Abstract
INTRODUCTION: Clinical outcomes of patients with myelodysplastic syndromes (MDS) and chronic myelomonocytic leukemia (CMML) are very heterogeneous. Although next generation sequencing has identified a number of somatic mutations in MDS and CMML the analysis of their prognostic impact has resulted in contradictory results. Although the number of cytogenetic abnormalities is a well-known prognostic factor in MDS, there is limited information evaluating the prognostic relevance of the number of mutations in this disease. Correlation of mutation data with patient outcomes and integration into clinical practice is still lacking.
METHODS: We conducted whole exome sequencing of 88 previously untreated patients with MDS and 26 with CMML. Exome capture hybrid was performed using Agilent SureSelect All Exon V4. Sequencing was performed with Illumina HiSeq 2000 and aligned to the hg19 human genome reference. Common virtual normal in house pool was used for somatic variant calling. Variant allele frequency (VAF) estimates were used to evaluate clonal and subclonal relationships within each individual sample. Clonal relationships were tested using Pearson chi-squared tests. Clinical and demographic data was obtained from clinical records. As a validation cohort, we studied 413 patients with untreated MDS and CMML with sequencing data obtained by use of a 28-gene next generation sequencing platform. The Kaplan-Meier produce limit methods were used to estimate the median overall survival (OS) and leukemia-free survival (LFS).
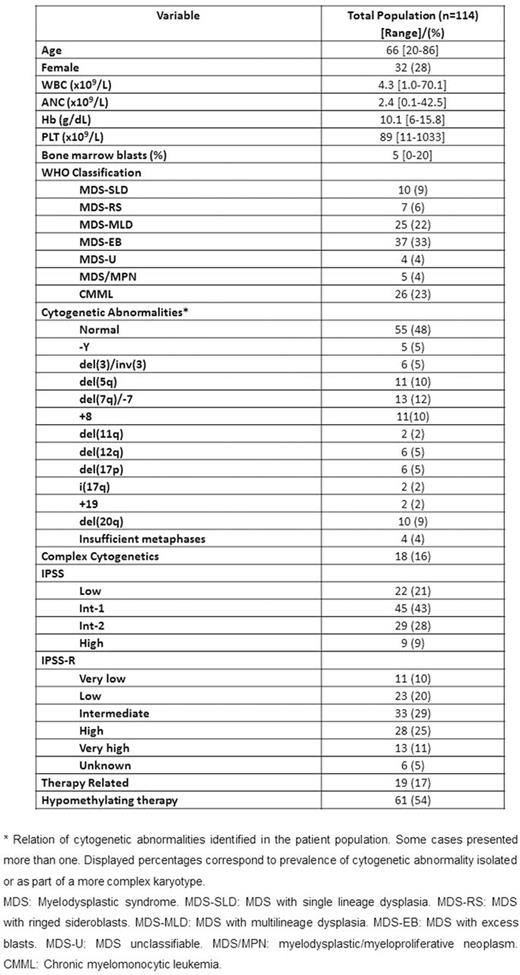
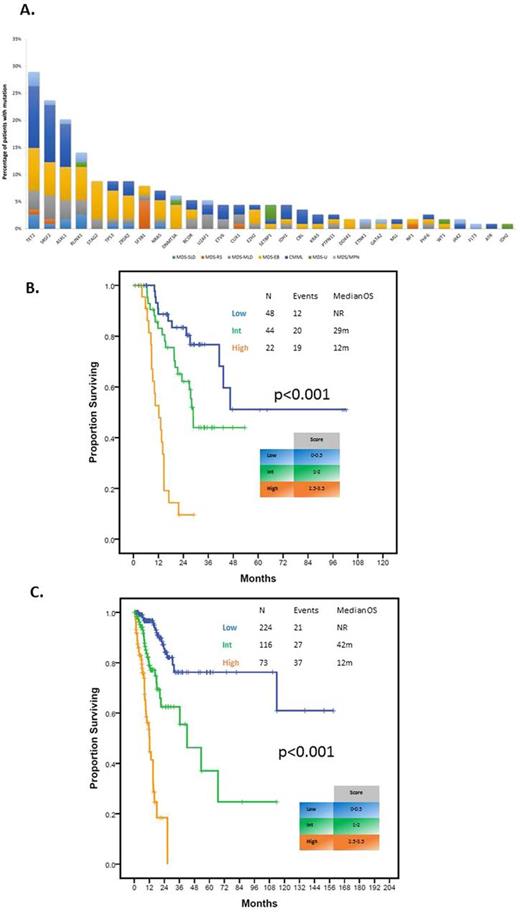
RESULTS: A total of 218 high-confidence driver mutations in 32 genes in 98 (86%) patients were identified. Patient characteristics are shown in Table 1. Median age was 66 (19-85). Median number of driver mutations was 2 (range 0-7). Most frequently detected mutations included TET2, SRSF2, ASXL1 and RUNX1 in >10% patients (Figure 1A). Median number of altered pathways was 2 (range 0-6) with mutations in TP53, splicing and DNA methylation tending to have higher VAF. Fifty-seven (50%) patients had at least 2 mutations and were evaluable for clonal heterogeneity testing. Among these patients, 32 (56%) were clonaly heterogeneous. The most frequent subclonal mutations included ASXL1 (31%), TET2 (25%) and RUNX1 (19%). Median follow up was 21.6 months (range 1-102 months). By univariate analysis, mutations in BCOR (HR 2.85, 95% CI 1.12-7.29, p=0.029), STAG2 (HR 2.45, 95% CI 1.04-5.80, p=0.041) and TP53 (HR 5.25, 95% CI 2.37-11.63, p<0.001) affected OS unfavorably and SF3B1 mutation affected OS favorably (median survival NR vs 28.7 months, p=0.023). Overall survival negatively correlated with increased number of mutations (p=0.02) with patients with 3 or more mutations having significantly worse outcomes (HR 1.94, 95% CI 1.10-3.41, p=0.022). Mutations in EZH2 (HR 8.49, 95% CI 1.85-39.02, p=0.006) and TP53 (HR 6.24, 1.63-23.86, p=0.007) as well as number of mutations predicted for shorter LFS (HR 1.50, 95% CI 1.06-2.12, p=0.021). Clonal heterogeneity was not associated with significant differences in OS or LFS. By multivariate analysis IPSS-R group (Intermediate: HR 1.45, 95% CI 0.56-3.77, p=0.448; High/Very High; HR 4.66, 95% CI 2.01-10.84, p<0.001), TP53 mutation (HR 3.12, 95% CI 1.30-7.49, p=0.011), and ³3 mutations (HR 2.51, 95% CI 1.32-4.76, p=0.005) retained their adverse impact on outcome. A new prognostic model integrating these variables separated patients into three risk groups (low, intermediate and high) with 3-year survival of 76, 62 and 0%, respectively (p<0.001) (Figure 1B), restaged 54 (47%) patients from IPSS-R and had increased stratification potential compared to IPSS-R (Dxy 0.46 vs 0.43; c-index 0.73 vs 0.71). Application of this model to the validation cohort rendered similar results (Figure 1C) with the new molecular model exhibiting higher discrimination potential for survival compared to IPSS-R (Dxy values 0.54 vs 0.51; c-index 0.77 vs 0.75).
CONCLUSIONS: Incorporation of mutation data into existing risk models improves prognostication of patients with MDS. Like cytogenetic abnormalities, the number of driver mutations have independent prognostic impact in MDS which seems almost equivalent to that of TP53mut. We believe that our study sets the basis for future multicenter studies aimed at developing a new prognostic model incorporating molecular data generated by use of a uniform sequencing technology.
Konopleva:Cellectis: Research Funding; Calithera: Research Funding. Jabbour:ARIAD: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Novartis: Research Funding; BMS: Consultancy. Daver:Sunesis: Consultancy, Research Funding; Karyopharm: Honoraria, Research Funding; Ariad: Research Funding; BMS: Research Funding; Kiromic: Research Funding; Pfizer: Consultancy, Research Funding; Otsuka: Consultancy, Honoraria. DiNardo:Celgene: Research Funding; Abbvie: Research Funding; Novartis: Research Funding; Agios: Research Funding; Daiichi Sankyo: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract