Abstract
Abstract 2389
Poster Board II-366
Children with Down syndrome (DS) have a 10 to 20-fold increased risk of developing acute lymphoblastic leukemia (ALL), and they have experienced poorer outcomes on recent major protocols worldwide. The cytogenetic abnormalities which are generally common in childhood ALL and contribute to risk-based treatment assignment are markedly less frequent in children with DS-ALL. Recently, activating mutations in Janus kinase 2 (JAK2) have been identified in approximately 20% of DS-ALL, and interstitial deletions involving cytokine receptor-like factor 2 (CRLF2) in approximately 50% of DS-ALL. Global gene expression profiling may provide insights into the biologic consequences of these molecular lesions. We performed microarray analysis of RNA from diagnostic bone marrow samples in 23 DS-ALL and 26 non-DS ALL cases using the Affymetrix Human Genome U133 Plus 2.0 array. CRLF2 expression was high in 10 of the 23 DS-ALL cases, 3 of which also bore JAK2 mutations, and in a single non-DS ALL case. Unsupervised hierarchical clustering analysis demonstrated clustering of non-DS ALL cases belonging to known cytogenetic subgroups such as E2A-PBX1, MLL rearrangement, and high hyperdiploidy. In contrast, neither DS-ALL cases overall nor the JAK2-mutated or high CRLF2 expressing cases formed a cohesive cluster. Supervised analysis identified 43 genes that were differentially expressed between CRLF2 high versus low cases with a false discovery rate <10%. Several of the most highly differentially expressed genes were validated by quantitative real-time PCR. These included three genes with high expression in CRLF2-high cases: chemokine (C-C motif) ligand 17 (CCL17) (p=0.01), V-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (YES1) (p=0.007), and Iroquois homeobox 2 (IRX2) (p=0.008); and one gene with expression inversely correlated with CRLF2 expression: dual specificity phosphatase 6 (DUSP6) (p=0.0015). Our findings suggest that DS-ALL does not form a single distinct biologic subgroup, but nearly half of DS-ALL cases are defined by high CRLF2 expression, a substantial enrichment for this lesion compared to its prevalence in non-DS ALL. Identification of downstream pathways may identify opportunities for targeted intervention, including interactions with other cytokines and activation of the JAK-STAT pathway.
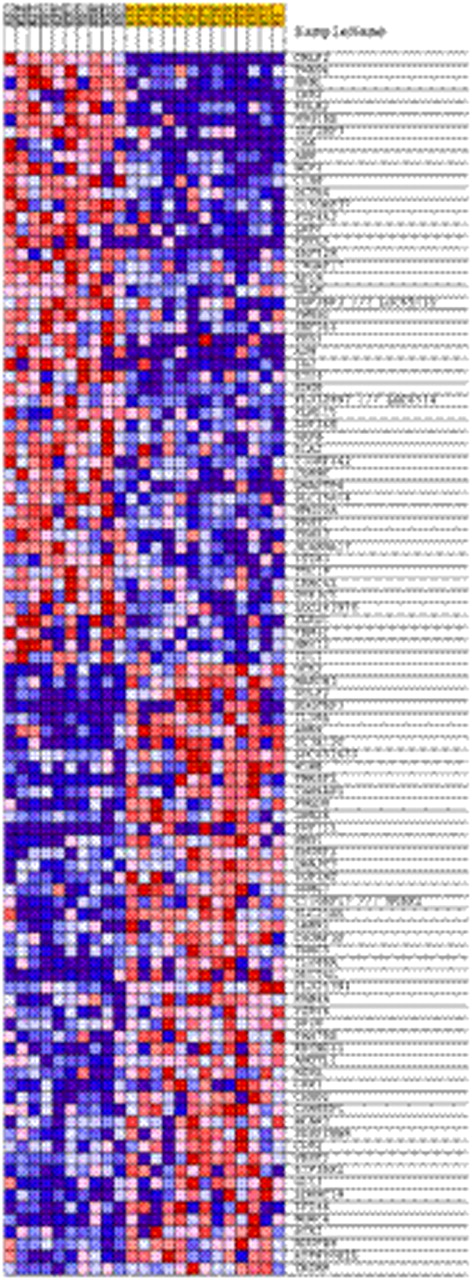
Gene expression signature of top differentially expressed genes in Down syndrome (DS) acute lymphoblastic leukemia (ALL) cases with high versus low CRLF2 expression. Each column indicates a case, with CRLF2-high cases depicted in gray and CRLF2-low cases in gold. Each row indicates one of the top 100 differentially expressed genes as determined by Gene Set Enrichment Analysis.
Gene expression signature of top differentially expressed genes in Down syndrome (DS) acute lymphoblastic leukemia (ALL) cases with high versus low CRLF2 expression. Each column indicates a case, with CRLF2-high cases depicted in gray and CRLF2-low cases in gold. Each row indicates one of the top 100 differentially expressed genes as determined by Gene Set Enrichment Analysis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.