Abstract
Therapy-related acute promyelocytic leukemia (t-APL) with t(15;17) translocation is a well-recognized complication of cancer treatment with agents targeting topoisomerase II. However, cases are emerging after mitoxantrone therapy for multiple sclerosis (MS). Analysis of 12 cases of mitoxantrone-related t-APL in MS patients revealed an altered distribution of chromosome 15 breakpoints versus de novo APL, biased toward disruption within PML intron 6 (11 of 12, 92% vs 622 of 1022, 61%: P = .035). Despite this intron spanning approximately 1 kb, breakpoints in 5 mitoxantrone-treated patients fell within an 8-bp region (1482-9) corresponding to the “hotspot” previously reported in t-APL, complicating mitoxantrone-containing breast cancer therapy. Another shared breakpoint was identified within the approximately 17-kb RARA intron 2 involving 2 t-APL cases arising after mitoxantrone treatment for MS and breast cancer, respectively. Analysis of PML and RARA genomic breakpoints in functional assays in 4 cases, including the shared RARA intron 2 breakpoint at 14 446-49, confirmed each to be preferential sites of topoisomerase IIα-mediated DNA cleavage in the presence of mitoxantrone. This study further supports the presence of preferential sites of DNA damage induced by mitoxantrone in PML and RARA genes that may underlie the propensity to develop this subtype of leukemia after exposure to this agent.
Introduction
The occurrence of acute promyelocytic leukemia (APL) as a second tumor (sAPL) frequently has been reported as a late complication of chemotherapy and/or radiotherapy (therapy-related APL [t-APL]), although sAPL cases arising in patients whose primary tumors were treated by surgery alone have also been described.1-3 The agents most often associated with development of t-APL induce DNA damage through targeting of topoisomerase II, with mitoxantrone, epirubicin, adriamycin, and etoposide being most commonly implicated.3,4 The latency period between chemotherapy exposure and the onset of t-APL is relatively short (< 3 years) and typically occurs without a preceding myelodysplastic phase.3,4
In a study by Mistry et al5 concerning molecular mechanisms underlying formation of the t(15;17) in t-APL, breakpoints in cases arising after mitoxantrone exposure for prior breast carcinoma were found to be clustered in an 8-bp region within PML intron 6; this corresponded in functional assays to a preferential site of mitoxantrone-induced topoisomerase II–dependent cleavage at position 1484. Although these findings highlighted the leukemogenic role of drug-induced DNA cleavage at specific sites in the genome, the precise mechanism by which secondary leukemias with balanced chromosomal translocations such as the t(15;17) in APL develop remains controversial.6-9 This is compounded by the fact that many patients have been exposed to multiple cytotoxic drugs often accompanied by radiotherapy, making it difficult to categorically ascribe the etiology of therapy-related acute myeloid leukemia (t-AML) in any given case.
Previous studies on t-AML have focused on patient populations that feasibly could have been enriched for persons at particular risk of leukemia, having already developed one form of cancer. Therefore, to investigate whether particular chemotherapeutic agents have a propensity to induce specific molecular subtypes of t-AML, it is of interest to study patients exposed to topoisomerase II targeting drugs used in the treatment of nonmalignant conditions, such as mitoxantrone in the management of multiple sclerosis (MS). MS is a putative autoimmune disease affecting the central nervous system for which mitoxantrone represents the latest in a long list of general immunosuppressive agents used in the treatment of this condition.10,11 In recent years, an increasing number of APL cases have been reported in MS patients treated with mitoxantrone.3,5,12-20 However, to date, no attempts have been made to systematically characterize translocation breakpoints in APL cases that developed in this setting.
In the present study, we analyzed at the genomic level the PML and RARA breakpoints of 14 patients who developed APL on a background of MS, including 12 who received mitoxantrone for their primary disease. Furthermore, we used functional cleavage assays to better elucidate the mechanisms underlying the formation of the t(15;17) in this setting.
Methods
Patients and samples
The main patient characteristics, including demographic data, MS type, and treatments received for MS, are reported in Table 1. Seven patients were diagnosed in 5 Italian institutions, 3 in 2 Spanish institutions, 3 in the United Kingdom, and the remaining patient in Austria. Analyses were undertaken after informed patient consent was obtained in accordance with the Declaration of Helsinki with ethical approval of University Tor Vergata of Rome and St Thomas' Hospital of London. Bone marrow samples were obtained at the time of diagnosis of APL. Mononuclear cells were collected after centrifugation on a Ficoll-Hypaque gradient and stored at −70°C as dry pellets. In all cases, APL diagnosis was confirmed at the genetic level by reverse-transcriptase polymerase chain reaction (RT-PCR) amplification of the PML-RARA hybrid gene.
Main clinical and molecular features of 14 sAPL patients
| Patient no. . | Age (y) at time of MS diagnosis . | Sex . | Primary disease . | Therapy of MS . | Latency between MS and APL, mo . | Latency between mitoxantrone and APL, mo . | Bcr subtype . | PML* breakpoint der(15)-der(17) . | RARA* breakpoint der(15)-der(17) . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of treatment . | Mitoxantrone schedule . | Total dose, mg . | |||||||||
| UPN 1 | 59 | F | SPMS | Mitoxantrone | 10 mg/m2 every 1 mo | 30 | 13 | 10 | 1 | 1496-97 1496-97 | 6104-05 6104-05 |
| UPN 2 | 43 | F | SPMS | Mitoxantrone | 10 mg/m2 every 2 mo | 35 | 192 | 37 | 1 | 1657-60 1657-60 | 14446-49 14446-49 |
| UPN 3 | 56 | F | PPMS | Mitoxantrone | 10 mg/m2 every 3 mo | 70 | 30 | 4 | 1 | 1485-87 1485-87 | 15266-68 15266-68 |
| UPN 4 | 24 | F | RRMS | Mitoxantrone | 12 mg/m2 every 2 mo | 147 | 150 | 6 | 1 | 1922-23 1922-23 | 12418-19 12418-19 |
| UPN 5 | 21 | F | RRMS | Mitoxantrone | 12 mg/m2 every 2 mo | 170 | 66 | 18 | 1 | 1150-51 1169-70 | 14082-83 14054-55 |
| UPN 6 | 53 | F | RRMS | Mitoxantrone | 10 mg/m2 (every 1 mo, 3 doses), 10 mg/m2 (every 3 mo, 7 doses) | 234 | 72 | 51 | 1 | 1483-84 1484-85 | 12909-10 12908-09 |
| UPN 7 | 25 | M | RRMS | Mitoxantrone | 12 mg/m2 (every 1 mo, 3 doses), 6 mg/m2 (every 3 mo, 5 doses) | 110 | 37 | 27 | 1 | 1165 1161 | 7974 7978 |
| UPN 8 | 44 | M | SPMS | Mitoxantrone | 1.9 mg/m2 every 1 mo | 100 | 204 | 29 | 1 | 1409-10 1429-30 | 718-19 675-76 |
| UPN 9 | 33 | M | PPMS | Mitoxantrone | 10 mg/m2 every 2 mo | 176 | 144 | 30 | 3 | 1286-87 1286-87 | 15386-87 15386-87 |
| UPN 10 | 26 | M | RRMS | INF beta | NA | NA | 18 | NA | 3 | 1117-22† | 14920-25 |
| UPN 11 | 37 | M | RRMS | Mitoxantrone | 13 mg monthly (5 doses) | 81 | 97 | 17 | 1 | 1483-86 1486-1506 | 11586-89 11683-84 |
| UPN 12 | 49 | M | RRMS | Prednisone | NA | NA | 216 | NA | 1 | 1489-91 1485 | 14785-87 14117 |
| UPN 13 | 45 | F | PPMS | Mitoxantrone | 8 mg/m2 every 1 mo | 64 | 120 | 60 | 1 | 1485-87 1486-88 | 11569-71 11569-71 |
| UPN 14 | 25 | F | SPMS | Mitoxantrone | 11.36 mg/m2 every 1 mo | 120 | 237 | 34 | 1 | 1488-89 1483-86 | 12038-39 12035-38 |
| Patient no. . | Age (y) at time of MS diagnosis . | Sex . | Primary disease . | Therapy of MS . | Latency between MS and APL, mo . | Latency between mitoxantrone and APL, mo . | Bcr subtype . | PML* breakpoint der(15)-der(17) . | RARA* breakpoint der(15)-der(17) . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of treatment . | Mitoxantrone schedule . | Total dose, mg . | |||||||||
| UPN 1 | 59 | F | SPMS | Mitoxantrone | 10 mg/m2 every 1 mo | 30 | 13 | 10 | 1 | 1496-97 1496-97 | 6104-05 6104-05 |
| UPN 2 | 43 | F | SPMS | Mitoxantrone | 10 mg/m2 every 2 mo | 35 | 192 | 37 | 1 | 1657-60 1657-60 | 14446-49 14446-49 |
| UPN 3 | 56 | F | PPMS | Mitoxantrone | 10 mg/m2 every 3 mo | 70 | 30 | 4 | 1 | 1485-87 1485-87 | 15266-68 15266-68 |
| UPN 4 | 24 | F | RRMS | Mitoxantrone | 12 mg/m2 every 2 mo | 147 | 150 | 6 | 1 | 1922-23 1922-23 | 12418-19 12418-19 |
| UPN 5 | 21 | F | RRMS | Mitoxantrone | 12 mg/m2 every 2 mo | 170 | 66 | 18 | 1 | 1150-51 1169-70 | 14082-83 14054-55 |
| UPN 6 | 53 | F | RRMS | Mitoxantrone | 10 mg/m2 (every 1 mo, 3 doses), 10 mg/m2 (every 3 mo, 7 doses) | 234 | 72 | 51 | 1 | 1483-84 1484-85 | 12909-10 12908-09 |
| UPN 7 | 25 | M | RRMS | Mitoxantrone | 12 mg/m2 (every 1 mo, 3 doses), 6 mg/m2 (every 3 mo, 5 doses) | 110 | 37 | 27 | 1 | 1165 1161 | 7974 7978 |
| UPN 8 | 44 | M | SPMS | Mitoxantrone | 1.9 mg/m2 every 1 mo | 100 | 204 | 29 | 1 | 1409-10 1429-30 | 718-19 675-76 |
| UPN 9 | 33 | M | PPMS | Mitoxantrone | 10 mg/m2 every 2 mo | 176 | 144 | 30 | 3 | 1286-87 1286-87 | 15386-87 15386-87 |
| UPN 10 | 26 | M | RRMS | INF beta | NA | NA | 18 | NA | 3 | 1117-22† | 14920-25 |
| UPN 11 | 37 | M | RRMS | Mitoxantrone | 13 mg monthly (5 doses) | 81 | 97 | 17 | 1 | 1483-86 1486-1506 | 11586-89 11683-84 |
| UPN 12 | 49 | M | RRMS | Prednisone | NA | NA | 216 | NA | 1 | 1489-91 1485 | 14785-87 14117 |
| UPN 13 | 45 | F | PPMS | Mitoxantrone | 8 mg/m2 every 1 mo | 64 | 120 | 60 | 1 | 1485-87 1486-88 | 11569-71 11569-71 |
| UPN 14 | 25 | F | SPMS | Mitoxantrone | 11.36 mg/m2 every 1 mo | 120 | 237 | 34 | 1 | 1488-89 1483-86 | 12038-39 12035-38 |
UPN indicates unique patient number (some details regarding UPNs 1, 4 and 5, and 7 were reported in references 20, 17, and 5, respectively); RRMS, relapsing-remitting multiple sclerosis; SPMS, secondary progressive multiple sclerosis; PPMS, primary progressive multiple sclerosis; and NA, not applicable.
Breakpoint locations are numbered according to the following GenBank accession numbers: PML intron 6 (bcr 1), S57791; PML intron 3 (bcr 3), S51489; and RARA intron 2, AJ297538.
No DNA was available to sequence the reciprocal RARA-PML in this case.
Amplification of DNA spanning possible break points (PML-RARA): long-range PCR and DNA sequencing
To determine the exact chromosomal breakpoint position in PML and RARA genes, genomic DNA extracted from APL blasts collected at diagnosis was amplified by a 2-step, long-range nested PCR method as reported elsewhere.5,21 Two forward and 8 reverse primers were designed for each step to cover the PML breakpoint region (bcr1 or bcr3, as previously known based on diagnostic RT-PCR results available for all cases) and the 16.9-kb-long RARA intron 2. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA). Samples were loaded in 96-well plates and covered with mineral oil. The amplified products were separated with a capillary electrophoresis-based system (CEQ 8000 Genetic Analysis System; Beckman Coulter, Fullerton, CA) using the “LFR1 Test” default run method and sequenced using appropriate primers.5,21 Rigorous procedures were used to reduce risk of PCR contamination,22 and genomic breakpoints were in all cases confirmed by PCR analysis of a fresh aliquot of DNA. Moreover, in 3 cases, breakpoint analyses were performed independently in parallel in the Rome and London laboratories, yielding identical results.
Amplification and sequence analysis of the reciprocal RARA-PML genomic breakpoint junction
Genomic RARA-PML was amplified using patient specific primers (designed on the basis of PML and RARA breakpoints) and fresh aliquots of DNA. In 13 cases, the reciprocal RARA-PML genomic breakpoint junction was sequenced, providing further confirmation of the t(15;17) translocation breakpoints at the genomic level. In one case (unique patient number [UPN] 10), no DNA was available to carry out sequencing of the reciprocal RARA-PML.
Alignment of sequenced nucleotides using BLAST algorithm
The patients' genomic PML-RARA junction sequences were aligned against normal PML (GenBank accession number S57791 for bcr1 and S51489 for bcr3) and RARA intron 2 (GenBank accession number AJ297538) nucleotides as a reference text input in BLAST/alignment program. The purpose of alignment was to identify any microhomologies between PML and RARA in the vicinity of the breakpoint.23
In vitro DNA cleavage assays
Human topoisomerase IIα was expressed in Saccharomyces cerevisiae24 and purified as described previously.25,26 Assays were performed as described previously.5 Briefly, having identified genomic junction sequences, regions of the normal homologs encompassing the breakpoint sites were amplified by PCR and subcloned into the pBluescript SKII(+) vector. The optimal insert size for the assay was 200 to 500 bp, with the breakpoint site located approximately 50 to 100 bp from the 5′ end of the insert. Substrates containing 25 ng of the normal homologs of the translocation breakpoints were 5′end-labeled (30 000 cpm) and incubated with 147 nM of human DNA topoisomerase IIα, 1 mM of ATP in the presence or absence of 20 μM mitoxantrone.5 In all cases, additional reactions were carried out to evaluate the heat stability of the covalent complexes formed. Cleavage complexes were irreversibly trapped by the addition of sodium dodecyl sulfate, and purified products were resolved in an 8% polyacrylamide-7.0 M of urea gel in parallel with dideoxy sequencing reactions primed at the same 5′-end, visualized by autoradiography, and quantified using PhosphoImager and IMAGEQUANT software (GE Healthcare, Little Chalfont, United Kingdom).
Results
Clinical features
As shown in Table 1, a total of 14 patients with APL developing in a background of MS were studied. The series included 12 cases exposed to a median total dose of 105 mg mitoxantrone (range, 30-234 mg), whereas 2 patients received other treatments for their primary disease (interferon-β in UPN 10 and corticosteroids in UPN 12). The median latency period between the first exposure to mitoxantrone and APL diagnosis was 28 months (range, 4-60 months). Patients were treated with all-trans retinoic acid and anthracycline-based chemotherapy, mostly using AIDA-like (all-trans retinoic acid + idarubicin) protocols27 (Table 2); however, UPN 13 died of cerebral hemorrhage within 3 hours of APL diagnosis before antileukemic therapy was started. The remaining 13 patients achieved hematologic and molecular remission. Of these, 11 remain in first molecular remission at a median follow-up of 10 months, whereas UPN 7 relapsed at 28 months and achieved second molecular remission after salvage therapy with arsenic trioxide, and UPN 1 died of cerebral hemorrhage while in remission after 7 months.
Main clinical features and treatment outcome of sAPL patients
| Patient no. . | Age (y) at time of APL diagnosis . | Sex . | APL characteristics . | Therapy of APL . | Outcome . | ||
|---|---|---|---|---|---|---|---|
| WBC, ×109/L . | PLT, ×109/L . | DIC, Yes/No . | |||||
| UPN 1 | 60 | F | 5.5 | 25 | Yes | PETHEMA APL 2005 | Died at 7 months due to cerebral hemorrhage |
| UPN 2 | 59 | F | 8.9 | 74 | Yes | GIMEMA AIDA protocol | CCR 21 mo |
| UPN 3 | 58 | F | 0.5 | 37 | No | PETHEMA APL 2005 | CCR 16 mo |
| UPN 4 | 36 | F | 0.8 | 74 | No | GIMEMA AIDA protocol | CCR 16 mo |
| UPN 5 | 26 | F | 1.1 | 30 | Yes | GIMEMA AIDA protocol | CCR 24 mo |
| UPN 6 | 59 | F | 1.1 | 58 | Yes | GIMEMA AIDA protocol | CCR 19 mo |
| UPN 7 | 28 | M | 0.7 | 5 | No | UK MRC protocol | Relapse at 28 mo |
| UPN 8 | 61 | M | 3.3 | 31 | No | PETHEMA APL 2005 | CCR 4 mo |
| UPN 9 | 45 | M | 13 | 12 | No | GIMEMA AIDA protocol | CCR 4 mo |
| UPN 10 | 27 | M | 33.7 | 11 | Yes | GIMEMA AIDA protocol | CCR 10 mo |
| UPN 11 | 45 | M | 0.6 | 66 | No | GIMEMA AIDA protocol | CCR 10 mo |
| UPN 12 | 67 | M | 1.1 | 9 | No | PETHEMA APL 2005 | CCR 9 mo |
| UPN 13 | 55 | F | 8.9 | 13 | Yes | NA | Died 3 h after APL diagnosis |
| UPN 14 | 45 | F | 72.2 | 40 | Yes | PETHEMA APL 2005 | CCR 1 mo |
| Patient no. . | Age (y) at time of APL diagnosis . | Sex . | APL characteristics . | Therapy of APL . | Outcome . | ||
|---|---|---|---|---|---|---|---|
| WBC, ×109/L . | PLT, ×109/L . | DIC, Yes/No . | |||||
| UPN 1 | 60 | F | 5.5 | 25 | Yes | PETHEMA APL 2005 | Died at 7 months due to cerebral hemorrhage |
| UPN 2 | 59 | F | 8.9 | 74 | Yes | GIMEMA AIDA protocol | CCR 21 mo |
| UPN 3 | 58 | F | 0.5 | 37 | No | PETHEMA APL 2005 | CCR 16 mo |
| UPN 4 | 36 | F | 0.8 | 74 | No | GIMEMA AIDA protocol | CCR 16 mo |
| UPN 5 | 26 | F | 1.1 | 30 | Yes | GIMEMA AIDA protocol | CCR 24 mo |
| UPN 6 | 59 | F | 1.1 | 58 | Yes | GIMEMA AIDA protocol | CCR 19 mo |
| UPN 7 | 28 | M | 0.7 | 5 | No | UK MRC protocol | Relapse at 28 mo |
| UPN 8 | 61 | M | 3.3 | 31 | No | PETHEMA APL 2005 | CCR 4 mo |
| UPN 9 | 45 | M | 13 | 12 | No | GIMEMA AIDA protocol | CCR 4 mo |
| UPN 10 | 27 | M | 33.7 | 11 | Yes | GIMEMA AIDA protocol | CCR 10 mo |
| UPN 11 | 45 | M | 0.6 | 66 | No | GIMEMA AIDA protocol | CCR 10 mo |
| UPN 12 | 67 | M | 1.1 | 9 | No | PETHEMA APL 2005 | CCR 9 mo |
| UPN 13 | 55 | F | 8.9 | 13 | Yes | NA | Died 3 h after APL diagnosis |
| UPN 14 | 45 | F | 72.2 | 40 | Yes | PETHEMA APL 2005 | CCR 1 mo |
DIC indicates disseminated intravascular coagulation; CCR, continuous complete remission; and NA, not applicable.
Location of t(15;17) translocation breakpoints within the PML and RARA loci
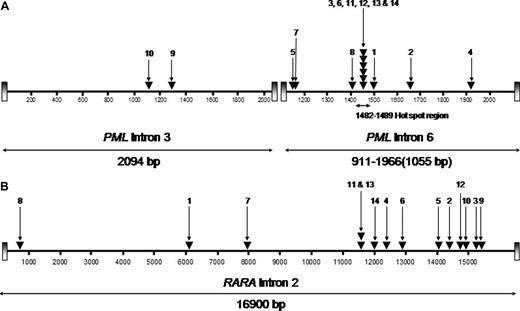
RT-PCR showed the bcr1 PML-RARA isoform (PML intron 6 breakpoint) in 12 cases, whereas in the remaining 2 cases the PML breakpoint fell within intron 3 (bcr3; Figure 1). This breakpoint distribution appeared skewed in favor of the bcr1 isoform, which previously has been reported to account for approximately 55% of unselected APL cases.28-30 Comparison of the breakpoint distribution in MS patients with mitoxantrone-related APL relative to a cohort of 1022 consecutive cases of newly diagnosed de novo APL from GIMEMA, PETHEMA, and United Kingdom MRC trials confirmed significant overrepresentation of involvement of PML intron 6 in the former group (11 of 12, 92% vs 622 of 1022, 61%: P = .035 by Fisher exact test). PML genomic breakpoints within intron 6 were found to fall between nucleotide positions 1482 and 1489 in 6 patients (UPNs 3, 6, 11, 12, 13, and 14; Figure 1A), coinciding precisely with the “hotspot” previously identified in t-APL after mitoxantrone treatment for breast cancer.5 Interestingly, one of these patients (UPN 12) had not received mitoxantrone therapy for MS. In the 2 patients (UPNs 9 and 10) with the bcr3 PML-RARA isoform, the breakpoints in PML intron 3 were detected between nucleotides 1286 and 1287 and 1117 through 1122, respectively (Figure 1A). The breakpoints within the RARA locus were distributed across intron 2 without particular clustering in any restricted small region (Figure 1B). However, one breakpoint (in UPN 2) mapped precisely to a breakpoint found in a case of t-APL arising after mitoxantrone therapy for breast cancer, studied previously by Mistry et al.5
Characterization of t(15;17) breakpoints within the PML and RARA loci. The location of breakpoints indicated by ▾ in the 14 patients (numbers correspond with UPNs in Tables 1 and 2) within the PML gene on chromosome 15 (A; bcr3 region and bcr1/2 region) and intron 2 of RARA on chromosome 17 (B) are shown. Breakpoint locations are numbered according to the following GenBank accession numbers: PML intron 6 (bcr 1), S57791; PML intron 3 (bcr 3), S51489; and RARA intron 2, AJ297538.23
Characterization of t(15;17) breakpoints within the PML and RARA loci. The location of breakpoints indicated by ▾ in the 14 patients (numbers correspond with UPNs in Tables 1 and 2) within the PML gene on chromosome 15 (A; bcr3 region and bcr1/2 region) and intron 2 of RARA on chromosome 17 (B) are shown. Breakpoint locations are numbered according to the following GenBank accession numbers: PML intron 6 (bcr 1), S57791; PML intron 3 (bcr 3), S51489; and RARA intron 2, AJ297538.23
Sequence analyses of the reciprocal RARA-PML fusion revealed a balanced translocation in 7 of 13 analyzed cases. Six patients showed size variable deletions and/or insertions at the breakpoint junction (Table 1). Microhomologies at the breakpoint junctions were indicative of DNA repair by the nonhomologous end-joining (NHEJ) pathway.5
t(15,17) translocation breakpoints are preferential sites for mitoxantrone-induced DNA cleavage by human topoisomerase IIα
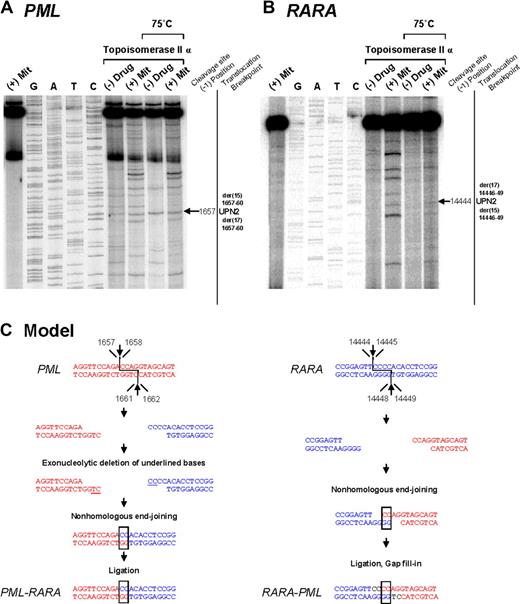
To investigate the mechanisms by which the t(15;17) chromosomal translocation may have been formed in MS patients treated with mitoxantrone, we evaluated topoisomerase IIα–mediated cleavage of the normal homologs of PML and RARA encompassing the respective breakpoints detected in 4 cases (UPNs 2, 7, 8, and 14) in the presence or absence of this agent. These included cases (ie, UPN 2 and UPN 14) in which the genomic breakpoint in the RARA or PML locus coincided with those reported previously in cases of t-APL arising in breast cancer patients treated with multiple DNA-damaging agents, including mitoxantrone.5 Few cleavage sites were observed in the absence of drug; however, bands of various sizes and intensities were observed in the presence of mitoxantrone in a topoisomerase IIα-dependent manner (Figures 2, 3 top panels). Cleavage bands that were significantly enhanced by mitoxantrone corresponding to the location of the observed genomic breakpoints in the PML and RARA loci were detected in each of the cases analyzed (Figures 2A,B, 3A,B; and data not shown). These bands remained detectable after heating, indicating stability of the cleavage complexes. In UPN 2, the case in which the RARA breakpoint was shared with a t-APL case that arose after mitoxantrone-containing breast cancer therapy,5 a functional site of mitoxantrone-induced cleavage by topoisomerase II was identified at position 14 444 (Figure 2B).
Investigation of t(15;17) translocation mechanism in UPN 2 by in vitro topoisomerase IIα DNA cleavage assay. Chromosomal breakpoint junctions were examined in an in vitro topoisomerase IIα cleavage assay using substrates containing PML (A) and RARA (B) translocation breakpoints in the APL case of UPN 2. Reactions in lane 1 were performed without DNA topoisomerase IIα and lanes 2 to 5 show dideoxy sequencing reactions. DNA cleavage reactions were performed in the presence of 147 nM of human DNA topoisomerase II alpha and in the absence (lanes 6 and 8) or presence of 20 μM mitoxantrone (lanes 7 and 9). Reactions in lanes 8 and 9 were incubated at 75°C to assess the heat stability of the cleavage products seen in lanes 6 and 7. In each case, the location of the relevant heat stable cleavage site is indicated by an arrow on the far right. (C) Native PML and RARA sequences are shown in red and blue, respectively. In the creation of the PML-RARA genomic fusion, processing includes exonucleolytic deletion to form a 2-base homologous overhang that facilitates repair via the error prone NHEJ pathway. In the creation of the reciprocal RARA-PML genomic fusion, 2-base homologies facilitate NHEJ repair, whereas in both instances polymerization of the relevant overhangs fills in any remaining gaps (shown black font).
Investigation of t(15;17) translocation mechanism in UPN 2 by in vitro topoisomerase IIα DNA cleavage assay. Chromosomal breakpoint junctions were examined in an in vitro topoisomerase IIα cleavage assay using substrates containing PML (A) and RARA (B) translocation breakpoints in the APL case of UPN 2. Reactions in lane 1 were performed without DNA topoisomerase IIα and lanes 2 to 5 show dideoxy sequencing reactions. DNA cleavage reactions were performed in the presence of 147 nM of human DNA topoisomerase II alpha and in the absence (lanes 6 and 8) or presence of 20 μM mitoxantrone (lanes 7 and 9). Reactions in lanes 8 and 9 were incubated at 75°C to assess the heat stability of the cleavage products seen in lanes 6 and 7. In each case, the location of the relevant heat stable cleavage site is indicated by an arrow on the far right. (C) Native PML and RARA sequences are shown in red and blue, respectively. In the creation of the PML-RARA genomic fusion, processing includes exonucleolytic deletion to form a 2-base homologous overhang that facilitates repair via the error prone NHEJ pathway. In the creation of the reciprocal RARA-PML genomic fusion, 2-base homologies facilitate NHEJ repair, whereas in both instances polymerization of the relevant overhangs fills in any remaining gaps (shown black font).
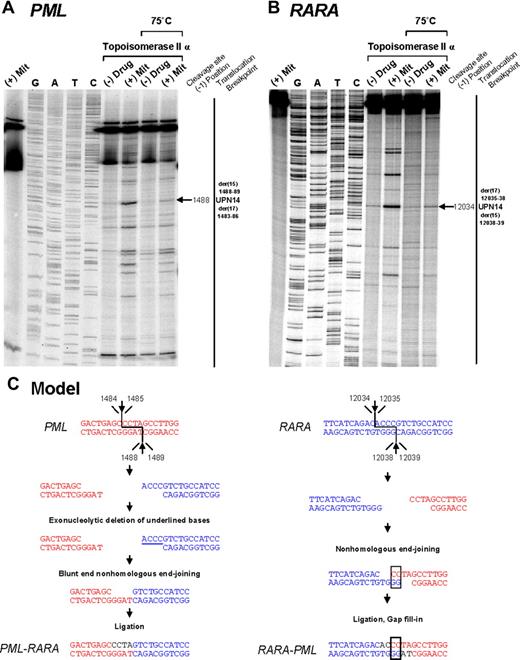
Investigation of t(15;17) translocation mechanism in UPN 14 by in vitro topoisomerase IIα DNA cleavage assay. DNA cleavage assays are shown for PML (A) and RARA (B) genomic breakpoint regions. For the PML assay, the reverse complement of the substrate containing the “hotspot” region between 1482 and 1489 described by Mistry et al5 was used. Lanes 1 to 9 of each cleavage assay are described in the legend to Figure 2. (C) Native PML and RARA sequences are shown in red and blue, respectively. In the creation of PML-RARA, processing includes exonucleolytic deletion and repair via the NHEJ pathway. In the creation of RARA-PML, 2-base homologies facilitate repair via the NHEJ pathway, whereas in both instances polymerization of the relevant overhangs fills in any remaining gaps (shown in black font).
Investigation of t(15;17) translocation mechanism in UPN 14 by in vitro topoisomerase IIα DNA cleavage assay. DNA cleavage assays are shown for PML (A) and RARA (B) genomic breakpoint regions. For the PML assay, the reverse complement of the substrate containing the “hotspot” region between 1482 and 1489 described by Mistry et al5 was used. Lanes 1 to 9 of each cleavage assay are described in the legend to Figure 2. (C) Native PML and RARA sequences are shown in red and blue, respectively. In the creation of PML-RARA, processing includes exonucleolytic deletion and repair via the NHEJ pathway. In the creation of RARA-PML, 2-base homologies facilitate repair via the NHEJ pathway, whereas in both instances polymerization of the relevant overhangs fills in any remaining gaps (shown in black font).
To provide further evidence that the region between positions 1482 and 1489 within PML intron 6 (which was involved in almost half the cases) is also a preferential site of mitoxantrone-induced DNA cleavage mediated by topoisomerase IIα, the reverse complement of the described PML substrate5 was used in the cleavage assay. A strong heat-stable cleavage band was detected in the presence of mitoxantrone at position 1488, which corresponds to the described functional cleavage at position 1484 on the upper strand5 (Figure 3A,C). Given that the chromosome 15 breakpoint in UPN 12 (in which there was no history of mitoxantrone exposure) also fell within this “hotspot,” it is interesting to note that a weak cleavage band was apparent in the presence of topoisomerase IIα in the absence of drug (Figure 3A lane 6). This finding suggests that the sequence may be a natural site of topoisomerase IIα–mediated cleavage that could be relevant to the etiology of APL in this case.
Based on sequence analysis of PML-RARA and reciprocal RARA-PML genomic breakpoints, the location of functional topoisomerase IIα cleavage sites in the vicinity of the breakpoints, and known mechanisms by which topoisomerase II induces double-strand breaks in DNA31 and their subsequent repair,6 it was possible to generate models as to how the t(15;17) chromosomal translocation could have been formed in the studied cases (Figure 2,3C). Type II topoisomerases introduce staggered nicks in DNA creating 5′-overhangs. In the models, repair of the overhangs in PML and RARA entails exonucleolytic digestion, pairing of complementary bases, and joining of DNA free ends by the NHEJ pathway, with template-directed polymerization to fill in any gaps.
Discussion
In this study on sAPL that developed after MS, we were able to identify a biased distribution of breakpoints in the PML gene that clustered in the same “hotspot” region previously identified in APL cases arising after treatment with mitoxantrone for breast cancer.5 In addition, we established in one patient who the breakpoint in RARA intron 2 at position 14446-49 coincided with a breakpoint identified by Mistry et al in 1 of 5 t-APL cases arising in breast cancer patients treated with the same agent.5 Given that intron 2 is almost 17 kb in length, such tight clustering of breakpoints between 2 different t-APL cases would be highly improbable to occur by chance. This observation strongly suggests that this is a preferential site of mitoxantrone-induced cleavage of DNA by topoisomerase IIα. The hypothesis is further supported by our functional in vitro data that show that this RARA site, together with the previously identified 8-bp “hotspot” region in PML intron 6, are preferential targets of mitoxantrone-induced DNA damage mediated by topoisomerase IIα.
Interestingly, of the 6 patients found to have PML breakpoints involving the “hotspot” region, one (UPN 12) did not receive mitoxantrone. Although mitoxantrone may significantly increase the chances of inducing DNA damage at this site, it is conceivable that this region represents a preferential site of cleavage by the native topoisomerase IIα and could in some instances act in concert with environmental or dietary agents that also target the enzyme.32-36 Accordingly, in the other case of sAPL that arose in the absence of mitoxantrone exposure (UPN 10), the in vitro DNA cleavage assay also revealed sites of cleavage in the PML and RARA substrates with topoisomerase II alone, which corresponded to the observed breakpoints (data not shown).
In some cases, the occurrence of short homologies of 1 or 2 nucleotides at the breakpoint region between the PML and RARA genes precluded precise assignment of the breakpoint within each respective gene. However, further investigation using the in vitro functional assays enabled the location of preferential sites of mitoxantrone-induced topoisomerase IIα–mediated DNA cleavage to be mapped. Taking into account the mechanisms by which type II topoisomerases induce double-strand DNA breaks31 and the processes mediating their repair, most probably involving the NHEJ pathway,5,30 it was possible to model the generation of the t(15;17) chromosomal translocation underlying the development of APL in these cases. Previous studies have established the presence of functional topoisomerase II cleavage sites at translocation breakpoints in MLL-associated t-AML, indicating that direct DNA damage coupled with aberrant repair by the NHEJ pathway is probably relevant to the formation of translocations that disrupt other genes that are commonly involved in t-AML.37-39
To the best of our knowledge, 20 cases of sAPL occurring in patients with MS have been reported to date,3,5,12-20 4 of which are included in the present series (UPNs 1, 4, 5, and 7) [5, 17, 20]. However, this is the first study to systematically analyze such cases at the genomic level. The incidence of sAPL arising in MS patients treated with mitoxantrone is not firmly established, as no systematic analysis has been undertaken to address this issue and only case reports have been published. On reviewing the records of 2336 MS patients treated with mitoxantrone, Voltz et al40 described 5 cases of t-AML and 2 sAPL as a case report. Ghalie et al41 assembled the records of 1378 patients treated with mitoxantrone in 3 MS studies and reported 2 patients who developed t-AML with an observed incidence proportion of 0.15% (95% confidence interval, 0.00%-0.40%). In addition to the reported 20 cases of sAPL, 8 cases of t-AML (non-M3) arising after mitoxantrone treatment for MS have been described, with the majority showing balanced translocations in their leukemic cells.40,42-46 Therefore, although it appears that an excess of sAPL cases are observed in the MS setting, the reasons underlying this phenomenon remain unclear at present and warrant further basic and epidemiologic investigation. It is unknown, for example, whether factors other than mitoxantrone may play a role in sAPL development in the context of MS. Finally, it would be important to assess prospectively the true incidence of APL development in the MS setting (with or without mitoxantrone). Considering the risk of leukemia development and cardiac toxicity, the Therapeutics and Technology Subcommittee of the American Academy of Neurology recently has recommended that mitoxantrone be reserved for patients with progressive MS who have failed other therapies.47
In conclusion, this study lends further support to the presence of preferential sites of DNA damage induced by mitoxantrone within PML intron 6 and suggests the existence of a further “hotspot” at the distal end of RARA intron 2. The susceptibility of these regions of the PML and RARA loci to topoisomerase IIα–mediated cleavage by mitoxantrone may underlie the propensity to develop this particular subtype of AML after exposure to this agent. Further studies are warranted to investigate whether MS patients have a particular predisposition to the development of sAPL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. Boggild for provision of clinical data, Jelena Jovanovic for performance of MRD analyses, and Mireia Camos for kindly providing the DNA from UPN 12.
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (F.L.C.) and Italian Ministry of Health (Progetto Integrato Oncologia), Leukemia Research Fund of Great Britain (A.N.M., D.G.), the National Institutes of Health (grant R01CA077683 to C.A.F.; and grant GM33944 to J.A.W.B., N.O.), and the Polish Ministry of Science and Education (grant PBZ KBN 107 P04 2004, to M.L.).
Authorship
Contribution: S.K.H. and A.N.M. performed the experiments, analyzed the data, and contributed to the manuscript; T.O. and M.L. assisted in experimental design and performance of experiments; A. Ledda, G.L.N., C.S., C.C., E.B., L.M., E.M., J.C., G.S., A. Lennard, J.E., M.T.V., and W.R.S. contributed to the samples, clinical data, and interpretation of results; J.A.W.B. and N.O. supplied vital reagents; D.G., M.A.S., C.A.F., and S.A. analyzed the data, critically reviewed the manuscript, and amended the final report; and F.L.C. and D.G. designed the study, supervised the research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Lo-Coco, Department of Biopathology, University Tor Vergata, Via Montpellier 1, 00133, Rome, Italy; e-mail: francesco.lo.coco@uniroma2.it.
References
Author notes
*S.K.H. and A.N.M. contributed equally to the experimental analyses and should be considered joint first authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal