Abstract
Hodgkin disease (HD) represents a malignant lymphoma in which the putative malignant Hodgkin and Reed-Sternberg cells are rare and surrounded by abundant reactive nonmalignant cells. It has been suggested that cytokines such as interleukin-6 (IL-6) are involved in the pathogenesis of the disease. The expression of the IL-6 receptor (IL-6R) complex and its link to the activation of signal transducers and activators of transcription (STAT) molecules in HD cell lines was investigated. Gel retardation and Western blot analyses revealed a high level of constitutively activated STAT3 in 5 of 7 HD cell lines, which could not be detected in Burkitt lymphoma cell lines. Different levels of IL-6R protein were measured in various HD cell lines: L428 and Dev cells were characterized by very low levels of gp80 and gp130, on KMH2 cells only gp130 but no gp80 was detected, whereas L540, L591, HDLM2, and L1236 were positive for both gp80 and gp130, suggesting a possible autocrine stimulation of STAT3. However, a further increase in STAT3 activation on IL-6 or IL-6/soluble IL-6R stimulation was not observed. Neutralizing monoclonal antibodies against IL-6, gp80, gp130, or both receptor subunits did not affect the proliferation or the constitutive activation of STAT molecules in HD cell lines. However, the tyrosine kinase inhibitor AG490 blocked the constitutive activation of STAT3 and inhibited spontaneous growth of HD tumor cells. The evidence suggests abnormal STAT signaling and growth regulation in Hodgkin cell lines.
Introduction
Hodgkin disease (HD) is characterized by a disruption of the lymph node architecture with low numbers of Hodgkin and Reed-Sternberg (H-RS) cells surrounded by an abundance of reactive nonmalignant cells.1,2 This histologic pattern of HD may be due to the production and release of cytokines from H-RS cells that are involved in the growth of the lymphoma cells and the interactions with bystander cells.3-5
Recently it has been shown that HD cell lines and primary H-RS cells express interleukin-6 (IL-6) and the α-chain of the IL-6 receptor (IL-6R; gp80).3,4,6 IL-6 is synthesized by a variety of cells on stimulation and acts on a wide range of different target cells to regulate cell growth, differentiation, or gene expression.7,8 IL-6 may act as an autocrine or paracrine growth factor for multiple myeloma, Epstein Barr virus (EBV)–immortalized B cells, and perhaps for H-RS cells, although this has not been proven yet for HD.3,7,9-13 IL-6 exerts its action via a cell surface receptor complex consisting of at least 2 subunits: the IL-6R gp80 and the signal transducer gp130.8,14 It has been shown that gp80 is not expressed on normal resting B cells and most Burkitt lymphoma (BL) cell lines15; gp130 is shared by a number of cytokines as a signal transducing receptor component that does not bind IL-6 by itself.7,8,16 The binding of IL-6 to gp80 leads to an association and dimerization of gp130, followed by the rapid activation of tyrosine kinases of the Janus family (Jak) and a subsequent activation of transcription factors of the signal transducers and activators of transcription (STAT) family.17-19 STAT proteins are latent cytoplasmic transcription factors that become activated by tyrosine phosphorylation in response to a number of cytokines. STATs become phosphorylated and translocate as homodimers and heterodimers to the nucleus, where they bind to defined DNA elements within the promoter region of target genes and activate their transcription.19-21 The activation by IL-6 is mainly associated with the tyrosine phosphorylation of STAT3.19,22 Besides this extracellular mechanism of STAT activation by cytokines evidence also suggests that intracellular events may influence the activation state of STAT proteins and render them independent of extracellular stimuli. Constitutive STAT activation has been reported by several authors for acute leukemias, multiple myeloma, or breast cancer.13 23-29
To further explore the putative role of IL-6 in HD, the expression of gp130 and the influence on the activation of STAT proteins in HD cell lines was analyzed. Neutralizing monoclonal antibodies against IL-6, gp80, gp130, or both receptor subunits did not affect the proliferation of the HD cells or the constitutive activation of STAT molecules. However, the tyrosine kinase inhibitor AG490 inhibited both the constitutive activation of STAT3 and the growth of Hodgkin cell lines in vitro.
Materials and methods
Cell lines
The HD cell lines L1236, L428, L540, HDLM2, Dev, and KMH2 were described previously.30 31 U266 is a multiple myeloma cell line; HepG2 is a hepatocyte-derived cell line; OciAML2 and NB4 are acute myeloid leukemia (AML) cell lines; DG75 is a BL cell line; and Jurkat is a T-cell line. All cell lines were obtained from the Deutsche Sammlung für Mikroorganismen (DSM, Braunschwieg, Germany). BL2, mutucl59, chepBL, and BL60 are BL cell lines (kindly provided by M. Falk, G. Lenoir, and J. Wolf). The cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum and glutamine, penicillin, and streptomycin at cell densities between 2 × 105 and 1 × 106 cells/mL. At 18 hours before stimulation, cells were washed and transferred to fresh medium (all cell culture reagents were obtained from Sigma-Aldrich, Deisenhofen, Germany). For stimulation with IL-6 or IL-6 and soluble IL-6R (sIL-6R), cells were washed twice with cold phosphate-buffered saline (PBS), resuspended in serum-free medium supplemented with 0.5% bovine serum albumin (BSA) and glutamine/penicillin/streptomycin at 5 × 105cells/mL, and incubated with the cytokine for 30 minutes before preparing nuclear extracts.
Cytokines
Human recombinant IL-6 was purchased from Life Technologies (Heidelberg, Germany) and used at a concentration of 250 U/mL. Recombinant sIL-6R was prepared as described previously and used at 1 μg/mL.32
Immunostaining of proteins
For Western blot analysis cells were harvested and washed with PBS. The cellular extracts were prepared in 2 × Laemmli buffer, boiled for 10 minutes, and chilled on ice. Proteins were separated on a discontinuous sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with a 7.5% acrylamide resolving gel, before electroblotting onto nitrocellulose filters (Hybond C extra, Amersham-Pharmacia, Freiburg, Germany). Equal loading of the gels was verified by Ponceau S staining. After incubation with blocking buffer for 1 hour, the blots were incubated with monoclonal mouse α-gp130 B-T12 (1 μg/mL) overnight at 4°C, washed 3 times with triethanolamine-buffered saline (TBS)/0.05% Tween and incubated with the peroxidase-coupled second antibodies (goat α-mouse horseradish peroxidase [HRP; Dako, Hamburg, Germany]).16The blots were developed using the enhanced chemiluminescence (ECL) system (Amersham-Pharmacia) according to the manufacturer's instructions.
Phosphorylated STAT3 proteins were detected by Western blot analysis separating 20 μg nuclear extracts on a discontinuous SDS-PAGE with a 10% acrylamide resolving gel, before electroblotting onto nitrocellulose filters (Hybond C extra). Equal loading of the gels was verified by Ponceau S staining (Biorad, München, Germany). After incubation with blocking buffer for 1 hour the blots were incubated with a rabbit polyclonal antibody directed against phospho-specific STAT3 (New England Biolabs, Schwalbach, Germany) at a dilution 1:1000 overnight at 4°C, washed 3 times with TBS/0.05% Tween, and incubated with the peroxidase-coupled second antibodies (goat α-rabbit HRP 1:2000 [New England Biolabs]). The blots were developed using the ECL system. After that the filters were stripped and reprobed with rabbit polyclonal antibody directed against STAT3 (New England Biolabs) at a dilution 1:1000 overnight at 4°C, washed 3 times with TBS/0.05% Tween, and incubated with the peroxidase-coupled secondary antibodies (goat α-rabbit HRP 1:2000 [New England Biolabs]) and developed using the ECL system.
Immunofluorescence analysis
For detection of cell surface antigen expression, 1 × 106 cells were washed twice in PBS supplemented with 0.5% BSA. Cells were incubated with monoclonal antibodies (5-10 μg/mL) for 10 minutes on ice in the dark. Cells were washed with PBS, 0.5% BSA, and stained with phycoerythrin (PE)–conjugated goat α-mouse Ig antibodies.33 34
Monoclonal antibodies used and their sources were as follows: α-gp130 (clones B-S12, B-R3), α-gp80 (clones BN12, B-R6), α-IL-6 (clones B-E8, B-F6) (Diaclone, Besancon, France); α-gp130 (clone AM64, Becton Dickinson, Heidelberg, Germany); goat α-mouse Ig-PE (Dako).16 Flow cytometry was performed using a FACScan (Becton Dickinson). Data are presented as specific mean linear fluorescence intensity after subtraction of background staining with isotype-matched control. Dead cells were excluded by propidium iodide staining.
IL-6–enzyme-linked immunosorbent assay
Cells (5 × 105/mL) were cultured for 24 hours in RPMI 1640 complete medium. The IL-6 content in the supernatants was measured by a specific enzyme-linked immunosorbent assay (ELISA) (Diaclone). The lower detection level of this ELISA was 3.5 pg/mL.
Analysis of cell proliferation
DNA synthesis of cultured L428 HD cells (1 × 105cells/well) was monitored by 3H-thymidine incorporation (0.4 μCi/well) (Amersham-Pharmacia) during the last 16 hours of culture in the absence or presence of the tyrphostin AG490 at 50 μM or 100 μM (Calbiochem, Schwalbach, Germany). The tyrphostin was solved in DMSO. The cells cultured in the absence of the tyrphostin were incubated with a corresponding amount of DMSO as control. Cells were harvested and analyzed by liquid scintillation counting (Beckmann, Düsseldorf, Germany). Data are presented as relative inhibition of proliferation plus SD of triplicate measurements. The proliferation of L428 cells in the presence of DMSO was taken as 100%. Parallel cultures were analyzed for STAT3 and STAT1 activation.
Gel retardation assays
Nuclear extracts were prepared as described.35 Protein concentrations were measured using the Biorad protein assay (Bradford, Biorad). A double-stranded mutated SIE (sis-inducible element)-oligonucleotide from the c-fospromoter (m67SIE) was labeled by filling in 5′ protruding ends with the Klenow enzyme, using (α-32P)dATP (3.000 Ci/mmol; 10 mCi/mL [Amersham-Pharmacia]). Nuclear extracts containing 5 μg protein were incubated with about 10 fmol (10 000 cpm) of probe in gel shift incubation buffer: 10 mM HEPES pH 7.8, 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM MgCl2, 10% glycerol, 5 mM dithiothreitol, 0.7 mM phenylmethylsulfonyl fluoride, 0.1 mg/mL poly(dI-C), and 1 mg/mL BSA for 10 minutes at room temperature. The protein-DNA complexes were separated on a 4.5% polyacrylamide gel containing 7.5% glycerol in 0.25-fold triethanolamine-borat-EDTA (TBE) at 20 V/cm for 4 hours. Gels were fixed in a water solution of 10% methanol and 10% acetic acid for 30 minutes, dried, and autoradiographed on Kodak-Biomax films (Sigma-Aldrich). Oligonucleotides used were purchased from Eurogentec (Serain, Belgium) with the following sequences (lower case letters indicate the cohesive ends of the double-stranded oligonucleotide used for Klenow labeling, the mutations are given in underlined italics): human c-fos promoter.
STAT1/3 (m67SIE): 5′ gatcCGGGAGGGATTTACGGGAAATGCTA;
STAT1/3M1: 5′ gatcCGGGAGGGATTTACGCTAGATGCTA;
STAT1/3M2: 5′ gatcCGGGAGGCTAGTACGGGAAATGCTA;
Supershift antibodies against STAT1 (clone E-23), STAT3 (clone H-190), STAT5 (clone G-2), and PEA1 (clone L-173; PEA1 is an unrelated transcription factor and used as negative supershift control) were purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Supershifts were performed by incubating the nuclear extracts with the corresponding antibody before adding the radiolabeled oligonucleotide to the binding reaction mixture and incubating them for 45 minutes at room temperature.
Results
STAT3 is constitutively activated in Hodgkin cells
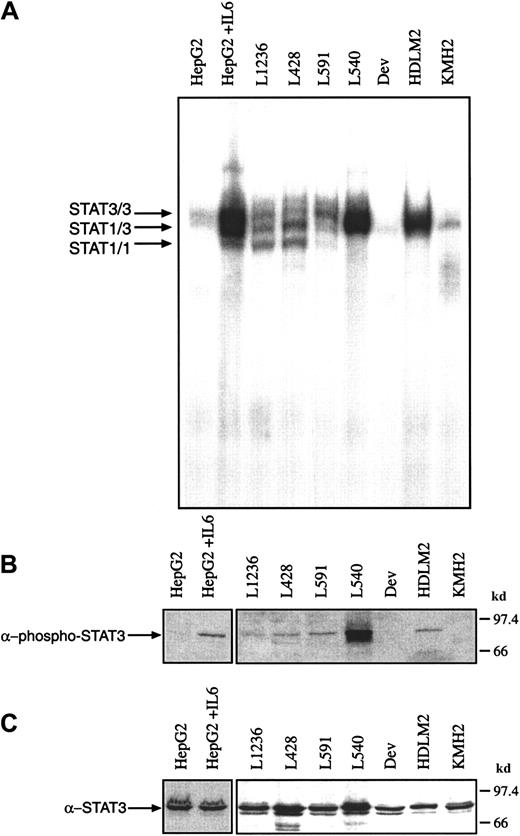
It has been shown previously that IL-6 and the IL-6R gp80 are expressed in H-RS cells. The binding of IL-6 to its receptor subunit gp80 is usually followed by an association of the IL-6/gp80 complex with 2 gp130 molecules. The formation of the high-affinity IL-6/IL-6R/gp130 complex is accompanied by the rapid activation of Janus kinases and STAT proteins. In some cases, however, it has been reported that STAT molecules are constitutively active in transformed cells. To analyze whether STAT proteins are constitutively activated in HD cell lines, gel retardation experiments (electrophoretic mobility shift assay [EMSA]) were performed. Nuclear extracts from untreated Hodgkin cells formed protein-DNA complexes with the promoter element of the c-fos gene as detected by EMSA (Figure1A). The intensity of the retarded bands varied between the different cell lines: L1236, L428, L591, L540, and HDLM2 cells were characterized by intense bands, whereas KMH2 and Dev cells showed only weak bands in the EMSA. In L428, L591, and L1236 cells the protein-DNA complexes comigrated with those induced by IL-6 in HepG2 hepatoma cells, previously identified as STAT3/3 and STAT3/1 and STAT1/1 homodimers and heterodimers. In L540 and HDLM2 cells the STAT3/3 homodimer was dominant. Repeated experiments showed that STAT3/3 homodimers were regularly detected, whereas STAT1/1 homodimers were not always visible in the EMSA.
Proteins from nuclear extracts from Hodgkin cell lines contain constitutively activated STAT3
. (A) Nuclear extracts (5 μg) of each cell line were analyzed in a gel retardation experiment using the radiolabeled STAT1/3 oligonucleotide. The DNA-protein complexes were separated by electrophoresis using a native 4.5% polyacrylamide gel as described in “Material and methods” and visualized by autoradiography. For comparison, nuclear extracts from unstimulated and IL-6–stimulated HepG2 cells were included in the study. Arrows show the position of STAT3 and STAT1 homodimers and heterodimers. (B) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3 and visualized by ECL. (C) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel B was reprobed with an antibody specific for STAT3 and the protein was visualized by ECL.
Proteins from nuclear extracts from Hodgkin cell lines contain constitutively activated STAT3
. (A) Nuclear extracts (5 μg) of each cell line were analyzed in a gel retardation experiment using the radiolabeled STAT1/3 oligonucleotide. The DNA-protein complexes were separated by electrophoresis using a native 4.5% polyacrylamide gel as described in “Material and methods” and visualized by autoradiography. For comparison, nuclear extracts from unstimulated and IL-6–stimulated HepG2 cells were included in the study. Arrows show the position of STAT3 and STAT1 homodimers and heterodimers. (B) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3 and visualized by ECL. (C) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel B was reprobed with an antibody specific for STAT3 and the protein was visualized by ECL.
The same Hodgkin cell lines were analyzed by Western blot experiments for the presence of phosphorylated and unphosphorylated STAT3 using corresponding specific antibodies (Figure 1B,C). Nuclear extracts (20 μg) from untreated Hodgkin cells were analyzed, demonstrating that only in those cells with specific protein-DNA complexes was phosphorylated STAT3 detected, whereas unphosphorylated STAT3 was expressed at comparable amounts in all HD cell lines analyzed (Figure1C).
In 2 representative HD cell lines, L428 (Figure2A) and L540 (Figure 2B), we tested the specificity of the protein-DNA interaction by competition with unlabeled oligonucleotides and with supershift assays. The incubation of nuclear extracts from L428 HD cells with an excess of unlabeled STAT1/3 oligonucleotides resulted in the disappearance of the STAT1 and STAT3 bands in EMSA (Figure 2A, lane 2), whereas in experiments with nuclear extracts from L540 HD cells the dominant STAT3/3 homodimer band disappeared (Figure 2B, lane 2). With oligonucleotide M1 containing point mutations in the core STAT binding site (“GGAA” to “CTAG”) no competition was detected (Figure 2A,B, lanes 3). Accordingly, when oligonucleotide M1 was used as the radiolabeled probe for gel retardation experiments no DNA-STAT complexes were formed (Figure 2A, lanes 5-8; Figure 2B, lanes 6-10). Mutations nearby or outside the core binding sequence for STAT1 and STAT3 in oligonucleotides M2 and M3 did not interfere with binding and competition was observed (Figure 2A,B, lanes 4; Figure 2B, lane 5). Antibodies directed against STAT1 or against STAT3 disrupted the respective DNA-protein complexes in nuclear extracts isolated from L428 cells (Figure 2A, lanes 9 and 10) and L540 cells (Figure 2B, lanes 11 and 12). The interaction of STAT3 with the corresponding site in the STAT1/3 oligonucleotide is very strong because the supershift antibody directed against STAT3 induced a supershift, but still there remained a DNA-protein complex visible (Figure 2B, lane 12). In these experiments in L428 cells the STAT1/1 homodimer-containing band was not as clearly visible as in the experiments shown in Figure 1. The antibodies directed against STAT5 or PEA1 used as specificity controls did not react with nuclear extracts from HD cell lines in EMSA (Figure 2A, lanes 11 and 12; Figure 2B, lanes 13 and 14). Taken together these experiments show that nuclear extracts from untreated HD cells contain activated STAT3 homodimers and heterodimers and to a much lower extent STAT1 homodimers, suggesting constitutive activation of these transcription factors in HD.
Activation of STAT molecules.
STAT molecules are constitutively activated in the HD cell lines L428 (A) and L540 (B). Analysis of the DNA-protein complexes formed between nuclear extracts from the L428 using radiolabeled oligonucleotides containing either the consensus or mutated STAT1/3 binding sites, by competition with corresponding unlabeled oligonucleotides. Supershift experiments were performed using antibodies directed against STAT1 and STAT3, and against STAT5 and PEA1 as negative controls. Nuclear extracts (5 μg) were analyzed in a gel retardation experiment using radiolabeled oligonucleotides. The nuclear extracts were incubated with the corresponding STAT1/3 (consensus) or STAT1/3-M1 (mutated GGAA to CTAG) oligonucleotide, the DNA-protein complexes were separated by electrophoresis using a native 4.5% polyacrylamide gel and visualized by autoradiography (Figure 1). Arrows show the position of STAT3 and STAT1 homodimers and heterodimers and supershifted bands; brackets and asterisks indicate the radiolabeled oligonucleotide used.
Activation of STAT molecules.
STAT molecules are constitutively activated in the HD cell lines L428 (A) and L540 (B). Analysis of the DNA-protein complexes formed between nuclear extracts from the L428 using radiolabeled oligonucleotides containing either the consensus or mutated STAT1/3 binding sites, by competition with corresponding unlabeled oligonucleotides. Supershift experiments were performed using antibodies directed against STAT1 and STAT3, and against STAT5 and PEA1 as negative controls. Nuclear extracts (5 μg) were analyzed in a gel retardation experiment using radiolabeled oligonucleotides. The nuclear extracts were incubated with the corresponding STAT1/3 (consensus) or STAT1/3-M1 (mutated GGAA to CTAG) oligonucleotide, the DNA-protein complexes were separated by electrophoresis using a native 4.5% polyacrylamide gel and visualized by autoradiography (Figure 1). Arrows show the position of STAT3 and STAT1 homodimers and heterodimers and supershifted bands; brackets and asterisks indicate the radiolabeled oligonucleotide used.
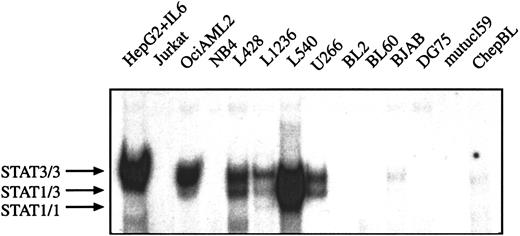
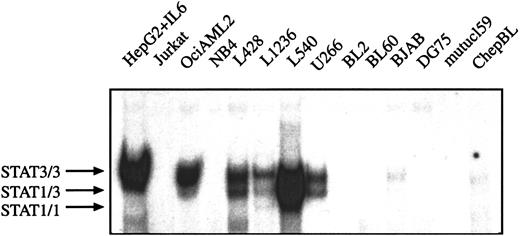
Comparison of STAT1 and STAT3 activation in Hodgkin, non-Hodgkin lymphoma, and acute leukemia cell lines
It has been described that myeloma and myeloid leukemia cell lines are characterized by constitutively activated STAT proteins.13 24 To analyze whether the constitutive STAT activation can also be observed in other hematopoetic malignant cells the DNA-protein interactions of nuclear extracts from HD, BL, multiple myeloma, and leukemia cell lines were compared in EMSA. The following cell lines were included in the analysis: chepBL and mutucl59 are EBV+ type I BL cell lines, BL60 is an EBV+ type II BL cell line, BL2 and DG75 are EBV− BL cell lines, and BJAB is derived from a BL lacking the myc-Ig translocation, which is typical for BL. Jurkat is an acute T-cell leukemia cell line. OciAML2 and NB4 are cell lines from AML and U266 from multiple myeloma. No specific activation of STAT1 and STAT3 was detected in BL cell lines, NB4 AML cells, and Jurkat T-leukemia cells. However, nuclear extracts from U266 multiple myeloma cells and OciAML2 cells contained DNA-binding activities similar to those detected in the various HD cells and IL-6–stimulated hepatoma cells (Figure3). Thus, constitutive activation of STAT3 (and STAT1) is not confined to HD cells and can also be observed in AML and myeloma cells, whereas in BL cells no activation of STAT1 and STAT3 was found.
STAT3 is constitutively activated in HD cell lines but not in BL cells lines.
Equal amounts of nuclear extracts of each cell line (as indicated) were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus or a mutated STAT1/3 binding site as described in “Material and methods.” HepG2 cells were stimulated with IL-6 (15 minutes, 250 U) before nuclear extracts were prepared. The position of STAT3 and STAT1 homodimers and heterodimers are shown by arrows.
STAT3 is constitutively activated in HD cell lines but not in BL cells lines.
Equal amounts of nuclear extracts of each cell line (as indicated) were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus or a mutated STAT1/3 binding site as described in “Material and methods.” HepG2 cells were stimulated with IL-6 (15 minutes, 250 U) before nuclear extracts were prepared. The position of STAT3 and STAT1 homodimers and heterodimers are shown by arrows.
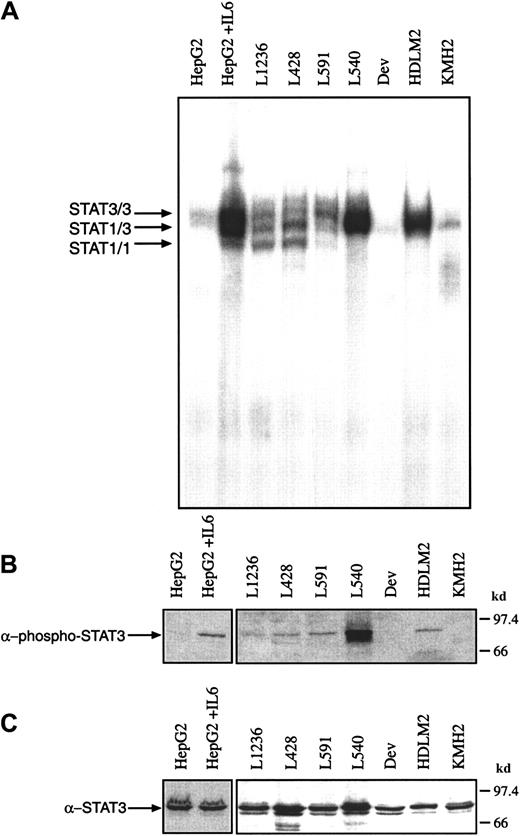
IL-6R (gp80) and gp130 are expressed on HD cell lines
Because IL-6 is produced by Hodgkin cell lines, the binding of IL-6 to its receptor could be responsible for STAT3 activation. IL-6R gp80 chains have been detected in Hodgkin cell lines and primary H-RS cells. The expression of gp130, which is essential for intracellular signaling, was analyzed by Western blot analysis using the α-gp130 antibody B-T12. The myeloma cell line U266, which served as a positive control for gp130 expression, and the HD cell lines KMH2, Dev, L540, and L591, expressed gp130, whereas L428, L1236, and HDLM2 were negative for gp130 expression in Western blot analysis (Figure4A). In the HD cell line KMH2 an additional faint band of higher mobility was detected suggesting the existence of a truncated gp130 molecule (Figure 4A, gp130*). Size differences of gp130 (Figure 4A, gray bar) may be due to different degrees of glycosylation.
Expression of gp130 and gp80 in HD cell lines.
(A) Immunoblot of gp130 expression in HD cell lines using the monoclonal antibody B-T12. Proteins isolated from 2 × 105 cells/lane were separated on a discontinuous SDS-PAGE with a 7.5% acrylamide resolving gel and electroblotted onto nitrocellulose filters. Specifically bound α-gp130 antibodies were detected using peroxidase-coupled secondary antibodies and ECL (unspecific bands [#]). gp130* is a faster migrating variant of gp130 detected in KMH2. The gray bar indicates the different migrations of gp130, which are probably due to differential glycosylation. (B) Expression of gp130 and gp80 on the surface of HD-derived cell lines visualized by FACS using the monoclonal antibody B-S12 for gp130 and B-R6 for gp80. Cells were incubated with unconjugated monoclonal antibodies and stained with goat-α-mouse-PE (GAM-PE) antibodies. Filled histograms indicate staining of gp130 or gp80; open histograms show staining with isotype control antibody.
Expression of gp130 and gp80 in HD cell lines.
(A) Immunoblot of gp130 expression in HD cell lines using the monoclonal antibody B-T12. Proteins isolated from 2 × 105 cells/lane were separated on a discontinuous SDS-PAGE with a 7.5% acrylamide resolving gel and electroblotted onto nitrocellulose filters. Specifically bound α-gp130 antibodies were detected using peroxidase-coupled secondary antibodies and ECL (unspecific bands [#]). gp130* is a faster migrating variant of gp130 detected in KMH2. The gray bar indicates the different migrations of gp130, which are probably due to differential glycosylation. (B) Expression of gp130 and gp80 on the surface of HD-derived cell lines visualized by FACS using the monoclonal antibody B-S12 for gp130 and B-R6 for gp80. Cells were incubated with unconjugated monoclonal antibodies and stained with goat-α-mouse-PE (GAM-PE) antibodies. Filled histograms indicate staining of gp130 or gp80; open histograms show staining with isotype control antibody.
The expression of gp130 was analyzed by flow cytometry with indirect staining (α-mouse-Ig-PE) using the monoclonal antibody B-S12, which binds to native surface gp130, but does not detect gp130 in Western blot.16 All HD cell lines except KMH2 expressed gp130 at low levels as compared to the myeloma cell line U266, which was brightly stained (Figure 4B). Using the directly conjugated α-gp130 antibody (AM64) the same results were obtained (data not shown). Differences between the expression levels of gp130 detected by Western blot analyses or fluorescence-activated cell sorting (FACS) are probably due to different amounts of total cellular gp130 and surface-expressed gp130.
The expression of gp130 was compared with the expression of IL-6R (gp80) α-chain (Figure 4B).4,37 The results of immunofluorescence staining for gp80 are in line with our previous reports showing the expression on all analyzed cell lines with the exception of KMH2.3 4 The recently established HD cell line L1236 also expressed gp80, but at a low level (Figure 4B). The results of Western blot and flow cytometry analyses are summarized in Table 1, which also provides IL-6 levels detected by ELISA. The data show that the IL-6R/gp80 and the signal transducer gp130 were expressed on most HD cell lines. Thus, it could be possible that the observed STAT3 activation in HD may be the result of the activation of the IL-6R complex.
Constitutive STAT3 activation in HD cell lines does not depend on gp130 or IL-6
Interleukin-6 together with sIL-6R is able to enhance IL-6–mediated signals by directing IL-6 to the gp130 signal transducer independently of the expression level of gp80 on the cell surface. To test whether IL-6 and sIL-6R can further enhance the observed STAT activity in HD cells, L540 and L1236 cells were incubated with IL-6 in the presence or absence of sIL-6R. No difference was observed between the DNA-binding activities of nuclear extracts prepared from cells incubated in the presence of exogenous IL-6 alone or together with sIL-6 receptor (Figure 5). Similar results were obtained for L428 cells (data not shown).
Activation of STAT3 in HD cell lines is not enhanced by IL-6.
The cell lines L540 (lanes 1-4) and L1236 (lanes 5-8) were cultured as described in “Materials and methods.” The cells were washed twice with serum-free medium and incubated with serum-containing medium (lanes 1 and 5), in 0.5% BSA-containing medium (lanes 2 and 6), in 0.5% BSA-containing medium with 250 U/mL IL-6 (lanes 3 and 7), or in 0.5% BSA-containing medium with 250 U/mL IL-6 and 1 μg/mL soluble recombinant IL-6R (lanes 4 and 8) for 30 minutes. Nuclear extracts (5 μg) of each cell line were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1 and STAT3 binding sites as described in the legend to Figure 1. Arrows indicate the position of STAT3 and STAT1 homodimers and heterodimers.
Activation of STAT3 in HD cell lines is not enhanced by IL-6.
The cell lines L540 (lanes 1-4) and L1236 (lanes 5-8) were cultured as described in “Materials and methods.” The cells were washed twice with serum-free medium and incubated with serum-containing medium (lanes 1 and 5), in 0.5% BSA-containing medium (lanes 2 and 6), in 0.5% BSA-containing medium with 250 U/mL IL-6 (lanes 3 and 7), or in 0.5% BSA-containing medium with 250 U/mL IL-6 and 1 μg/mL soluble recombinant IL-6R (lanes 4 and 8) for 30 minutes. Nuclear extracts (5 μg) of each cell line were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1 and STAT3 binding sites as described in the legend to Figure 1. Arrows indicate the position of STAT3 and STAT1 homodimers and heterodimers.
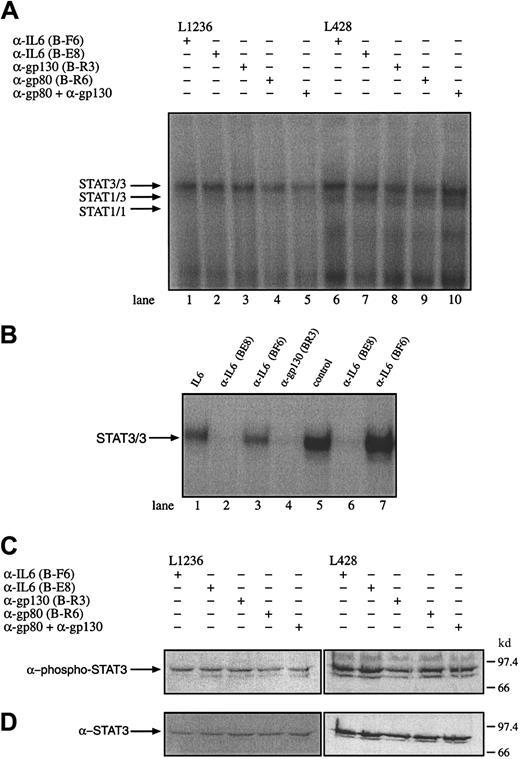
To test whether IL-6, gp80, or gp130 is involved in the observed STAT3 activation or may influence the proliferation of the HD cells, the cell lines L428, L540, and L1236 were incubated with α-IL-6 (nonneutralizing B-F6 and neutralizing B-E8 antibodies), α-gp80 (B-R6), or antagonistic α-gp130 antibodies (B-R3). The effect of these antibodies of interrupting the constitutive activation of STAT3 was analyzed by gel retardation and Western blot experiments. Nuclear extracts from untreated and antibody-treated cells were analyzed. Neutralizing α-IL-6 and α-gp130 antibodies but not nonneutralizing α-IL-6 antibodies were able to block STAT3 activation in HepG2 cells treated with 250 U/mL exogenous IL-6 or IL-6 containing the supernatant of L428 (Figure 6B). However, the DNA-STAT interaction was not affected by the neutralization of IL-6, gp80, and gp130 or gp80 together with gp130 in the tested HD cell lines L428, L1236 (Figure 6A), and L540 (data not shown). Similar results were obtained in Western blot experiments, which revealed no reduction of phosphorylated STAT3 molecules by incubation of the cells with α-IL-6 or α-IL-6R antibodies (Figure 6C,D). The addition of IL-6 to KMH2 cells was not associated with an activation of STAT3 as analyzed by gel shift experiments (data not shown). These observations indicate that the activation of STAT3 in Hodgkin cells is independent of IL-6 or cytokines sharing the receptor subunit gp130 as signal transducing molecule.
Constitutive activation of STAT3 in HD cell lines is independent of IL-6, the IL-6R gp80, or gp130.
(A) The cell lines L1236 (lanes 1-5) and L428 (lanes 6-10) were cultured as described in “Materials and methods.” The cells were incubated for 3 days with antibodies against IL-6 (lanes 1 and 6; α-IL-6 B-F6 nonneutralizing, lanes 2 and 7; α-IL-6 B-E8 neutralizing), gp130 (lanes 3 and 8; α-gp130 B-R3), gp80 (lane 4 and 9; α-gp80 B-R6), or gp80 together with gp130 (lanes 5 and 10). Nuclear extracts (5 μg) of each probe were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1 and 3 binding sites as described in the legend to Figure 1. The position of STAT3 and STAT1 homodimers and heterodimers is indicated by arrows. (B) Nuclear extracts from HepG2 cells incubated with IL-6 (250 U/mL) in the absence of an antibody (lane 1), in the presence of neutralizing α-IL-6 (B-E8) antibodies (lane 2), in the presence of nonneutralizing α-IL-6 (B-F6) antibodies (lane 3) and antagonistic α-gp130 (B-R3) antibodies (lane 4) were analyzed for STAT activation. The ability to neutralize IL-6 in the supernatant of L428 cells producing 260 pg/mL IL-6 within 24 hours was tested by incubating L428 cells with neutralizing α-IL-6 (B-E8) or nonneutralizing α-IL-6 (B-F6) antibodies. The L428 supernatants were taken after 20 hours. The corresponding supernatants were incubated with HepG2 cells (lanes 5-7). Nuclear extracts (5 μg) of each probe were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT3 binding sites as described in the legend to Figure 1. The position of STAT3/3 homodimers is indicated by an arrow. (C) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3, and visualized by ECL. (D) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel C was stripped and reprobed with an antibody specific for STAT3 and the protein was visualized by ECL.
Constitutive activation of STAT3 in HD cell lines is independent of IL-6, the IL-6R gp80, or gp130.
(A) The cell lines L1236 (lanes 1-5) and L428 (lanes 6-10) were cultured as described in “Materials and methods.” The cells were incubated for 3 days with antibodies against IL-6 (lanes 1 and 6; α-IL-6 B-F6 nonneutralizing, lanes 2 and 7; α-IL-6 B-E8 neutralizing), gp130 (lanes 3 and 8; α-gp130 B-R3), gp80 (lane 4 and 9; α-gp80 B-R6), or gp80 together with gp130 (lanes 5 and 10). Nuclear extracts (5 μg) of each probe were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1 and 3 binding sites as described in the legend to Figure 1. The position of STAT3 and STAT1 homodimers and heterodimers is indicated by arrows. (B) Nuclear extracts from HepG2 cells incubated with IL-6 (250 U/mL) in the absence of an antibody (lane 1), in the presence of neutralizing α-IL-6 (B-E8) antibodies (lane 2), in the presence of nonneutralizing α-IL-6 (B-F6) antibodies (lane 3) and antagonistic α-gp130 (B-R3) antibodies (lane 4) were analyzed for STAT activation. The ability to neutralize IL-6 in the supernatant of L428 cells producing 260 pg/mL IL-6 within 24 hours was tested by incubating L428 cells with neutralizing α-IL-6 (B-E8) or nonneutralizing α-IL-6 (B-F6) antibodies. The L428 supernatants were taken after 20 hours. The corresponding supernatants were incubated with HepG2 cells (lanes 5-7). Nuclear extracts (5 μg) of each probe were analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT3 binding sites as described in the legend to Figure 1. The position of STAT3/3 homodimers is indicated by an arrow. (C) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3, and visualized by ECL. (D) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel C was stripped and reprobed with an antibody specific for STAT3 and the protein was visualized by ECL.
The proliferation of these cell lines could not be inhibited in the presence of neutralizing antibodies against IL-6, gp80, and gp130 as measured by 3H-thymidine incorporation after 5 days of antibody incubation (data not shown). This is in line with earlier experiments in which we found that the growth of HD cells, which produce IL-6 and express the IL-6R gp80, could not be inhibited by α-IL-6 antibodies and further suggests that cytokines sharing the gp130 receptor subunit are not involved in proliferation of Hodgkin cell lines.4
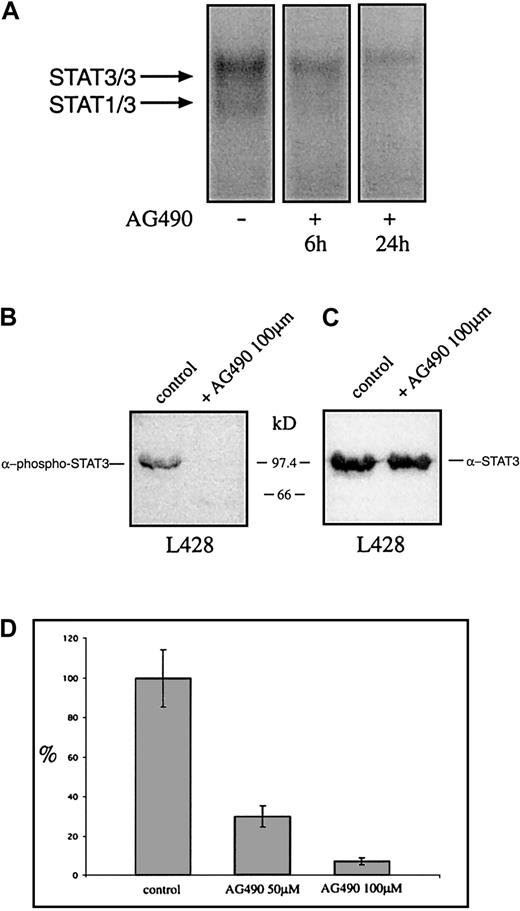
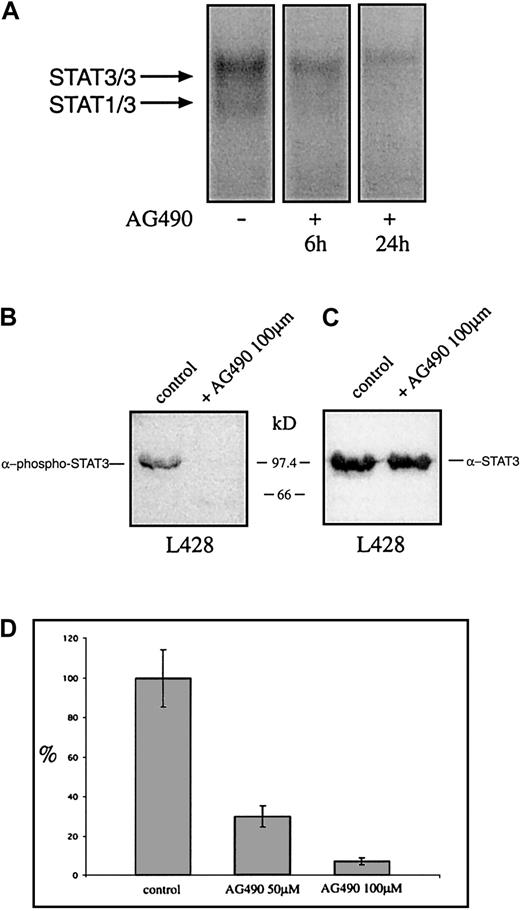
The tyrphostin AG490 inhibits STAT3 activation and growth of the Hodgkin cell line L428
Recently it was shown that tyrphostin AG490 is a Jak2 inhibitor leading to inhibition of constitutively activated STAT3 molecules in acute leukemias and multiple myeloma. Thus it was analyzed whether AG490 affects the constitutive activation and the proliferation of HD cell lines. Cells were incubated with AG490 and analyzed for STAT3 activation 6 and 24 hours after incubation with AG490. Nuclear extracts from untreated HD cells formed protein-DNA complexes with the promoter element of the c-fos gene as detected by EMSA, whereas the AG490-treated cells showed drastically reduced amounts of specific protein-DNA complexes (Figure7A). The use of specific STAT3-phosphotyrosine antibodies demonstrates that 24 hours after incubation with the tyrphostin the phosphorylation of tyrosines on STAT3 was completely blocked, whereas STAT3 protein level was unaltered (Figure 7B,C). After 3 days the proliferation was tested. As measured by 3H-thymidine incorporation, the proliferation of L428 cells was completely inhibited by AG490 (Figure 7D). Thus the Jak-inhibitor tyrphostine AG490 blocked the constitutive activation of STAT3 and inhibited spontaneous growth of HD tumor cells.
The tyrphostin AG490 inhibits constitutive activation of STAT3 and proliferation of the HD cell line L428.
(A) Inhibition of STAT3-DNA binding by the tyrphostin AG490. L428 cells were cultured as described in “Materials and methods.” The cells were incubated in 0.1% DMSO (control, nuclear extract isolated after 24 hours) or 50 μM AG490 (nuclear extract isolated after 6 and 24 hours). Five micrograms of protein of each probe was analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1/3 binding sites as described in the legend to Figure 1. The positions of the STAT3/3 homodimer and the STAT1/3 heterodimer are indicated by arrows. (B) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3, and visualized by ECL. (C) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel B was stripped and reprobed with an antibody specific for STAT3 and the protein was visualized by ECL. (D) Inhibition of proliferation of L428 cells by the tyrphostin AG490. The cells were incubated for 3 days in 0.1% DMSO (control) or 50 μM or 100 μM AG490. Sixteen hours before harvest 0.4 μCi/well3H-thymidine was added to the cells. The results are shown as relative inhibition of proliferation. The proliferation of L428 cells was not affected by DMSO.
The tyrphostin AG490 inhibits constitutive activation of STAT3 and proliferation of the HD cell line L428.
(A) Inhibition of STAT3-DNA binding by the tyrphostin AG490. L428 cells were cultured as described in “Materials and methods.” The cells were incubated in 0.1% DMSO (control, nuclear extract isolated after 24 hours) or 50 μM AG490 (nuclear extract isolated after 6 and 24 hours). Five micrograms of protein of each probe was analyzed in a gel retardation experiment using radiolabeled oligonucleotides containing the consensus STAT1/3 binding sites as described in the legend to Figure 1. The positions of the STAT3/3 homodimer and the STAT1/3 heterodimer are indicated by arrows. (B) Western blot analysis of phosphorylated STAT3. Nuclear extracts (20 μg) of each cell line were analyzed in a 10% SDS-polyacrylamide gel, transferred to a nylon membrane, incubated with an antibody specific for phosphorylated STAT3, and visualized by ECL. (C) Western blot analysis of unphosphorylated STAT3. The same nylon membrane as in panel B was stripped and reprobed with an antibody specific for STAT3 and the protein was visualized by ECL. (D) Inhibition of proliferation of L428 cells by the tyrphostin AG490. The cells were incubated for 3 days in 0.1% DMSO (control) or 50 μM or 100 μM AG490. Sixteen hours before harvest 0.4 μCi/well3H-thymidine was added to the cells. The results are shown as relative inhibition of proliferation. The proliferation of L428 cells was not affected by DMSO.
Discussion
Biochemical and molecular analysis of HD is still hampered by the fact that very low numbers of H-RS cells are surrounded by a variety of reactive cells.2,34 Recent analyses revealed that H-RS cells are derived from B cells in the germinal center and are characterized by rearranged, somatically hypermutated Iggenes that are frequently nonfunctional.2 The mechanism responsible for the oncogenic transformation is still unknown. Interestingly, H-RS cells express a variety of cytokines and cytokine receptors that may regulate interactions between tumor cells and reactive cells.3 Constitutive expression of growth factors may lead to unlimited proliferation in certain tumor cells. We and other groups have previously shown that IL-6 and IL-6R gp80 are expressed both in HD cell lines and in primary H-RS cells and IL-6, and perhaps additional cytokines may be involved in the pathogenesis of HD.1 3-6
The aim of this study was to evaluate whether IL-6 signal transduction is important for the growth of HD cell lines. Using a panel of well-characterized HD cell lines we identified constitutively activated DNA-binding proteins in these cells. Competition experiments with unlabeled oligonucleotides and supershift assays revealed that the DNA-binding proteins in the nuclear extracts from HD cell lines contain STAT3 and, at lower levels, STAT1.
The HD cell lines differ in expression levels of gp80, gp130, and IL-6. However, incubation with IL-6 in the presence or absence of sIL-6R did not increase the DNA-binding activity of STAT factors. Activation of STAT was independent of endogenous IL-6 as shown by neutralization experiments. Thus, STAT activation was already high without stimulation by exogenous factors. In addition antibodies against IL-6, IL-6R, or gp130 did not inhibit the DNA-binding activity of STAT3. This reveals that IL-6–type cytokines such as IL-6, IL-11, leukemia inhibitory factor, oncostatin M, ciliary neurotrophic factor, and cardiotrophin are not involved in the observed constitutive activation of STAT3 in HD cells.8
It is unlikely that soluble factors from the culture medium are responsible for the observed STAT activation because incubation of the cells in starvation medium did not reduce STAT activity. We formally cannot exclude that HD cells produce a factor-mediating activation of a receptor resulting in STAT activation, although exchange of the conditioned medium with fresh medium did not alter STAT activation (data not shown).
Interleukin-6 is the major survival factor for multiple myeloma tumor cells where STAT3 is constitutively activated. In the multiple myeloma cell line U266 blocking of the IL-6R signaling from Janus kinases to STAT3 inhibits Bcl-x expression and induces apoptosis, providing evidence that constitutively activated STAT3 contributes to the pathogenesis of multiple myeloma.13 It has been hypothesized that genes regulated by STAT3 act cooperatively in cell cycle progression and prevention of apoptosis in BAF-BO3 cells.55,56 However, in a melanoma cell line the opposite effect was detected in that IL-6 was involved in STAT-dependent growth inhibition by up-regulating p27/Kip1.57 Furthermore, evidence suggests that gp130 can induce opposite pathways at least in certain cells.58,59 However it has been reported by several authors that constitutively activated STAT molecules can be detected in acute leukemic cells and bone marrow mononuclear cells from patients with multiple myeloma.13,24,25,28,38 In a recent report it has been shown in addition that the substitution of 2 cysteines in STAT3 led to a spontaneous activation that by itself can mediate cellular transformation.39 Here we show that STAT proteins are constitutively activated in HD. This deregulated STAT activation could be due either to gp130-independent activation of Jak or other tyrosine kinases.40-42 The deregulation of the Jak/STAT pathway has been recently described in a T-acute lymphocytic leukemia (ALL) line, in which a t(9;12) chromosomal translocation led to a TEL-Jak2 fusion protein. TEL-mediated oligomerization of TEL-Jak2 resulted in the constitutive activation of its tyrosine kinase activity and conferred cytokine-independent proliferation to the IL-3–dependent Ba/F3 cell line.27
Recent studies indicated that intracellular events may influence the activation of STAT proteins, thereby rendering the cells to grow independently of extracellular stimuli. It has been described that src can activate STAT directly without Jak.43-46 Other proto-oncogenes like Eyk, Bmx, v-abl, or the bcr-abl fusion protein can also activate the Jak/STAT pathway using bypass mechanisms to activate cytokine signal transduction pathways and promote transformation.26,47-49 Thus, oncogenes not yet identified could be responsible for the activation of STAT molecules in HD. HD cell lines as well as primary H-RS cells so far analyzed indeed show a heterogeneous pattern of proto-oncogene expression and gene translocations including aberrant c-fes transcripts.50 51
Alternatively, inhibitors of STAT activation such as SOCS and PIAS may be absent or suppressed in HD cells. Thus, the analysis of potential mutations in proteins involved in the activation or inactivation of STAT proteins, or the identification of translocations leading to constitutive STAT activating kinases, may be an object of further investigation in HD. Recently it was shown that the Jak2 gene is amplified in Hodgkin cells, which could be responsible for constitutive STAT3 activation.60 However, we could not demonstrate the overexpression or enhanced activation of Jak2 compared to other Janus kinases (data not shown).
Recently it was shown that HD cells contain constitutively activated nuclear factor (NF)-κB, another transcription factor activated by a number of cytokines.52,53 This constitutively active NF-κB is most likely the result of mutated IκB-α, which is unable to bind NF-κB and thus to inactivate this transcription factor.54 It was suggested that constitutive activation of NF-κB contributes to the pathogenesis of HD by preventing apoptosis in HD cells.52 54 Our findings now provide evidence that other transcription factors like STAT3 are constitutively activated in HD cells and perhaps involved in the transformation of Hodgkin cells.
We have shown that in the HD cell line L428 the constitutively activated STAT3 could be inhibited by the tyrphostin AG490, which also inhibits the proliferation of the cells. Recently it has been shown that AG490 selectively blocks STAT activation in primary pre-B ALL, cutaneous T-cell lymphomas, and the myeloma cell line U266. In these cell lines AG490 also inhibited the proliferation of the tumor cells indicating a link between cell proliferation and constitutively activated STAT molecules.13,25,28,39 AG490 is a protein tyrosine kinase inhibitor that inhibits Jak2 and perhaps other kinases. We analyzed the activation of several Jaks in HD cell lines using Western blot analyses. Our preliminary results show that a number of Janus kinases are phosphorylated in HD cell lines independent of whether they contain activated STAT3 or not (data not shown). However, STAT3 could also be activated by different mechanisms including the activation of src kinases or the lack of STAT3 inhibitors. The fact that the STAT3 activation is inhibited by a tyrosine kinase inhibitor makes it less probable that a mutation might be the reason for constitutive activation.39
In conclusion, we have identified constitutively activated STAT3 in Hodgkin cell lines. The observed STAT activation may render H-RS cells to grow independently of STAT-activating factors and support the notion that activated STAT proteins participate in oncogenic transformation.
We would like to acknowledge A. Jox and G. Mosialos for helpful discussions on the manuscript and H. Straub for technical assistance.
Supported by grants from the Deutsche Forschungsgemeinschaft within the Sonderforschungsbereich 502 (Cologne), the Köln Fortune Program–Faculty of Medicine University of Cologne and the Sonderforschungsbereich 542 (Aachen).
D.K. and U.H. contributed equally to this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dieter Kube, Eberhard-Karls-Universität, Institut für Tropenmedizin, Sektion für Humanparasitologie, Wilhelmstraße 27, D-72074 Tübingen, Germany; e-mail:dieter.kube@uni-tuebingen.de.


![Fig. 4. Expression of gp130 and gp80 in HD cell lines. / (A) Immunoblot of gp130 expression in HD cell lines using the monoclonal antibody B-T12. Proteins isolated from 2 × 105 cells/lane were separated on a discontinuous SDS-PAGE with a 7.5% acrylamide resolving gel and electroblotted onto nitrocellulose filters. Specifically bound α-gp130 antibodies were detected using peroxidase-coupled secondary antibodies and ECL (unspecific bands [#]). gp130* is a faster migrating variant of gp130 detected in KMH2. The gray bar indicates the different migrations of gp130, which are probably due to differential glycosylation. (B) Expression of gp130 and gp80 on the surface of HD-derived cell lines visualized by FACS using the monoclonal antibody B-S12 for gp130 and B-R6 for gp80. Cells were incubated with unconjugated monoclonal antibodies and stained with goat-α-mouse-PE (GAM-PE) antibodies. Filled histograms indicate staining of gp130 or gp80; open histograms show staining with isotype control antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/3/10.1182_blood.v98.3.762/5/m_h81511331004.jpeg?Expires=1766262629&Signature=AFAebnsN0XZD4-3bvpOcgX3JqCE3BkkBIdzY-tagOgreYTXIsFMghTFf7eLAS641o6DnvDRHx1X6UWlSxSwyYsoy~FcBvObD9~qkvOJ1iL3TPAIBxXtiWyNULTffYRy2JQvPQxGBMVlep3uR~enlo92QlXzQpPo-2xq9qesSUcWQ17K48OAMttZj7fvuefG2GpTCY2TJpvvwr7c4wxzVNilRrTr9eO~jO~x14sQay~UnkVUncnWx-ALu8NUquAolQ0bNg5~pJTxcRuuj13ah9n0E~6XTQJ9uV8-K8kwFkZzEykTchtlfC4-oBzoxUcJP7kfoByHQYLNnrd1wY7gpog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Fig. 4. Expression of gp130 and gp80 in HD cell lines. / (A) Immunoblot of gp130 expression in HD cell lines using the monoclonal antibody B-T12. Proteins isolated from 2 × 105 cells/lane were separated on a discontinuous SDS-PAGE with a 7.5% acrylamide resolving gel and electroblotted onto nitrocellulose filters. Specifically bound α-gp130 antibodies were detected using peroxidase-coupled secondary antibodies and ECL (unspecific bands [#]). gp130* is a faster migrating variant of gp130 detected in KMH2. The gray bar indicates the different migrations of gp130, which are probably due to differential glycosylation. (B) Expression of gp130 and gp80 on the surface of HD-derived cell lines visualized by FACS using the monoclonal antibody B-S12 for gp130 and B-R6 for gp80. Cells were incubated with unconjugated monoclonal antibodies and stained with goat-α-mouse-PE (GAM-PE) antibodies. Filled histograms indicate staining of gp130 or gp80; open histograms show staining with isotype control antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/3/10.1182_blood.v98.3.762/5/m_h81511331004.jpeg?Expires=1766550343&Signature=gxhhqVQOyeWOs7hDgLi6ZNf7JPi2~0sNmBFiwqGVSRngIWY4Y8fdunxSaGjo~KMzaMfvMe41jqt2o89vsWXKbykSl5nWmcptjZBW5FbdlZ-UdV~6W2UXRhQCzTaXFdHeEroSqDzm-y2XvIRc-bKwE28gWNSNBmzdlcfwnx87SL9s8uqyv8rnAZKoddKLxClx3vcwETGi843f3HYNdbW34PwpDHU8XrHcOKUQoFyJlWnNIS8e26r2gckJZnMVNcXF7PEpt2RVNTftbLoLUwBac93afndXk1ZX9ntUKZ8MoIBe8Ry8TaLCFUbdV0aeITlQQ~oa8-ZUQgNqH92YSrluKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)