Angiogenesis has a critical role in the pathophysiology of multiple myeloma (MM); however, the molecular mechanisms underlying this process are not completely elucidated. The new tumor-suppressor gene inhibitor of growth family member 4 (ING4) has been recently implicated in solid tumors as a repressor of angiogenesis. In this study, we found that ING4 expression in MM cells was correlated with the expression of the proangiogenic molecules interleukin-8 (IL-8) and osteopontin (OPN). Moreover, we demonstrate that ING4 suppression in MM cells up-regulated IL-8 and OPN, increasing the hypoxia inducible factor-1α (HIF-1α) activity and its target gene NIP-3 expression in hypoxic condition. In turn, we show that the inhibition of HIF-1α by siRNA suppressed IL-8 and OPN production by MM cells under hypoxia. A direct interaction between ING4 and the HIF prolyl hydroxylase 2 (HPH-2) was also demonstrated. Finally, we show that ING4 suppression in MM cells significantly increased vessel formation in vitro, blunted by blocking IL-8 or OPN. These in vitro observations were confirmed in vivo by finding that MM patients with high IL-8 production and microvascular density (MVD) have significantly lower ING4 levels compared with those with low IL-8 and MVD. Our data indicate that ING4 exerts an inhibitory effect on the production of proangiogenic molecules and consequently on MM-induced angiogenesis.
Introduction
Angiogenesis has a critical role in the pathophysiology and progression of multiple myeloma (MM) supporting the growth and survival of MM cells.1,,,–5
The angiogenic process in MM is sustained mainly by the overexpression of proangiogenic factors directly by MM cells including VEGF,6 angiopoietin-1 (ANG-1),7 osteopontin (OPN),8 and interleukin-8 (IL-8).9 Nevertheless, the molecular mechanisms underlying the regulation of angiogenesis in MM have not been completely elucidated.
The new candidate tumor-suppressor gene inhibitor of growth family member 4 (ING4) has been recently implicated in solid tumors as a repressor of tumor growth and angiogenesis through the association with NF-κB. ING4 is a nuclear factor expressed in all normal tissues and markedly reduced in glioblastoma cells and head and neck squamous cell carcinoma, with levels inversely correlated with tumor grade.10,11 Inhibition of ING4 expression strongly promotes the growth of glioma cells in vivo, whereas its overexpression leads to growth inhibition through ING4's capability to interact with p65 subunit of NF-κB.10
Interestingly, it has been also shown that tumors lacking ING4 showed increased vascularization compared with ING4-expressing tumors.12 Moreover ING4 down-regulated the angiogenic-related molecules including IL-8 and the hypoxia inducible factor-1α (HIF-1 α) activity in hypoxic condition through the involvement of HIF prolyl hydroxylase 2 (HPH-2)10,13 In turn, the role of hypoxia has been recently highlighted in the promotion of angiogenesis.14
The expression of ING4 by MM cells, as well as its potential role in MM-induced angiogenesis, has never been investigated. In this study, we evaluated the expression of ING4 in malignant MM cells and the potential relationship between ING4 and the production of proangiogenic molecules by MM cells in normoxic and hypoxic conditions, as well as its relationship with the “in vitro and in vivo” angiogenesis.
Patients, materials, and methods
Cells and cell culture conditions
Cell lines.
Human myeloma cell lines (HMCLs) XG-6, XG-1, and JJN3 were obtained from Dr Bataille (Nantes, France). U266 was obtained from the American Type Culture Collection (Rockville, MD). OPM2 and RPMI-8226 were purchased from DSM (Braunschweig, Germany). ARP-1 and H929 were generously received from Dr Shaughnessy's laboratory (Little Rock, AR).
Cell cultures.
HMCLs were incubated in RPMI medium at 10% FCS (Invitrogen Life Technologies, Milan, Italy) and maintained with or without IL-6 (3 ng/mL; Endogen Woburn, MA). In a series of experiments, HMCLs were incubated with the HPH-2 inhibitor dymethyloxaloylglycine (DMOG, 0.2 mM; Cayman Chemical, Ann Arbor, MI) or vehicle in the presence or absence of hypoxic condition (1% O2, 5% CO2) or treated with the hypoxic mimetic CoCl2 at 100 μM (Sigma Aldrich, St Louis, MO) or vehicle for 12 hours. Similarly, HMCLs, 24 hours after siRNA transfection or 48 hours after lentiviral infection, were incubated in the presence or absence of hypoxic condition.
siRNA transfection.
HMCLs were transfected as previously described.8 In brief, HMCLs were washed twice with serum-free Opti-MEM (Gibco BRL, Paisley, United Kingdom) and resuspended to a final concentration of 2 × 107 cells/mL in Opti-MEM (Gibco BRL). Subsequently, 0.4 mL of cell suspension was mixed either with 1 nmol of a smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or/and HIF-1α or HPH-2 or with a nonspecific control siRNA (siRNA Cy) (Dharmacon Tech, Lafayette, CO) and electroporated in a 0.4-cm cuvette using the Gene Pulser electroporation apparatus (Bio-Rad Laboratories, Hercules, CA) using a single-pulse protocol.8 The viability of HMCLs after the transfection was evaluated by flow cytometry (Becton Dickinson, San Jose, CA).
Overexpression of ING4 in HMCLs.
Plasmids containing the wild-type ING4 coding region were a generous gift from Dr Greco (Department of Experimental Oncology, National Cancer Institute, Milan, Italy) and Dr Harris (Laboratory of Human Carcinogenesis, National Cancer Institute, Bethesda, MD). The amplified ING4 cDNA sequence was cloned into the pWPI lentiviral vector (kindly provided by Dr Didier Trono, National Center for Competence in Research, Switzerland). Recombinant lentivirus was produced by transient transfection of 293T cells following a standard protocol.15 In brief, crude virus was concentrated by ultracentrifugation at 90 000g for 90 minutes. Viral titers were determined by measuring the amount of HIV-1 p24 antigen by enzyme-linked immunosorbent assay (ELISA; NEN Life Sciences, Boston, MA). A 99% transduction efficiency of HMCLs (RPMI-8226 and JJN3) was achieved with 3000 ng lentiviral p24 particles/106 cells as evaluated by green fluorescence protein (GFP) expression using flow cytometry.
In vitro angiogenesis assay.
In vitro angiogenesis was assessed by Angio-kit obtained from TCS Biologicals (Buckingham, United Kingdom), as previously described. In brief, cells were stimulated with VEGF (2 ng/mL, positive control) or suramin (20 μM negative control) or with conditioned media (CM) of HMCLs/D-MEM at 10% FBS (ratio, 1:2) transfected with siRNA anti-ING4 or siRNA Cy in the presence or absence of blocking anti-OPN mAb (Assay Designs, Ann Arbor, MI) or anti–IL-8 blocking mAb (R&D Systems, Minneapolis, MN) or anti-IgG control Ab.
At day 11, cells were fixed and stained using an anti-CD31 Ab (TCS Biologicals) according to the instructions provided. To measure the formation of the capillary network, the number of connections among 3 or more capillary-like structures was considered and plates were quantified by a computerized image analysis (TCS Biologicals).
Patients
We studied a total of 50 patients (median age, 66 years; range, 53-73 years) with newly diagnosed MM (stages I-III). BM aspirates and bone biopsies were obtained from iliac crest of all patients after informed consent was obtained in accordance with the Declaration of Helsinki. Approval was obtained from the Institutional Review Board of the Azienda Ospedaliero-Universitaria de Parma (Parma, Italy). Eleven patients with monoclonal gammopathy of unknown significance (MGUS) were also included in the study. CD138+ plasma cells obtained from reactive tonsils have been used as healthy controls. Both normal and malignant CD138+ plasma cells were purified with an immunomagnetic method using anti-CD138 mAb-coated microbeads (magnetic-activated cell sorting [MACS]; Miltenyi Biotec, Bergisch-Gladbach, Germany). Only samples with purity of more than 90% were tested. Fresh MM cells were analyzed immediately after purification.
BM plasma was obtained from BM aspirate (5 mL) of MM patients (n = 35) treated with EDTA to prevent clotting after centrifugation.
RNA isolation and reverse-transcriptase–polymerase chain reaction amplification
For reverse-transcription–polymerase chain reaction (RT-PCR) analysis, total cellular RNA was extracted from cells using RNeasy mini kit (Qiagen, Valencia, CA). RNA (1 μg) was reverse transcribed using the Superscript RnaseH-reverse transcriptase kit according to the manufacturer's protocol (Invitrogen Life Technologies). cDNAs were amplified by PCR with specific primer pairs. PCR reactions were performed in a thermal cycler (MiniCycler; MyResearch, Watertown, MA) for 30 cycles. The following specific primer pairs have been used: ING4: forward (F), 5′-ATTGCCTTTGTCACCAGGTC-3′ and reverse (R), 5′-AAAGGACCCTCCCTCTGAAA-3′; VEGF: F, 5′-CGAAGTGGTGAAGTTCATGGATG-3′ and R, 5′-TTCTGTTCAGTCTTTCCTGGTGAG-3′; IL-8: F, 5′-TACTCCAAACCTTTCCACCC-3′ and R, 5′-AACTTCTCCACAACCCTCTG-3′; ANG-1: F, 5′-GGAAGTCTAGTTTCCAAAGAGGC-3′ and R, 5′-CTTTATCCCATTCAGTTTTCCATG-3′; OPN: F, 5′-ATATGATGGCCGAGGTGATA-3′ and R, 5′-GACCTCAGAAGATGCACTAT-3′; HIF-1α: F, 5′-CTCAAAGTCGGACAGCCTCA-3′ and R, 5′-CCCTGCAGTAGGTTTCTGCT-3′; NIP-3: F, 5′-CCTGGGTAGAACTGCACTTCAGCAAT-3′ and R, 5′-TCATGACGCTCGTGTTCCTCATGCA-3′; GAPDH: F, 5′-CAACGGATTTGGTCGTATTG-3′ and R, 5′-GGAAGATGGTGATGGGATTT-3′.
Annealing temperatures used were as follows: ING4: 56°C; VEGF: 66°C; IL-8: 64°C; ANG-1: 58°C, OPN: 60°C, HIF-1α: 59°C, NIP-3: 68°C; and GAPDH: 58°C. Product size were as follows: ING-4: 465 bp; VEGF: 375 bp; IL-8: 158 bp; ANG-1: 429 bp; OPN: 475 bp; HIF-1α: 454 bp; and GAPDH: 209 bp.
Pictures of the electrophoresed cDNAs were recorded with a digital DC 120 Kodak camera (Rochester, NY) and quantified, as previously described.7
Real-time quantitative PCR
Real-time PCR was performed by adding 2 of 20 μL cDNA, obtained from 1 μg RNA, to a universal master Mix (Applied Biosystem, Applera, Milan, Italy), primers and TaqMan probes, as previously described.16 Primers and probes were synthesized and purchased from Applied Biosystem. The sequence of the primers and probes used was as follows: ING4: F, 5′-CCAGGTCTCCTATGGAGAGATGA-3′ and R, 5′-CCCGAGGCTTGGTTGTCA-3′; probe: 6-FAM-ACCCTGATTGTTCCATTGAGTGGTTCCA-MGB; VEGF: F, 5′-TGCTCTCACCTCCACCATGCCAA-3′ and R, 5′-TGATGATTCTGCCCTCCTCCTTC-3′; probe: 6-FAM-TGGTCCCAGGCTGCACCCATGGC-MGB; IL-8: F, 5′-CTCTTGGCAGCCTTCCTGATT-3′ and R, 5′-TATGCACTGACATCTAAGTTCTTTAGCA-3′; probe: 6-FAM-CTTGGCAAAACTGCACCTTCACACAGA-MGB; ANG-1: F, 5′-GCAACTGGAGCTGATGGACACA-3′ and R, 5′-CATCTGCACAGTCTCTAAATGGT-3′; probe: 6-FAM-CAATCTTTGCACTAAAGAAGGTGTTTTACT-MGB; NIP-3: F, 5′-GCAGGGCTCCTGGGTAGAA-3′ and R, 5′-AACCGAGGCTGGAACGCT-3′; probe: 6-FAM-TGCACTTCAGCAATAATGGGAACGGG-MGB.
For OPN, HIF-1α, and HPH-2 real-time PCR, the TaqMan Gene Expression Assays HS00167093-M1, HS00936379-M1, and HS00254392-M1, respectively, were used according to the manufacturer's protocols. To normalize for differences in RNA quality and reverse-transcription efficiency, we applied the comparative Ct method using the endogenous reference gene ABL.16–17 The relative ING4 mRNA quantification was performed as previously described16,18 by the ΔCt method (ΔCt = mean CT ING4 − mean CT ABL).
ΔΔCt was evaluated as the difference between the ΔCt of a sample and the ΔCt of the control. The fold change in ING4 mRNA expression (n-fold) was calculated as 2−ΔΔCt. The same procedures were used for IL-8, OPN, VEGF, ANG-1, HIF-1α, and HPH-2.
Statistical analysis was performed using SPSS for Mac software (version 10.0; SPSS, Chicago, IL). The independent 2-tailed t test was used for comparison ING4 and the angiogenic molecule expression in MM patients. The correlation between ING4 and the angiogenic molecules was analyzed by a linear regression.18
Gene expression profiling and microarray analysis
Gene expression profiling, using the Affymetrix U133Plus2.0 microarray (Santa Clara, CA), was performed, as previously described.19 Affymetrix U133Plus2.0 microarrays were preprocessed using GCOS1.1 software (Affymetrix) and normalized using conventional GCOS1.1 scaling.
NF-κB and HIF-1α activity
HIF-1α and NF-κB activation in nuclear extracts was evaluated using an ELISA-based method (TransAM HIF-1 and TransAM NF-κB family kits; Active Motif, Carlsbad, CA) according to the manufacturer's procedures.
Western blot analysis
Western blot analysis was performed as previously described.8 Nuclear extracts were prepared using the Nuclear Extraction kit (Active Motif) according to the manufacturer's protocol. Nuclear extracts (40 μg) were tested. p29ING4 protein was detected using a polyclonal anti-p29ING4 by Rockland (Gilbertsville, PA). A polyclonal goat anti–HIF-1α Ab (R&D Systems) was used to detect HIF-1α, whereas histone H1 mAb was used as internal control (Upstate, Lake Placid, NY). HPH-2 protein was detected using an anti–HPH-2 polyclonal Ab (Novus Biological, Littleton, CO).
Coimmunoprecipitation assay
Coimmunoprecipitation assay was performed using a commercially available kit (Active Motif). In brief, nuclear lysates of HMCLs, infected with ING4 lentiviral vector, were incubated in the presence of anti–HPH-2 rabbit polyclonal Ab or anti-ING4 goat polyclonal Ab overnight at 4°C. Antibody-protein complexes were precipitated with protein A–agarose beads, washed extensively with lysis buffer, and eluted with SDS loading buffer. Either coimmunoprecipitated ING4 or HPH-2 was detected by Western blot analysis using anti–HPH-2 and anti-ING4 Abs, respectively.
ELISAs
Soluble IL-8, OPN, and VEGF were detected in the CM of myeloma cells and BM plasma from MM patients using ELISAs purchased from R&D Systems, according to the manufacturer's protocols. Cytokine levels in the CM were normalized to the number of cells at the end of culture period.
BM angiogenesis evaluation
Bone biopsies were obtained in a subgroup of 28 MM patients. Blood vessels were detected in 3-μm sections of 4% formalin and B5 solution–fixed paraffin-embedded biopsies. Angiogenesis was measured as density of microvessels (capillaries and small venules) by staining endothelial cells using anti-CD34 monoclonal Ab (working dilution: 1:50, clone QBEnd/10; NeoMarkers, Fremont, CA) as previously described.7 The mean of vascular density (MVD) and the mean number of microvessels in each biopsy were expressed on the basis of the total number of microvessel transversal sections (capillaries and/or venules) per the total cellular area (in mm2) and per the total number of fields (400× magnification), respectively.
Results
Expression of ING4 in human myeloma cells and its correlation with proangiogenic molecules
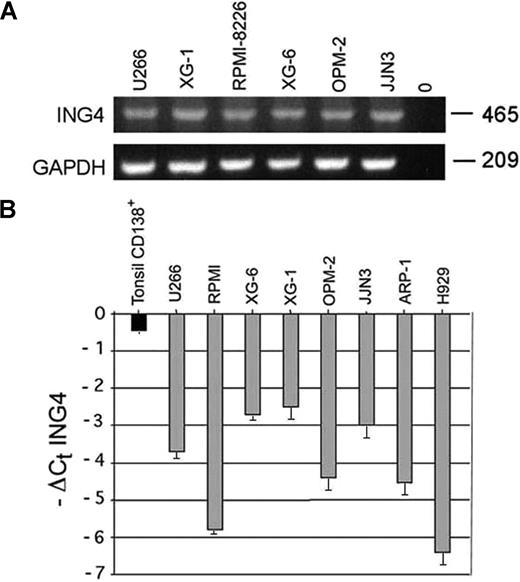
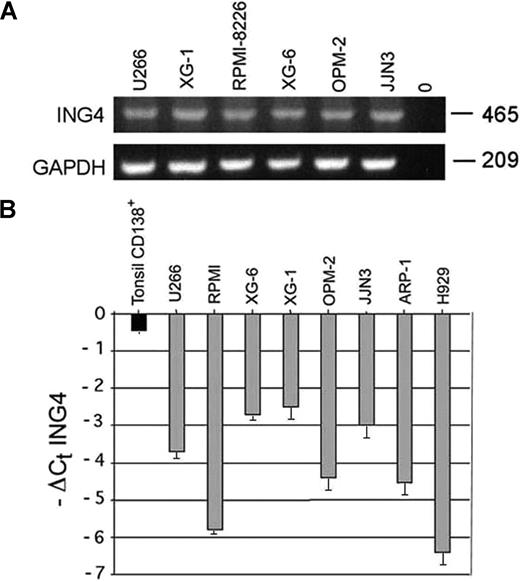
The presence of ING4 mRNA was observed in all HMCLs tested (Figure 1A). By real-time PCR we found that ING4 mRNA levels were significantly reduced in HMCLs compared with normal CD138+ tonsil plasma cells with a median −ΔCt ING4 of −3.2 versus −0.45 (Figure 1B). The nuclear p29ING4 protein level was consistently reduced in HMCLs compared with controls (data not shown). A significant negative correlation was found between ING4 mRNA levels and p65 nuclear activity (R = −0.83; Pearson 2-tailed test: P = .005) in HMCLs, as reported in Table 1.
ING4 expression by human myeloma cell lines (HMCLs). ING4 mRNA expression was evaluated by RT-PCR in several HMCLs (U266, XG-1, RPMI-8226, XG-6, OPM-2, JJN3). GAPDH was used as internal control (A). ING4 mRNA levels were quantified by real-time PCR in HMCLs. Data are expressed (graphs) as −ΔCt ING4: −(mean Ct ING4 − mean Ct ABL) plus or minus SD of triplicate measures. Purified tonsil CD138+ plasma cells were used as healthy control (B).
ING4 expression by human myeloma cell lines (HMCLs). ING4 mRNA expression was evaluated by RT-PCR in several HMCLs (U266, XG-1, RPMI-8226, XG-6, OPM-2, JJN3). GAPDH was used as internal control (A). ING4 mRNA levels were quantified by real-time PCR in HMCLs. Data are expressed (graphs) as −ΔCt ING4: −(mean Ct ING4 − mean Ct ABL) plus or minus SD of triplicate measures. Purified tonsil CD138+ plasma cells were used as healthy control (B).
Thereafter, we correlated ING4 level in HMCLs with the expression of proangiogenic molecules by quantitative PCR. A significant negative correlation was demonstrated between ΔCt ING4 and ΔCt IL-8 (R = −0.84, Pearson 2-tailed test: P = .008) and between ΔCt ING4 and ΔCt OPN (R = − 0.64, P = .05), suggesting that HMCLs with lower ING4 expression have significantly higher IL-8 and OPN mRNA levels. On the other hand, any significant correlation was not observed between ING4 mRNA and ANG-1 or VEGF mRNA expression (P = NS) (data not shown).
Effect of ING4 suppression on the production of angiogenic molecules by human myeloma cells
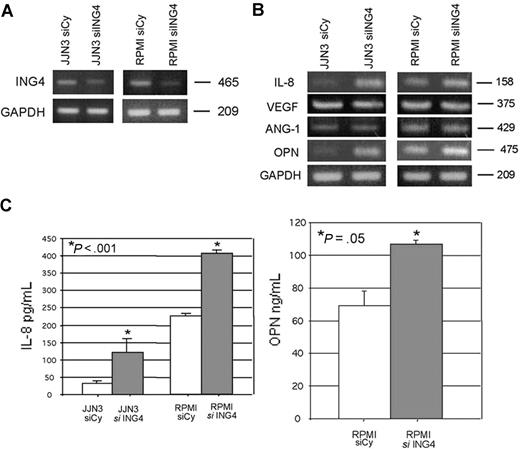
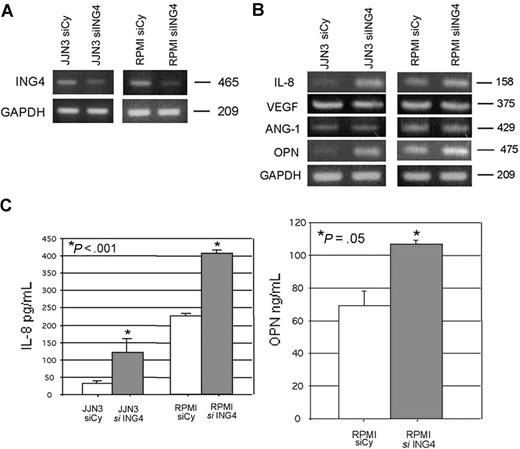
To demonstrate the potential relationship between ING4 and the production of proangiogenic molecules, we suppressed ING4 expression in HMCLs by siRNA. The transfection with ING4 siRNA, but not with the nonspecific control siRNA, suppressed ING4 mRNA as shown for JJN3 and RPMI-8226 (Figure 2A), leading to an increase of the p65 NF-κB nuclear binding activity (44% ± 4.6%, P = .03). Subsequent to ING4 suppression, we found that IL-8 mRNA was increased (Figure 2B; Table 2). Similarly, we found that OPN mRNA expression was increased in HMCLs transfected with ING4 siRNA, as shown in Figure 2B and Table 2. On the contrary ANG-1 and VEGF mRNA expression was not modified by ING4 siRNA transfection (Figure 2B; Table 2). Data were confirmed at the protein level by the finding that ING4 suppression in HMCLs significantly increased IL-8 (mean ± SD) (JJN3: 126 ± 4.3 vs 365 ± 14.3 pg/mL; RPMI-8226: 151 ± 4.1 vs 320 ± 6.2 pg/mL; P < .001) and OPN protein (RPMI-8226: 68.9 ± 11.0 vs 106.7 ± 2.3 ng/mL; P = .05) secretion (Figure 2C) but not VEGF and ANG-1 (data not shown). The potential effect of ING4 suppression on gene expression profiling of myeloma cells was investigated by microarray in RPMI-8226. As reported in Table 3, other than IL-8 and OPN, the up-regulation of a series of proangiogenic molecules was observed in RPMI-8226 transfected with siRNA anti-ING4 as compared to the same cells transfected with the nonspecific siRNA.
Effect of ING4 suppression in HMCLs on the production of the proangiogenic molecules. JJN3 and RPMI-8226 were transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or with a nonspecific control siRNA (Cy). After 24 hours, cells were harvested for ING4 evaluation by RT-PCR (A). The expression of the proangiogenic molecules IL-8, VEGF, ANG-1, and OPN was evaluated by RT-PCR in both JJN3- and RPMI-8226–transfected cells (B). After 48 hours, IL-8 (JJN3 and RPMI-8226) and OPN (RPMI-8226) levels were measured in the conditioned media of transfected cells by ELISA. Graphs represent the mean plus or minus SD of IL-8 and OPN levels normalized for the number of cells (C).
Effect of ING4 suppression in HMCLs on the production of the proangiogenic molecules. JJN3 and RPMI-8226 were transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or with a nonspecific control siRNA (Cy). After 24 hours, cells were harvested for ING4 evaluation by RT-PCR (A). The expression of the proangiogenic molecules IL-8, VEGF, ANG-1, and OPN was evaluated by RT-PCR in both JJN3- and RPMI-8226–transfected cells (B). After 48 hours, IL-8 (JJN3 and RPMI-8226) and OPN (RPMI-8226) levels were measured in the conditioned media of transfected cells by ELISA. Graphs represent the mean plus or minus SD of IL-8 and OPN levels normalized for the number of cells (C).
Effect of ING4 suppression in myeloma cells on angiogenic molecules in hypoxic condition: role of HIF-1α
We have further investigated the role of ING4 in the production of angiogenic molecules by MM cells in hypoxic condition.
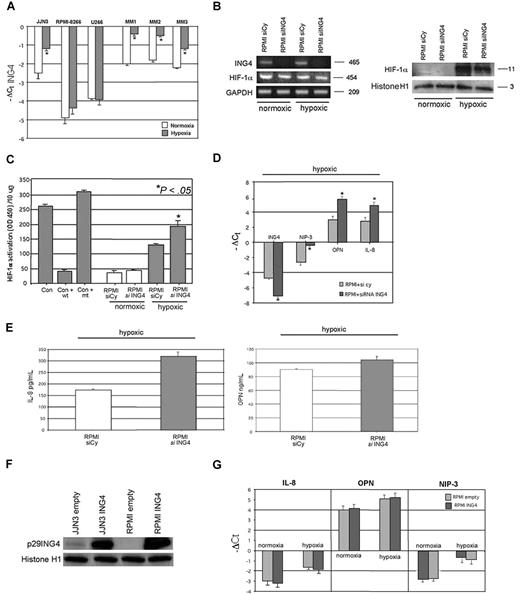
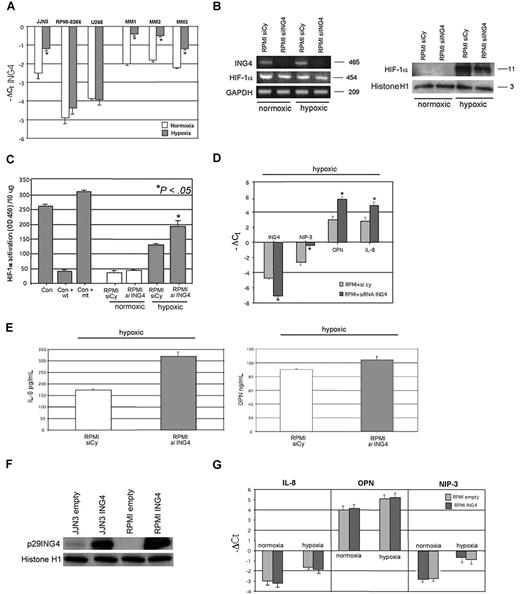
First we found that hypoxia increased ING4 mRNA level in JJN3 but not in RPMI-8226 and U266 (Figure 3A), whereas a significant up-regulation of ING4 expression was observed in almost all MM patients tested, as reported for 3 representative ones (Figure 3A). The hypoxic treatment of HMCLs significantly up-regulated the protein and activity levels of HIF-1α without modifying its mRNA expression and increased the levels of the angiogenic molecules (JJN3: IL-8: + 106% ± 12%, P < .001; OPN: + 40% ± 8%, P = .05; RPMI-8228: IL-8: + 50% ± 11%, P < .001; OPN: + 34% ± 6%, P = .005).
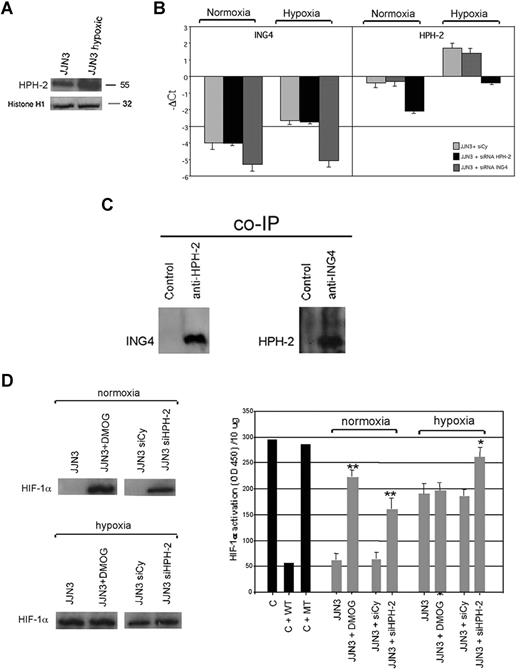
Effects of ING4 suppression and overexpression in HMCLs on IL-8 and OPN production and HIF-1α activity under hypoxic condition. HMCLs and fresh purified CD138+ MM cells obtained from 3 MM patients were incubated in the presence or absence of hypoxic conditions for 12 hours. ING4 mRNA expression was evaluated by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value of 3 independent experiments (*P < .05) (A). RPMI-8226 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or with a nonspecific control siRNA (Cy) and after 24 hours incubated in the presence or absence of hypoxic condition (1% O2, 5% CO2 atmosphere or CoCl2 treatment) for 12 hours. Thereafter, ING4 and HIF-1α mRNA expression was evaluated by RT-PCR (B left panel). HIF-1α protein level was checked by Western blot in nuclear extracts (B right panel). HIF-1α activity was quantified in RPMI-8226–transfected cells by a transcriptional factor assay kit, as described in “NF-κB and HIF-1α activity.” Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (Con). Graphs represent the mean plus or minus SD of HIF-1α activity normalized for 10 μg protein analyzed of 2 independent experiments performed in triplicate (OD = optical density) (C). The mRNA levels of ING4, NIP-3, OPN, and IL-8 in RPMI-8226–transfected cells incubated in hypoxic condition were quantified by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (D). Aliquots of conditioned media of RPMI-8226–transfected cells incubated in hypoxic condition were tested by ELISA. Graphs represent the mean plus or minus SD of IL-8 and OPN levels in 2 independent experiments measured in triplicate (*P < .05) (E). Both JJN3 and RPMI-8226 were infected by empty lentiviral vector, used as control, or ING4 overexpression vector, as described in “Cells and cell culture conditions; siRNA transfection.” p29ING4 protein was detected after 72 hours by Western blot analysis (F). RPMI-8226 cells overexpressing ING4 or the empty vector were incubated in the presence or absence of hypoxic condition for 12 hours and then evaluated for IL-8, OPN, and NIP-3 mRNA expression by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (G).
Effects of ING4 suppression and overexpression in HMCLs on IL-8 and OPN production and HIF-1α activity under hypoxic condition. HMCLs and fresh purified CD138+ MM cells obtained from 3 MM patients were incubated in the presence or absence of hypoxic conditions for 12 hours. ING4 mRNA expression was evaluated by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value of 3 independent experiments (*P < .05) (A). RPMI-8226 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or with a nonspecific control siRNA (Cy) and after 24 hours incubated in the presence or absence of hypoxic condition (1% O2, 5% CO2 atmosphere or CoCl2 treatment) for 12 hours. Thereafter, ING4 and HIF-1α mRNA expression was evaluated by RT-PCR (B left panel). HIF-1α protein level was checked by Western blot in nuclear extracts (B right panel). HIF-1α activity was quantified in RPMI-8226–transfected cells by a transcriptional factor assay kit, as described in “NF-κB and HIF-1α activity.” Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (Con). Graphs represent the mean plus or minus SD of HIF-1α activity normalized for 10 μg protein analyzed of 2 independent experiments performed in triplicate (OD = optical density) (C). The mRNA levels of ING4, NIP-3, OPN, and IL-8 in RPMI-8226–transfected cells incubated in hypoxic condition were quantified by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (D). Aliquots of conditioned media of RPMI-8226–transfected cells incubated in hypoxic condition were tested by ELISA. Graphs represent the mean plus or minus SD of IL-8 and OPN levels in 2 independent experiments measured in triplicate (*P < .05) (E). Both JJN3 and RPMI-8226 were infected by empty lentiviral vector, used as control, or ING4 overexpression vector, as described in “Cells and cell culture conditions; siRNA transfection.” p29ING4 protein was detected after 72 hours by Western blot analysis (F). RPMI-8226 cells overexpressing ING4 or the empty vector were incubated in the presence or absence of hypoxic condition for 12 hours and then evaluated for IL-8, OPN, and NIP-3 mRNA expression by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (G).
ING4 suppression by siRNA did not modify either HIF-1α mRNA expression by RPMI-8226 (Figure 3B) or HIF-1α nuclear protein level (Figure 3B) but significantly increased its nuclear activity in hypoxic condition (Figure 3C). Consistently we observed a significant increase of the HIF-1α target gene NIP-3 at transcriptional level by quantitative PCR (Figure 3D). Similarly, IL-8 and OPN mRNA (Figure 3D) and protein secretion (Figure 3E) were up-regulated by ING4 inhibition in RPMI-8226 under hypoxic condition. Nevertheless, ING4 lentiviral overexpression in HMCLs (Figure 3F) did not modify IL-8, OPN, and NIP-3 mRNA expression both in normoxic and hypoxic conditions, as shown in RPMI-8226 (Figure 3G).
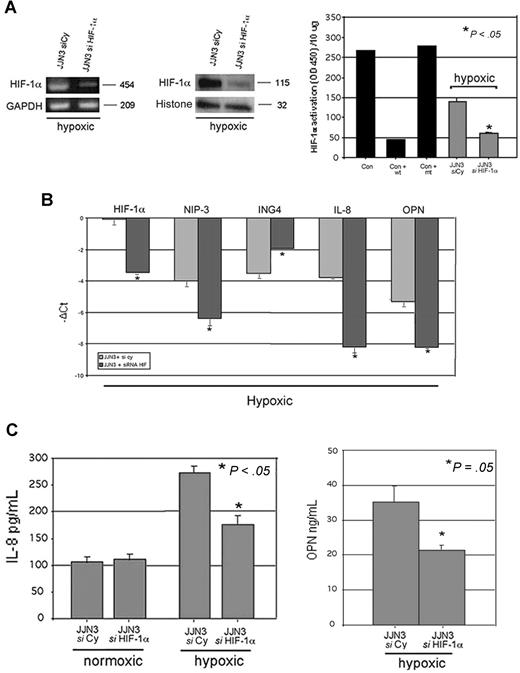
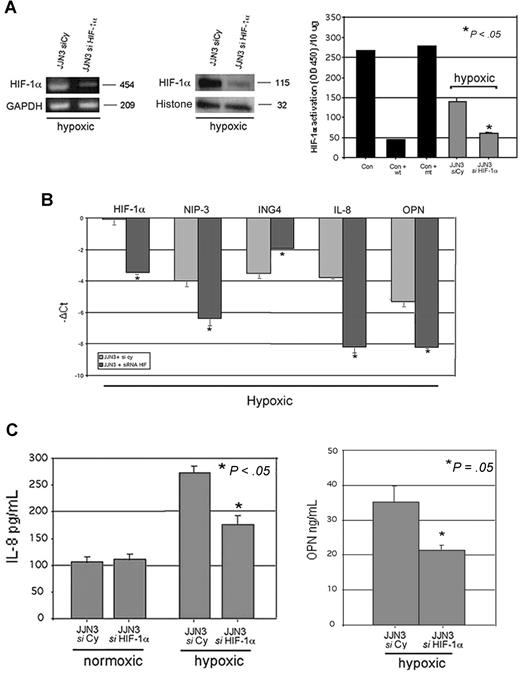
The potential relationship between ING4 and HIF-1α system was further analyzed by blocking HIF-1α expression by specific siRNA in HMCLs. Both RPMI-8226 and JJN3 were transfected with siRNA anti–HIF-1α with higher efficiency of transfection in JJN3. The inhibition of HIF-1α by siRNA led to the suppression of HIF-1α mRNA and protein expression as well as HIF-1α activity (Figure 4A). Subsequent to HIF-1α suppression, IL-8 production by JJN3 was inhibited in hypoxic condition but not in normoxic one (Figure 4B). Similarly, OPN production was suppressed in JJN3 in hypoxic condition (Figure 4B).
Effect of HIF-1α suppression on ING4 and the angiogenic-related molecules in hypoxic condition. JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against HIF-1α or with a nonspecific control siRNA (Cy). After 24 hours, cells were incubated in the presence or absence of hypoxic condition (1% O2, 5% CO2 atmosphere or CoCl2 treatment) for 12 hours. HIF-1α mRNA expression was evaluated by RT-PCR, whereas HIF-1α protein level and activity were detected by Western blot and ELISA, respectively, as described in “Western blot analysis” and “ELISAs.” Graphs represent the mean plus or minus SD of 2 independent experiments measured in triplicate (OD = optical density). Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (Con) (A). Aliquots of conditioned media of JJN3 transfected cells were tested by ELISA to detect IL-8 and OPN levels. Graphs represent the mean plus or minus SD of IL-8 or OPN levels in 2 independent experiments measured in triplicate (B). JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against HIF-1α or ING4 or HIF-1α plus ING4 or with a nonspecific control siRNA (Cy) and incubated after 24 hours in the presence or absence of hypoxic condition. Thereafter ING4, IL-8, OPN, HIF-1α, and NIP-3 mRNA expression was quantified by real-time PCR. Data are expressed as mean −ΔCt plus or minus SD as described in “Real-time quantitative PCR” (C).
Effect of HIF-1α suppression on ING4 and the angiogenic-related molecules in hypoxic condition. JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against HIF-1α or with a nonspecific control siRNA (Cy). After 24 hours, cells were incubated in the presence or absence of hypoxic condition (1% O2, 5% CO2 atmosphere or CoCl2 treatment) for 12 hours. HIF-1α mRNA expression was evaluated by RT-PCR, whereas HIF-1α protein level and activity were detected by Western blot and ELISA, respectively, as described in “Western blot analysis” and “ELISAs.” Graphs represent the mean plus or minus SD of 2 independent experiments measured in triplicate (OD = optical density). Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (Con) (A). Aliquots of conditioned media of JJN3 transfected cells were tested by ELISA to detect IL-8 and OPN levels. Graphs represent the mean plus or minus SD of IL-8 or OPN levels in 2 independent experiments measured in triplicate (B). JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against HIF-1α or ING4 or HIF-1α plus ING4 or with a nonspecific control siRNA (Cy) and incubated after 24 hours in the presence or absence of hypoxic condition. Thereafter ING4, IL-8, OPN, HIF-1α, and NIP-3 mRNA expression was quantified by real-time PCR. Data are expressed as mean −ΔCt plus or minus SD as described in “Real-time quantitative PCR” (C).
To investigate whether the effect of ING4 on IL-8 and OPN production in hypoxic condition is mediated by HIF-1α activation, we have performed ING4 down-regulation in the context of HIF-1α suppression by a double transfection using siRNA anti-ING4 and anti–HIF-1α both in normoxic and hypoxic conditions. In normoxic condition, ING4 inhibition retained the stimulatory effect on IL-8 and OPN expression also in the presence of HIF-1α suppression, whereas HIF-1α inhibition had no effect on IL-8 and OPN expression (Figure 4C). On the other hand, in hypoxic condition, we found that HIF-1α inhibition blunted the stimulatory effect of ING4 down-regulation on IL-8 and OPN expression (Figure 4C).
Relationship between ING4 and HPH-2 in HMCLs
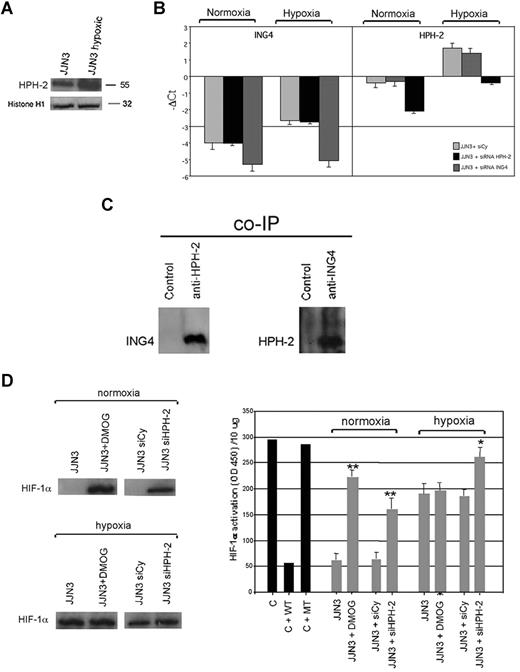
Because recent data indicate that the effect of ING4 on HIF-1α regulation could be mediated by HPH-2, we have investigated its expression and role in HMCLs. First we showed that HPH-2 is present at nuclear level in HMCLs and up-regulated by hypoxia, as shown for JJN3 (Figure 5A). In addition, we checked whether HPH-2 and ING4 expression might be reciprocally regulated and found that ING4 suppression by siRNA did not modify HPH-2 mRNA expression both in normoxic and hypoxic conditions. Similarly, HPH-2 inhibition by siRNA had no effect on ING4 mRNA expression (Figure 5B). On the other hand, we demonstrated the direct interaction between ING4 and HPH-2 at nuclear level by coimmunoprecipitation in JJN3 overexpressing ING4 (Figure 5C). The potential role of ING4/HPH-2 interaction on HIF-1α protein level and activity was also investigated. In normoxic condition, we found that the inhibition of both HPH-2 expression by siRNA and activity by DMOG up-regulated HIF-1α protein and activity (Figure 5D). In hypoxic condition, the block of HPH-2 activity by DMOG did not affect HIF-1α protein level and activity. On the other hand, similar to that observed with ING4 suppression (Figure 3B,C), the inhibition of HPH-2 expression by siRNA did not modify HIF-1α protein level but significantly increased its activity (Figure 5D) as also demonstrated by the increase of NIP-3 expression (−ΔCt NIP-3: HPH-2 siRNA vs nonspecific siRNA: −0.9 vs −2).
Relationship between ING4 and HPH-2 in JJN3. JJN3 was treated with or without hypoxic condition for 12 hours and then nuclear extracts were evaluated for HPH-2 expression by Western blot analysis. Histone H1 was used as internal control (A). JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or HPH-2 or with a nonspecific control siRNA (Cy). After 24 hours, cells were incubated in the presence or absence of hypoxic condition for 12 hours. ING4 and HPH-2 mRNA expression was quantified by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (B). Nuclear lysates of JJN3 overexpressing ING4 were immunoprecipitated with anti–HPH-2 Ab or anti-IgG control and with anti-ING4 Ab or anti-IgG control. ING4 coimmunoprecipitated with HPH-2 and HPH-2 coimmunoprecipitated with ING4 were detected by Western blot analysis using anti-ING4 and anti–HPH-2 Abs, respectively (C). JJN3 was pretreated with the HPH-2 inhibitor DMOG (0.2 mM) for 2 hours or transfected by siRNA against HPH-2 or with a nonspecific control siRNA (Cy) and then incubated with or without hypoxic condition for 12 hours. HIF-1α protein level was detected in nuclear lysates by Western blot analysis (D left). HIF-1α activity was quantified by a transcriptional factor assay kit as described in “NF-κB and HIF-1α activity.” Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (C). Graphs represent the mean plus or minus SD of HIF-1α activity normalized for 10 μg protein analyzed of 2 independent experiments performed in triplicate (OD = optical density; *P = .01; **P = .001; D right).
Relationship between ING4 and HPH-2 in JJN3. JJN3 was treated with or without hypoxic condition for 12 hours and then nuclear extracts were evaluated for HPH-2 expression by Western blot analysis. Histone H1 was used as internal control (A). JJN3 was transfected by electroporation with 1 nmol smart pool double-stranded RNA oligonucleotides (siRNA) against ING4 or HPH-2 or with a nonspecific control siRNA (Cy). After 24 hours, cells were incubated in the presence or absence of hypoxic condition for 12 hours. ING4 and HPH-2 mRNA expression was quantified by real-time PCR. Graphs represent the mean plus or minus SD of −ΔCt value (B). Nuclear lysates of JJN3 overexpressing ING4 were immunoprecipitated with anti–HPH-2 Ab or anti-IgG control and with anti-ING4 Ab or anti-IgG control. ING4 coimmunoprecipitated with HPH-2 and HPH-2 coimmunoprecipitated with ING4 were detected by Western blot analysis using anti-ING4 and anti–HPH-2 Abs, respectively (C). JJN3 was pretreated with the HPH-2 inhibitor DMOG (0.2 mM) for 2 hours or transfected by siRNA against HPH-2 or with a nonspecific control siRNA (Cy) and then incubated with or without hypoxic condition for 12 hours. HIF-1α protein level was detected in nuclear lysates by Western blot analysis (D left). HIF-1α activity was quantified by a transcriptional factor assay kit as described in “NF-κB and HIF-1α activity.” Nuclear extracts of COS-7 treated with CoCl2 in the presence or absence of wild-type (wt) or mutated (mt) competitor oligonucleotides were tested as control (C). Graphs represent the mean plus or minus SD of HIF-1α activity normalized for 10 μg protein analyzed of 2 independent experiments performed in triplicate (OD = optical density; *P = .01; **P = .001; D right).
Involvement of ING4 in myeloma-induced angiogenesis in vitro
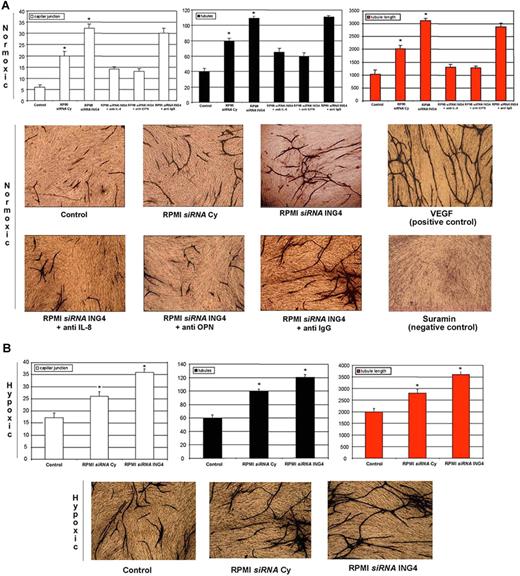
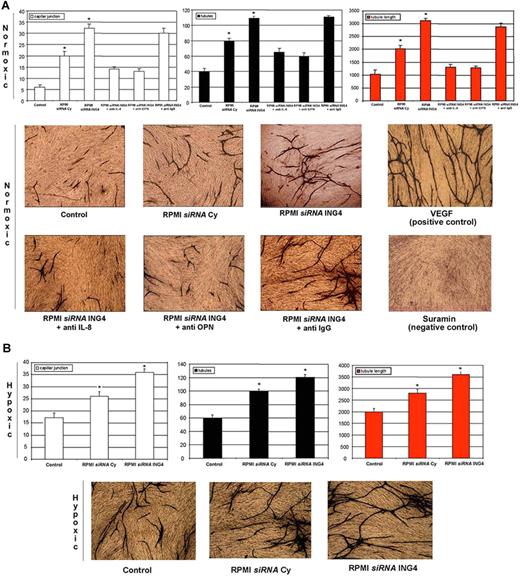
The potential role of ING4 as angiogenic repressor was tested in an experimental in vitro model of angiogenesis. In this model, we found, as expected, that CM of RPMI-8226 transfected with a specific siRNA increased vessel formation evaluated as capillary junctions, tubules, and tubule length compared with the nonconditioned medium used as control to validate the system. Consistent with the angiogenic role of ING4, we found that CM of RPMI-8226 transfected with siRNA against ING4 significantly increased vessel formation compared with CM of RPMI-8226 transfected with siRNA Cy (Figure 6A). The presence of either blocking anti–IL-8 or anti-OPN Abs but not isotypic anti-IgG Ab significantly blunted the stimulatory effect of the CM obtained from RPMI-8226 transfected with ING4 siRNA (Figure 6A).
ING4 involvement in MM-induced in vitro angiogenesis. Using an angiogenic in vitro assay (ANGIOkit TCS Biologicals, Buckingham, United Kingdom), endothelial-like cells were stimulated with the conditioned media (dilution 1:2) of RPMI-8226 previously transfected with a nonspecific control siRNA (siRNA Cy) or anti-ING4 siRNA and incubated in normoxic (A) or hypoxic (B) conditions. Blocking anti–IL-8 or anti-OPN or nonspecific anti-IgG Abs were added in the conditioned media of RPMI-8226 transfected with siRNA anti-ING4 incubated in normoxic condition. VEGF and suramin treatments were used as positive and negative controls, respectively. Every 3 days the medium was replaced, and after 13 days cells were fixed and stained with anti-CD31 Ab according to the manufacturer's protocol. (Images were obtained on a Nikon Eclipse TE300 microscope at 10×/0.13 NA using a DS-US digital slight at 4×/0.12 NA objective lens. Original magnification, 10×). Vessel formation was quantified as described in “Cells and cell culture conditions; In vitro angiogenesis assay.” Graphs on the left, middle, and right represent the mean plus or minus SD of the number of capillary junctions, tubules, and the tubule length, respectively, of 2 independent experiments tested twice (control represents D-MEM medium nonconditioned by HMCLs; *P < .05).
ING4 involvement in MM-induced in vitro angiogenesis. Using an angiogenic in vitro assay (ANGIOkit TCS Biologicals, Buckingham, United Kingdom), endothelial-like cells were stimulated with the conditioned media (dilution 1:2) of RPMI-8226 previously transfected with a nonspecific control siRNA (siRNA Cy) or anti-ING4 siRNA and incubated in normoxic (A) or hypoxic (B) conditions. Blocking anti–IL-8 or anti-OPN or nonspecific anti-IgG Abs were added in the conditioned media of RPMI-8226 transfected with siRNA anti-ING4 incubated in normoxic condition. VEGF and suramin treatments were used as positive and negative controls, respectively. Every 3 days the medium was replaced, and after 13 days cells were fixed and stained with anti-CD31 Ab according to the manufacturer's protocol. (Images were obtained on a Nikon Eclipse TE300 microscope at 10×/0.13 NA using a DS-US digital slight at 4×/0.12 NA objective lens. Original magnification, 10×). Vessel formation was quantified as described in “Cells and cell culture conditions; In vitro angiogenesis assay.” Graphs on the left, middle, and right represent the mean plus or minus SD of the number of capillary junctions, tubules, and the tubule length, respectively, of 2 independent experiments tested twice (control represents D-MEM medium nonconditioned by HMCLs; *P < .05).
Similarly, we found that CM of RPMI-8226 transfected with ING4 siRNA incubated in hypoxic condition stimulated vessel formation compared with RPMI-8226 transfected with siRNA Cy (Figure 6B).
ING4 expression in MM patients: relationship with angiogenic-related molecules and BM angiogenesis
In vitro data were extended in vivo in 50 MM patients. First, we found by real-time PCR that CD138+ MM cells had significantly reduced levels of ING4 mRNA compared with normal plasma cells (median −ΔCt ING4: −3.7 vs −0.45; P = .01) with a mean n-fold expression of 0.11 normalized to controls, whereas the difference in ING4 expression compared with MGUS subjects (median −ΔCt ING4: −2.7) did not reach a statistical significance (P = .1, NS).
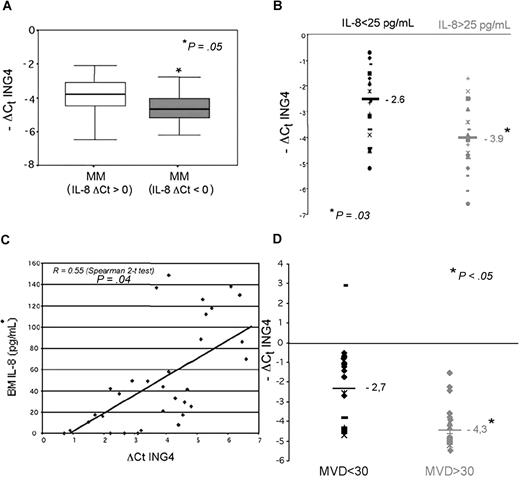
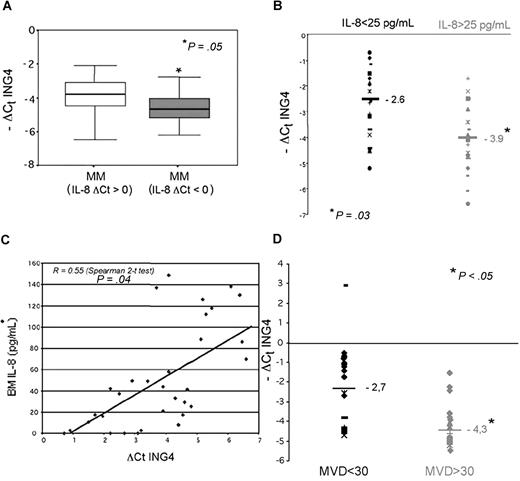
We observed a relationship between ING4 and IL-8 mRNA levels in CD138+ MM cells by quantitative PCR and found that patients with higher IL-8 mRNA levels (ΔCt IL-8: < 0) have significantly lower ING4 mRNA levels (Figure 7A). Consequently, we found that IL-8 median level was significantly higher in BM plasma of MM patients compared with control subjects (30.5 vs 25.2 pg/mL; P = .04) and that MM patients with higher BM IL-8 levels (> 25 pg/mL) have significantly lower ING4 mRNA levels (median −ΔCt ING4: −3.9 vs −2.6; P = .03; Figure 7B). A significant correlation was consistently observed between BM IL-8 plasma level and ING4 mRNA expression in purified MM cells (R = 0.55; Spearman 2-tailed test: P = .04; Figure 7C). Similarly, we found that MM patients positive for OPN mRNA and protein expression had lower ING4 levels compared with those who were negative (median −ΔCt ING4: −3.6 vs −1.8; P = .05).
ING4 expression in MM patients: relationship with IL-8 and BM angiogenesis. ING4 mRNA expression by CD138+ MM cells was evaluated in MM patients (n = 50) in relationship with IL-8 mRNA expression. Box plots represent the median −ΔCt ING4 with the 25th to 75th percentiles in MM patients with low levels of IL-8 mRNA (IL-8 ΔCt: > 0) and high IL-8 mRNA levels (IL-8 ΔCt: < 0) (A). ING4 mRNA levels in CD138+ MM were evaluated in relationship with bone marrow (BM) IL-8 plasma levels measured by ELISA in a subgroup of 35 MM patients. Plots represent the −ΔCt value of ING4 in MM patients with low and high BM IL-8 levels (B). The potential relationship between BM IL-8 levels and ΔCt ING4 values was analyzed in MM patients by the Spearman 2-tailed test analysis (C). BM angiogenesis was evaluated in bone biopsies of 28 MM patients as described in “BM angiogenesis evaluation.” Plots represent the −ΔCt value of ING4 in MM patients with low and high microvascular density (MVD) (D).
ING4 expression in MM patients: relationship with IL-8 and BM angiogenesis. ING4 mRNA expression by CD138+ MM cells was evaluated in MM patients (n = 50) in relationship with IL-8 mRNA expression. Box plots represent the median −ΔCt ING4 with the 25th to 75th percentiles in MM patients with low levels of IL-8 mRNA (IL-8 ΔCt: > 0) and high IL-8 mRNA levels (IL-8 ΔCt: < 0) (A). ING4 mRNA levels in CD138+ MM were evaluated in relationship with bone marrow (BM) IL-8 plasma levels measured by ELISA in a subgroup of 35 MM patients. Plots represent the −ΔCt value of ING4 in MM patients with low and high BM IL-8 levels (B). The potential relationship between BM IL-8 levels and ΔCt ING4 values was analyzed in MM patients by the Spearman 2-tailed test analysis (C). BM angiogenesis was evaluated in bone biopsies of 28 MM patients as described in “BM angiogenesis evaluation.” Plots represent the −ΔCt value of ING4 in MM patients with low and high microvascular density (MVD) (D).
In agreement with our previously published evidence,7–8 we considered MM patients as having high BM angiogenesis if they had a median MVD value superior to 30 and a median number of vessels per field (400 ×) superior to 6. Accordingly, we found that MM patients with high BM MVD (> 30) showed significantly lower ING4 mRNA levels in purified CD138+ MM cells compared with those with low MVD (< 30; median −ΔCt: −4.3 vs −2.7; Mann-Whitney 2-tailed: P = .04; Figure 7D). Consistently, a significant negative correlation between ING4 mRNA levels in MM cells and MVD was found in our cohort of MM patients (Pearson chi-square: P = .04; Cochran linear trend: P = .02).
Similarly, we found that MM patients with a higher number of microvessels per field (400×) have significantly lower ING4 levels (data not shown).
Discussion
In the present study, we analyzed the expression of the tumor-suppressor gene ING4 in myeloma cells and its potential relationship with the angiogenic process in MM.
ING4 is a new suppressor gene expressed in normal tissues that acts as a repressor of angiogenesis, but it is not directly involved in the regulation of the cell cycle.10,–12,20 Data obtained in several human cancer cell lines have demonstrated that ectopic expression of ING4 suppressed the loss of contact inhibition, but had no direct effect on cellular proliferation.11,20 ING4 expression is significantly reduced in tumor cells compared with normal tissues, as demonstrated in glioblastoma and in head and neck squamous cell carcinomas.10,12 In line with these observations, our study showed by real-time PCR that ING4 mRNA expression is reduced in MM cells compared with normal plasma cells. On the other hand, we failed to find a statistically significant difference in the median ING4 mRNA level of expression by CD138 plasma cells between MGUS and all MM patients even if ING4 levels were higher in MGUS versus MM patients. This observation, which could be due to the relatively small number of MGUS subjects included in the study, is not completely surprising as few genes differently expressed between MGUS and MM patients are reported.21–22 Furthermore, any significant difference in the expression of proangiogenic molecules, such as VEGF by CD138 plasma cells, was not reported between MGUS and MM patients.23 A similar behavior was observed for ING4 expression in this study even if ING4 levels were found to be higher in MGUS subjects compared with MM with high BM angiogenesis.
The mechanisms by which ING4 expression is reduced in tumor cells compared with normal ones are under investigation. Deletion of the ING4 locus in the chromosome 12p13 has been observed in 10% to 20% of breast cancer cells11 ; loss of heterozygosity in 66% of head and neck squamous cell carcinomas,12 but not in glioblastoma cells, has also been reported. Different splice variants in the nuclear localization signal (NLS) region of ING4 have also recently been shown to have a different suppressive effect on p21WAF activation.24 However, using denaturing high-performance liquid chromatography and microsatellite analysis, we failed to find mutations or loss of heterozygosity in several screened HMCLs; in addition, no significant difference was observed between normal plasma cells and MM cells in terms of ING4 NLS splicing variants (D.M. et al, unpublished data, March 2006). Moreover, a recent report has identified novel ING4 spliced variant mRNAs encoding proteins devoid of different portions in both normal and tumoral cells.25 Further studies in MM cells will be necessary to clarify if the difference in ING4 mRNA expression that we have observed between normal and malignant plasma cells could be due to the presence of these different splice isoforms.
Ongoing experiments are currently focused on the mechanisms responsible for ING4 tumor suppression. In the glioblastoma model, it has been shown that ING4 physically interacts with p65 (RelA) subunit of NF-κB–suppressing NF-κB–responsive genes.10 Similarly in our study, a correlation between ING4 and p65 NF-κB activity was demonstrated in HMCLs, confirmed by a finding that ING4 suppression in HMCLs cells stimulated NF-κB–related p65 activity. Clearly, other than the block of NF-κB activity, other mechanisms could be involved, such as the recently described capability of ING4 to bind p53 and enhance its activity.26,27
Much of the evidence indicates that ING4 exerts its growth-suppressive effect mainly through the inhibition of angiogenesis. In the mouse xenograft model, it has been shown that human glioblastoma cells, which have decreased expression of ING4, grow significantly faster and have high vascularization, suggesting that ING4 may act as a repressor of angiogenesis.10 As known in MM, an increase of BM angiogenesis has been demonstrated to be due to the capacity of MM cells to stimulate vessel formation through the production of proangiogenic factors as VEGF, ANG-1, OPN, and IL-8.6,,–9 The angiogenic switch that occurs within myeloma cell infiltration into the BM also correlates with the progression of MM,3 suggesting that vessel formation is critically involved in the pathophysiology of MM; however, the molecular mechanism underlying the angiogenic switch in MM is not completely clear. Different approaches were used in this study to investigate the potential relationship of ING4 with the proangiogenic molecules produced by MM cells. First we compared ING4 levels with those of the main angiogenic molecules produced by myeloma cells by a quantitative PCR. Then we evaluated the effect of ING4 suppression in myeloma cells on the production of proangiogenic molecules. Among several proangiogenic factors, we found that ING4 negatively correlates with IL-8 and OPN expression and regulates their production by myeloma cells; on the other hand, we failed to find a direct relationship between ING4 and VEGF or ANG-1. This observation was confirmed by the finding that ING-4 suppression in myeloma cells up-regulates both IL-8 and OPN but not VEGF and ANG-1. Similarly, it has been previously reported that ING4 suppression in glioblastoma cells increases IL-8 secretion, but had no effect on VEGF secretion. However, we failed to observe an inhibitory effect on IL-8 and OPN production in HMCLs overexpressing ING4, suggesting that ING4 is necessary but not sufficient for the regulation of these 2 molecules. Similarly, ING4 overexpression in glioblastoma cells has been reported to have a slight inhibitory effect on IL-8 production.10
IL-8 (also termed CXCL8) is a chemokine that exerts a potent angiogenic activity, binding the receptors CXCR1 and CXCR2 present on endothelial cells.28,29 Previous evidence indicates that myeloma cells, as well as BM stromal cells9,30,31 directly produce IL-8. Tumor cell expression of IL-8 has been linked to the metastatic potential in many solid tumors.28,32 In MM cells, IL-8 expression has been correlated with aberrant CD28 expression and consequently with MM progression and extramedullary localizations.9,30
Other than IL-8, we show that ING4 may be involved in the regulation of OPN production. OPN is a multifunctional bone matrix protein able to stimulate endothelial cell survival and migration, acting as a potent proangiogenic factor in vivo.33,–35 Recently, we have shown that myeloma cells produce OPN, in part through the activation of the Runx2/Cbfa1 gene, which acts as proangiogenic factor in MM patients.8 A correlation between OPN secretion and BM angiogenesis was also demonstrated.8 In this study, we found that ING4 regulates OPN production by myeloma cells, suggesting the potential involvement of the NF-κB pathway other than Runx2/Cbfa1 in OPN production. It has been recently demonstrated that the oncosuppressor gene BRMS1 consistently regulates OPN in breast cancer cell line by abrogating NF-κB activation.36
Other that IL-8 and OPN, other molecules could be regulated by ING4. By microarray analysis, we found that ING4 suppression in HMCLs also stimulated the expression of CXCL12 (SDF-1), heparin binding EGF-like growth factor (HBEGF), and the endothelial cell growth factor 1 (ECGF1), which are recognized as potent proangiogenic factors in cancer cells37,–39 and regulated at least in part through an NF-κB–dependent mechanism.40
Because it has been recently suggested that ING4 represses the activation of HIF-1α in hypoxic condition,13,41 the potential role of ING4 in the angiogenic response and HIF-1α activation has been also investigated in this study. We show that hypoxia stimulates the production of IL-8 and OPN by myeloma cells and that ING4 suppression further increases their secretion, indicating that ING4 represses these molecules also in hypoxia. The induction of IL-8 and OPN by hypoxia in myeloma cells has never been reported, however a stimulatory effect by hypoxia was demonstrated on IL-8 secretion by endothelial cells as well as on OPN by tumor cells.42,43
Within tumors, the availability of O2 is often limited, and hypoxia represents the trigger to induce tumor cells to overexpress the proangiogenic molecules.44 Low O2 is normally present in the BM, indicating that MM cells in the early phase of the disease grow in hypoxic microenvironment that triggers the angiogenic switch and the production of angiogenic molecules that precedes the progression of the disease.45 HIF-1α is highly regulated by hypoxia and acts as master regulator of hypoxia-responsive genes in cancer.46,47 Many genes are regulated by HIF-1α, including proangiogenic molecules such as VEGF and angiopoietins, as well as IL-8 and OPN, linking HIF-1α activation to the angiogenic process.44 In turn, it has been demonstrated that HIF-1α induction or overexpression in several tumor cell lines may correlate with the tumor progression.46,47 Our data indicate that HIF-1α is induced by hypoxia in myeloma cells and that ING4 has a suppressive effect on HIF-1α as demonstrated by the finding that the block of ING4 in myeloma cells increases HIF-1α activity and its target gene NIP-3 expression. Interestingly, we demonstrate that HIF-1α regulates the angiogenic-related molecule expression because the suppression of HIF-1α in hypoxic condition significantly reduced IL-8 and OPN production by MM cells and blunted the stimulatory effect of ING4 on IL-8 and OPN production, suggesting that HIF-1α activation was critical in the regulation of IL-8 and OPN by ING4 in hypoxic condition.
The potential mechanisms involved in ING4 regulation of HIF-1α system in hypoxia were also investigated, showing that ING4 directly interacts with the HIF-1α–regulating key molecule HPH-2. As known, HPH-2, a HIF prolyl hydroxylase enzyme, is responsible for the O2-dependent hydroxylation of HIF-1α, leading to its proteosomal degradation in normoxic condition.41,48,49 On the other hand, in hypoxia HPH-2 is up-regulated and regulates HIF-1α activity rather than its stability.41,48,49 Consistently, we found that the inhibition of HPH-2 expression by siRNA, rather than the inhibition of its hydroxylation activity by DMOG, increased HIF-1α activity but not its stability. Recent data suggest that the HPH-2 effect on HIF-1α activity in hypoxia could be due to the recruitment of ING4.13,41 In line with this hypothesis, we show that both ING4 and HPH-2 are up-regulated under hypoxia in HMCLs and directly associated, suggesting that both molecules are necessary in the negative regulation of HIF-1α activity. This hypothesis was also confirmed by the finding that the single suppression of ING4 or HPH-2 by siRNA was sufficient for the loss of their inhibitory effect on HIF-1α activity. Consistently, the single overexpression of ING4 in HMCLs did not lead to a further increase of their suppressive regulation of HIF-1α.
The involvement of ING4 as an angiogenic repressor in MM was further confirmed in an established in vitro angiogenic model showing that the CM of MM cells with suppressed ING4 levels significantly stimulates vessel formation in normoxic and hypoxic condition. This stimulatory effect was blunted, in normoxic condition, by blocking anti-OPN or anti–IL-8 Abs, confirming that the repressive effect of ING4 on MM-induced angiogenesis was mediated by the regulation of IL-8 and OPN production.
Finally, our in vitro evidence was supported in vivo in a cohort of MM patients, in which a negative correlation between ING4 and either the related proangiogenic molecules or BM angiogenesis was found. In line with this evidence, we have previously reported that a significant correlation exists between OPN expression and BM angiogenesis in MM patients.8 These observations support the hypothesis that ING4 may act as angiogenic repressor also in vivo, as previously demonstrated in glioblastoma,10 suggesting that a reduced ING4 expression in myeloma cells may contribute to the increased BM angiogenesis that occurs in MM patients. The link between ING4 and angiogenesis in MM patients suggests that this oncosuppressor gene may be potentially involved in the progression of MM.
In conclusion, our data indicate that the tumor-suppressor gene ING4, as reported for other tumors, is involved in the angiogenic switch and potentially in MM progression exerting an inhibitory effect on the production of the proangiogenic molecules IL-8, OPN, and HIF-1α in hypoxic condition. This evidence supports the hypothesis that ING4 acts as a suppressor of myeloma-induced angiogenesis and that it is involved in the pathophysiology of MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Associazione Italiana Contro le Leucemie (AIL) Parma section.
We thank Alessandra Leporati, Gennari Dirce, and Zanardi Giuliana for the technical support.
Authorship
Contribution: S.C. provided molecular biology experiments, lentiviral infection of myeloma cells, microarray experiments and analysis, and cell cultures, and provided critical review of the manuscript; S.T. performed real-time PCR experiments and contributed to molecular biology experiments; F.M. performed cell culture experiments, MM cell purification, ELISA, and Western blot; P.L. performed siRNA transfection experiments; G.D. contributed to the construction of the lentiviral vectors and cell culture experiments; D.M. contributed to the molecular biology experiments and lentiviral vectors; C. Mancini evaluated bone marrow angiogenesis; M.L. performed immunohistochemistry on bone biopsies; L.M. contributed to siRNA transfection; L.R. contributed to molecular biology experiments; S.B. contributed to the characterization and purification of MM patients; L.F. contributed to performing Western blot analysis; C. Miranda contributed to the preparation of lentiviral vectors and molecular biology experiments; M.L. provided the critical revision of the paper and the analysis of real-time quantitative PCR; T.M.N. contributed to molecular biology experiments; A.N. provided a critical revision of the paper; A.G. contributed to the preparation of lentiviral vectors; M.M. provided vital reagents and recruited MM patients; A.B. contributed to the transfection experiments by siRNA; V.R. provided vital reagents, recruited MM patients, and contributed to the critical revision of the paper; N.G. designed the research, performed cell culture experiments, provided the recruitment of MM patients, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicola Giuliani, Chair of Hematology and BMT Center, Department of Internal Medicine and Biomedical Science, University of Parma, via Gramsci 14, 43100 Parma, Italy; e-mail:nicola.giuliani@unipr.it.