Abstract
To study the impact of oncogenic K-Ras on T-cell leukemia/lymphoma development and progression, we made use of a conditional K-RasG12D murine knockin model, in which oncogenic K-Ras is expressed from its endogenous promoter. Transplantation of whole bone marrow cells that express oncogenic K-Ras into wild-type recipient mice resulted in a highly penetrant, aggressive T-cell leukemia/lymphoma. The lymphoblasts were composed of a CD4/CD8 double-positive population that aberrantly expressed CD44. Thymi of primary donor mice showed reduced cellularity, and immunophenotypic analysis demonstrated a block in differentiation at the double-negative 1 stage. With progression of disease, approximately 50% of mice acquired Notch1 mutations within the PEST domain. Of note, primary lymphoblasts were hypersensitive to γ-secretase inhibitor treatment, which is known to impair Notch signaling. This inhibition was Notch-specific as assessed by down-regulation of Notch1 target genes and intracellular cleaved Notch. We also observed that the oncogenic K-Ras-induced T-cell disease was responsive to rapamycin and inhibitors of the RAS/MAPK pathway. These data indicate that patients with T-cell leukemia with K-Ras mutations may benefit from therapies that target the NOTCH pathway alone or in combination with inhibition of the PI3K/AKT/MTOR and RAS/MAPK pathways.
Introduction
RAS proteins are small GTPases that cycle between an active (GTP-bound) and inactive (GDP-bound) state. GDP-GTP cycling is regulated by (1) guanine-nucleotide-exchange factors (GEFs) that stimulate the release of bound nucleotides followed by passive binding of cytoplasmic GTP; and (2) by GTPase-activating proteins (GAPs), stimulating intrinsic RAS GTPase activity.1-3 Ligand stimulation of receptor tyrosine kinases creates intracellular docking sites followed by recruitment of a multiprotein complex to the inner leaflet of the plasma membrane, activation of GEFs and several RAS effector pathways, including mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K), phospholipase Cϵ (PLC-ϵ) and Ral-GEF.4-7 These pathways are involved in the regulation of proliferation, cell survival, cytoskeletal organization, and differentiation. The RAS gene family consists of 3 members, KRAS, NRAS, and HRAS, which differ in their functional properties with respect to posttranslational modification, subcellular localization, activation of downstream effectors, and their role in development. Of note, the occurrence of RAS gene mutations in human cancers is highly variable with respect to tissue type and isoform.8-11
Somatic NRAS, HRAS, or KRAS mutations are detected in approximately 30% of all human cancers predominantly affecting codons 12, 13, and 61.11 In hematologic malignancies, activating RAS mutations are found in 10% to 15% of acute myeloid leukemias (AMLs), 25% of juvenile myelomonocytic leukemias (JMMLs), and 10% to 15% of acute lymphoblastic leukemias (B- and T-ALL).11-14 In addition, RAS germ line mutations or germ line mutations affecting other molecules in the RAS signaling pathways (eg, SHP2) have been detected in human developmental disorders associated with JMML (eg, neurofibromatosis type 1, Noonan and Costello syndromes; reviewed by Schubbert et al15 ). RAS can also be activated by gain-of-function mutations in upstream tyrosine kinases, including FLT3 internal tandem duplications, c-KIT point mutations, or TEL/ABL1 and BCR/ABL1 fusion genes.16-18
Mutations affecting signal transduction pathways have also been reported in T-ALL and include the NUP214-ABL fusions, accounting for approximately 5% to 10% of cases, and NRAS and KRAS mutations in approximately 10% and less than 2% of patients, respectively.11,14,19 In addition, RAS proteins have been reported to be activated in approximately 50% of T-ALLs.20 To investigate the effect of oncogenic K-Ras in the hematopoietic system, we used a model for conditional expression of this allele that has been previously described.21-23 Mx1-Cre/+KrasG12D/+ mice develop a rapidly fatal myeloproliferative disease.22,23 K-RasG12D–expressing cells show increased proliferation and hyperresponsiveness to cytokine stimulation, but the mutant allele is not sufficient to cause AML.24
Ras is required for proper thymocyte development and for the process of positive selection of these cells in the thymus.25-27 However, development of thymic lymphomas/leukemias has only been observed in mice expressing viral/oncogenic H-Ras that is not detected in human T-cell disease, and only rarely in animals expressing oncogenic K-Ras from its endogenous promoter.22,28,29 Furthermore, most murine T-ALL models have investigated the effects of aberrant expression or loss of transcription factors or molecules involved in cell-cycle regulation, but not of GTPases or tyrosine kinases.30-34 Therefore, the impact of oncogenic K-Ras on the initiation and maintenance of T-cell disorders has not been fully clarified.
In addition to the contribution of mutations that activate signal transduction pathways, in T-ALL, the NOTCH pathway has been shown to play a pivotal role in development of T-ALL in humans and in murine models of disease. The NOTCH1 gene encodes a transmembrane receptor expressed on hematopoietic stem cells and T cells. On binding to its ligands Delta1 or Jagged1, the NOTCH receptor undergoes proteolytic cleavage, leading to γ-secretase–mediated release of intracellular NOTCH (ICN). ICN translocates to the nucleus and activates transcription of NOTCH1 target genes, such as HES1 (Hairy/enhancer of split) and Deltex (DTX1; reviewed by Grabher et al35 ). Activating NOTCH1 mutations affecting the extracellular heterodimerization domain and/or the C-terminal PEST domain have been detected in more than 50% of human T-ALLs.36-38 The high prevalence of NOTCH1 mutations provides a rationale for targeting the NOTCH signaling path-way in T-cell malignancies. As a promising therapeutic strategy, γ-secretase inhibitors (GSIs), initially developed for Alzheimer disease, have been shown to inhibit proteolytic cleavage of NOTCH1 and induce a G1/G0 cell-cycle arrest in human T-ALL cell lines.36
We observed that, when we transplanted whole BM cells from polyinosinic-polycytidylic acid (pI-pC)–induced Mx1-Cre/+K-RasG12D/+ animals into syngeneic wild-type recipients, recipient mice developed a highly penetrant and aggressive T-cell disease, with lymphoblastic cells expressing CD4 and CD8, but also CD44 and pre–T-α. Immunophenotypic analysis of donor-derived, preleukemic thymocytes revealed a block in differentiation at the DN1 stage that was associated with increased apoptosis. Of note, these tumors were found to harbor secondary mutations in the PEST domain of Notch1. Treatment of tumor-derived cell lines with minimal doses of γ-secretase inhibitor led to inhibition of cell proliferation and induction of apoptosis.
Methods
Mouse strains
Lox-stop-lox (LSL)-K-RasG12D mice were crossed to C57BL/6 for at least 8 generations. To obtain double-transgenic mice, LSL-K-RasG12D/+ mice were crossed to C57BL/6 Mx1-Cre/+ mice (hereafter called KM mice). For all experiments, LSL-K-RasG12D/+ mice were used as controls (hereafter called control). For induction of Cre expression, 3- to 4-week-old mice were injected with a single dose (20 mg/kg) of pI-pC (GE Healthcare, Little Chalfont, United Kingdom). Genotyping and verification of Cre-mediated recombination were performed as described previously.21
All mice were housed in microisolator cages, monitored daily for evidence of disease, and killed when moribund. All experiments were conducted with the ethical approval of the Harvard Medical Area Standing Committee on Animals.
Bone marrow transplantation assay
For competitive bone marrow transplantation (BMT) assays, mice were killed 8 to 10 days after pI-pC injection, bone marrow (BM) cells were harvested from femurs and tibias, and single-cell suspensions were prepared. BM cell suspensions derived from K-RasG12D/+Mx1Cre/+ mice or K-RasG12D/+ (CD45.2 at the CD45 locus) were mixed with bone marrow single-cell suspensions derived from wild-type B6SJL/C57BL6 mice at a 1:1 or 5:1 ratio (CD45.1/CD45.2 at the CD45 locus). The mixed cell population was resuspended in Hank's balanced salt solution at a concentration of 0.8 × 106 cells/mL or 2.4 × 106 cells/mL, respectively, and 0.5 mL was injected into the lateral tail vein of wild-type B6.SJL recipient animals (CD45.1 at the CD45 locus), which were lethally irradiated (2 × 550 cGy).
Histopathology
Histopathologic analysis was performed at the Dana-Farber/Harvard Cancer Center Specialized Histopathology Services Core facility. In brief, tissues were fixed for at least 72 hours in 10% neutral buffered formalin (Sigma-Aldrich, St Louis, MO), dehydrated in alcohol, cleared in xylene, and infiltrated with paraffin on an automated processor (Leica Microsystems, Deerfield, IL). Tissue sections (4 μm thick) were placed on charged slides, deparaffinized in xylene, rehydrated through graded alcohol solutions, and stained with hematoxylin and eosin. Images of histological slides were obtained on a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) equipped with a SPOT RT color digital camera, model 2.1.1 (Diagnostic Instruments, Sterling Heights, MI), and analyzed in Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Flow cytometric analysis
Thymus and spleen tissue was homogenized and passed through a cell strainer (BD Biosciences Discovery Labware, Bedford, MA) to generate a single-cell suspension: BM mononuclear cells were flushed from hind leg bones with RPMI (Cambrex, East Rutherford, NJ) + 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), lysed on ice with red blood cell lysis solution (Gentra Systems, Minneapolis, MN), and washed in phosphate-buffered saline + 2% FBS. Cells (106) were blocked with Fc-block (BD Biosciences PharMingen, San Diego, CA) for 10 minutes and stained in phosphate-buffered saline + 2% FBS for 30 minutes on ice with the following monoclonal antibodies at a 1:100 dilution: CD3, CD4, CD8, CD25, CD44, TCR-β, or TCT-γ/δ (BD Biosciences, San Jose, CA). All analyses were gated on live cells based on propidium iodide staining. Cells were subsequently analyzed using a 4-color FACSCalibur cytometer or a FACSAria cytometer (BD Biosciences).
Apoptosis assays were performed by staining freshly harvested thymocytes or cells cultured in RPMI + 10% FBS + 1% penicillin/streptomycin supplemented with or without interleukin-2 (IL-2) and IL-7 for CD4 and CD8 followed by annexin V and 7-amino-actinomycin D staining according to the manufacturer's protocol (BD Biosciences PharMingen).
A minimum of 10 000 events was acquired and analyzed using FlowJo software (BD Biosciences).
TCR rearrangement PCR and RT-PCR
For PCR analysis, total RNA was extracted from 5 × 106 thymocytes according to the RNeasy Mini Kit (Qiagen, Valencia, CA) protocol, and 5 μg of total RNA was reverse-transcribed using the superscript III cDNA synthesis kit (Invitrogen). Each PCR reaction consisted of 2.5 μL of resultant cDNA and 20 pmol of different combinations of TCR-β–specific and TCR-α–specific primers as described elsewhere.39
For specific amplification of RAG1, RAG2, pre–T-α, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the following primers were used: RAG1-for 5′-ACCCGATGAAATTCAACACCC-3′; RAG1-rev 5′- CTGGAACTACTGGAGACTGTTCT-3′; RAG2-for 5′-CTCCCACCTCTTCGTTATCCA-3′; RAG2-rev 5′-GCTCATTGTTTGGTGTTTTCCC-3′; pTα-for 5′-CGTCA-GGTGTCAGGCTCTAC-3′; pTα-rev 5′-GTGAAGGCGTCTAGGGCAC-3′; GAPDH-for 5′-AGCCTCGTCCCGTAGACAAAA-3′; GAPDH-rev 5′-TG-GCAACAATCTCCACTTTGC-3′.
DNA resequencing of Notch1
Genomic DNA was isolated from 5 × 106 tumor cells using the DNA Blood Mini Kit (Qiagen). Amplification of Notch1 exons 26, 27, and 34 was performed as described previously.40 Amplification products were sequenced in both directions at the Biopolymers Facility (Harvard Medical School, Boston, MA).
Western blotting
A total of 5 × 106 cells of the primary T-ALL cell line 470 were treated with γ-secretase inhibitor XXI or PD98059 (Calbiochem, San Diego, CA) as indicated, and cells were lysed as previously described.41 Protein lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes as previously described.41
The following antibodies were used for Western blot analysis: anticleaved Notch1 (Val1744), anti-PARP, antiphospho-p42/44, anti-p42/44 (Cell Signaling Technology, Danvers, MA), and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA).
Tissue culture
Cell lines established from secondary transplant mice were propagated in RPMI 1640 + 10% FBS, 1% penicillin/streptomycin supplemented with 2 ng/mL of IL-2 and 5 ng/mL of IL-7 (PeproTech, Rocky Hill, NJ) or without cytokines. For growth inhibition assays, cells were seeded at a density of 106/mL in cytokine-conditioned medium with various concentrations of PD98059, rapamycin, or γ-secretase inhibitor XXI (Calbiochem), and the number of viable cells was determined after 48 hours using the CellTiter 96AQueous One Solution Proliferation Assay (Promega, Madison, WI).
Quantification of Notch target gene expression
Primary T-ALL cells were treated with GSI as indicated, and RNA extraction and cDNA synthesis were performed. The levels of Hes1, Dtx1, and Hey1 expression were assessed by quantitative RT-PCR (EZ RT-PCR Core Reagents; Applied Biosystems, Foster City, CA) following the manufacturer's recommendations. Expression levels of Hes1 (Mm00468601_m1 assay; Applied Biosystems), Dtx1 (Mm00492297_m1 assay; Applied Biosystems), and Hey1 (Mm00468865_m1 assay; Applied Biosystems) were normalized to GAPDH (assay no. 4352932E; Applied Biosystems).
Statistical analysis
Survival analysis was performed using the Kaplan-Meier method. Comparisons between 2 groups were performed using the unpaired Student t test. A P value of less than .05 was considered significant. Statistical computations were performed using GraphPad Prism software, version 4.0c (GraphPad, San Diego, CA).
Results
K-RasG12D induces an aggressive T-cell leukemia/lymphoma
Expression of K-RasG12D was induced by injecting 3- to 4-week-old C57BL/6 K-RasG12D/+Mx1-Cre/+ mice (hereafter called KM mice) with as single dose of pI-pC (20 mg/kg). Eight days after injection, mice were killed and whole BM cells were collected. Expression of oncogenic K-Ras was confirmed by excisional PCR analysis. At this time point, the animals showed evidence of a myeloproliferative disease (MPD) with mild leukocytosis, slightly increased spleen weight, and a prominent Mac-1–positive fraction in peripheral blood and BM. To investigate the effect of oncogenic K-Ras on the hematopoietic stem cell compartment, we performed a competitive transplantation assay by admixing CD45.2 donor cells with CD45.1/CD45.2 double-positive (DP) helper cells in a 1:1 or 5:1 ratio, and injecting into lethally irradiated CD45.1 recipient mice (Figure 1A). We observed that the majority of mice died of an aggressive T-cell disease with a latency of 114 or 107 days, respectively (Figure 1B). Surviving mice were analyzed at week 16 after transplantation. Of these, 2 animals developed a mild MPD only, whereas 2 mice developed both an MPD-like disease and a thymic lymphoma (data not shown). Negative controls with pI-pC–treated, single-transgenic LSL-Kras G12D/+ mice (hereafter called control) showed no evidence of K-Ras–mediated disease.
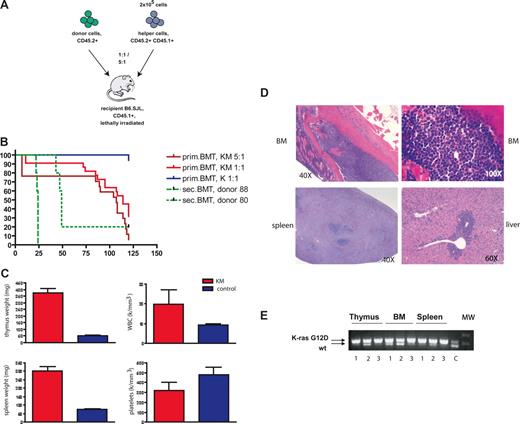
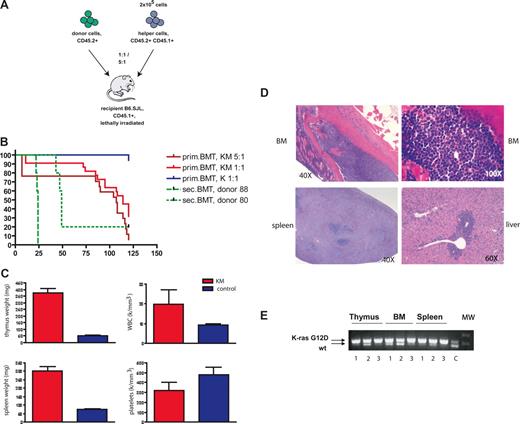
Mice transplanted with K-RasG12D–expressing bone marrow cells develop an aggressive T-cell disease. (A) Schematic illustration of the bone marrow transplantation strategy. (B) Kaplan-Meier survival plot of mice transplanted with BM cells derived from K-RasG12D/+Mx1-Cre+ (KM) and K-RasG12D/+ (K) together with 2 × 105 helper cells or with 106 cells derived from the thymi of 2 different diseased animals (donors 9988 and 9980; n = 5 each). Cumulative survival was plotted against days after BMT. KM 5:1 (n = 17) and KM 1:1 (n = 22) developed an aggressive T-cell disease with a median survival of 107 and 114 days, respectively. Sixteen of 17 K-mice were healthy during observation; 1 mouse died of a spontaneous thymic lymphoma. (C) Hematologic and pathologic data of diseased mice. KM mice have increased weight of thymus and spleen, elevated weight blood cell counts, and mild thrombocytopenia. (D) Histopathologic sections of BM (first row, original magnification 40×, left, original magnification 100×, right; hematoxylin and eosin), spleen (bottom left, original magnification 40×; hematoxylin and eosin), and liver (bottom right, original magnification 60×; hematoxylin and eosin) from a representative mouse with T-ALL. Diseased mice show infiltration of lymphoblasts in BM and spleen with destruction of normal architecture as well as extramedullary hematopoiesis in liver. (E) Cre-mediated activation of the oncogenic K-Ras allele. PCR for WT and activated (Δ) K-Ras allele demonstrates the presence of the activated K-Ras allele in thymus, bone marrow, and spleen of 3 representative patients with T-ALL. C indicates control DNA from an patient with KM+-induced MPD; MW, molecular weight marker.
Mice transplanted with K-RasG12D–expressing bone marrow cells develop an aggressive T-cell disease. (A) Schematic illustration of the bone marrow transplantation strategy. (B) Kaplan-Meier survival plot of mice transplanted with BM cells derived from K-RasG12D/+Mx1-Cre+ (KM) and K-RasG12D/+ (K) together with 2 × 105 helper cells or with 106 cells derived from the thymi of 2 different diseased animals (donors 9988 and 9980; n = 5 each). Cumulative survival was plotted against days after BMT. KM 5:1 (n = 17) and KM 1:1 (n = 22) developed an aggressive T-cell disease with a median survival of 107 and 114 days, respectively. Sixteen of 17 K-mice were healthy during observation; 1 mouse died of a spontaneous thymic lymphoma. (C) Hematologic and pathologic data of diseased mice. KM mice have increased weight of thymus and spleen, elevated weight blood cell counts, and mild thrombocytopenia. (D) Histopathologic sections of BM (first row, original magnification 40×, left, original magnification 100×, right; hematoxylin and eosin), spleen (bottom left, original magnification 40×; hematoxylin and eosin), and liver (bottom right, original magnification 60×; hematoxylin and eosin) from a representative mouse with T-ALL. Diseased mice show infiltration of lymphoblasts in BM and spleen with destruction of normal architecture as well as extramedullary hematopoiesis in liver. (E) Cre-mediated activation of the oncogenic K-Ras allele. PCR for WT and activated (Δ) K-Ras allele demonstrates the presence of the activated K-Ras allele in thymus, bone marrow, and spleen of 3 representative patients with T-ALL. C indicates control DNA from an patient with KM+-induced MPD; MW, molecular weight marker.
All recipient animals in the competitive transplantation assay showed a significant increase in thymus size and increased spleen weight (Figure 1C). In addition, 75% of mice had elevated white blood cell counts and thrombocytopenia (Figure 1C). Histopathologic examination showed diffuse infiltration of lymphoblastic cells into BM, spleen, and liver, as well as into extrahematopoietic organs, such as kidney, lung, and brain (Figure 1D; data not shown). K-RasG12D expression was stably maintained as assessed by excision PCR analysis of cells derived from infiltrated thymus, BM, and spleen (Figure 1E).
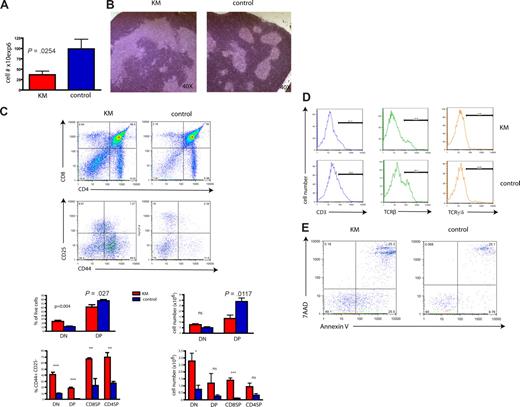
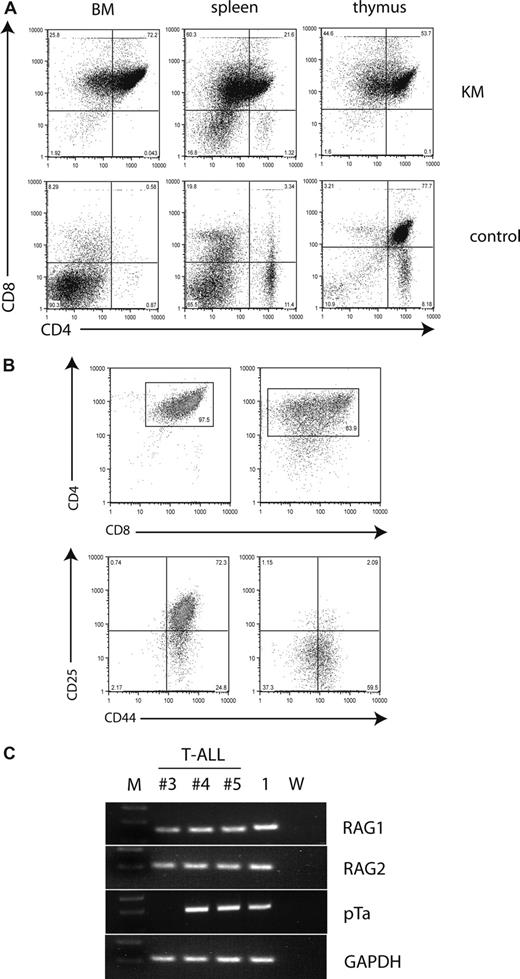
Immunophenotypic analysis of disease showed that tumor cells in the thymus, BM, and spleen were DP for CD4 and CD8 (Figure 2A). However, some malignant thymocytes had low levels of CD4 expression. To further characterize the phenotype of the observed T-cell disease, we analyzed the expression of the early T-cell markers CD25 and CD44. CD4+CD8+ tumor cells in KM animals expressed high levels of CD44 with or without coexpression of CD25 (Figure 2B). Expression of CD3, CD62L, and TCR-β was heterogeneous in the tumor samples analyzed, and no expression of TCR-γ/δ or CD24 was detected (data not shown). In addition, we analyzed tumor samples for expression of RAG1, RAG2, and pre–T-α using RT-PCR analysis and found that all samples expressed RAG1 and RAG2, and 2 of 3 expressed pre–T-α (Figure 2C). In summary, KM tumors were positive for CD4 and CD8, displayed persistent expression of CD44+/−CD25, and expressed RAG1, RAG2, and pre–T-α, indicative of a block in differentiation at the CD4+CD8+ stage with persistent expression of CD44+/−CD25.
Malignant T cells are CD4/CD8 double-positive and aberrantly express early markers of T-cell development. (A) Flow cytometric analysis of single-cell suspensions of BM, spleen, and thymus of a representative diseased mouse demonstrates a CD4/CD8 double-positive population in all 3 tissues, with some cells becoming single-positive for CD8. The percentages of cells are indicated in each quadrant. (B) Flow cytometric analysis of single-cell suspensions of the thymus from 2 diseased mice demonstrates variable phenotypes. Plots were gated on live cells and stained for CD4 and CD8 (top panel). The gated tumor cell population was analyzed for the expression of CD25 and CD44 (bottom panel). The percentages of cells of interest are indicated. (C) T-ALL cells express RAG1, RAG2, and pre–T-α. Tumor and control cDNA was amplified using primer for RAG1, RAG2, pre–T-α, and GAPDH, electrophoresed through a 2% agarose gel and stained with ethidium bromide. M indicates molecular weight marker; 1, KM donor 1; and W, water.
Malignant T cells are CD4/CD8 double-positive and aberrantly express early markers of T-cell development. (A) Flow cytometric analysis of single-cell suspensions of BM, spleen, and thymus of a representative diseased mouse demonstrates a CD4/CD8 double-positive population in all 3 tissues, with some cells becoming single-positive for CD8. The percentages of cells are indicated in each quadrant. (B) Flow cytometric analysis of single-cell suspensions of the thymus from 2 diseased mice demonstrates variable phenotypes. Plots were gated on live cells and stained for CD4 and CD8 (top panel). The gated tumor cell population was analyzed for the expression of CD25 and CD44 (bottom panel). The percentages of cells of interest are indicated. (C) T-ALL cells express RAG1, RAG2, and pre–T-α. Tumor and control cDNA was amplified using primer for RAG1, RAG2, pre–T-α, and GAPDH, electrophoresed through a 2% agarose gel and stained with ethidium bromide. M indicates molecular weight marker; 1, KM donor 1; and W, water.
K-RasG12D–induced T-ALL/lymphomas are oligoclonal or polyclonal and associated with secondary Notch1 mutations
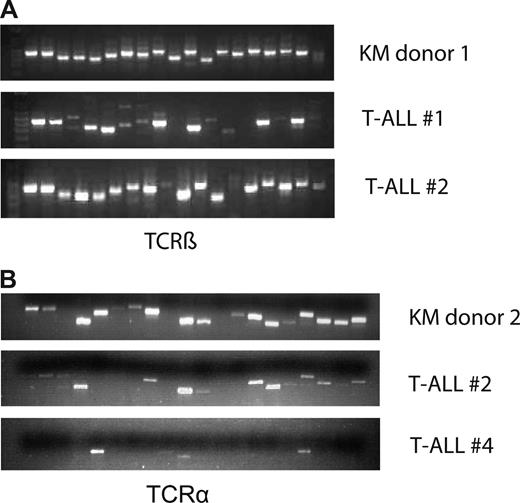
To determine whether the T-cell disease was clonal or polyclonal in origin, we performed a PCR-based assay on cDNA obtained from bulk tumor cells using primer pairs specific for each of the 19 or 20 possible variable/constant region junctions at the murine TCR-β or TCR-α locus, respectively. Whereas all 19 TCR-β junctions could be amplified from primary oncogenic KM thymocytes, we observed a variable loss of specific Vβ-Cβ combinations with concomitant predominance of others (Figure 3A). In contrast, whereas 18 of 20 TCR-α junctions could be detected in primary donor thymocytes, 6 (T-ALL #2) and 16 (T-ALL #4) specific Vα-Cα combinations were lost in the tumor samples (Figure 3B). Therefore, tumor cells either express pre–T-α or have a reduced number of TCR-α rearrangement, consistent with a block at the DP stage. Together, these results indicate that K-RasG12D induces a polyclonal and, in some cases, an oligoclonal T-cell disorder that requires the acquisition of additional genetic alterations for malignant transformation.
T-cell disease is oligoclonal. Analysis of TCR-β rearrangement. Thymocyte/tumor cDNA was amplified using primer pairs specific for each of the 19 and 20 possible variable/constant region junctions at the murine TCR-β locus (A) and TCR-α (B) locus, respectively. The samples were electrophoresed through a 1.8% agarose gel and stained with ethidium bromide. Top line represents KM donor; middle and bottom lines, 2 different tumor samples.
T-cell disease is oligoclonal. Analysis of TCR-β rearrangement. Thymocyte/tumor cDNA was amplified using primer pairs specific for each of the 19 and 20 possible variable/constant region junctions at the murine TCR-β locus (A) and TCR-α (B) locus, respectively. The samples were electrophoresed through a 1.8% agarose gel and stained with ethidium bromide. Top line represents KM donor; middle and bottom lines, 2 different tumor samples.
To determine whether the primary disease was transplantable, we injected tumor cells derived from the thymi of diseased animals into the tail vein of sublethally irradiated secondary recipients. The disease was readily transplantable into most recipients; however, latency and penetrance were different for independent donor cells (Figure 1B). Moreover, we were only able to establish suspension cell lines from spleen or BM of mice transplanted with cells from certain donors (eg, no. 9988), but not from mice transplanted with cells from donor no. 9980 or cells derived from primary diseased animals. These results suggest that, although the first genetic event in progression to T-ALL (ie, expression of oncogenic K-Ras) is identical among all animals, additional and disparate mutations are required, which in turn impart biologic differences in the behavior of the malignant clone.
We next sequenced the Notch1 gene for mutations in the heterodimerization or PEST domains that have been previously observed in both human and murine models of T-ALL. No mutations were detected at the heterodimerization cleavage site, but approximately 50% of murine tumors harbored PEST domain mutations (Table 1; GenBank accession number NM_008741). This finding is in concordance with previously reported murine T-ALL models bearing genetic alterations in Notch1, in which PEST domain mutations are more frequent than heterodimerization domain mutations.40
Oncogenic K-Ras induces a differentiation block at stage DN1 in primary donor mice
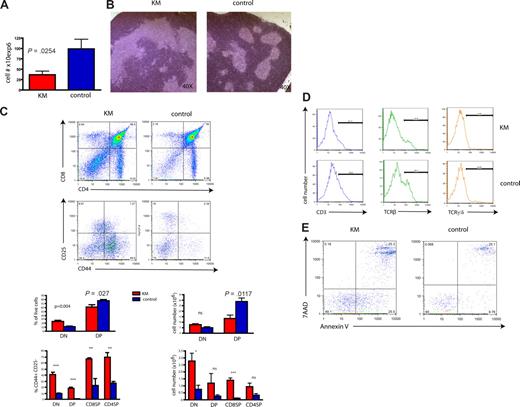
To further explore the mechanism by which BM from a mouse with an MPD-like disease can result in a T-cell leukemia/lymphoma on transplantation, we investigated the effect of expression of oncogenic K-Ras in primary animals. KM mice, analyzed 8 to 10 days after pI-pC injection, showed a significant reduction in thymic cellularity (Figure 4A). Furthermore, histopathology demonstrated lymphocyte-depleted thymi with distorted thymic architecture compared with littermate controls (Figure 4B). Analysis for expression of CD4 and CD8 showed a significant decrease of the DP cell fraction in KM thymi compared with controls (Figure 4C), probably accounting for the observed decrease of total thymic cell numbers. In contrast, we observed a mild increase of CD4/CD8 double-negative (DN) cells, suggesting an arrest at this stage of T-cell development. To further define the stage of the differentiation block, we analyzed the expression of the early T-cell markers CD25 and CD44. Most primitive, intrathymic, DN (CD4−CD8−) precursors progress from DN1 (CD44+CD25−) to DN2 (CD44+CD25+), DN3 (CD44−CD25+) and DN4 (CD44−CD25−).42 Indeed, CD44/CD25 staining demonstrated that the majority of cells within the DN population were arrested at the DN1 stage (Figure 4C). Of note, single-positive (SP) CD8 or CD4, as well as DP cells, showed higher expression of CD44 than control cells (Figure 4C). In addition, KM DN cells had lower expression of CD3 (10.5% vs 19.8%), TCR-β (22% vs 35.5%), and TCR-γ/δ (4.2 vs 5.3%), all markers for later stages of differentiation and commitment to the T-cell lineage (Figure 4D).
Primary, oncogenic K-Ras–expressing mice have a severe block in differentiation during early T-cell development. (A) Decreased thymic cellularity in oncogenic K-Ras–expressing mice (KM). Total thymocyte number of 4-week-old KM mice is reduced (P = .025) compared with control mice (8 days after pI-pC induction). (B) Histopathologic sections of the thymus of a primary mouse expressing oncogenic K-Ras (left) compared with normal control (right), 8 days after pI-pC induction (original magnification 40×; hematoxylin and eosin) show a perturbed thymic architecture with an increase in stromal tissue in the former. Shown is 1 representative example. (C) Expression of oncogenic K-Ras is associated with a decrease of the DP population and an increase of DN1 cells. Single-cell suspension from thymi of 4-week-old KM (n = 7) and control (n = 7) mice, 8 days after pI-pC induction, were prepared and stained for CD4 and CD8. CD25 and CD44 expression was analyzed gating on DN, DP, CD8 single-positive (SP), and CD4 SP populations. Compared with normal controls, DP cells were significantly reduced (P = .027, shown as percentage of live cells and P = .012 for total cell numbers), whereas DN1 and CD8 SP populations were significantly increased. Shown is 1 representative experiment of 7 independent experiments (top panel), and bar diagrams represent combined results (bottom: left, percentage of live cells; right, total cell number). DN indicates double-negative; DP, double-positive; and SP, single-positive. (D) DN cells of KM mice show reduced expression of CD3, TCR-β, and TCR-γ/δ. Histograms were gated on the DN population of thymocytes derived from KM or control mice. Shown is a representative example of 6 for each experiment. (E) Primary thymocytes exhibit enhanced apoptosis. DN-gated cells were analyzed with 7AAD and annexin V staining. Data are a representative of 3 independent experiments.
Primary, oncogenic K-Ras–expressing mice have a severe block in differentiation during early T-cell development. (A) Decreased thymic cellularity in oncogenic K-Ras–expressing mice (KM). Total thymocyte number of 4-week-old KM mice is reduced (P = .025) compared with control mice (8 days after pI-pC induction). (B) Histopathologic sections of the thymus of a primary mouse expressing oncogenic K-Ras (left) compared with normal control (right), 8 days after pI-pC induction (original magnification 40×; hematoxylin and eosin) show a perturbed thymic architecture with an increase in stromal tissue in the former. Shown is 1 representative example. (C) Expression of oncogenic K-Ras is associated with a decrease of the DP population and an increase of DN1 cells. Single-cell suspension from thymi of 4-week-old KM (n = 7) and control (n = 7) mice, 8 days after pI-pC induction, were prepared and stained for CD4 and CD8. CD25 and CD44 expression was analyzed gating on DN, DP, CD8 single-positive (SP), and CD4 SP populations. Compared with normal controls, DP cells were significantly reduced (P = .027, shown as percentage of live cells and P = .012 for total cell numbers), whereas DN1 and CD8 SP populations were significantly increased. Shown is 1 representative experiment of 7 independent experiments (top panel), and bar diagrams represent combined results (bottom: left, percentage of live cells; right, total cell number). DN indicates double-negative; DP, double-positive; and SP, single-positive. (D) DN cells of KM mice show reduced expression of CD3, TCR-β, and TCR-γ/δ. Histograms were gated on the DN population of thymocytes derived from KM or control mice. Shown is a representative example of 6 for each experiment. (E) Primary thymocytes exhibit enhanced apoptosis. DN-gated cells were analyzed with 7AAD and annexin V staining. Data are a representative of 3 independent experiments.
We next assessed apoptosis in KM versus control thymus cells using single-cell suspensions and cultured cells either in media supplemented with IL-7 and IL-2 for 4 hours or directly stained them for CD4, CD8, and annexin V. Compared with control cells, KM thymocytes showed increased apoptotic cell death in the DN and the DP compartments (Figure 4E). This increase in apoptosis was dependent on total thymocyte number as no difference in apoptosis was observed in cells derived from thymi with cell numbers comparable with those of control animals. These differences were probably the result of different stages of tumor progression at the time point of analysis; however, we could not clearly distinguish between K-Ras induced-, cell-intrinsic apoptotic cell death versus extrinsic causes, such as atrophy of thymus tissue.
K-RasG12D–expressing cells are sensitive to γ-secretase inhibitors
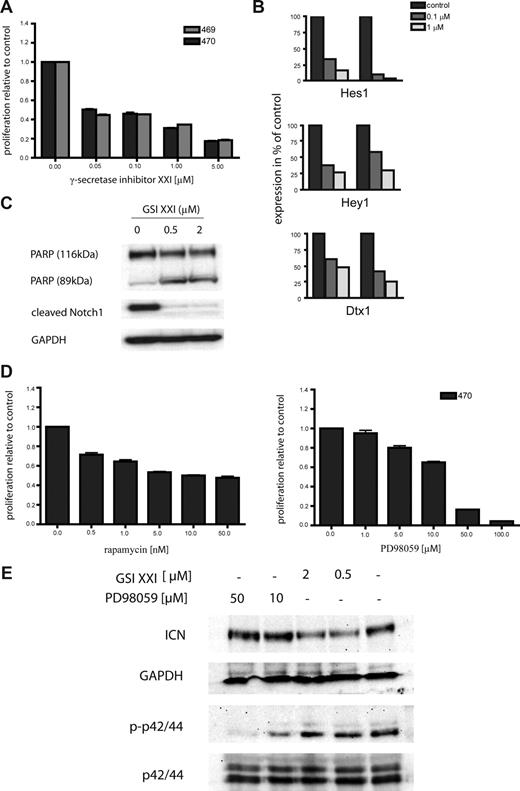
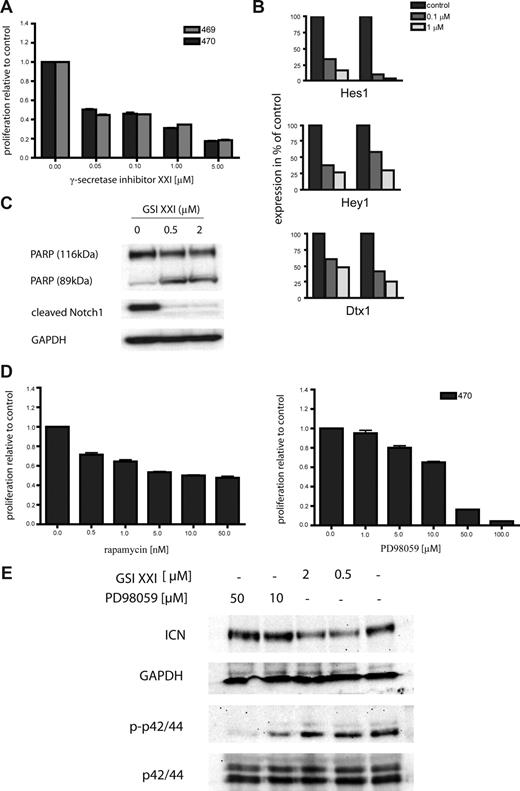
To determine the sensitivity of K-Ras–induced T-cell leukemia to inhibitor therapy, we treated several cell lines with small molecule inhibitors of Notch signaling. These included 2 independent cell lines, designated 469 and 470, derived from diseased secondary transplant recipient no. 9988 that harbored a Notch1 mutation in the PEST domain. Both primary cell lines were sensitive to inhibition of Notch signaling at concentrations as low as 0.05 μM (Figure 5A). The decrease in proliferation was the result of increased apoptotic cell death as demonstrated by PARP cleavage and a dose-dependent increase in annexin V–positive cells (Figure 5C; data not shown). To demonstrate γ-secretase inhibitor induced suppression of Notch function, we analyzed the expression of Notch downstream targets after inhibitor treatment. Expression of Hes1, Hey1, and Dtx1 were decreased in a dose-dependent manner in both cell lines (Figure 5B). In addition, expression of cleaved Notch1 was decreased after treatment with γ-secretase inhibitor XXI, as shown by Western blotting (Figure 5B). To investigate the effects of inhibition of Ras downstream pathways, we also treated the primary cell line 470 with the MEK inhibitor PD98059. Treatment for 48 hours caused a dose-dependent inhibition of proliferation of the 470 cell line (Figure 5D right). Finally, as both Notch1 and Ras signaling pathways affect PI3K signaling,5,43 we treated 470 cells with rapamycin. Again, inhibitor treatment resulted in a dose-dependent suppression of cellular proliferation (Figure 5D left). As survival of the leukemic cells was dependent on both the Ras-MAPK pathway and Notch signaling, we were interested in determining whether inhibition of either pathway influenced the activation status of the other. Therefore, we treated 470 cells with GSI XXI and PD98059 for 6 hours and performed Western blot analysis. As shown in Figure 5E, inhibition of Notch signaling caused down-regulation of ICN but no suppression of phospho-p42/44. Similarly, treatment with PD98059 had no effect on expression of ICN but strongly inhibited p42/44 phosphorylation. Together, these data indicate that progression of T-ALL initiated by oncogenic K-Ras is potentiated by acquisition of Notch1 mutations that confer sensitivity to treatment with γ-secretase inhibitors, as well as to inhibition of downstream effector of the RAS/MAPK pathway.
Oncogenic K-Ras–driven T-cell disease is hypersensitive toward Notch1 inhibitor therapy. (A) Primary oncogenic K-Ras–expressing T-ALL cells and the human cell line CCRF-CEM were grown in RPMI media without cytokines and treated with the γ-secretase inhibitor XXI as indicated. Cell viability was measured after 48 hours, and the proportion of viable cells relative to the control (no inhibitor) was plotted. Experiments were performed in triplicate. Values are mean plus or minus SEM. (B) Inhibition of Notch1 signaling caused reduced expression of known Notch1 target genes. Primary cells were treated as indicated, and expression of Hes1, Hey1, and Dtx1 mRNA levels was determined by RQ-PCR. Each experiment was performed in duplicate. (C) Inhibition of Notch1 results in cleavage of PARP (top panel) and a decrease of ICN levels (middle panel). Equal loading was confirmed by measuring GAPDH protein levels. GSI indicates γ-secretase inhibitor. (D) The primary oncogenic K-Ras expressing T-ALL cell line 470 and the human cell line CCRF-CEM were grown in RPMI-media without cytokines and treated with rapamycin and PD98059 as indicated. Cell viability was measured after 48 hours, and the proportion of viable cells relative to the control (no inhibitor) was plotted. Experiments were performed in triplicate. Values are mean plus or minus SEM. (E) Primary 470 T-ALL cells were treated with GSI XXI or PD98059 for 6 hours as indicated. Protein lysates were extracted, and immunoblot analysis was performed using antiphosph-p42/44 and anti-ICN antibodies. Equal loading was confirmed by measuring p42/44 and GAPDH protein levels, respectively. GSI indicates γ-secretase inhibitor.
Oncogenic K-Ras–driven T-cell disease is hypersensitive toward Notch1 inhibitor therapy. (A) Primary oncogenic K-Ras–expressing T-ALL cells and the human cell line CCRF-CEM were grown in RPMI media without cytokines and treated with the γ-secretase inhibitor XXI as indicated. Cell viability was measured after 48 hours, and the proportion of viable cells relative to the control (no inhibitor) was plotted. Experiments were performed in triplicate. Values are mean plus or minus SEM. (B) Inhibition of Notch1 signaling caused reduced expression of known Notch1 target genes. Primary cells were treated as indicated, and expression of Hes1, Hey1, and Dtx1 mRNA levels was determined by RQ-PCR. Each experiment was performed in duplicate. (C) Inhibition of Notch1 results in cleavage of PARP (top panel) and a decrease of ICN levels (middle panel). Equal loading was confirmed by measuring GAPDH protein levels. GSI indicates γ-secretase inhibitor. (D) The primary oncogenic K-Ras expressing T-ALL cell line 470 and the human cell line CCRF-CEM were grown in RPMI-media without cytokines and treated with rapamycin and PD98059 as indicated. Cell viability was measured after 48 hours, and the proportion of viable cells relative to the control (no inhibitor) was plotted. Experiments were performed in triplicate. Values are mean plus or minus SEM. (E) Primary 470 T-ALL cells were treated with GSI XXI or PD98059 for 6 hours as indicated. Protein lysates were extracted, and immunoblot analysis was performed using antiphosph-p42/44 and anti-ICN antibodies. Equal loading was confirmed by measuring p42/44 and GAPDH protein levels, respectively. GSI indicates γ-secretase inhibitor.
Discussion
In this study, we transplanted whole BM cells expressing oncogenic K-RasG12D from its endogenous promoter into syngeneic recipients. We had intended to study the effects of expressing oncogenic K-Ras in the hematopoietic stem cell compartment. However, our ability to reliably measure long-term reconstitution at 16 weeks after BMT was limited because of early development of an aggressive T-cell leukemia/lymphoma with infiltration of BM, spleen, and nonhematopoietic organs, whereas a minority of mice developed a myeloproliferative disease. This is in contrast to donor K-RasG12D/Mx1-Cre–expressing transgenic mice that, on induction with pI-pC, develop myeloproliferative disease as the most prominent hematopoietic phenotype.22,23
Nonetheless, this observation provided an opportunity to assess the role of oncogenic K-Ras in a murine model system that is genotypically and phenotypically similar to human cases of T-ALL associated with activating mutations in KRAS. We observed a significant decrease of total thymocyte numbers that was accompanied by impaired differentiation at the CD4−CD8− DN stage in primary animals. This is similar to other murine T-ALL models including (1) mice with aberrant expression of Tal1/Scl with or without coexpression of LMO1/2, (2) E2A-deficient mice, or (3) Tal1/E2A+/− or Tal1/HEB+/− transgenic mice.30,32,33,44-46 However, as each of these latter models is induced by aberrant expression of transcription factors, it is intriguing that a similar T-ALL phenotype is observed in our K-RasG12D model with a constitutively activated GTPase. Our observations indicate that mutated K-Ras can act as an initiating event in pathogenesis of T-ALL and give rise to a phenotype that includes impaired T-cell differentiation and is nearly identical to those initiated by aberrant expression of transcription factors. Although this is somewhat unexpected given the association with oncogenic Ras alleles and hyperproliferation, a K-RasG12D–induced block in differentiation has also been described during erythropoiesis.47,48 In this model system using the same allele, K-RasG12D, cells arrest at the transition from Ter119-negative erythroid progenitors (CD71-positive or -negative) toward Ter119-positive mature erythrocytes in the fetal liver, as well as during adult erythropoiesis.47,48
In addition to the block in T-cell differentiation, we also observed increased apoptotic cell death accompanied with reduced thymic cellularity in primary donor mice. Transplantation of this preleukemic population causes a fatal lymphoid disease in wild-type recipients, which is only rarely seen in primary mice, probably because of the rapid onset and the fatal course of the myeloid disease in these animals.22 As Mx1-driven Cre-recombinase expression is also observed in some mesenchymal cells, it is tempting to speculate that the thymic/hematopoietic microenvironment contributes to establishing this preleukemic phenotype, in accord with recent reports demonstrating an essential role of the microenvironment in the development of MPD in RB or RAR-γ knockout mice.49,50 At least stroma-derived signals might be involved in the development of the preleukemic state. Beyond this step, thymic cells are independent of oncogenic K-Ras–expressing stromal cells, as transplantation of BM cells into wild-type recipients results in full malignant transformation. On the other hand, as we transplanted whole BM cells, it is also possible that oncogenic K-Ras–expressing hematopoietic stem cells or common lymphoid progenitors (CLPs) are the cell of origin in our tumor model. These cells could then migrate to the thymus and become fully transformed with potential support from the normal thymic environment. In addition, analysis of the few remaining DN cells derived from thymic lymphomas showed a similar block of differentiation as observed in primary donor mice (data not shown), suggesting a block of differentiation independent of the stromal microenvironment.
The long latency of approximately 100 days and clonal evolution of T-cell lymphomagenesis in KM recipients indicates that additional mutations are required for malignant transformation. As in human disease, Notch1 alterations have been observed in several mouse models of T-ALL.40 We detected Notch1 mutations exclusively in the PEST domain of the Notch1 gene in approximately 50% of KM recipient animals. Thus, acquisition of Notch1 mutations appears to be a disease progression allele in this model, as in other models of T-ALL. However, because only 50% of tumors were positive for Notch1 mutations, the acquisition of additional mutations affecting Notch signaling or other pathways are likely and could include mutations affecting the FBW7 locus or p16INK4A locus, TAL1/SCL, LMO1/2, HOX family members, or other Notch family members.34,51-53
However, the fact that K-Ras–initiated T-ALL with acquisition of Notch1 mutations phenocopies T-ALL initiated by transcription factors, such as TAL1/SCL, which also acquire Notch1 mutations, suggests the possibility that these latter tumors also contain activating mutations in the RAS/MAPK or related signal transduction pathways. Indeed, approximately 5% of cases of human T-ALL have NUP214-ABL fusions that may coexist with Notch mutations. This model system provides further support for strategies to comprehensively search for mutations in signal transduction pathways, such as RAS and NUP214-ABL, which could represent therapeutic targets in T-ALL. Furthermore, this model also demonstrates that K-Ras–initiated tumors, like other tumors with activating mutations in Notch1, are sensitive to inhibition of γ-secretase. Indeed, in comparison with other models of T-ALL,36,54 the 2 cell lines tested here were remarkably sensitive to γ-secretase inhibitors, with induction of apoptotic cell death at concentrations as low as 50 nM.
There is also reason to consider targeting of the PI3K/AKT pathway in this model because Notch itself has been reported to regulate the pathway54 and because oncogenic K-Ras is an upstream effector of PI3K/AKT signaling. Indeed, we observed that pharmacologic inhibition of mTOR or Erk1/2 resulted in suppression of cell growth in cell lines derived from diseased KM mice. In addition, oncogenic RAS signaling has been shown to activate wild-type NOTCH1 in human foreskin fibroblasts (BJ) and human embryonic kidney epithelial cells (HEK) expressing the human telomerase reverse transcriptase subunit (hTERT), SV40 oncoproteins, and HRAS V12 and NOTCH1 signaling is required to maintain this neoplastic phenotype.55 Therefore, inhibition of NOTCH1 signaling alone, in combination with rapamycin, or with FTI/MEK1/2 inhibitors, might represent a promising strategy for T-ALL patients harboring both RAS and NOTCH1 mutations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Gilliland laboratory for support and helpful discussions, and Drs David A. Tuveson and Tyler E. Jacks for providing us with the conditional murine K-RasG12D model.
This work was supported by National Institutes of Health grants CA66996 and CA105423 (D.G.G.) and the Leukemia & Lymphoma Society (D.G.G.). T.K. is a fellow of the German Research Foundation. D.G.G. is a Doris Duke Charitable Foundation Distinguished Clinical Scientist and an Investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: T.K. designed and performed experiments and wrote the manuscript; M.G.C. and C.S. performed research and analyzed data; J.L., D.S.L., and J.E.H. provided technical assistance; S.F. and B.H.L. analyzed data; and D.G.G. designed and analyzed experiments and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Kindler, 3rd Medical Department, Johannes Gutenberg-University Mainz, Langenbeckstrasse 1, 55131 Mainz, Germany; e-mail: thomas.kindler@ukmainz.de.