Abstract
Leukemic cells from bone marrow (BM) of 17 infants and 127 children with newly diagnosed ALL, as well as fetal liver and BM and normal infant BM samples, were analyzed for presence of a t(4;11) translocation using standard cytogenetic techniques and expression of an MLL-AF4 fusion transcript using standard reverse transcriptase-polymerase chain reaction (RT-PCR) assays as well as nested RT-PCR that is 100-fold more sensitive than standard RT-PCR. Overall, 9 of 17 infants and 17 of 127 noninfant pediatric ALL patients were positive for expression of MLL-AF4 fusion transcripts, as determined by standard and/or nested RT-PCR assays. None of theMLL-AF4+ cases were positive for E2A-PBX1or BCR-ABL fusion transcript expression. Although 8 of 9MLL-AF4+ infants had cytogenetically detectable t(4;11)(q21;q23), 15 of the 17 MLL-AF4+noninfants were t(4;11)−. Infants withMLL-AF4+ ALL had poor outcomes, whereas non-infant MLL-AF4+/t(4;11)−patients had favorable outcomes similar toMLL-AF4− patients. Notably, MLL-AF4transcripts also were detected by nested RT-PCR in 4 of 16 fetal BMs, 5 of 13 fetal livers, and 1 of 6 normal infant BMs, but not in any of the 44 remission BM specimens from pediatric ALL patients. Our results provide unprecedented evidence that MLL-AF4 fusion transcripts can be present in normal hematopoietic cells, indicating that their expression is insufficient for leukemic transformation of normal lymphocyte precursors.
© 1998 by The American Society of Hematology.
CHROMOSOMAL TRANSLOCATIONS found in childhood and adult acute lymphoblastic leukemia (ALL) may result in the production of chimeric fusion proteins with leukemogenic potential.1,2 The prototype of such translocations, t(9;22)(q34;q11), is associated with high risk in B-lineage ALL and results in the production of BCR-ABL fusion mRNAs and proteins.3,4 Similarly, the t(4;11)(q21;q23) translocation, a hallmark of infant ALL, results in the fusion of the MLL gene on chromosome 11 and the AF4 gene on chromosome 4.5-9 The presence of a t(4;11) translocation is associated with unfavorable presenting features such as very high white blood cell (WBC) count,10,11 and the majority of t(4;11)+infant ALL patients have an extremely poor prognosis.12-16Recently, the identification of in utero rearrangements have implicated the MLL gene in the leukemogenesis of infant ALL.17
The MLL gene encodes a protein that contains regions with homology to the Drosophila trithorax gene,18,19including A-T hook and transcriptional repression domains located 5′ to the MLL-AF4 breakpoint, a zinc finger homology domain located at the breakpoint, and a transcriptional activation domain located 3′ to the breakpoint.20,21 The A-T hook domain has been shown to mediate binding of MLL to cruciform DNA, irrespective of sequence. The AF4 gene encodes a protein with serine- and proline-rich regions located near the AF4 breakpoint and a nuclear localization signal domain located 3′ to the breakpoint.19 22 Thus, translocations between MLLand AF4 that disrupt these functional activities may alter expression of genes regulated by MLL or AF4, or may endow novel fusion proteins with transcriptional regulatory activity.
Although fusion transcripts from both reciprocal derivative chromosomes are found in leukemic cells with t(4;11)(q21;q23) translocations, it is unclear which of the two products is leukemogenic.19,23However, clustering of breakpoints found in various 11q23 rearrangements, heterogeneity of breakpoints in 11q23 fusion partners, and loss of the translocated MLL region telomeric to 11q23 have lead to the hypothesis that the der(11) fusion product, which is encoded by the 5′ portion of the MLL gene, represents the biologically more relevant potential oncogenic fusion protein.23-26
Rubnitz et al,27 as well as Chen et al28 and Griesinger et al,29 have shown the presence of MLLgene rearrangements in infant ALL patients with or without cytogenetically detectable t(4;11) or other structural chromosomal aberrations involving 11q23. These studies were interpreted to indicate that the frequency of molecular 11q23 rearrangements andMLL-AF4 fusion transcripts was greater than that reported using cytogenetic analyses alone, and suggested the importance of molecular-based techniques for screening ALL patients.
Recently, however, new questions concerning the clinical significance of malignancy-associated fusion transcripts have arisen as a result of several reports that documented their occurrence in normal or nonleukemic cells. For example, Limpens et al30,31 and other groups,32-35 using highly sensitive polymerase chain reaction (PCR) techniques, have demonstrated the presence of lymphoma-associated t(14;18)(q32;q21)36,37 in benign follicular hyperplasias and normal blood of healthy individuals. Similarly, Biernaux et al38 recently reported that theBCR-ABL fusion transcript, which is thought to be derived from the t(9;22) translocation associated with high-risk ALL, was present in hematopoietic cells of healthy individuals. Thus, the relationship between the translocation and transformation in lymphoid malignancies requires further investigation.
These observations led us to examine primary leukemic cells from ALL patients, fetal tissues, and normal infant bone marrows (BMs) for the presence of MLL-AF4 fusion transcripts. Our results show thatMLL-AF4 fusion transcripts, detectable by a sensitive nested reverse transcriptase (RT)-PCR assay, are frequently generated in patients whose cells lack cytogenetically detectable t(4;11) and that expression of MLL-AF4 fusion transcripts is not a significant prognostic factor for these patients. Notably, MLL-AF4 fusion transcripts were also detected by nested PCR in normal hematopoietic cells from fetal tissues and infant BMs. These results suggest that the presence of MLL-AF4 fusion transcripts may not be sufficient for neoplastic transformation of lymphocyte precursors and add to recent evidence that leukemia-associated gene rearrangements can occur in normal hematopoiesis throughout the lifespan of an individual, thereby providing only a potentially contributory but not final transforming event.
MATERIALS AND METHODS
Patient samples and cell lines.
The patient population included 17 infants and 127 children with newly diagnosed ALL treated on the Children's Cancer Group (CCG) 1800 series protocols between December 1993 and December 1994 for whom complete cytogenetic, immunophenotypic, and clinical data were obtained. Also included were 44 children with ALL in remission. Most of the children included in the present analysis had standard-risk ALL. Diagnosis of ALL was based on morphological, biochemical, and immunological features of the leukemic cells, including lymphoblast morphology on Wright-Giemsa–stained BM smears, positive nuclear staining for terminal deoxynucleotidyl transferase (TdT), and cell-surface expression of two or more lymphoid differentiation antigens, as previously described.39 Surplus cells from diagnostic BM specimens were used for molecular genetic studies. In two cases, 10 sequential BM specimens were examined for MLL-AF4 fusion transcript expression by nested RT-PCR. Informed consent was obtained from parents, patients, or both, as deemed appropriate, according to the Department of Health and Human Services guidelines. The RS4;11 cell line harboring the t(4;11) translocation was obtained from John H. Kersey (University of Minnesota) and served as a positive control inMLL-AF4 RT-PCR assays.
Fetal tissues and normal infant BMs.
Human fetal livers (N = 13) and fetal BMs (N = 16) from prostaglandin-induced human abortuses of gestational age 15 to 22 weeks were used according to the guidelines of the University of Minnesota Committee on the Use of Human Subjects in research for secondary use of pathological or surgical tissue. Normal BM specimens were obtained from infants (N = 6) who were either BM donors in the context of sibling BM transplantation or had been examined for suspicious cells in the blood but were found to have a normal BM without any evidence of leukemia.
Immunophenotyping.
Mononuclear cell fractions comprised primarily of leukemic cells were isolated from diagnostic BM aspirate samples by centrifugation on Ficoll-Hypaque density gradients, as previously described.39 Immunophenotyping was performed centrally in the CCG ALL Biology Reference Laboratory by indirect immunofluorescence and flow cytometry using monoclonal antibodies (MoAbs) reactive with the following differentiation antigens: CD2, CD3, CD5, CD7, CD10, CD19, and CD34, as previously described.39 Patients were classified as B-cell precursor ALL if ≥30% of their leukemic cells were positive for CD19 and <30% of their leukemic cells were positive for CD2, CD5, or CD7.
Cytogenetic analysis.
Cytogenetic analysis of leukemic cells was performed by local institutions at diagnosis before initiation of therapy. Banded chromosomes were prepared from unstimulated peripheral blood or direct and 24-hour cultured preparations of fresh BM, as described previously.12,40 Chromosome abnormalities were designated using the 1995 International System for Human Cytogenetics Nomenclature.41 Abnormal clones were defined as two or more metaphase cells with identical structural chromosomal abnormalities or extra chromosomes, or three or more metaphase cells with identical missing chromosomes.
RT-PCR.
All PCR assays were performed centrally in the Children's Cancer Group ALL Biology Reference Laboratory, as described.42-44Briefly, total cellular RNA was extracted from cells using the RNeasy total RNA isolation kit (Qiagen, Santa Clarita, CA), and 20% of the total RNA sample was used for cDNA synthesis with Moloney murine leukemia virus (MMLV) reverse transcriptase (GIBCO-BRL, Gathersburg, MD) in the presence of dNTPs (= reaction mixture 1). For amplification, cDNA products were denatured, diluted in PCR buffer containing oligonucleotide primers and Amplitaq DNA polymerase (Perkin Elmer Cetus Corp, Norwalk, CT; = reaction mixture 2), and subjected to 35 cycles in a DNA thermal cycler as described.42,43 Primers were as follows: MLL-AF4, 5′-AGAGCAGAGCAAACAGAA-3′ and 5′-GCTGAGAATTTGAGTGAG-3′; E2A-PBX1, 5′-GCCAGCCAGGCACCCTCCC-3′ and 5′-GTTGTCCAGCCGCATCAGCT-3′; and BCR-ABL, 5′-TCCGAGGCCACCATCGTGGGCGTCGGC-3′ and 5′-TGTGATTATAGCCTAAGACCCGGAG-3′. For increased sensitivity, nested PCR11,45,46 was performed using the primers 5′-AAGTGGCTCCCCCGCCCAAGTAT-3′ and 5′-TTGGGTTACAGAACTGACATG-3′ for MLL-AF4 and 5′-CCAACGATGGCGAGGGCGCCT-3′ and 5′-CGAGCGGCTTCACTCAGACC-3′ for BCR-ABL. PCR products were separated by electrophoresis in 1.2% agarose, transferred to nylon membranes, and hybridized with the oligonucleotide probes specific for internal sequences of E2A-PBX1 orBCR-ABL as described.44 For the MLL-AF4fusion transcript, a specific AF4 oligonucleotide probe (5′-TAGGGAAAGGAAACTTGGATG-3′) was used. Reactions conducted in the absence of added mRNA substrate served as negative controls. Reactions conducted with RNA isolated from the cell line RS4;11, which harbors the t(4;11) translocation, and from a patient (unique patient number [UPN] 100), with t(9;22)+ ALL, were used as positive controls for MLL-AF4 and BCR-ABL fusion transcripts, respectively. Strict precautions were used to prevent cross-contamination of samples and negative controls were included at the RNA extraction as well as PCR amplification steps.47 To prevent carryover of amplified cDNA sequences, we prepared our samples in a dedicated room separated from the room in which the PCR reactions were performed. UV germicidal lamps were used in biosafety hoods to quickly damage any DNA left on exposed surfaces, making it unsuitable for subsequent amplification. Separate sets of supplies and pipetting devices were dedicated for sample preparation and for setting up reactions. Deionized water, buffer solutions, disposable pipette tips, and microcentrifuge tubes were autoclaved. We divided reagents into aliquots to minimize the number of repeated samplings necessary. All reagents used in the PCR reactions were prepared, divided, and stored in an area that is free of the PCR-amplified products. Similarly, oligonucleotides used for amplification were synthesized and purified in a PCR product-free environment. Individuals involved in PCR reactions were required to wear gloves and change them frequently. The barrel of pipetting devices may become contaminated with aerosols containing sample RNA, leading to cross-contamination of samples. To prevent this, we used pipettes with aerosal-resistant tips. We mixed reagents before dividing them into aliquots. All PCR reagents were combined into a “premixture,” which was then pipetted into reaction tubes containing RNA/cDNA. Nonsample components, such as premixed dNTPs, primers, buffer, and enzyme, were added to the reaction tubes before sample cDNA (Molecular Bio-Products, San Diego, CA). Negative controls included PCR products from RNA-free reaction mixture 1 plus reaction mixture 2 (= negative control 1) and reaction mixture 1 containing RNA from RS4;11 cell line plus DNA polymerase-free reaction mixture 2 (= negative control 2). RNA integrity was confirmed by PCR amplification of the cABL mRNA, which is ubiquitously expressed in human hematopoietic cells using the primers 5′-TTCAGCGGCCAGTAGCATCTGACTT-3′ and 5′-TGTGATTATAGCCTAAGACCCGGAG-3′. In sensitivity assays of the RT-PCR assay for MLL-AF4 fusion transcript expression, varying numbers (1 to 107) of RS4;11 cells were mixed with cells from an Epstein-Barr virus (EBV)-transformed lymphoblastoid cell line to yield 0.00001% to 100%MLL-AF4+ cells in a cell population containing a total of 107 cells. RNA subtracted from those 107 cell samples were analyzed by standard versus nested RT-PCR.
Molecular characterization of MLL gene rearrangements.
Total cellular DNA extracted from BM leukemic cells, fetal liver/BM, or normal infant BM (= test samples) and unrearranged control DNA isolated from normal peripheral blood lymphocytes were analyzed by Southern blotting using standard techniques. DNA was digested using the restriction endonucleases BamI-II, EcoRI, andHindIII. Fragments were electrophoretically separated on 0.8% agarose gels, transferred to nylon membranes, and hybridized with the radiolabeled MLL-specific probes PS/4, 98.40, and 4.2E.16 28 Membranes were washed under high stringency conditions and exposed to Kodak XAR-5 film (Eastman Kodak, Rochester, NY).
MLL-AF4 sequence analysis.
MLL-AF4 cDNA from RT-PCR reaction mixtures was purified with the QIAquick PCR purification kit (Qiagen). Purified PCR products were cloned into the pCR II sequencing vector using the TA Cloning kit (Invitrogen, San Diego, CA). Clones (24/patient) were sized byEcoRI restriction analysis and two to four clones were randomly chosen for sequencing. The cloned PCR products were purified with a Qiagen plasmid isolation kit and sequenced automatically with the Thermosequenase sequencing kit (Amersham, Arlington Heights, IL) and the ALF Sequencer (Pharmacia, LKB Biotech, Piscataway, NJ). The sequences were compared with the published MLL-AF4 and the t(4;11) translocation breakpoint region sequences obtained through Bionet (Bionet Accession codes HUMMLLAF44F, HSMLLAF4F, and S67825).
Statistical analysis.
Event-free survival (EFS) for ALL patients with or withoutMLL-AF4 fusion transcripts was analyzed using life table methods and associated statistics. Events included induction failure (nonresponse to therapy or death during induction), leukemic relapse at any site, death during remission, or second malignant neoplasm, whichever occurred first. Patients not experiencing an event at the time of EFS analysis were censored at the time of their last contact. Life table estimates were calculated by the Kaplan-Meier (KM) procedure, and the standard deviation of the life table estimate was obtained using Greenwood's formula.48 An approximate 95% confidence interval can be obtained from the life table estimate ± 1.96 SDs. Life table comparisons of EFS outcome pattern for patient groups generally used the log rank statistic,49 50 andP values for life table comparisons are based on the pattern of outcome across the entire period of patient follow-up.51
RESULTS
Detection of MLL-AF4 in infants and children with ALL by standard as well as nested RT-PCR.
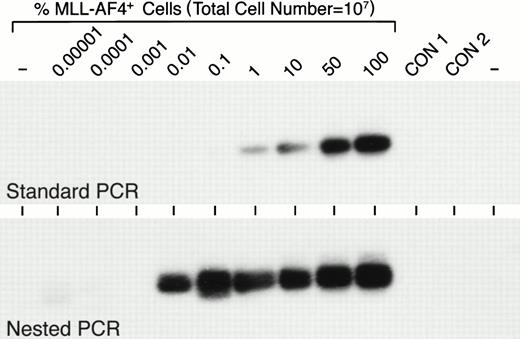
A balanced t(4;11)(q21;q23) translocation is frequently observed in infant ALL.9,11,14,15 Of 56 infants with newly diagnosed ALL entered on the most recent CCG infant ALL protocol, 21 (37.5%) had a t(4;11).13 By comparison, t(4;11) is a very rare cytogenetic abnormality in childhood ALL. There were only 8 t(4;11)+ patients (0.6%) among 1,322 children with newly diagnosed ALL entered on all CCG-1800 series protocols and only 2 of 653 (0.3%) children with standard-risk ALL entered on CCG-1881 and CCG-1891 risk-adjusted protocols had a t(4;11) (N.H., F.M.U., unpublished data, June 1998). In a recent study we used standard RT-PCR assays to examine primary leukemic cells from 642 children with ALL for the expression of MLL-AF4 fusion transcript and found MLL-AF4 expression only in 0.7% of the patient population, which excluded infants.44 Since Downing et al11 reported that standard PCR was not sensitive enough and nested PCR analyses were required for the detection ofMLL-AF4 fusion transcripts in 26% of t(4;11) ALL cases, we decided to examine primary leukemic cells for the expression of t(4;11)-specific MLL-AF4 fusion transcripts using both standard as well as nested RT-PCR assays. When we compared the detection levels achieved by standard versus nested PCR assays, we found that standard PCR was capable of detecting 1% MLL-AF4+ cells among 107 cells, whereas nested PCR was 100-fold more sensitive, allowing the detection of 0.01%MLL-AF4+ cell contamination among 107cells (Fig 1).
Sensitivity of standard and nested RT-PCR assays for detection of MLL-AF4 fusion transcript-positive cells. Varying numbers (1 to 107) of MLL-AF4+ RS4;11 cells were mixed with cells from an EBV-transformed lymphoblastoid B-cell line to yield 0.00001% to 100%MLL-AF4+ cells in a test cell population containing a total of 107 cells. RNA was subtracted from these 107 cell samples and analyzed by standard RT-PCR as well as nested RT-PCR for MLL-AF4positivity, as described in Materials and Methods. CON, control.
Sensitivity of standard and nested RT-PCR assays for detection of MLL-AF4 fusion transcript-positive cells. Varying numbers (1 to 107) of MLL-AF4+ RS4;11 cells were mixed with cells from an EBV-transformed lymphoblastoid B-cell line to yield 0.00001% to 100%MLL-AF4+ cells in a test cell population containing a total of 107 cells. RNA was subtracted from these 107 cell samples and analyzed by standard RT-PCR as well as nested RT-PCR for MLL-AF4positivity, as described in Materials and Methods. CON, control.
We first examined leukemic cells from 17 infants with ALL forMLL-AF4 expression. For comparison, cells were also examined for expression of t(1;19)-specific E2A-PBX1, and t(9;22)-specific BCR-ABL fusion transcripts. While 6 cases (35%) were determined to be MLL-AF4+ by standard PCR, 9 cases (53%) were found to beMLL-AF4+ by nested PCR (Table 1). Data for all 17 infants is summarized in Table 2. Cytogenetic analysis showed balanced t(4;11)(q21;q23) in 8 infants, normal diploid karyotypes in 2 infants, as well as t(1;19)(q23;p13), t(11;15)(q23;q26), and t(7;11)(q22;q23) in 1 infant each. Three additional infants had complex rearrangements and 1 infant was hyperdiploid without structural abnormalities. Both standard and nested RT-PCR results generally were in accord with the cytogenetics data. Specifically, all 6 standard PCR+ infants also had a cytogenetically detectable t(4;11). Of the additional 3 infants determined to beMLL-AF4+ by nested PCR only, 2 had a t(4;11) and 1 was normal diploid (Table 1 and Table 2). The E2A-PBX1 andBCR-ABL fusion transcripts were observed only in cells from infants who were t(1;19)+ and t(9;22)+, respectively. Infants with other translocations, including t(7;11) and t(11;15), did not express MLL-AF-4 fusion transcripts in their leukemic cells (Table 2).
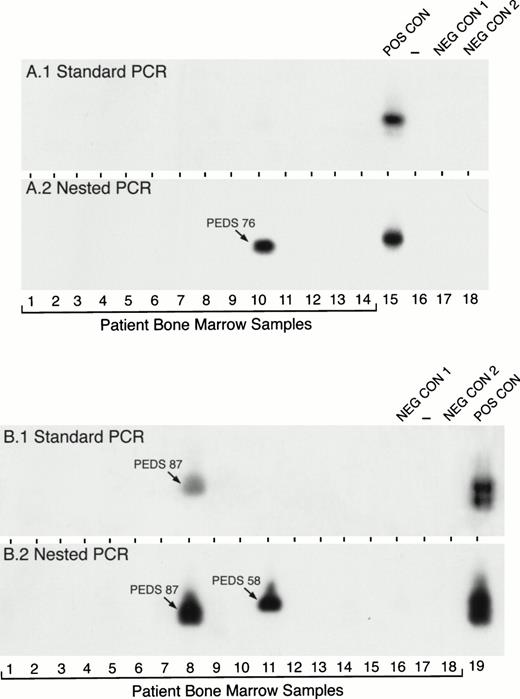
In contrast to the high expression frequency of MLL-AF4 in infant ALL, standard PCR detected MLL-AF4 positivity in only 1 of 127 (0.8%) pediatric ALL cases. However, MLL-AF4 fusion transcripts were found in leukemic cells from 17 (13%) patients when nested PCR assays were applied (Table 1). Figure 2 depicts representative experiments illustrating the detection of MLL-AF4 fusion transcripts in t(4;11) ALL by standard and nested PCR as well as detection ofMLL-AF4 expression in non-t(4;11) ALL patients by nested PCR. RNA was reextracted from cryopreserved cells in 10 of the 15 MLL-AF4+ non-t(4;11) ALL cases and reanalyzed by nested RT-PCR to confirm the initial results shown in Table 1 and Fig 2. As shown in Fig 3, each of these 10 non-t(4;11) cases were MLL-AF4+ when retested by nested PCR.
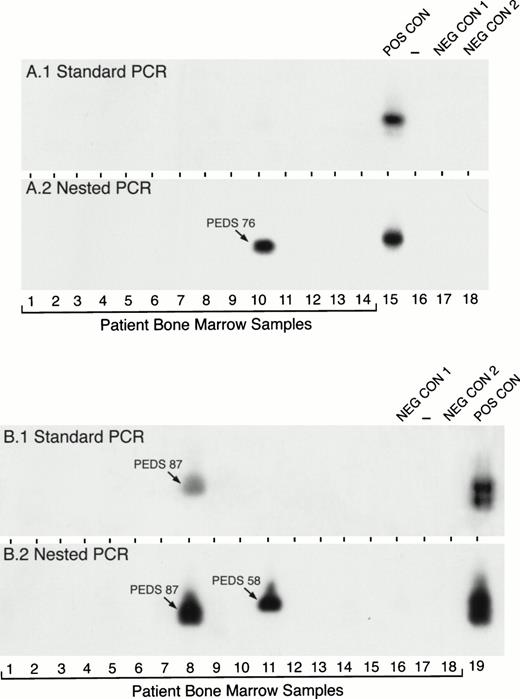
Detection of MLL-AF4 fusion transcripts in pediatric ALL BM specimens by standard and nested RT-PCR. RNA samples from primary leukemic cells of 32 (A, 14 patients; B, 18 patients) newly diagnosed pediatric ALL patients were examined forMLL-AF4 fusion transcript expression by standard RT-PCR (A1 and B1) and nested RT-PCR (A2 and B2). PEDS 76 shown in A.2 was a normal diploid case (see Tables 3 and 6). PEDS 87 shown in B.1. and B.2 was a hyperdiploid t(4;11) ALL case and PEDS 58 shown in B.2. was a hyperdiploid non-t(4;11) case with del(11)(q23) (see Tables 3 and 6). Amplified mRNA from the RS4;11 cell line was used as a positive control (POS CON). Negative controls were PCR products from RNA-free reaction mixture 1 plus reaction mixture 2 (NEG CON 1) and reaction mixture 1 plus DNA polymerase-free reaction mixture 2 (NEG CON 2).
Detection of MLL-AF4 fusion transcripts in pediatric ALL BM specimens by standard and nested RT-PCR. RNA samples from primary leukemic cells of 32 (A, 14 patients; B, 18 patients) newly diagnosed pediatric ALL patients were examined forMLL-AF4 fusion transcript expression by standard RT-PCR (A1 and B1) and nested RT-PCR (A2 and B2). PEDS 76 shown in A.2 was a normal diploid case (see Tables 3 and 6). PEDS 87 shown in B.1. and B.2 was a hyperdiploid t(4;11) ALL case and PEDS 58 shown in B.2. was a hyperdiploid non-t(4;11) case with del(11)(q23) (see Tables 3 and 6). Amplified mRNA from the RS4;11 cell line was used as a positive control (POS CON). Negative controls were PCR products from RNA-free reaction mixture 1 plus reaction mixture 2 (NEG CON 1) and reaction mixture 1 plus DNA polymerase-free reaction mixture 2 (NEG CON 2).
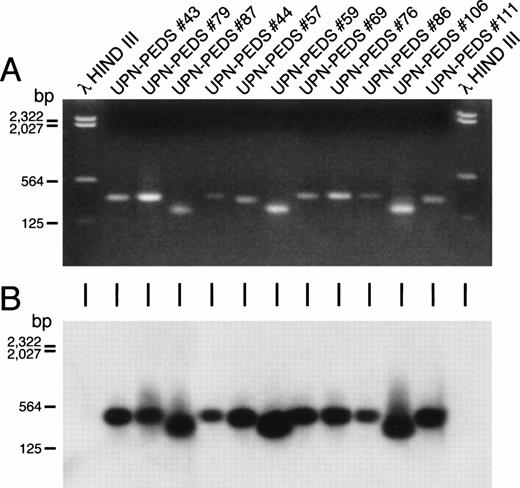
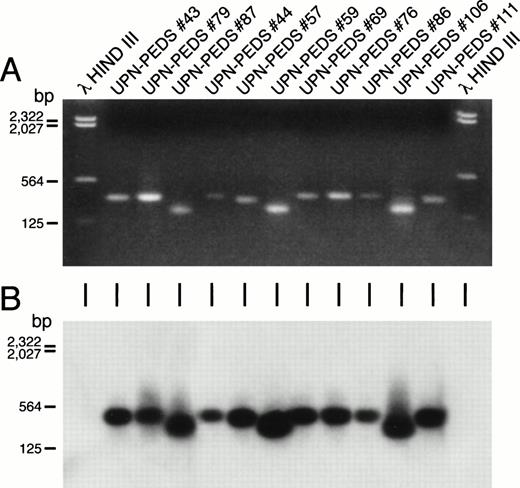
Detection of MLL-AF4 fusion transcripts in non-t(4;11) leukemic cells by PCR. Ethidium bromide–stained gel (A) and Southern blot (B) of the PCR reaction products from ALL patients who lack cytogenetically detectable t(4;11). First and last lanes contain molecular size markers. UPN-PEDS 69, 76, 86, and 106 had normal diploid karyotypes. UPN-PEDS 79 had a pseudodiploid karyotype. UPN-PEDS 43, 44, 57, and 59 had hyperdiploid karyotypes with structural abnormalities. UPN-PEDS 111 had hyperdiploid karyotypes without structural abnormalities. UPN-PEDS 87, a patient with t(4;11), was included as a positive control (see Tables 3 and 6).
Detection of MLL-AF4 fusion transcripts in non-t(4;11) leukemic cells by PCR. Ethidium bromide–stained gel (A) and Southern blot (B) of the PCR reaction products from ALL patients who lack cytogenetically detectable t(4;11). First and last lanes contain molecular size markers. UPN-PEDS 69, 76, 86, and 106 had normal diploid karyotypes. UPN-PEDS 79 had a pseudodiploid karyotype. UPN-PEDS 43, 44, 57, and 59 had hyperdiploid karyotypes with structural abnormalities. UPN-PEDS 111 had hyperdiploid karyotypes without structural abnormalities. UPN-PEDS 87, a patient with t(4;11), was included as a positive control (see Tables 3 and 6).
Of the 1,322 children treated on the CCG-1800 series protocols for whom centrally acceptable cytogenetic data were available, 399 (30.2%) were normal diploid, 74 (5.6%) were hypodiploid, 368 (27.8%) were pseudodiploid, and 481 (36.4%) were hyperdiploid (N.H., F.M.U., unpublished data, June 1998). Similarly, of the 127 children included in the present study, 49 (38.6%) were normal diploid, 3 (2.4%) were hypodiploid, 33 (26.0%) were pseudodiploid, and 42 (33.1%) were hyperdiploid. There were only 2 patients with t(4;11) (1 pseudodiploid and 1 hyperdiploid case); standard PCR detected MLL-AF4 expression in 1 of these cases, whereas both cases were positive by nested PCR (Table 1). Notably, 15 of the remaining 125 non-t(4;11) patients (12%), including 3 pseudodiploid, 6 normal diploid, and 6 hyperdiploid cases, were found to be nested PCR positive for MLL-AF4 fusion transcript expression even though standard PCR was negative in each of these 15 cases. Cytogenetic and PCR data as well as the age and WBC count at presentation for the 17 pediatric ALL patients who expressed MLL-AF4 fusion transcripts are detailed in Table 3. None of these 17 MLL-AF4 positive cases were positive forE2A-PBX1 (standard PCR) or BCR-ABL expression (standard and nested PCR). In summary, 17 of 127 noninfant pediatric ALL patients were positive for expression of MLL-AF4 fusion transcripts, yet only 2 of these patients had cytogenetically detectable t(4;11)(q21;q23). Thus, MLL-AF4 fusion transcripts were found in leukemic cells from patients lacking t(4;11) translocations.
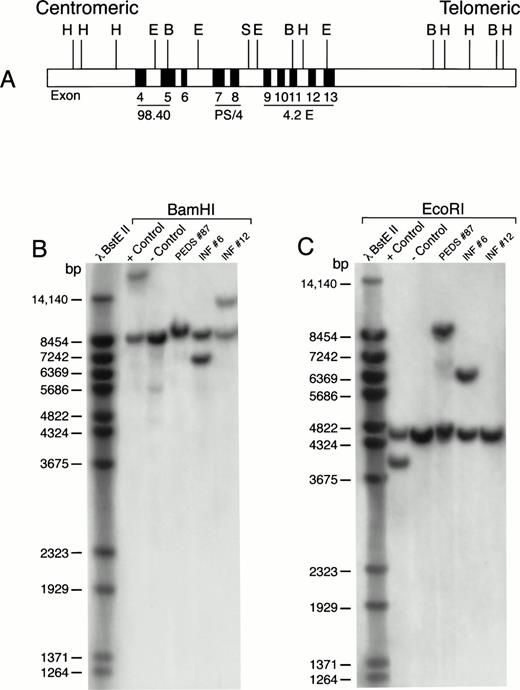
We next used Southern blot analyses to examine HindIII-,EcoRI-, or BamHI-digested genomic DNA of leukemic cells from a subset of standard PCR−, nested PCR+ patients with MLL-AF4 fusion transcripts for the presence of MLL gene rearrangements. As shown in Table 4 and Fig 4, cells from INF-6 and PEDS-87, two t(4;11)+, standard PCR−, nested PCR+ ALL patients, had MLL gene rearrangements occurring in the HindIII, EcoRI, and BamHI fragments as detected by the PS/4, 98.4, or 4.2E probes. Similarly, cells from PEDS-27, PEDS-44, and PEDS-5, three of the five t(4;11)−, standard PCR−, nested PCR+ ALL patients, had MLL gene rearrangements (Table 4, Fig 5). These results are consistent with the notion that in some ALL cases with MLLgene rearrangements and MLL-AF4 positivity, the level of expression for MLL-AF4 fusion transcripts in leukemic cells may be so low that a nested PCR is required for their detection. By comparison, the absence of MLL gene rearrangements in some patients with MLL-AF4 positivity (eg, PEDS-59 and PEDS-106 in Table 4) indicates the existence of a very small subpopulation (detectable by nested PCR only) of leukemic cells expressing theMLL-AF4 fusion transcript.
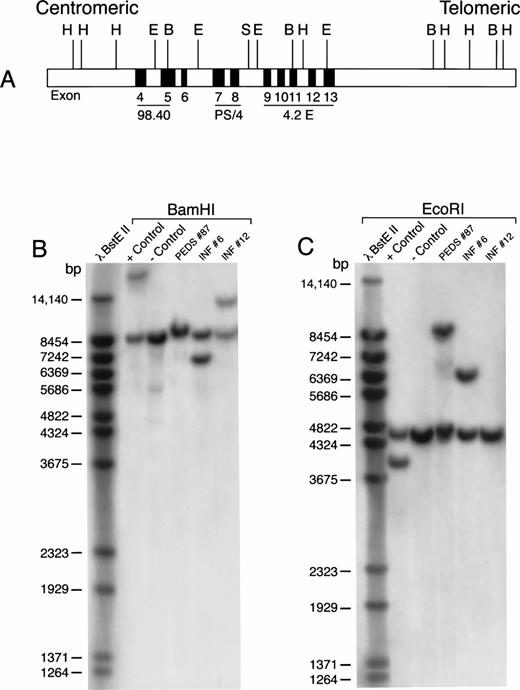
MLL rearrangement in leukemic cells from patients with ALL. Genomic DNA was digested with BamHI or EcoRI restriction enzymes, separated by electrophoresis, blotted to nylon membranes, and hybridized with the radiolabeled P/S4 probe. (A) Probes for analysis of MLL gene rearrangement. Partial restriction map of the 11q23 region containing the MLL gene. Approximate positions of MLL exons 4 through 13 (of 21 exons) are shown as gray boxes. Areas mapped by probes P/S4, 98.40, and 4.2E are indicated. Abbreviations: H, HindIII; E, EcoRI; B, BamHI; S, Sac I. (B) BamHI digest. (C) EcoRI digest. RS4;11 cells were used as a positive control and normal peripheral lymphocytes served as a negative control. bp, base pairs.
MLL rearrangement in leukemic cells from patients with ALL. Genomic DNA was digested with BamHI or EcoRI restriction enzymes, separated by electrophoresis, blotted to nylon membranes, and hybridized with the radiolabeled P/S4 probe. (A) Probes for analysis of MLL gene rearrangement. Partial restriction map of the 11q23 region containing the MLL gene. Approximate positions of MLL exons 4 through 13 (of 21 exons) are shown as gray boxes. Areas mapped by probes P/S4, 98.40, and 4.2E are indicated. Abbreviations: H, HindIII; E, EcoRI; B, BamHI; S, Sac I. (B) BamHI digest. (C) EcoRI digest. RS4;11 cells were used as a positive control and normal peripheral lymphocytes served as a negative control. bp, base pairs.
MLL rearrangement in leukemic cells, normal BM, and fetal liver. Genomic DNA was digested with EcoRI, separated by electrophoresis, blotted to nylon membranes, and hybridized with the radiolabeled probes. (A) P/S4 probe. (B) 98.40 probe. RS4;11 cells were used as a positive control and normal peripheral lymphocytes served as a negative control. FL7 was isolated from a fetus age 19 gestational weeks. NBM4 was a BM sample from a normal infant BM donor age 6 months. UPN-PEDS no. 91 was a male age 17.7 years with newly diagnosed t(4;11) ALL. UPN-PEDS no. 5 was a female age 2.9 years with newly diagnosed normal diploid ALL. bp, base pairs.
MLL rearrangement in leukemic cells, normal BM, and fetal liver. Genomic DNA was digested with EcoRI, separated by electrophoresis, blotted to nylon membranes, and hybridized with the radiolabeled probes. (A) P/S4 probe. (B) 98.40 probe. RS4;11 cells were used as a positive control and normal peripheral lymphocytes served as a negative control. FL7 was isolated from a fetus age 19 gestational weeks. NBM4 was a BM sample from a normal infant BM donor age 6 months. UPN-PEDS no. 91 was a male age 17.7 years with newly diagnosed t(4;11) ALL. UPN-PEDS no. 5 was a female age 2.9 years with newly diagnosed normal diploid ALL. bp, base pairs.
Prognostic significance of MLL-AF4 fusion transcripts in t(4;11)− pediatric ALL patients.
Eight of the 9 infants with MLL-AF4+ ALL had cytogenetically detectable t(4;11) and presented with very high WBC; all 9 MLL-AF4+ ALL infants were CD10− (Table 2). In accordance with previous reports, outcome among the MLL-AF4+ infants was poor, with only 2 of 9 patients surviving disease-free longer than 12 months after diagnosis despite intensive chemotherapy and/or BM transplantation (data not shown). Unlike MLL-AF4+infants, who were CD10−, all but 1 of theMLL-AF4+ children were CD10+, and the majority of these children (13 of 15) were ages 2 to 9.9 years (Table3).
The 15 MLL-AF4+/t(4;11)− patients were similar to fusion transcript−(MLL-AF4−/BCR-ABL−/E2A-PBX1−= PCR−) patients with respect to important presenting clinical characteristics, including age, WBC count, organomegaly, risk classification, and modal chromosome number.
The majority of patients in both groups were less than 10 years of age, had WBC counts <50,000/μL, and were assigned to CCG protocols for low- or intermediate-risk ALL, which corresponds to standard risk according to NCI classification.52
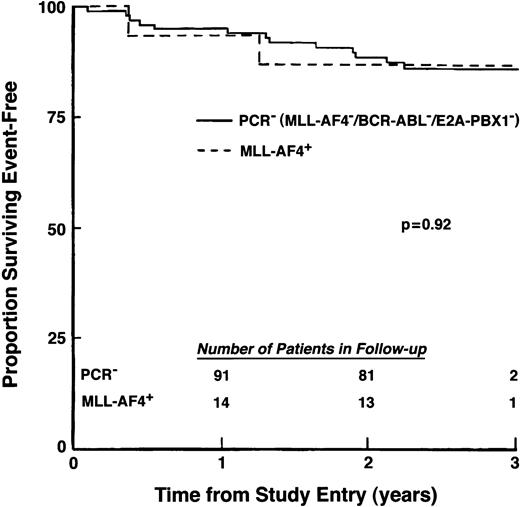
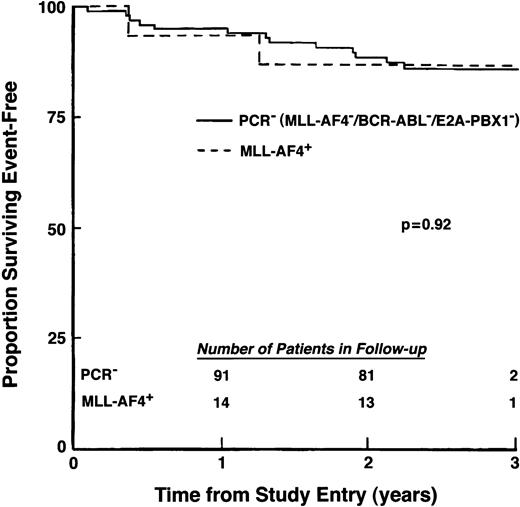
All MLL-AF4+/t(4;11)− patients and the majority (96%) of PCR− patients achieved remission after induction chemotherapy. Outcome forMLL-AF4+/t(4;11)− patients also was similar to that of the 97 MLL-AF4−, BCR-ABL−, E2A-PBX1− patients (Fig 6), with both groups achieving excellent EFS at 2 years of follow-up (86.7%, SD = 8.8% v88.4%, SD = 3.3%, respectively). Thus, among this subset of primarily standard-risk pediatric ALL patients, presence of an MLL-AF4fusion transcript in the absence of a cytogenetically detectable t(4;11) translocation was not associated with poor outcome.
EFS for patients with t(4;11) negative/MLL-AF4+ ALL. Percentage of patients surviving event-free was compared for 15MLL-AF4+ patients who lacked cytogenetically detectable t(4;11) and 97 patients who showed no evidence for MLL-AF4, BCR-ABL, or E2A-PBX1fusion transcripts. (Inset) Number of patients in follow-up at indicated times.
EFS for patients with t(4;11) negative/MLL-AF4+ ALL. Percentage of patients surviving event-free was compared for 15MLL-AF4+ patients who lacked cytogenetically detectable t(4;11) and 97 patients who showed no evidence for MLL-AF4, BCR-ABL, or E2A-PBX1fusion transcripts. (Inset) Number of patients in follow-up at indicated times.
Expression of MLL-AF-4 fusion transcripts in normal infant BM and fetal hematopoietic cells.
The presence of MLL-AF-4 fusion transcripts in leukemia cells without a cytogenetically detectable t(4;11) translocation prompted the hypothesis that MLL-AF4 fusion transcripts may be similarly expressed in rare populations of normal lymphohematopoietic cells. To test this hypothesis, we used RT-PCR to determine ifMLL-AF4 fusion transcripts were present in fetal BMs and fetal livers (gestational age, 15 to 22 weeks), as well as BM samples from normal infants.
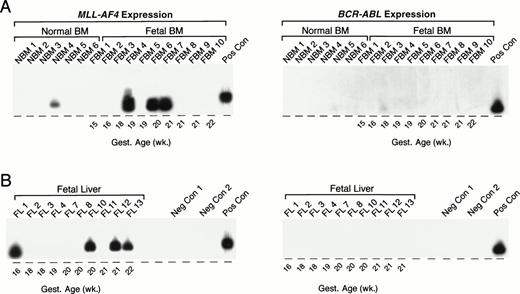
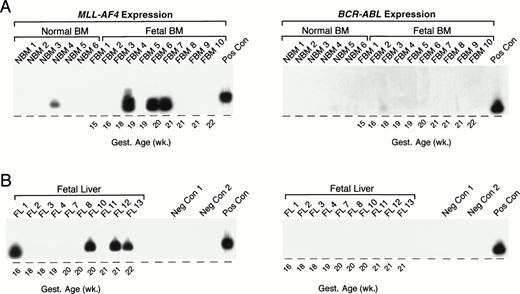
As summarized in Table 5 and illustrated in Fig 7, an MLL-AF4 fusion transcript was detected by nested (but not standard) PCR in 1 (NBM 4) of 6 (17%) normal infant BM samples. MLL-AF4 transcripts were detected by nested (but not standard) PCR in 4 of 16 (25%) fetal BMs: FBM 4, 19 weeks; FBM 6, 20 weeks; FBM 7, 21 weeks; FBM 11, 18 weeks. Representative samples are shown in Fig 7A. MLL-AF4 also was detected in 5 of 13 fetal livers: FL 1, 16 weeks gestational age; FL 14, 19 weeks gestational age; FL 10, 20 weeks gestational age; FL 12, 21 weeks gestational age; FL 13, 22 weeks gestational age; but not in any of the 44 remission BM specimens from pediatric ALL patients (Table 5). Nested PCR data from representative fetal liver samples are shown in Fig 7B. Unlike MLL-AF4 fusion transcripts, BCR-ABL fusion transcripts were not expressed in any of the normal infant BM, fetal BM, or fetal liver samples by standard or nested PCR (Fig 7).
Detection of MLL-AF4 fusion transcripts in normal hematopoietic cells by Southern blot analysis. (A) PCR products from normal infant and fetal BM. (B) PCR products from fetal liver. PCR products were transferred to nylon membranes and hybridized as described in Materials and Methods with 32P-labeled oligonucleotide detection probes for MLL-AF4 andBCR-ABL. Amplified mRNA from the RS4;11 and patient UPN100, with t(9;22)+ ALL, were used as positive controls (Pos Con) for MLL-AF4 and BCR-ABL fusion transcripts, respectively. Negative controls (Neg Con) were PCR products from RNA-free reaction mixture 1 plus reaction mixture 2 (negative control 1) and reaction mixture 1 plus DNA polymerase-free reaction mixture 2 (negative control 2). Gest., gestational.
Detection of MLL-AF4 fusion transcripts in normal hematopoietic cells by Southern blot analysis. (A) PCR products from normal infant and fetal BM. (B) PCR products from fetal liver. PCR products were transferred to nylon membranes and hybridized as described in Materials and Methods with 32P-labeled oligonucleotide detection probes for MLL-AF4 andBCR-ABL. Amplified mRNA from the RS4;11 and patient UPN100, with t(9;22)+ ALL, were used as positive controls (Pos Con) for MLL-AF4 and BCR-ABL fusion transcripts, respectively. Negative controls (Neg Con) were PCR products from RNA-free reaction mixture 1 plus reaction mixture 2 (negative control 1) and reaction mixture 1 plus DNA polymerase-free reaction mixture 2 (negative control 2). Gest., gestational.
As expected, MLL gene rearrangements were detected in leukemic cells from both of the t(4;11)+, standard PCR+, nested PCR+ cases (ie, INF-12 and PEDS-91) examined. In contrast, no MLL gene rearrangements were detected by Southern blot analysis in any of the DNA samples from normal cells with nested PCR positivity for MLL-AF4 fusion transcript expression (Table 4, Fig 5). The nested PCR positivity of these standard PCR− samples is likely caused by the presence of a small subpopulation of normal cells expressingMLL-AF4 fusion transcripts rather than very low levelMLL-AF4 expression in the majority of the cells.
Molecular characterization of MLL-AF4 fusion transcripts in leukemic and normal cells.
The predominant amplification products of the MLL-AF4 nested RT-PCR assays from 2 t(4;11)+ infants, 1 t(4;11)− infant, 2 t(4;11)+ children, and 13 t(4;11)− children with ALL, as well as 5 fetal livers and 2 fetal BMs, were characterized by sequence analyses. These analyses confirmed that the PCR-generated products in each of the 25 cases were MLL-AF4 fusion transcripts and showed common splicing variants in the various samples assayed. MLL-AF4sequence analysis data for these 25 samples are summarized in Table 6. In 3 of 4 t(4;11) leukemia cases, sequence analysis indicated that the PCR products resulted from fusions between MLL exon 6 and AF4 exon a, whereas the PCR product in the remaining case was the result of an MLL exon 7/AF4 exon b fusion. Among the 14 MLL-AF4+non-t(4;11) cases, 4 had MLL exon 6/AF4 exon a fusion transcripts, 7 had MLL exon 7/AF4 exon a fusion transcripts, and 3 had MLL exon 7/AF4 exon b fusion transcripts. By comparison, the nested PCR products from 7 fetal liver (N = 5)/fetal BM (N = 2) specimens resulted from fusions betweenMLL exon 6 and AF4 exon a (N = 4) or MLL exon 7 and AF4 exon b (N = 3).
DISCUSSION
We have examined the expression of MLL-AF4 fusion transcripts in pediatric and infant ALL patients with or without cytogenetically detectable t(4;11)(q21;q23), as well as in normal BM cells and fetal tissues using standard and nested RT-PCR assays. Overall,MLL-AF4 transcripts were detected by nested PCR in 9 of 17 infant ALL patients and 17 of 127 childhood ALL patients. Cytogenetically detectable t(4;11) translocations were found in 8 of the 9 MLL-AF4+ infants, but in only 2 of 17MLL-AF4+ children with ALL. Sequence analysis confirmed that the PCR-generated products were MLL-AF4 fusion transcripts and revealed common splicing variants in the various samples assayed. Notably, MLL-AF4 transcripts were found in 9 of 29 fetal tissues and 1 of 6 normal infant BM samples.
Although previous investigators15,27-29 have shown thatMLL-AF4 fusion transcripts were present in infant and adult ALL patients who were cytogenetically t(4;11)−, to our knowledge this is the first report to document the expression ofMLL-AF4 fusion transcripts in noninfant pediatric ALL patients lacking cytogenetic evidence of t(4;11). Interestingly, the expression of MLL-AF4 fusion transcripts in these patients was not associated with high-risk features or poor treatment outcome. Behm et al16 recently reported that MLL rearrangements confer poor outcome in pediatric ALL patients regardless of age. However, all patients in the study by Behm et al had MLLrearrangements in concert with a cytogenetically detectable 11q23 abnormality. Numerous investigators have documented the poor prognosis of ALL patients with t(4;11),12-16 although in one study the subset of patients 1 to 9 years of age with t(4;11) was distinct in having a favorable outcome.53
The observation that MLL-AF4 fusion transcripts are present in normal hematopoietic tissues raises crucial questions regarding the significance of MLL-AF4 rearrangements in leukemogenesis of ALL. Rearrangements between the 11q23 and 4q21 loci may occur in normal cells where their biological impact may be dependent on additional leukemogenic events. Altered expression or loss of expression of genes other than MLL and AF4 could be involved in both leukemogenesis and the poor prognosis of t(4;11)+ patients.
A crucial role for MLL in mammalian development has been shown by disruption of 11q23 in transgenic mice.54MLL+/− mice exhibited retarded growth, hematologic abnormalities, and disrupted segmental development of the axial skeleton, and MLL−/− was lethal to embryonic development. Although these studies clearly indicate that disruption of normal MLL expression throughout the organism can have deleterious effects, the effects of MLL-AF4 gene fusions restricted to blood-forming tissues in normal individuals or t(4;11)− leukemia patients may be much less dire. Further studies of MLL-AF4+ ALL patients and normal individuals will be required to confirm this hypothesis.
Consistent with our finding of MLL-AF4 fusion transcripts in normal tissues, recent reports by others have documented similar findings for other lymphoid malignancy-associated gene fusion products.BCL2-JH fusions, which were thought to arise via t(14;18)(q32;q21) in follicular cell lymphoma and diffuse large-cell lymphoma,36,37 have now been detected in peripheral blood cells from healthy individuals.31,33-35BCL2-JH fusions also were reported to occur in benign hyperplastic lymphoid tissues,30,32 as well as in all hematopoietic lineages of patients with B-cell non-Hodgkin's lymphoma (NHL).55 In addition, the BCR-ABL fusion transcript, which is thought to occur as a result of the t(9;22) translocation that represents a significant adverse risk factor in pediatric ALL,3,4,56 was recently identified in the blood of normal individuals38 as well as in a Ph− case of chronic myelogenous leukemia.57 In our current analysis, BCR-ABLfusions were detected in two ALL patients lacking classic t(9;22) translocations, but not in any of the normal tissues examined. It also is noteworthy that the gene fusion partners AF9 andENL, which are translocated in the 11q23 rearrangements t(9;11) and t(11;19), like AF4, encode proteins with serine- and proline-rich regions and nuclear localization signals.19,22,58 59 In light of our current findings, it would be of interest to determine if MLL-AF9 andMLL-ENL fusion transcripts are present in normal cells or leukemic cells lacking cytogenetically detectable t(9;11) and t(11;19).
In view of the findings described above, the contribution ofMLL-AF4 and other fusion transcripts toward leukemic transformation of lymphopoietic precursors requires further investigation. Furthermore, the utility of sensitive PCR-based detection of these fusion transcripts as sole indicators of minimal residual disease60 or for potential screening of cord blood before transplant should be reconsidered given their presence in nonleukemic cells.
ACKNOWLEDGMENT
The authors are grateful to Dr James Downing (St Jude Children's Research Hospital, Memphis, TN) for his generous and comprehensive assistance in establishing the RT-PCR assays in the CCG ALL Biology Reference Laboratory.
Supported in part by Department of Health and Human Services grants, including CCG Chairman's Grant Nos. CA-13539 and CA-60437 from the National Cancer Institute. F.M.U. is a Stohlman Scholar of the Leukemia Society of America and Parker Hughes Chair in Oncology.
Presented in part at the 39th Annual Meeting of the American Society of Hematology, December 5-9, 1997, San Diego, CA.
Address reprint requests to Fatih M. Uckun, MD, PhD, Hughes Institute, 2665 Long Lake Rd, Suite 330, St Paul, MN 55113.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.