Abstract
In multiple myeloma (MM), previous studies showed that mutations of the p53 gene are rare events in patients with newly diagnosed disease, but it is not known whether deletions of p53 are of any significance in MM. To address this question, we used interphase fluorescence in situ hybridization (FISH) with a DNA probe specific for the p53 locus at 17p13 and investigated bone marrow plasma cells from 72 patients with MM (59 patients = 81.9% before therapy). By FISH, deletions of p53, which were found to be predominantly monoallelic, were detected in 32.8% and 54.5% of patients with newly diagnosed and relapsed MM, respectively. Karyotypes from six of the patients with a p53 deletion by FISH showed a structural abnormality of 17p in only one of them. Additional FISH studies including a distal-17p probe (specific for theD17S34 locus) provided evidence for an interstitial deletion on 17p resulting in loss of p53 hybridization signals in myeloma cells. Among all 59 patients with newly diagnosed MM, presence of a p53 deletion was associated with stage III (P = .054), but not with other laboratory and clinical parameters. Patients with a p53 deletion had significantly shorter survival time compared with those without a deletion, both from the time of diagnosis (median 13.9v 38.7 months; P < .0001) and from the time of initiation of induction treatment consisting of conventional dose chemotherapy (median 15.9 months v median not reached at 38 months; P < .0002). On stepwise multivariate regression analysis, presence of a p53 deletion was the most significant independent parameter predicting for shortened survival (P = .002). We conclude that a p53 gene deletion, which can be identified by interphase FISH in almost a third of patients with newly diagnosed MM, is a novel prognostic factor predicting for short survival of MM patients treated with conventional-dose chemotherapy.
© 1998 by The American Society of Hematology.
THE p53 TUMOR suppressor gene, which maps to 17p13,1 has been shown to be implicated in the control of normal cellular proliferation, differentiation, and apoptosis.2 Inactivation of p53 due to mutation or allelic loss has been observed in many solid tumors, and results suggested a relation between presence of a p53 mutation and tumor development or progression.3,4 In addition, alterations of the p53 gene have been reported to be of prognostic relevance in non–small cell lung cancer5 as well as in breast cancer.6 In hematological malignancies, mutations of p53 generally occur at a lower frequency compared with solid tumors,7 but when present they appear to identify prognostically relevant subgroups of patients with acute myelogenous leukemia, myelodysplastic syndromes, and aggressive non-Hodgkin's lymphomas.8,9 In contrast, multiple myeloma (MM) is one of the few neoplasms in which p53 mutations are very rare at the time of diagnosis. Overall, p53 mutations were found to occur in fewer than 20% of patients with MM, which usually had advanced and aggressive forms of the disease including plasma cell leukemia.10-15 It was concluded from these studies that p53 mutations represent a late event in the progression of MM and have only limited value as a prognostic factor in MM.
p53 function may also be compromised by presence of a gene deletion, which has been shown in a recent study of patients with chronic lymphocytic leukemia (CLL).16 In this B-cell malignancy, deletions of p53 were predictive for nonresponse to purine-analogs like fludarabine and for short survival. In MM, chromosomal abnormalities leading to loss of 17p were only infrequently reported in conventional cytogenetic studies of MM.17-22 However, cytogenetic studies by means of metaphase analysis are limited in MM because of the low proliferative capacity of myelomatous plasma cells in vitro. We and others have previously shown that detection of chromosomal abnormalities in MM is improved by use of interphase fluorescence in situ hybridization (FISH), and that chromosomal aneuploidy is present in the majority of patients with MM.23-26
Thus, the aims of the present investigation were twofold: First, because deletions of 17p13 have not yet been studied systematically in MM, we investigated the incidence of p53 deletions using interphase FISH with a p53-specific DNA probe. Second, we addressed the question whether presence of a p53 deletion was of any clinical and prognostic significance in MM.
MATERIALS AND METHODS
Patients.
Seventy-four bone marrow (BM) samples from 72 consecutive, nonselected patients with MM (median age 63 years, range 34 to 87) were studied. There were 59 patients at diagnosis, 4 patients during induction chemotherapy, and 9 patients at relapse; 2 patients were studied both at initial presentation and at relapse. Main characteristics of patients are summarized in Table 1. When patients had symptomatic and/or progressive disease, they received conventional-dose chemotherapy as induction treatment: Melphalan-based regimens (melphalan/prednisone, vincristine/melphalan/cyclophosphamide/prednisone ± carmustine) were administered in 36 patients (5 of them concomitantly received interferon-α), 5 patients were treated with VAD (vincristine/doxorubicin/dexamethasone), and 1 patient received high-dose glucocorticoids. Response to treatment was defined as a sustained decrease (for longer than 4 weeks) of the paraprotein-level to less than 50% of the pretreatment value; in addition, achievement of a response required normalization of a preexisting hypercalcemia and no increase in the number and/or size of lytic bone lesions.27 One patient not responding to melphalan/prednisone was subsequently treated with high-dose melphalan (140 mg/m2) followed by autologous stem cell support.
Characteristics of 72 Patients With MM
| Feature . | n . | % . |
|---|---|---|
| Age: >60 yr | 52 | 72.2 |
| Sex: | ||
| Male | 29 | 40.3 |
| Female | 43 | 59.7 |
| Before therapy | 59 | 81.9 |
| Ig-subtype: | ||
| IgG | 47 | 65.3 |
| IgA | 20 | 27.8 |
| Bence-Jones only | 2 | 2.8 |
| Biclonal | 2 | 2.8 |
| Non-secretory | 1 | 1.3 |
| Stage (Durie & Salmon): | ||
| I | 13 | 18.1 |
| II | 12 | 16.7 |
| III | 47 | 65.2 |
| Creatinine >2 mg/dL | 8 | 11.1 |
| β2-Microglobulin >6 mg/L | 11 | 15.3 |
| LDH >240 U/L | 13 | 18.1 |
| Feature . | n . | % . |
|---|---|---|
| Age: >60 yr | 52 | 72.2 |
| Sex: | ||
| Male | 29 | 40.3 |
| Female | 43 | 59.7 |
| Before therapy | 59 | 81.9 |
| Ig-subtype: | ||
| IgG | 47 | 65.3 |
| IgA | 20 | 27.8 |
| Bence-Jones only | 2 | 2.8 |
| Biclonal | 2 | 2.8 |
| Non-secretory | 1 | 1.3 |
| Stage (Durie & Salmon): | ||
| I | 13 | 18.1 |
| II | 12 | 16.7 |
| III | 47 | 65.2 |
| Creatinine >2 mg/dL | 8 | 11.1 |
| β2-Microglobulin >6 mg/L | 11 | 15.3 |
| LDH >240 U/L | 13 | 18.1 |
Cells.
BM aspirates were obtained from the posterior iliac crest or sternum during standard diagnostic procedures and were collected in a heparinized syringe. Informed consent according to institutional guidelines was obtained for the use of an aliquot of the samples for research purposes. After dilution with phosphate-buffered saline (PBS), mononuclear BM cells were separated by density gradient centrifugation over Ficoll-Hypaque (density = 1.077; Sigma Inc, St Louis, MO). Mononuclear cells were washed twice with PBS, treated with Carnoy's fixative (methanol/glacial acetic acid 3:1 [vol/vol]), and stored at −20°C or −80°C.
Specimens used as controls consisted of peripheral blood (PB) (n = 4) and BM samples (n = 9) from healthy individuals. Reactive plasma cells were obtained from the PB of a patient with severe sepsis (nucleated PB cells consisted of 40% plasma cells).
Slides with normal metaphases derived from phytohemagglutinin-stimulated lymphocytes were obtained from Vysis, Inc (Downers Grove, IL).
Interphase FISH studies.
DNA probes specific for the p53-locus on 17p13 (directly conjugated with SPECTRUM-Orange), for the Rb-1 locus on 13q14 (directly conjugated with SPECTRUM-Orange), and for the centromeric region of chromosome 17 (directly conjugated with SPECTRUM-Green) were purchased from Vysis, Inc. A probe hybridizing to the D17S34 locus (labeled with digoxigenin), which maps distal of p53 on 17p,28 was obtained from Oncor (Gaithersburg, MD). An anti-digoxigenin antibody tagged with rhodamine (Oncor) was used as secondary reagent to visualize the hybridized D17S34 probe. Hybridization was performed as detailed in a previous report.29 To control for sufficient hybridization conditions, experiments were always performed as dual-color hybridizations combining the chromosome 17-centromeric probe with either the p53 or the D17S34 probe. Since none of the myeloma specimens was characterized by loss of the chromosome 17 centromere, presence of ≥2 signals with this probe in more than 90% of nuclei was considered as evidence for high hybridization efficiency. Only slides fulfilling these criteria were scored, and at least 200 cells with the appearance of plasma cells were evaluated.
Images were captured using a cooled charge coupled device (CCD) camera (Photometrics, Tucson, AZ) mounted on a Zeiss Axioplan-2 immunofluorescence microscope (Zeiss, Jena, Germany) and linked to an Apple Macintosh computer (Apple Comp Corp, Cuperdino, CA). Prints were obtained with a Phaser 440 Tektronix color printer (Tektronix, Inc, Wilsonville, OR).
Metaphase cytogenetics.
Mononuclear cells after Ficoll-Hypaque density separation were washed twice and subsequently cultured at 2 × 106/mL in α-MEM:McCoy's 5A (1:1, vol/vol) or RPMI1640 medium supplemented with 20% heat-inactivated fetal calf serum. Growth factors (granulocyte-macrophage colony-stimulating factor, 100 U/mL; interleukin-6 [IL-6], 250 U/mL; IL-10, 25 U/mL; 10% B- and T-cell growth supplement, obtained from Origen [Origen Inc, Rockville, MD]), either alone or in combination, were added to parallel cultures. Harvesting and spreading of metaphases on slides was performed using standard techniques, followed by the tristain protocol established by Schweizer.30,31 Metaphases were analyzed using the semi-automated Genevision 221 karyotyping system (Applied Imaging, Carlsbad, CA). Chromosomal aberrations were defined and described according to the International System for Cytogenetic Nomenclature.32
Statistical analysis.
Statistical evaluation of results included Fisher's exact test, χ2-test, t-test, and the nonparametric Kruskal-Wallis test. Odds ratios were determined according to Mantel-Haenszel. Kendall's tau coefficients were used in testing correlations. Survival time, measured both from the date of diagnosis and the date of start of induction treatment, was calculated using Kaplan-Meier estimates. Differences between survival curves were analyzed by means of Breslow and Mantel-Cox tests. Multivariate analysis of survival time data was performed using the proportional hazards regression model of Cox. Stepwise evaluation of maximum partial likelihood ratio was used to identify prognostic factors.
RESULTS
Control hybridizations.
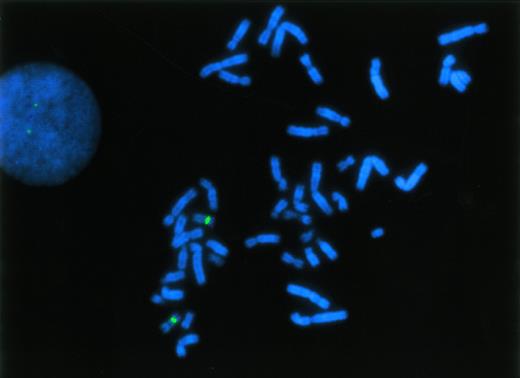
To establish the cut-off level for presence of a p53 deletion by interphase FISH, hybridizations were performed with PB (n = 9) and BM specimens (n = 4) from healthy individuals. As with the myeloma specimens, simultaneous hybridizations with probes specific for the chromosome 17 centromere and the p53 locus were performed (Fig1). In normal specimens, we observed fewer than two hybridization signals per nucleus with the p53 probe in 3.24% ± 1.79% (mean ± standard deviation [SD]) of nuclei. Of note, there was no significant difference between normal PB and BM samples. In addition, similar results were obtained with reactive plasma cells (PB plasmocytosis in a patient with severe sepsis) exhibiting one p53 hybridization signal in 3.4% of cells (407 cells evaluated). Thus, the cut-off level for the presence of a p53 deletion was set at 8.6% (mean + 3 SD of the frequency of control cells with only one p53 hybridization signal).
FISH analysis of the p53 gene at 17p13 using a specific DNA probe (red fluorescent signals), hybridized simultaneously with a chromosome 17-centromeric probe (green fluorescent signals). Probes were applied to normal, phytohemagglutinin-stimulated lymphocytes and a metaphase spread as well as an interphase nucleus showing a normal hybridization pattern are displayed.
FISH analysis of the p53 gene at 17p13 using a specific DNA probe (red fluorescent signals), hybridized simultaneously with a chromosome 17-centromeric probe (green fluorescent signals). Probes were applied to normal, phytohemagglutinin-stimulated lymphocytes and a metaphase spread as well as an interphase nucleus showing a normal hybridization pattern are displayed.
Deletions of p53 in MM by interphase FISH.
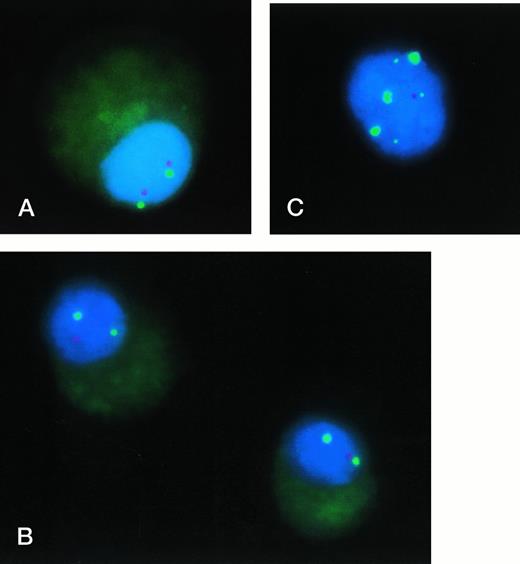
In 25 of 74 BM samples (33.8%) from patients with MM, interphase FISH analysis of plasma cells provided evidence for a p53 gene deletion (Fig2). Among 59 patients with newly diagnosed MM, a deletion of p53 was detected in 19 of them (32.8%); at relapse, a p53 deletion was observed in 6 of 11 patients (54.5%). None of the 4 patients studied during induction chemotherapy exhibited a p53 deletion. Two patients, who had normal findings for p53 by FISH at diagnosis, were again studied at relapse, and a deletion of p53 was detected in one of them. The percentage of plasma cells exhibiting a p53 deletion ranged between 17.8% and 95.0% (median, 49.1%). In 17 of the 25 patients, deletions of p53 were monoallelic, whereas in 7 patients loss of both p53 alleles was observed in 20% to 40% of the deleted plasma cell population. In one patient with relapsed MM, a biallelic deletion of p53 was found.
Interphase FISH analysis of the p53 gene in MM: (A) Plasma cell from a patient without p53 deletion: Two hybridization signals with both probes (chromosome 17 centromere, green signals; p53, red signals) can be recognized. (B) Plasma cells from patient 3/FJ (Table 2) exhibiting a monoallelic deletion of p53 (probes as in A). (C) Interphase cell of patient 2/NN (compare Table 2: trisomy 17 by metaphase cytogenetics) hybridized with 3 probes: Chromosome 17 centromere (labeled with SPECTRUM-Green; large green signals), p53 (labeled with SPECTRUM-Orange; red signal), and distal 17p (D17S34 locus, labeled with digoxigenin and visualized with anti-digoxigenin-fluorescein isothiocyanate; small green signals). Presence of three signals for chromosome 17 centromere andD17S34, but only one signal for p53 indicating an interstitial deletion on 17p involving the p53 locus.
Interphase FISH analysis of the p53 gene in MM: (A) Plasma cell from a patient without p53 deletion: Two hybridization signals with both probes (chromosome 17 centromere, green signals; p53, red signals) can be recognized. (B) Plasma cells from patient 3/FJ (Table 2) exhibiting a monoallelic deletion of p53 (probes as in A). (C) Interphase cell of patient 2/NN (compare Table 2: trisomy 17 by metaphase cytogenetics) hybridized with 3 probes: Chromosome 17 centromere (labeled with SPECTRUM-Green; large green signals), p53 (labeled with SPECTRUM-Orange; red signal), and distal 17p (D17S34 locus, labeled with digoxigenin and visualized with anti-digoxigenin-fluorescein isothiocyanate; small green signals). Presence of three signals for chromosome 17 centromere andD17S34, but only one signal for p53 indicating an interstitial deletion on 17p involving the p53 locus.
Gain of signals with the chromosome 17 centromeric probe was observed in 10 patients (13.3%), and in three of these patients, a deletion of p53 was found. Of note, also in these patients fewer than two hybridization signals with the p53-specific probe were observed (compare Fig 2C), indicating the presence of a true deletion and not just relative loss of p53. In the remaining 7 patients, gain of chromosome 17 centromere was paralleled by a gain of hybridization domains for the p53 gene.
Comparison of metaphase cytogenetics and interphase FISH.
Conventional cytogenetic studies were performed in 23 patients, and clonal chromosomal abnormalities were detected in 10 patients (43.5%). In 7 of these 23 patients, interphase FISH provided evidence for a p53 deletion in plasma cells. By metaphase cytogenetics, however, chromosome 17 was found to be structurally abnormal in only one patient (Table 2, patient 1/GN): One chromosome 17 was found to be derivative with loss of material from both arms, and on the second chromosome 17 additional material was found distal of band 17p13 (add17p13). In patient 2/NN, metaphase analysis indicated presence of trisomy 17 (without structural abnormalities involving chromosome 17), which corresponds to the finding of three signals by FISH with the chromosome 17 centromeric probe; however, only one p53 signal was observed by interphase FISH. In four patients, normal results for chromosome 17 were obtained by metaphase analysis, and in patient 7, only diploid metaphases were found.
Metaphase Cytogenetic Results in Seven Patients With a p53 Gene Deletion by Interphase FISH
| Patient . | Karyotype . | Interphase FISH . | ||
|---|---|---|---|---|
| No. 17-Centromere . | p53 . | D17S34 . | ||
| 1/GN | Hypodiploid (38-45),XX,der(1),del(1)(p12),−4,inv(6)(p12p25)/−6,del(10)(q21),−10, t(11;?) (q10;?),dic(16;?)(p13;?),add(17)(p13),der(17), add(22)(p13)/trc(22;?;?)(p13;?;?),+3mar[cp15]/46,XX[5] | 2 Signals | 1 Signal | 2-3 Signals |
| 2/NN | 52,XY,+t(1;16)(p10;p10,+3,+5,+ins(7;?)(p21;?),+9,+del(11)(q13),−13,−14, +15,−16,+17,+18,+19,+21[cp5]/106,idemx2[1]/46,XY[12] | 3 Signals | 1 Signal | 3 Signals |
| 3/FJ | 50,XX,del(1)(p34),+dic(1;18)(p12;q21),+t(1;16)(p10;p11),+add(3)(p22), der(4),+5,del(6)(q13q25),−13,−16,+19,−20,+21,−22[5]/46,XX[11] | 2 Signals | 1 Signal | 2 Signals |
| 4/EB | 53,XX,+3,+5,+7,+9,del(11)(q23),del(13)(q13q32),+15,+19,+mar[1]/46,XY[61] | 2 Signals | 1 Signal | 2 Signals |
| 5/RB | 40,XY,+t(1;16)(p10;p10)x2,−5,−10,−10,−13,−14,−16,−16,−21[1]/45,X, −Y[13]/46,XY[49] | 2 Signals | 1 Signal | 2 Signals |
| 6/RE | Hypodiploid(36-45),X,− X,dic(1;15)(q10;q26),t(2;10)(q14;p13),add(3)(q29), add(7)(p22),−8,var del(11)(q11/q14),der(12),−13,add(16)(q24),+2dicmar[cp20] | 2 Signals | 1 Signal | 2 Signals |
| 7/MS | 46,XX[19] | 2 Signals | 1 Signal | 2 Signals |
| Patient . | Karyotype . | Interphase FISH . | ||
|---|---|---|---|---|
| No. 17-Centromere . | p53 . | D17S34 . | ||
| 1/GN | Hypodiploid (38-45),XX,der(1),del(1)(p12),−4,inv(6)(p12p25)/−6,del(10)(q21),−10, t(11;?) (q10;?),dic(16;?)(p13;?),add(17)(p13),der(17), add(22)(p13)/trc(22;?;?)(p13;?;?),+3mar[cp15]/46,XX[5] | 2 Signals | 1 Signal | 2-3 Signals |
| 2/NN | 52,XY,+t(1;16)(p10;p10,+3,+5,+ins(7;?)(p21;?),+9,+del(11)(q13),−13,−14, +15,−16,+17,+18,+19,+21[cp5]/106,idemx2[1]/46,XY[12] | 3 Signals | 1 Signal | 3 Signals |
| 3/FJ | 50,XX,del(1)(p34),+dic(1;18)(p12;q21),+t(1;16)(p10;p11),+add(3)(p22), der(4),+5,del(6)(q13q25),−13,−16,+19,−20,+21,−22[5]/46,XX[11] | 2 Signals | 1 Signal | 2 Signals |
| 4/EB | 53,XX,+3,+5,+7,+9,del(11)(q23),del(13)(q13q32),+15,+19,+mar[1]/46,XY[61] | 2 Signals | 1 Signal | 2 Signals |
| 5/RB | 40,XY,+t(1;16)(p10;p10)x2,−5,−10,−10,−13,−14,−16,−16,−21[1]/45,X, −Y[13]/46,XY[49] | 2 Signals | 1 Signal | 2 Signals |
| 6/RE | Hypodiploid(36-45),X,− X,dic(1;15)(q10;q26),t(2;10)(q14;p13),add(3)(q29), add(7)(p22),−8,var del(11)(q11/q14),der(12),−13,add(16)(q24),+2dicmar[cp20] | 2 Signals | 1 Signal | 2 Signals |
| 7/MS | 46,XX[19] | 2 Signals | 1 Signal | 2 Signals |
Interphase FISH analysis of the D17S34 locus in MM.
Because interphase FISH, but not metaphase chromosome analysis, indicated loss of material from 17p, we performed additional hybridization studies using a distal 17p DNA probe specific for theD17S34 locus. D17S34 represents the most distal genetically mapped region of the chromosome 17p arm.27 In normal specimens, 96.7% ± 0.4% of nuclei showed two hybridization signals for D17S34, and only one hybridization domain was observed in 2.9% ± 0.5% of cells (cutoff level for loss ofD17S34: 4.4%). All 25 myeloma patients, who were found to have a p53 gene deletion by FISH, were studied with the D17S34 probe by interphase FISH (simultaneous hybridizations with the chromosome 17 centromeric probe). In only 3 of these 25 patients, a deletion ofD17S34 was observed by FISH (Table3), and the concomitant loss of hybridization signals for both p53 and D17S34 strongly suggests a deletion of the 17p arm. However, in all other patients (deletion of the p53 gene, but presence of the D17S34 locus) an interstitial deletion involving 17p13 should be the abnormality leading to the observed loss of p53 hybridization signals. To test this hypothesis, we performed simultaneous hybridizations with three probes (chromosome 17-centromere, p53, D17S34) to interphase nuclei of patient 2/NN (trisomy 17 by metaphase cytogenetics, but deletion of p53 by interphase FISH; compare Table 2): As illustrated in Fig 2C, a normal hybridization pattern with presence of hybridization signals for all three probes was observed for only one chromosome 17, whereas both remaining chromosomes 17 were characterized by loss of the p53 locus in the presence of hybridization signals for chromosome 17-centromere andD17S34. Taken together, these results indicate that deletion of the p53 gene in MM is mainly due to an interstitial deletion of a presumably narrow chromosomal region on 17p, and that this abnormality remains unrecognizable by metaphase chromosome analysis.
Interphase FISH Results of Three Patients With Loss of Distal-17p (D17S34 Locus)
| Patient . | Signal Distribution by Dual-Color FISH . | % Cells . | No. of Cells Evaluated . |
|---|---|---|---|
| 8/HP | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 35.0 | 305 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 26.4 | 308 | |
| 9/LW | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 95.0 | 208 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 64.0 | 307 | |
| 10/EK | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 42.6 | 252 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 30.7 | 209 |
| Patient . | Signal Distribution by Dual-Color FISH . | % Cells . | No. of Cells Evaluated . |
|---|---|---|---|
| 8/HP | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 35.0 | 305 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 26.4 | 308 | |
| 9/LW | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 95.0 | 208 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 64.0 | 307 | |
| 10/EK | 2 Signals [chromosome 17 centromere]/1 signal [p53] | 42.6 | 252 |
| 2 Signals [chromosome 17 centromere]/1 signal [D17/S34] | 30.7 | 209 |
Correlation of p53 deletions with laboratory and clinical findings.
Analyzing data from all 59 patients with newly diagnosed MM, we addressed the question whether or not the p53 status by FISH was correlated with clinical and major prognostic parameters. Regarding the Durie & Salmon stage, patients with a p53 deletion were predominantly at stage III (p53 deletion in 38.5% of stage III patients v12.5% of patients at stages I and II; P = .054). However, there was no significant correlation of p53 status with sex, age, type of the paraprotein, serum creatinine, hemoglobin, platelet count, serum lactat dehydrogenase-level, C-reactive protein, or beta-2 microglobulin (B2M). In 27 of the patients with newly diagnosed MM, results from interphase FISH studies of chromosomal region 13q14 were available. Loss of 13q14 was observed in 15 patients (55.6%); however, we did not observe an association between deletion of p53 and loss of 13q14 (contingency correlation r = .246,P < .1).
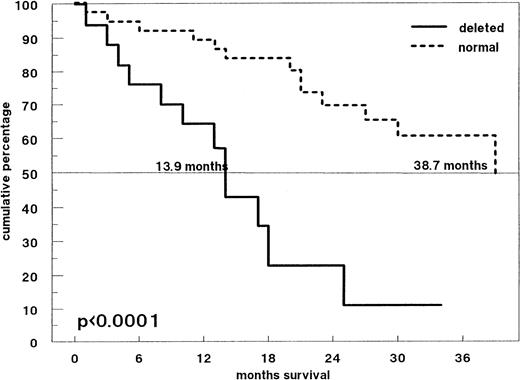
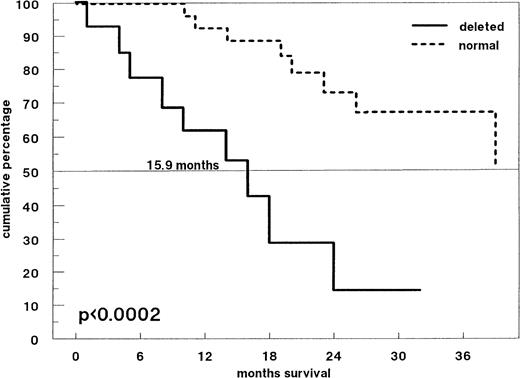
Figure 3 shows the survival probabilities from the time of diagnosis of all 59 previously untreated patients: Patients with and without p53 deletions had significantly different overall survival times (median 13.9 months and 38.7 months, respectively; P < .0001). Forty-two patients received induction treatment with conventional-dose chemotherapy. Response to treatment was observed in 26 patients (61.9%). Among the 14 patients with a p53 deletion by FISH, 42.9% responded compared with 71.4% of patients without p53 deletion (P = .078). However, survival time from the time of initiation of induction chemotherapy was significantly shorter for patients with a p53 deletion (Fig4; P < .0002). Among patients with a p53 deletion, only one patient survived longer than 30 months: This patient was a 36-year-old woman who did not respond to melphalan/prednisone, but achieved a partial remission after high-dose melphalan and autologous PB stem cell support.
Survival probabilities from the time of diagnosis in 59 patients with newly diagnosed MM. The solid line represents patients with a p53 deletion by FISH (n = 19), the dotted line those without a p53 deletion (n = 40). The median survival times of both groups are significantly different (P < .0001).
Survival probabilities from the time of diagnosis in 59 patients with newly diagnosed MM. The solid line represents patients with a p53 deletion by FISH (n = 19), the dotted line those without a p53 deletion (n = 40). The median survival times of both groups are significantly different (P < .0001).
Survival probabilities from the time of start of induction therapy (conventional-dose chemotherapy) in 42 patients with newly diagnosed MM. Patients with a p53 deletion (n = 14; solid line) have a significantly shorter survival time compared with MM patients without a p53 deletion (n = 28; dotted line) (P < .0002).
Survival probabilities from the time of start of induction therapy (conventional-dose chemotherapy) in 42 patients with newly diagnosed MM. Patients with a p53 deletion (n = 14; solid line) have a significantly shorter survival time compared with MM patients without a p53 deletion (n = 28; dotted line) (P < .0002).
When comparing noncensored survival data from 52 myeloma patients who had either been observed for more than 12 months or had died within the first year after diagnosis, deletion of p53 was found to be associated with a more than fivefold risk to die within 12 months from diagnosis, as compared with patients with normal findings for p53 (Table4).
Survival of 52 Patients Observed Longer Than 12 Months or Until Death
| . | Survival <12 mos . | Survival >12 mos . |
|---|---|---|
| Normal p53 | 10.8% | 89.2% |
| Deletion of p53 | 40.0% | 60.0% |
| Odds ratio (95% CI) | 5.50 (1.36-22.18) | |
| Significance | P < .02 | |
| . | Survival <12 mos . | Survival >12 mos . |
|---|---|---|
| Normal p53 | 10.8% | 89.2% |
| Deletion of p53 | 40.0% | 60.0% |
| Odds ratio (95% CI) | 5.50 (1.36-22.18) | |
| Significance | P < .02 | |
Abbreviation: CI, confidence interval.
Univariate linear regression identified deletion of p53, elevated serum-creatinine, elevated B2M, age over 65, stage III, and a high percentage of BM plasma cells as prognostic factors for shortened survival. On stepwise multivariate regression analysis, presence of a p53 deletion proved to be the best prognostic factor: It contributed more than twice as much to accurate prediction of survival than advanced stage, which remained the second independent prognostic factor (Table 5).
Results of Stepwise Maximum Partial Likelihood Ratio Test (Cox Model)
| Step No. . | Variable . | DF . | Log Likelihood . | Improvement . | Global . | ||
|---|---|---|---|---|---|---|---|
| χ2 . | P Value . | χ2 . | P Value . | ||||
| 0 | (Maximum) | −61.204 | |||||
| 1 | p53 Deletion | 1 | −56.526 | 9.36 | .002 | 11.95 | .001 |
| 2 | Myeloma stage | 2 | −54.422 | 4.21 | .040 | 14.50 | .001 |
| % Plasma cells | 2.13 | .144 | NS | ||||
| Age | 1.41 | .236 | NS | ||||
| β2-Microglobulin | 0.37 | .543 | NS | ||||
| Serum creatinine | 0.23 | .631 | NS | ||||
| Step No. . | Variable . | DF . | Log Likelihood . | Improvement . | Global . | ||
|---|---|---|---|---|---|---|---|
| χ2 . | P Value . | χ2 . | P Value . | ||||
| 0 | (Maximum) | −61.204 | |||||
| 1 | p53 Deletion | 1 | −56.526 | 9.36 | .002 | 11.95 | .001 |
| 2 | Myeloma stage | 2 | −54.422 | 4.21 | .040 | 14.50 | .001 |
| % Plasma cells | 2.13 | .144 | NS | ||||
| Age | 1.41 | .236 | NS | ||||
| β2-Microglobulin | 0.37 | .543 | NS | ||||
| Serum creatinine | 0.23 | .631 | NS | ||||
Abbreviation: NS, not significant.
DISCUSSION
In this study we report that deletions of the p53 gene are detectable by interphase FISH in a significant proportion of patients with MM. Specifically, plasma cells from 32.8% of patients with newly diagnosed MM carry a p53 deletion, and the incidence of this abnormality increases to 54.5% in patients with relapsed MM. Our results indicate that deletions of 17p involving the p53 gene locus are much more common than previously assumed based on findings by metaphase cytogenetics. Evidence that 17p may be deleted in MM was also obtained in a recent molecular cytogenetic study of 13 MM cell lines and 8 tumor specimens including 5 cases of plasma cell leukemia: By means of comparative genomic hybridization, underrepresentation of region 17p11.2-p13 was a recurrent finding.33 The higher frequency of p53 gene deletions by FISH compared with banding analysis can only in part be explained by the fact that metaphase cytogenetics remains normal or noninformative in the majority of patients with newly diagnosed MM despite the finding of chromosomal aneuploidy by interphase FISH.23 Of note, we also observed p53 deletions by FISH in cases with abnormal cytogenetics, but apparently normal findings for chromosome 17 or 17p. Although it may be difficult to identify bands on metaphase chromosomes from lymphoid malignancies,34 the most likely explanation for the failure of detecting this deletion by banding studies is the presence of a physical deletion of 17p on the submicroscopical level. By additional hybridization experiments we have obtained evidence that the D17S34 locus distal on 17p is present in all but three cases with a p53 deletion by FISH. Thus, it is our conclusion that loss of p53 hybridization signals is predominantly due to an interstitial deletion that presumably is too small to be detected on metaphase chromosomes. In agreement with this conclusion, there is increasing evidence that FISH is a more sensitive method than banding analysis to detect deletions of narrow chromosomal regions. For example, FISH was shown to identify deletions of 9p involving theCDKN2 gene (p16INK4a/MTS1) in acute lymphoblastic leukemia,35,36 and of 12p in acute leukemias,37-41 both in the absence of microscopically visible deletions on metaphase chromosomes.
Our findings by FISH show that in the majority of patients with MM, p53 deletions are monoallelic. Although we do not have data on p53 mutations in our group of patients, we consider the coincidence of p53 deletions and mutations as a rare event in newly diagnosed MM because of the low reported frequency of p53 mutations at this stage of the disease.10-15 This is in contrast to observations in many other solid and hematological malignancies, where inactivation of p53 most commonly occurs through point mutation in one p53 allele, and deletion of the second one.7 Currently, we cannot answer the question whether p53 function in myeloma cells is compromised already by presence of a monoallelic deletion or by any additional mechanism. For example, inactivation of p53 may occur by interaction with other cellular proteins, particularly the murine double minute 2 (MDM2) protein.42 In MM cell lines, overexpression of MDM2 as well as binding of MDM2 to p53 has recently been shown.43 However, it is not known if this is also true for myeloma cells isolated directly from patients, and if suppression of p53 function by MDM2 may be facilitated in case of a p53 gene deletion.
With respect to clinical correlations, our study shows that presence of a p53 deletion in plasma cells from patients with MM was associated with significantly shortened survival. This specific chromosomal abnormality proved to be the most powerful prognostic factor of all variables investigated. Stepwise Cox analysis revealed two independent prognostic factors for shortened survival, namely deletion of p53 and advanced stage. Other known indicators of poor prognosis, like elevated B2M, impaired renal function, and advanced age, could not improve the prognostic power of these two factors.
The main reason for short survival of myeloma patients with a p53 deletion by FISH was poor response to chemotherapy, which in this group of patients was administered at conventional doses. The outcome was independent of the type of primary treatment whether it was melphalan-based or VAD. The association of a p53 deletion with poor response is reminiscent of what has been observed in patients with B-CLL in whom presence of a p53 deletion was correlated with nonresponse to otherwise very effective purine-analogs like fludarabine.16 Obviously, patients with p53 deletions show poor response to various unrelated chemotherapeutic agents. Because many cytotoxic drugs act through induction of apoptosis, and apoptosis requires a functional p53, it has been suggested that in tumor cells with impaired p53 function, apoptosis may not occur leading to resistance to chemotherapeutic agents.44 Alternatively, an altered p53 gene may lose its function as ‘guardian of the genome,’45,46 and, as a consequence, complex cytogenetic abnomalities may develop. These genetic changes could then be the true reason for resistance to treatment. Indeed, although karyotypes obtained from MM patients are usually complex, some specific chromosomal abnormalities were recently shown to be indicative for shortened event-free and overall survival, even after an intensive therapeutic approach including two autotransplants: Partial or complete loss of chromosome 13, aberrations involving 11q, and presence of translocations were identified as such unfavorable cytogenetic features.47 48 With respect to loss of 13q, which was found in 15 of 27 newly diagnosed patients of this series, we did not observe a correlation with p53 deletions. However, this does not exclude presence of any other unfavorable cytogenetic abnormality present in myeloma patients with a p53 gene deletion, but a comprehensive FISH analysis of prognostically relevant chromosomal regions in these patients is in progress.
We conclude that in contrast to p53 mutations, allelic loss of p53 is not uncommon in newly diagnosed MM and is predictive for short survival of patients treated with conventional-dose chemotherapy. It remains to be determined whether or not the unfavorable outcome of myeloma patients with a p53 deletion can be improved by high-dose treatment including autologous stem cell support. In case of prolonged survival of these high-risk patients after intensive therapy, molecular cytogenetics performed at diagnosis could contribute to identification of an unfavorable group of MM patients benefiting significantly from a risk-adapted therapeutic approach.
Supported by grants from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (P-10893-MED, P-12432-MED) and the Austrian National Bank (Jubiläumsfonds 5839).
Address reprint requests to Johannes Drach, MD, University of Vienna, First Department of Internal Medicine, Division of Clinical Oncology, Währinger Gürtel 18-20, A-1090 Vienna, Austria; e-mail:johannes.drach@akh-wien.ac.at.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal