself was too low to analyze reliably by surface plasmon resonance and radiolabeling abrogates its binding to uPAR, we turned to competition-inhibition experiments to explore the mechanism of action of the kringle in greater detail. To do so, we first compared the capacity of scuPA to be displaced by ATF (amino acid 1-135) and its GFD (amino acids 4-43) from its binding to immobilized suPAR measured by surface plasmon resonance. Binding of ATF to suPAR was more stable than was GFD. ATF added at 200-fold molar excess totally displaced scuPA binding, whereas only about 50% of scuPA was displaced by GFD at the same molar ratio (Figure 3). In contrast, binding of ΔK-scuPA was totally displaced by the same fold molar excess of both GFD and ATF (Figure 4). LMW-uPA and ΔGFD-scuPA served as negative controls and showed no detectable binding to suPAR, as expected (data not shown). This result strongly suggests the kringle helps to stabilize the binding of scuPA to suPAR by providing a second low-affinity binding site beside the GFD or by interacting with the GFD whether it is part of the intact molecule or of its amino-terminal fragment. Because these studies were performed using soluble uPAR, we then compared the capacity of ATF and GFD to inhibit the binding of 125I-scuPA to GPI-anchored uPAR expressed on WT CHO cells. Again, binding of 125I-scuPA was inhibited almost totally by ATF at concentrations at which GFD inhibited labeled ligand by only 60% (Figure 5), a figure in close agreement to the 50% inhibition by GFD seen by SPR. However, GFD totally inhibited uPA binding at higher concentrations, affirming this fragment retained its capacity to bind to uPAR.
Effect of GFD and ATF on binding of WT-scuPA to suPAR measured as surface plasmon resonance. Binding of scuPA to suPAR was measured using a BIA X optical Biosensor (Biacore). WT-suPAR was coupled to a CM5-research grade sensor chip. (A) Buffer was injected for 3 minutes (no wash) followed by a 3-minute injection of 2 nM scuPA with a 3-minute wash delay (black); 400 nM GFD was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA for 3 minutes with a 3-minute wash delay (gray). (B) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was then injected for 3 minutes (no wash) followed by a mixture of 400 nM ATF plus 2 nM scuPA for 3 minutes with a 3-minute wash delay (gray). Mixtures of scuPA with GFD or ATF were incubated for 30 minutes prior to injection. Data were collected at a flow rate of 30 μL/min with the data collection rate set to “high” (5 data points per second).
Effect of GFD and ATF on binding of WT-scuPA to suPAR measured as surface plasmon resonance. Binding of scuPA to suPAR was measured using a BIA X optical Biosensor (Biacore). WT-suPAR was coupled to a CM5-research grade sensor chip. (A) Buffer was injected for 3 minutes (no wash) followed by a 3-minute injection of 2 nM scuPA with a 3-minute wash delay (black); 400 nM GFD was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA for 3 minutes with a 3-minute wash delay (gray). (B) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was then injected for 3 minutes (no wash) followed by a mixture of 400 nM ATF plus 2 nM scuPA for 3 minutes with a 3-minute wash delay (gray). Mixtures of scuPA with GFD or ATF were incubated for 30 minutes prior to injection. Data were collected at a flow rate of 30 μL/min with the data collection rate set to “high” (5 data points per second).
The effect of GFD and ATF on ΔK-scuPA binding to suPAR measured as surface plasmon resonance. ΔK-scuPA binding to recombinant suPAR was monitored as described in Figure 2. (A) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM GFD was then injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (gray). (B) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was then injected for 3 minutes (no wash) followed by a mixture of 400 nM ATF plus 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (gray). Mixtures of ΔK-scuPA with GFD or ATF were incubated for 30 minutes prior to injection. Data were collected at a flow rate of 30 μL/min with the data collection rate set to “high” (5 data points per second).
The effect of GFD and ATF on ΔK-scuPA binding to suPAR measured as surface plasmon resonance. ΔK-scuPA binding to recombinant suPAR was monitored as described in Figure 2. (A) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM GFD was then injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (gray). (B) Buffer was injected for 3 minutes (no wash) followed by an injection of 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was then injected for 3 minutes (no wash) followed by a mixture of 400 nM ATF plus 2 nM ΔK-scuPA for 3 minutes with a 3-minute wash delay (gray). Mixtures of ΔK-scuPA with GFD or ATF were incubated for 30 minutes prior to injection. Data were collected at a flow rate of 30 μL/min with the data collection rate set to “high” (5 data points per second).
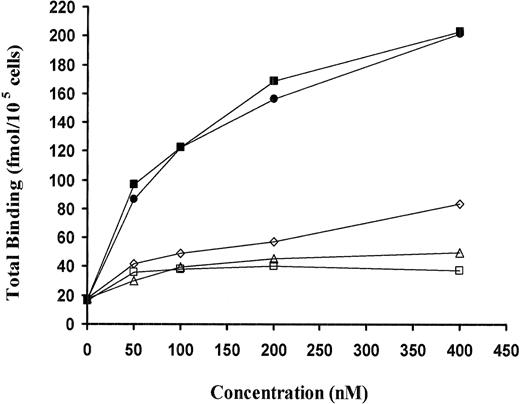
Inhibition of binding of WT-scuPA to uPAR by variant scuPA. (A) Binding of 125I-scuPA to CHO cells line expressing GPI-anchored uPAR was measured in the absence and presence of scuPA variants. CHO cells were prechilled for 30 minutes and washed twice with binding buffer (PBS, 1% BSA). Cells were then incubated with 125I-scuPA (10 nM) in the presence of the indicated concentrations (0.012 to 800 nM) of GFD (□) or ATF 1-135 (▪) for 1 hour at 4°C. The cells were washed 4 times with binding buffer, and the cell-associated ligand was eluted with glycine, pH 2.8, and counted. The mean ± SD of 3 experiments is shown.
Inhibition of binding of WT-scuPA to uPAR by variant scuPA. (A) Binding of 125I-scuPA to CHO cells line expressing GPI-anchored uPAR was measured in the absence and presence of scuPA variants. CHO cells were prechilled for 30 minutes and washed twice with binding buffer (PBS, 1% BSA). Cells were then incubated with 125I-scuPA (10 nM) in the presence of the indicated concentrations (0.012 to 800 nM) of GFD (□) or ATF 1-135 (▪) for 1 hour at 4°C. The cells were washed 4 times with binding buffer, and the cell-associated ligand was eluted with glycine, pH 2.8, and counted. The mean ± SD of 3 experiments is shown.
The kringle of uPA contributes to scuPA-suPAR enzymatic activity
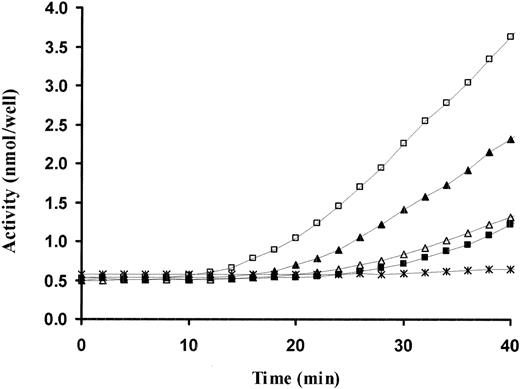
As a second, independent approach, we next examined the role of the kringle in suPAR-mediated activation of scuPA. We have previously observed that the plasminogen activator activity of the scuPA-suPAR complex requires persistent contact between the reactants and is dissipated when scuPA is displaced from suPAR by ATF.19 We therefore compared the capacity of GFD, ATF, and a catalytically inactive full-length uPA variant (scuPA-Ser356Ala) to compete with WT-scuPA for binding to suPAR and thereby inhibit plasminogen activator activity. ATF and scuPA-Ser356Ala added at 100-fold molar excess clearly inhibited the plasminogen activator activity of WT-scuPA-suPAR, whereas GFD at the same molar excess caused complete inhibition at the initial time points, again confirming the integrity of the fragment, but caused only partial inhibition after the first 20 minutes (Figure 6). Of note, the activity of scuPA-suPAR was reduced by ATF to the same level as scuPA alone, as previously reported,19 whereas the activity of both scuPA and scuPA-suPAR was even lower in the presence of 100-fold excess scuPA-Ser356Ala.50,51 The finding that ATF is a more effective/durable inhibitor of scuPA-suPAR activity than GFD suggests that the kringle contributes to the scuPA-suPAR interaction but does not exclude potential effects on the evolution of the catalytic site or an interaction with plasminogen that prevents its conversion to plasmin.
Inhibition of scuPA-suPAR-mediated plasminogen activator activity by scuPA variants. Plasminogen activation was measured using a chromogenic assay. scuPA (2 nM) alone (▪) or complexed with 4 nM suPAR (□) was incubated in the absence and presence of 100-fold molar excess of GFD (▴), ATF 1-135 (▵), or scuPA-S356A (*). Glu-plasminogen (20 nM) and plasmin chromogenic substrate (500 μM) were added, and the optical density at 405 nm was measured continuously over time at 37°C. The data shown are from 1 experiment representative of 3 so performed.
Inhibition of scuPA-suPAR-mediated plasminogen activator activity by scuPA variants. Plasminogen activation was measured using a chromogenic assay. scuPA (2 nM) alone (▪) or complexed with 4 nM suPAR (□) was incubated in the absence and presence of 100-fold molar excess of GFD (▴), ATF 1-135 (▵), or scuPA-S356A (*). Glu-plasminogen (20 nM) and plasmin chromogenic substrate (500 μM) were added, and the optical density at 405 nm was measured continuously over time at 37°C. The data shown are from 1 experiment representative of 3 so performed.
The uPA kringle promotes interdomain cooperativity in suPAR
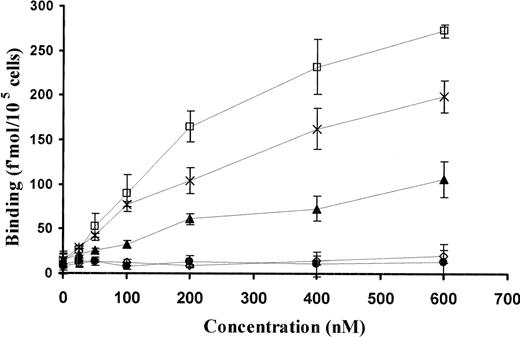
To investigate the mechanism by which the kringle cooperates with GFD to stabilize binding of uPA to its receptor in greater detail, the capacity of WT and variant uPAs to form a ternary complex with isolated domains of suPAR was examined.25 We have previously reported that WT-scuPA in solution at concentrations above those used to measure binding to intact uPAR is capable of forming a ternary complex with soluble domain I of uPAR and cell-associated or soluble domains II+III, a measure of receptor interdomain cooperativity. This interdomain cooperativity was confirmed using soluble domain I and domain II+III and CHO cells expressing GPI-anchored scuPA.26 ATF and the full-length molecule showed a similar ability to form a ternary complex with the isolated domains of uPAR (Figure 7). In contrast, GFD alone or in presence of isolated kringle was unable to form a ternary complex with the isolated receptor domains, although it inhibited the binding scuPA to intact uPAR (Figure 5). Moreover, the ability of ΔK-scuPA to form a ternary complex with the receptor fragments was greatly impaired relative to the wild-type molecule and was only slightly above the levels seen with LMW-uPA (135-411) or ΔGFD-scuPA, a variant lacking the GFD but containing the kringle linked to LMW-uPA, both of which are unable to bind to uPAR. These data indicate that the GFD must be present for the kringle to generate interdomain cooperativity within uPAR.
Binding of sDI to DII+DIII-uPAR requires GFD and kringle domains. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-anchored uPAR DII+DIII was measured in the presence of the indicated (0 to 600 nM) concentrations of WT-scuPA (□), GFD (○), ATF 1-135 (×), ΔGFD-scuPA (•), and ΔK-scuPA (▴). Cells were incubated at 4°C for 1 hour, washed 4 times with binding buffer, and the cell-bound ligand was eluted with glycine, pH 2.8, and counted. The mean ± SD of 3 experiments is shown.
Binding of sDI to DII+DIII-uPAR requires GFD and kringle domains. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-anchored uPAR DII+DIII was measured in the presence of the indicated (0 to 600 nM) concentrations of WT-scuPA (□), GFD (○), ATF 1-135 (×), ΔGFD-scuPA (•), and ΔK-scuPA (▴). Cells were incubated at 4°C for 1 hour, washed 4 times with binding buffer, and the cell-bound ligand was eluted with glycine, pH 2.8, and counted. The mean ± SD of 3 experiments is shown.
We then asked whether the kringle reconstituted interdomain cooperatively in uPAR through direct binding to the receptor. No specific binding of recombinant kringle to suPAR was observed using surface plasmon resonance measurements made at concentrations as high as 1 μM or to cell-associated uPAR using a radiolabeled ligand binding assay (not shown). Moreover, ΔGFD-scuPA did not bind to suPAR or cell-associated uPAR, nor did it express enzymatic activity in the presence of suPAR (data not shown). These data support the concept that the kringle enhances stability of the complex through an effect on GFD, although the possibility that a receptor-binding epitope is not exposed in the isolated kringle cannot be excluded.
Interaction between the kringle and GFD promotes receptor binding
The data to this point suggest that the kringle supports the GFD to induce formation of a more stable complex with uPAR. This presupposes that the kringle and GFD can interact with one another and that the kringle and GFD may mutually constrain their flexibility and/or relative orientation in the wild-type molecule. To begin to examine this possibility, we aligned the kringles from the 15 available nuclear magnetic resonance (NMR) structures of ATF.52 Four of these structures (chosen at random) are shown in Figure 8. When the kringles from all structures are closely aligned, the position of the GFD is seen to be quite variable with respect to the kringle in 3-dimensional space. Based on this observation, we posited that residues 43-49 that connect the GFD with the kringle form a functional unit that is highly flexible and enables new and stable interactions to develop between these 2 domains. In view of our observation that both domains are required to optimize complex stability, we further postulated that such interactions might be important for binding to uPAR. If so, perturbations in this region might interfere with kringle-GFD interactions that we postulate occur in the wild-type molecule.
NMR structures of ATF. Shown are 4 structures representative of the 15 NMR published structures of ATF.55 Each structure is displayed in a separate color. Residues 105-110 (PDNRRR) are rendered as space-filling models. Of note is the variability in the spatial relationship between the GFD and kringle domains.
NMR structures of ATF. Shown are 4 structures representative of the 15 NMR published structures of ATF.55 Each structure is displayed in a separate color. Residues 105-110 (PDNRRR) are rendered as space-filling models. Of note is the variability in the spatial relationship between the GFD and kringle domains.
We were unable to measure the binding of isolated kringle to isolated GFD directly by plasmon resonance. It could be argued therefore that the integrity of the isolated GFD fragment was compromised, although the same fragment inhibits scuPA to uPAR-expressing CHO cells (Figure 5). Nevertheless, to test this hypothesis and to address this issue, we produced a series of uPA variants designed to disengage the proposed cooperation between the native GFD and the native kringle within the full-length molecule. To do so, we first analyzed a uPA variant in which the 7 amino acids in this putative linker region were converted to alanine (scuPA-43-49Ala; Table 1) to generate an α-helical structure.53 scuPA-43-49Ala showed an even lower affinity for suPAR than did ΔK-scuPA and was incapable of reconstituting the isolated uPAR domains (Figure 9). The same results were seen when amino acids 45-DKSK-48 (designated ΔLeu45-48-scuPA) in the linker were deleted or when a proline was introduced at position 48 (scuPA-Lys48Pro) to limit the flexibility between the GFD and the kringle. In contrast, a conservative mutation in the linker (scuPA-Ser47Gly) had no effect on scuPA binding or its capacity to reconstitute the uPAR domains (Figure 9). GFD totally blocked the binding of scuPA-Lys48Pro to suPAR, as was seen with ΔK-scuPA, whereas scuPA-Ser47Gly behaved like the wild-type molecule (Figures 10, 3, and 4).
Binding of sDI to DII/DIII-uPAR mediated by the scuPA linker mutants. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-DII-DIII of uPAR was measured in the presence of the indicated (0 to 400 nM) concentrations of WT-scuPA (▪), scuPA-43-49Ala (▵), ΔLeu43-45-scuPA (□), scuPA-Lys48Pro (⋄), or scuPA-Ser47Gly (•). Binding was measured as described in the legend to Figure 7. The data shown are from a single experiment representative of 2 so performed.
Binding of sDI to DII/DIII-uPAR mediated by the scuPA linker mutants. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-DII-DIII of uPAR was measured in the presence of the indicated (0 to 400 nM) concentrations of WT-scuPA (▪), scuPA-43-49Ala (▵), ΔLeu43-45-scuPA (□), scuPA-Lys48Pro (⋄), or scuPA-Ser47Gly (•). Binding was measured as described in the legend to Figure 7. The data shown are from a single experiment representative of 2 so performed.
Effect of GFD and ATF on the binding of scuPA linker mutants to suPAR measured as surface plasmon resonance. Binding of scuPA-Lys48Pro (A-B) and scuPA-Ser47Gly (C-D) to recombinant suPAR was monitored as described in the legends to Figures 2 and 5. (A,C) Buffer was injected for 3 minutes (no wash) followed by 2 nM scuPA-Lys48Pro (A) or scuPA-Ser47Gly (C) for 3 minutes with a 3-minute wash delay (black); 400 nM GFD was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA mutant for 3 minutes with a 3-minute wash delay (gray). (B,D) Buffer was injected for 3 minutes (no wash) followed by 2 nM scuPA-Lys48Pro (B) or scuPA-Ser47Gly (D) for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA mutant for 3 minutes with a 30-minute wash delay (gray).
Effect of GFD and ATF on the binding of scuPA linker mutants to suPAR measured as surface plasmon resonance. Binding of scuPA-Lys48Pro (A-B) and scuPA-Ser47Gly (C-D) to recombinant suPAR was monitored as described in the legends to Figures 2 and 5. (A,C) Buffer was injected for 3 minutes (no wash) followed by 2 nM scuPA-Lys48Pro (A) or scuPA-Ser47Gly (C) for 3 minutes with a 3-minute wash delay (black); 400 nM GFD was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA mutant for 3 minutes with a 3-minute wash delay (gray). (B,D) Buffer was injected for 3 minutes (no wash) followed by 2 nM scuPA-Lys48Pro (B) or scuPA-Ser47Gly (D) for 3 minutes with a 3-minute wash delay (black); 400 nM ATF was injected for 3 minutes (no wash) followed by a mixture of 400 nM GFD plus 2 nM scuPA mutant for 3 minutes with a 30-minute wash delay (gray).
Identification of kringle epitopes involved in stabilization of receptor binding
It can be argued that removing the entire kringle domain from scuPA might have a nonspecific impact on the binding of the remaining domains to uPAR. To address this possibility, point mutations were introduced into the native kringle to identify the portions required for cooperative interaction with the GFD within the full-length molecule. We began by examining the contribution of amino acids 105-110 in the kringle, which have been implicated previously in binding to heparin and PAI-1.40,50,54,55 This portion of the kringle is found in apposition to the GFD in several of the published NMR structures, consistent with the flexibility of the ATF. To examine the involvement of this region within the full-length molecule, we generated a scuPA-kringle variant (designated scuPA-Lys105-110Ala) in which each of the charged amino acids implicated in heparin binding were mutated to alanine. Binding of scuPA-Lys105-110Ala to suPAR, like ΔK-scuPA (Figure 4), was comparably inhibited by ATF and GFD (data not shown), unlike the binding of WT-scuPA (Figure 3). ΔK-scuPA and scuPA-Lys105-110Ala each required a 5- to 10-fold higher concentration to inhibit binding by 50% (IC50) to CHO-uPAR than WT-scuPA (not shown). Moreover, scuPA-Lys105-110Ala, like ΔK-scuPA, was unable to fully reconstitute the isolated fragments of suPAR (Figure 11). In contrast, mutation of amino acids 136-143, immediately C-terminal to the kringle (ΔCP-scuPA), had no effect on scuPA binding to uPAR. Variants containing 1, 2, or 3 of the charged amino acids mutated to alanine (scuPA-Lys110Ala, scuPA-Lys108-110Ala, and scuPA-Lys107-109Ala) were approximately 80% as effective as WT-scuPA or the variant scuPA-Lys107Ala. These data indicate that both the net positive charge and specific residues in this region of the kringle contribute to stability of the scuPA-suPAR complex.
Binding of sDI to DII+DIII-uPAR in the presence of scuPA kringle mutants. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-anchored uPAR DII+DIII was measured in the presence of the indicated (0 to 400 nM) concentrations of WT-scuPA (▪), scuPA-Lys105-110Ala (□), scuPA-Lys107-109Ala (▴), scuPA-Lys108-110Ala (×), scuPA-Lys110Ala (○), and scuPA-Lys107Ala (•). Binding was measured as described in the legend to Figure 7. The data shown are from a single experiment representative of 2 so performed.
Binding of sDI to DII+DIII-uPAR in the presence of scuPA kringle mutants. Binding of 125I-sDI (50 nM) to 293 cells expressing GPI-anchored uPAR DII+DIII was measured in the presence of the indicated (0 to 400 nM) concentrations of WT-scuPA (▪), scuPA-Lys105-110Ala (□), scuPA-Lys107-109Ala (▴), scuPA-Lys108-110Ala (×), scuPA-Lys110Ala (○), and scuPA-Lys107Ala (•). Binding was measured as described in the legend to Figure 7. The data shown are from a single experiment representative of 2 so performed.
Discussion
The diverse physiologic and pathophysiologic activities attributed to urokinase/urokinase receptor system demonstrate that these proteins develop complex pathways of cross-talk with other cellular partners. The capacity of uPAR, which lacks a transmembrane domain, to interact with low density lipoprotein related receptor (LRP),56 various integrins,8,57 and integrin ligands7,37 and the modulation of several of these interactions by uPA4,31,58 suggests the uPA-uPAR complex can adopt multiple conformations, which, in turn, govern its biologic behavior.8 The process by which the uPA-uPAR complex assumes these additional and diverse conformations and activities is largely unknown. It is clear, however, that such dynamic biologic activities are unlikely to be explained by a simple binding process mediated exclusively through a limited portion of the growth factor domain.28-33
The results of the present study show that the kringle contributes to the interaction of uPA with uPAR and that this contribution is dependent on the GFD. This conclusion is based on several findings. First, a deletion mutant of scuPA lacking the kringle (ΔK-scuPA), while showing a similar association constant, dissociates more rapidly from soluble uPAR than does its wild-type counterpart. This impact was readily apparent with a more prolonged dissociation phase than has been explored previously9 and with suPAR oriented on the sensor surface in a homogeneous manner using a monoclonal nonblocking antibody that simulated the cellular context, allowing less direct influence by the dextran surface.59 Second, the amino-terminal fragment (ATF) of scuPA, which contains the GFD linked to the kringle, inhibits binding of full-length scuPA to suPAR more effectively than does the GFD fragment alone. Third, whereas the apparent Kd's of ATF and full-length scuPA calculated from the initial “on-rate” an “off-rates” are nearly identical,28 ATF but not GFD is able to displace full-length scuPA from suPAR upon incubation. This is consistent with some reports, but not others, in which differences between the affinities of full-length scuPA and its fragments to uPAR have been noted.16,28,31,36,60 Fourth, ATF and full-length scuPA reconstitute the separate domains of suPAR into a stable unit, whereas GFD does not. Fifth, deletion of the kringle almost completely abrogates the capacity of uPA to form a ternary complex with isolated domains of uPAR, a more stringent test of interdomain cooperativity caused by the greater flexibility and degree of freedom of the fragments than when they are part of the intact receptor.26
This latter finding suggests a new paradigm for understanding the uPA-uPAR interaction. It is known that interdomain cooperation26,61,62 is needed for optimal binding of uPA (ie, neither the isolated domains nor combinations of any 2 domains of uPAR binds uPA with high affinity). These data indicate that similar cooperation among the domains of uPA is required to optimize binding to uPAR. Although the kringle participates in the binding of uPA to uPAR, the isolated kringle itself did not bind to the receptor and nor did it inhibit the binding of scuPA to suPAR, indicating that the effect of the kringle requires the presence of the GFD. This is similar to the requirement for domains II and III of uPAR for high-affinity binding of uPA to domain I. We postulate that this difference indicates that the kringle in the full-length molecule serves a secondary function (ie, to stabilize the GFD-mediated binding to uPAR). In accordance with this postulate, kinetic analysis of the optical biosensor data shows that ΔK-scuPA binds to suPAR with a similar apparent “on-rate” as the wild-type molecule, but with an increased apparent “off-rate,” supporting the concept that the role of the kringle is to stabilize the scuPA-uPAR interaction once formed.
There are several potential mechanisms by which the kringle may contribute to the stabilization of binding. The kringle may interact with GFD within scuPA. This interaction may help to maintain the GFD in an optimal configuration for binding to uPAR. An alternative, but not mutually exclusive, possibility is an “induced-fit” model in which initial binding of GFD to uPAR generates conformational changes within scuPA and/or uPAR that expose additional receptor binding sites in the GFD, which require the kringle for their expression. Lastly, binding of GFD may induce binding sites in uPAR for the kringle itself.
The results of our experiments provide support for the first of these proposed mechanisms. The NMR structure of ATF suggests the GFD and the kringle can assume multiple orientations with respect to each other, rotating around a linker sequence comprising amino acids 43-49.52 Mutations introduced into this region designed to generate a more orderly α-helical conformation, to shorten the linker, or to distort its flexibility by introducing a proline at position 48 each resulted in variants that showed a dissociation rate constant comparable to ΔK-scuPA. Moreover, like ΔK-scuPA, binding of each of these linker variants to suPAR was inhibited comparably by GFD and ATF, unlike WT-scuPA, which required the GFD to be linked to the kringle to be inhibited. Thus, as observed with deletion of the kringle itself, mutations in the linker sequence disengage the cooperation between the GFD and kringle and cause the binding of scuPA to its receptor to depend solely on the GFD. These results involving point mutations within the full-length molecule also exclude the possibility that these results are simply due to perturbation in the function of the isolated GFD fragment or a destructive effect on the interaction of scuPA with suPAR as a result of domain deletion.
Although these results provide additional support for the hypothesis that the kringle helps to stabilize the interaction between GFD and uPAR, they neither exclude a direct interaction of the kringle or the linker region itself with uPAR nor an interaction of the linker region with the GFD that extends its β sheet strand.63 Additional experiments will be required to elucidate the relative contribution of the 2 mechanisms. Regardless of the mechanism, these added contacts are likely to play role in pairing of the uPA-uPAR complex with its various transmembrane adapters implicated in cell signaling, cell proliferation, cell adhesion, and cell migration.8,9,37,56,64-69 Strategies to interrupt the cooperative interactions between the GFD and kringle may permit selective interruption of uPAR-mediated proteolysis versus uPAR-mediated interaction with specific cellular partners.
Prepublished online as Blood First Edition Paper, July 24, 2003; DOI 10.1182/blood-2003-03-0949.
Supported by grants HL60169, HL66442, and HL67381 from the National Institutes of Health (D.B.C. and A.A.H.) and a Beginning Grant-In-Aid Award 0060251U from the Mid-Atlantic Division of the American Heart Association (K.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank G. Cohen and R. Eisenberg from the University of Pennsylvania School of Dental Medicine for the use of the BIACORE X.

![Figure 1. Dissociation of radiolabeled WT- and ΔK-scuPA from soluble uPAR. suPAR-coated wells were incubated with 125I-WT-scuPA (5 [▾], 10 [▴], and 20 nM [▪]) or 125I-ΔK-scuPA (5 [▿], 10 [▵], and 20 nM [□]) for 2 hours at 22°C. Wells were washed 4 times, and the released radioactive ligand was counted at the indicated times. The lines represent the best fit to the single exponential decay model as determined using GraphPad software.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/10/10.1182_blood-2003-03-0949/6/m_h82235250001.jpeg?Expires=1767726609&Signature=skAaTnPr3MGZ5UXlE8yQGfxbV1Q-BtQkLGEQb49iHx-y7Uw58bBFrP4jaoFgvXk31Xyn9tIjMAV59l05KGLvfq1HWbuCgPQTlQSPDeQUde8fNoexdmVf8SGrC5KWAv1TvI1DKxoHldJVs7iEy0Ku7hVPKxQ1Pm8vLfCRMd6O6RaAIhuT2cR73Q9ihHTR-Z-MyelAtzYTokDWrgDqb3Np5EjGvnHkL~DknIXRkzSKxT3Urq2h2IEWNz0ukAjT4hduj4jt3VFzBTCiea9K4H7~Cxk3X16I4~4bMfLMmA~pRbmHLobJvv8L2ehDZG1TaAAi7lQSiX7SlEB0ZhgHheD~xw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal