Abstract
With the goal of creating a resource for in-depth study of myelopoiesis, we have executed a 2-pronged strategy to obtain a complementary DNA (cDNA) clone set enriched in hematopoietic genes. One aspect is a library subtraction to enrich for underrepresented transcripts present at early stages of hematopoiesis. For this, a hematopoietic cDNA library from primary murine bone marrow cells enriched for primitive progenitors was used as tester. The subtraction used 10 000 known genes and expressed sequence tags (ESTs) as driver. The 2304 randomly picked clones from the subtracted cDNA libraries represent 1255 distinct genes, of which 622 (50%) are named genes, 386 (30%) match uncharacterized ESTs, and 247 (20%) are novel. The second aspect of our strategy was to complement this subtracted library with genes known to be involved in myeloid cell differentiation and function. The resulting cDNAs were arrayed on polylysine-coated glass slides. The microarrays were used to analyze gene expression in primary and cultured murine bone marrow–derived progenitors. We found expression of various types of genes, including regulatory cytokines and their receptors, signal transduction genes, and transcription factors. To assess gene expression during myeloid differentiation, we examined patterns of change during induced differentiation of EML cells. Several hundred of the genes underwent fluctuations in expression level during myeloid cell differentiation. The complete database, accessible on the World Wide Web at http://yale130132115135.med.yale.edu/, allows for retrieval of information regarding these genes. Our microarray allows for genomewide expression analysis of myeloid stem cells, which will help in defining the regulatory mechanisms of stem cell differentiation.
Introduction
Acute myeloid leukemia (AML) remains a highly lethal malignancy requiring novel therapeutic strategies.1,2 An integral component of the AML phenotype is the loss of the capacity to differentiate into mature myeloid cells. Consequently, a major focus of research in this area has been on the molecular mechanisms controlling normal myeloid differentiation. One conclusion of this work is that differential expression of key regulatory genes in hematopoietic stem cells controls their differentiation into mature cell types, including erythrocytes, platelets, neutrophils, monocytes, eosinophils, and basophils. A detailed understanding of the gene expression patterns throughout hematopoietic differentiation obtained by means of messenger RNA (mRNA)–expression profiling and bioinformatics can provide valuable insights into this complex process and will perhaps lead to novel treatment approaches for AML. We are interested in the patterns of gene expression at early stages of myeloid commitment and differentiation. Previous studies have identified a small but diverse group of genes that are down-regulated during this process, includingCD34,3ckit,4Jagged2,5mpl,6sca-1,7SCL,8GATA-1 and GATA-2,8Flt-1,9Notch,10Ap-1,11Mzf-1,12C/ebp,13 andSTATs.14 Up-regulated genes include Pu.115 and others.16However, there are likely to be many more genes, some known and some yet to be identified, involved in the molecular events of differentiation.17 18
To better understand the interacting pathways and networks involved in hematopoiesis, we decided to employ the genomewide strategy of gene expression profiling using complementary DNA (cDNA) microarrays.19 A requisite component of this technology is inclusion of potentially critical genes on the array. With the aim of developing a cDNA microarray for use in the study of gene expression during early myelopoiesis, we constructed a subtracted cDNA library derived from sorted hematopoietic progenitor cells, and we complemented this set of genes with a set of available clones known to be important to myelopoiesis. This clone set was sequenced, characterized, and then spotted onto glass slides to create a microarray for analyzing the profile of gene expression during early steps in myelopoiesis. We have employed this microarray to assess expression in primary hematopoietic precursors and to analyze changes in gene expression during induced differentiation of the myeloid progenitor cell line EML.
Materials and methods
Reagents
The α-[32P]deoxycytidine 5′triphosphate (α-[32P]dCTP) (3000 Ci/mmol [111 TBq/mmol]) was purchased from Amersham Pharmacia Biotech (Buckinghamshire, United Kingdom), and restriction enzymes were purchased from New England Biolab (Beverly, MA). Iscoves modified Dulbecco medium (IMDM) (Life Technologies, Rockville, MD) supplemented with 20% horse serum was the culture medium used throughout. Recombinant human stem cell factor (SCF) was purchased from Peprotech (Rocky Hills, NJ). Rhodamine123 and Hoechst 33342 dyes were from Molecular Probes (Eugene, OR). Plasmid vectors λ ZipLox and pSport were from Life Technologies (Rockville, MD).
Cell lines
The EML and EPRO cell lines20 were generously contributed by S. Tsai. EML cells were maintained in IMDM supplemented with 20% horse serum, 15% BHK/MKL–conditioned medium (containing SCF), 1% l-glutamine, 1% penicillin/streptomycin/amphotericin (P/S/A), and 1% nonessential amino acids. EPRO cells were maintained in IMDM supplemented with 20% horse serum, 10% HM-5–conditioned medium (containing granulocyte-macrophage colony-stimulating factor), 1%l-glutamine, 1% P/S/A. Cell lines were cultured at 37°C in 5% CO2. EML cells were induced to differentiate into myeloid cells with 10 μM all trans-retinoic acid (ATRA) and 5% WEHI-conditioned medium as a source of interleukin-3 (IL-3).
cDNA library for subtraction
The lineage−rhodamineLowHoechstLow(Lin−rhodamineLowHoechstLow) (LRH) library was derived by means of previously described techniques,21,22 and its construction has been described.23 Briefly, primary bone marrow cells were depleted of lineage-committed cells and then further enriched for primitive cells by fluorescent-activated cell sorting for cells with low-level staining with rhodamine123 and Hoechst 33342 dyes. From 30 mice, 5000 cells were obtained, from which a directionally cloned cDNA library was created in the lambda vector λ ZipLox (Gibco BRL) asSalI-EagI fragments, in such a way that the 5′ end of the cDNA was adjacent to the SalI site and the 3′ end was adjacent to the EagI site. The original library had an initial plating complexity of 1.44 × 107clones.23
The LRH library was converted to single-stranded DNA (ssDNA) by in vivo excision via cre-mediated excision and filamentous phage rescue as described.24 Briefly, we electroporated 50 ng library DNA into competent Escherichia coli DH5 alpha F′ bacteria, F′ φ80 ΔlacZΔM15 Δ(lacZYA-argF) U169deoR recA1 endA1 hsdR17 (r, m) phoA supE44λ-thi-1 gyrA96relA1. We incubated the transformed bacteria in 100 mL 2 × YT broth at 30°C on an orbital shaker for 1 hour and then added 150 μL ampicillin (50 mg/mL) and 1 mL 20% glucose. Bacteria were grown overnight until OD600 = 0.1 (OD indicates optical density), followed by incubation at 37°C for 1 hour until OD600 = 0.2. The culture was superinfected with M13 KO7 helper phage and then cultured an additional 2 hours. After eliminating bacteria by centrifugation filamentous phage particles were precipitated with the addition of 4 g polyethylene glycol and 2.92 g NaCl into 100 mL solution and incubation at 4°C for 16 hours. Particles were collected by centrifugation; the phage DNA was purified with phenol-chloroform extraction; and the final product was dissolved in TE (10 mM Tris-HCl pH 7.9, 1 m MEDTA). The ssDNA was confirmed by digestion with mung-bean nuclease. This yielded approximately 25 mg single-stranded phage DNA composing the entire LRH library of cDNAs.
Prior to hybridization, the single-stranded library DNA was purified by means of hydroxyapatite (HAP) column chromatography to eliminate double-stranded DNAs (dsDNAs). First, 10 μg library ssDNA was digested with PvuII, and then it was applied to a 10-mL HAP column. The flowthrough (7 to 8 mL), representing the ssDNA, was collected and concentrated by means of Qiagen (Valencia, CA) spin columns following the manufacturer's protocol. DNA eluted from the Qiagen spin column was precipitated and resuspended in 5 μL double-distilled water (ddW). The ssDNA was confirmed by digestion with mung-bean nuclease.
Preparation of driver DNA
The DNA driver pool was prepared with 10 000 mouse cDNA clones that were a gift from Research Genetics (Huntsville, AL). These clones were derived from mouse testes, kidney, diaphragm, skin, lung, brain, heart, and whole embryonic fetus; mouse melanoma; embryonic carcinoma; and mouse macrophages. The inserts were amplified by polymerase chain reaction (PCR) with Expand high-fidelity PCR system (Invitrogen, Carlsbad, CA) under the following conditions: 94°C, 7 min for 1 cycle; 20 cycles at 94°C for 1 minute, 55°C for 2 minutes, and 72°C for 3 minutes; and a final extension of 7 minutes at 72°C. The PCR products were purified by phenol-chloroform extraction and checked by ethidium bromide–stained agarose gel electrophoresis. All inserts were combined to make the driver pool by transfer of 2 μL from each PCR product.
Subtraction of cDNA library
Subtraction of the LRH library was performed essentially as described by Bonaldo et al24 with minor modifications. The hybridizations of library ssDNA and pooled driver DNA were performed in 20 μL volume hybridization buffer (50% formamide, 0.12M NaCl, and 1% sodium dodecyl sulfate [SDS]) with 2.5 μg driver DNA and 50 ng tracer ssDNA from cDNA library at 30°C for 110.4 hours (Cot = 50; here, Co is substrate concentration of total DNA in solution, and t is hybridization time at 30°C). To block hybridization via the vector and poly(adenylic acid) (poly[A]) tail sequences, blocking oligonucleotides were designed (Table 1) and were included in the hybridization at a concentration of 2 μg/μL for blocking vector homology sequence and 0.5 μg/μL for blocking poly(A) tail sequence. Following hybridization, DNA molecules remaining single stranded were purified by HAP chromatography and were concentrated to 11 μL volume as described above. The ssDNA was converted into dsDNA in vitro by transferring the ssDNA into premixed reaction solution, (5 μL sequenase buffer [5×] and 1 μL M13 forward primer [1 μg/mL]), heating at 65°C for 5 minutes, then 37°C for 3 minutes, adding 2 μL deoxynucleoside 5′-triphosphates (10 mM each), 1 μL dithiothreitol (DTT) (0.1 M), and 1 μL sequenase (5 U/μL) into reaction solution, incubating at 37°C for 30 minutes, and then purifying the dsDNA with phenol-chloroform extraction.
The resultant dsDNA was transformed into E coli DH10α, which was then plated on Luria-Bertani broth/ampicillin agar plates. The total number of clones were calculated. An aliquot of the subtracted LRH library was submitted to Lawrence Livermore National Laboratory (Livermore, CA) for transformation, plating, and robotic picking of colonies into 96-well plates as part of the Cancer Genome Anatomy Project (National Institutes of Health, Bethesda, MD). A separate aliquot of the libraries was amplified as a population and used to prepare DNA.
DNA preparation and sequencing
Plasmid DNA was prepared in 96-well plates. The clones derived from the subtracted cDNA libraries were grown in 96-well plates, and plasmids were isolated by the alkaline-lysis method. The final plasmid was dissolved in 100 μL ddW. (Note: for sequencing purposes, we used 20 μL plasmid directly, and for arraying, we further purified the plasmid DNA with a 96-well filter plate. The picked clones were sequenced with single-pass automated sequences by the W. M. Keck Facility at Yale University (New Haven, CT) and/or the Genome Sequencing Center at Washington University Medical School (St Louis, MO) with the use of an M13AEK forward primer (5′ CAA AAG GGT CAG TGC TG 3′), which primes synthesis at the 3′ of clones. Some clones were also sequenced from the 5′ end with the use of the T7 promoter primer (5′ TAA TAC GAC TCA CTA TAG GG 3′). The M13/pUC reverse primer (AGC GGA TAA CAA TTT CAC ACA GGA) for 5′ termini was used to confirm LRH novel sequences.
Sequence editing and analysis
Because some sequencing primers contained common vector sequence, we first removed vector sequences from the sequences with CodonCode-Cross_Match software (http://www.codoncode.com). FASTA formatted DNA sequences were compared with known nucleotide sequences with the use of the Blast algorithm in batches of 3228 sequences and the use of the blastall program (BLASTN and BLASTX programs25) installed in a Dell Workstation with a Linux operating system. Three publicly accessible databases were searched: Genbank nonredundant (nr) nucleotide, database for expressed sequence tags (dbESTs), and Genbank nr protein. Internal redundancy within our clone set was determined by comparison of each sequence against our own database. Categorization of sequence homology was based on the following criteria: exact match to known named mouse genes (threshold score exceeding 200) or protein, or near-identity to a known gene or protein from a species other than mouse (usually either human or rat); EST only (no extensive homology to any published or characterized protein, but identity to ESTs from mouse, rat, or human); or novel (no extensive homology to any nucleotide or protein sequence in these public databases). Sequence data from 5′ and 3′ sequence reads were assembled with the use of the PHRAP software package (http://www.phrap.org/) kindly provided by Phil Green (Washington University). Protein motifs within the assembled sequences were identified by converting the DNA sequence to open reading frame using the ORF analysis program (http://curagen.com/) (CuraGen, New Haven, CT) and then performing domain searches with Pfam, ProDom, Prosite, and Prints software programs (http://curagen.com/) (CuraGen). Cutoff parameters for match selection wereP < .05; identities exceeded 40%, and positives exceeded 50%.
Southern hybridization
Five micrograms of library DNA was double-digested with restriction enzymes BamHI and EcoR1, fractionated on 0.8% agarose gel, and transferred to nylon membranes.29 Hybridization probe DNAs were cut with restriction enzyme, gel-purified, and labeled with random primer DNA labeling. The labeled probes were purified with Sephadex G-50 Quick Spin Column (Boehringer Mannheim, Germany), and Southern blot analysis was performed according to standard methods.
Preparation of DNA samples for arraying
Bacterial cultures were grown overnight in 96-well culture plates (Qiagen), and plasmid DNA was prepared as described above. The cDNA inserts were amplified by means of PCR (96-Well GeneAmp PCR System 9700) (Perkin Elmer–Applied Biosystems, Foster City, CA) in 96-well plates with the use of M13 AEK forward and reverse primers (1 μM) for amplification. The PCR reaction was carried out in 100 μL solution of 1 mM deoxyadenosine 5′-triphosphate (dATP), dCTP, deoxyguanosine 5′-triphosphate (dGTP), and deoxyribothymidine 5′-triphosphate (dTTP); 1.5 mM MgCl2; and 2.5 U Taq polymerase in 96-well plate with the following cycles: 5 cycles of 94°C for 50 seconds, 55°C for 1 minute, and 72°C for 1.5 minutes; followed by 30 cycles of 94°C for 30 seconds, 56°C for 1 minute, and 72°C for 1.5 minutes; and then 1 cycle of 72°C for 10 minutes. Resulting PCR products were purified with the use of a 96-well glass-fiber filter (MAFB NOB) (Millipore, Bedford, MS) according to the manufacturer's user manual. The purity and yield were approximated by running the purified PCR products on a 0.8% agarose gel. The DNAs were prepared for arraying by transferring 5 μL to 384-well plates and adding SSC to a final concentration of 3 ×. Glass slides were prepared for printing and arrayed by the Yale Microarray Facility (http://info.med.yale.edu/wmkeck/dna_arrays.htm) with the use of a GeneMachines (San Carlos, CA) Omnigene Arrayer. After printing, the slides were postprocessed as described by P. Brown and J. DeRisi (http://www.microarrays.org.protocols.html).
RNA preparation, probe labeling, and hybridization
Total RNA was prepared by means of Trizol reagent (Life Technologies). Microarray slide hybridization was performed as follows. The cDNA probes were synthesized by reverse transcription with oligo deoxythymidine (dT) as primer, incorporating allyl amine-deoxyuridine triphosphate (aa-dUTP) (Sigma, St Louis, MO) into synthesized cDNA. Reactions were performed in 100 μL reaction with the following final concentrations: 85.8 μg/mL oligo dT primer; 0.5 mM each dATP, dCTP, and dGTP; 0.2 mM aa-dUTP; 0.3 mM dTTP; 10 mM DTT; and 1280 U MLV reverse transcriptase (SuperscriptII) per milliliter. Coupling of cyanine-3 (cy-3) or cy-5 dyes to aa-dU–modified cDNAs was done with the use of NHS-ester cy-3 or cy-5 dye (Pharmacia, Piscataway, NJ) by incubation at 25°C for 1 hour in subdued light. Hybridization of fluorescently labeled probes to glass slides was performed with hybridization buffer (50% deionized formamide, 12.5% SSPE, 0.625% SDS, 1.5 × Denhardt reagent with blockers [0.5 μg/μL mouse Cot 1 DNA, 0.1 μg/μL poly(A) (15A), and 0.2 μg/μL yeast transfer RNA] at 42°C for 18 to 24 hours. After hybridization, the slides were washed first with 1 × SSC, 0.1% SDS, at 25°C for 15 minutes, then with 0.2 × SSC, 0.1% SDS, and finally with 0.2 × SSC. The slides were scanned with a GSI Lumonics (Packard, Billerica, MA) or Axon (Axon Instruments, Union City, CA) laser scanner. Analysis of the fluorescent hybridization signal of microarray slide was performed with Quantarray (Packard) or Genepix software (Axon Instruments, Union City, CA), and the data were analyzed by means of Microsoft Excel. Further clustering of data was performed with the use of Genespring software (Silicon Genetics, Redwood City, CA).
Northern blot analysis
Northern hybridization was carried out following standard methods. Total RNA (10 μg) was electrophoresed on a 1% agarose/formaldehyde gel and was blotted onto Hybond-N nylon membranes (Amersham Pharmacia Biotech) followed by UV cross-linking. DNA probes were labeled with random primers, and the hybridization was performed at 65°C for 16 hours. Signals on the washed filter were visualized by autoradiography.
Results
Creation of a subtracted myeloid cDNA library that is enriched for low-abundance transcripts
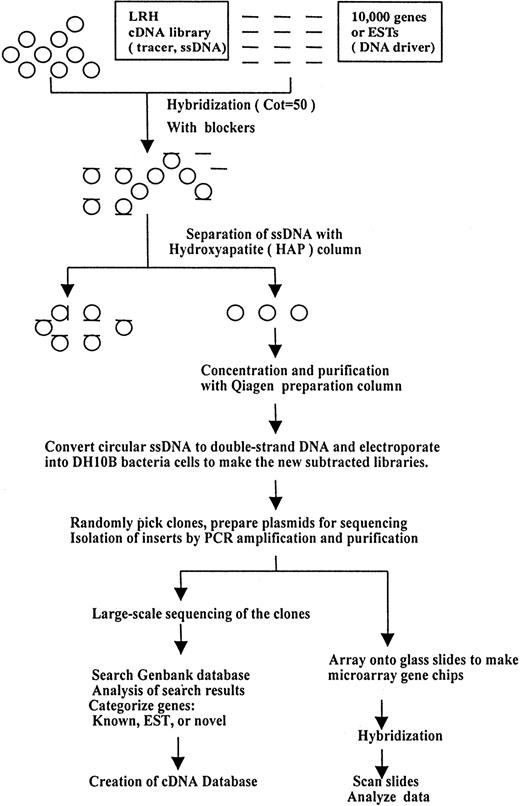
With the long-term goal of fully characterizing changes in gene expression during the early stages of myelopoiesis, we wanted to develop a cDNA microarray that was enriched in genes expressed in primitive hematopoietic cells and early committed myeloid cells. We took a 2-pronged approach to achieve this: one prong was to create a subtracted cDNA library from an early hematopoietic library; the second was to complement this set with available genes known to be involved in myelopoiesis. As starting points for the library subtractions, we used cDNA library LRH, derived from primary bone marrow samples that were sorted for early progenitors by flow cytometry (Degar et al23). The initial complexity of the LRH cDNA library was 1.44 × 107 clones. Subtraction of the libraries was performed by using as driver a pool of 10 000 mouse Integrated Molecular Analysis of Genomes and Their Expression (IMAGE) Consortium cDNA clones that were derived from several different mouse organ cDNA libraries (Figure 1). Following subtraction, we obtained 1 × 106 total clones; the complexity within this population is not known.
Schematic for production of the normalized hematopoietic progenitor cDNA library and construction of microarray gene chips.
At top, single-strand circles represent the starting cDNA library produced by means of filamentous phage rescue. The ssDNA was hybridized with driver (10 000 IMAGE consortium cDNAs) with appropriate blocking oligonucleotides. The fraction that remains single stranded (flowthrough from HAP column) was converted to double-strand circles, electroporated into DH10Bα, and propagated under ampicillin selection to generate an amplified normalized cDNA library. Large-scale sequencing of clones was performed with the use of the M13AEK forward primer. To make the myeloid-specific gene chips, the sequenced cDNA clones were amplified by PCR, purified, and printed onto polylysine-coated glass slides.
Schematic for production of the normalized hematopoietic progenitor cDNA library and construction of microarray gene chips.
At top, single-strand circles represent the starting cDNA library produced by means of filamentous phage rescue. The ssDNA was hybridized with driver (10 000 IMAGE consortium cDNAs) with appropriate blocking oligonucleotides. The fraction that remains single stranded (flowthrough from HAP column) was converted to double-strand circles, electroporated into DH10Bα, and propagated under ampicillin selection to generate an amplified normalized cDNA library. Large-scale sequencing of clones was performed with the use of the M13AEK forward primer. To make the myeloid-specific gene chips, the sequenced cDNA clones were amplified by PCR, purified, and printed onto polylysine-coated glass slides.
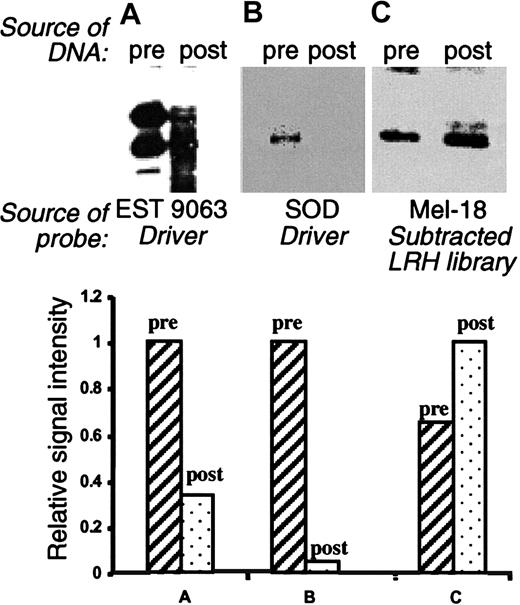
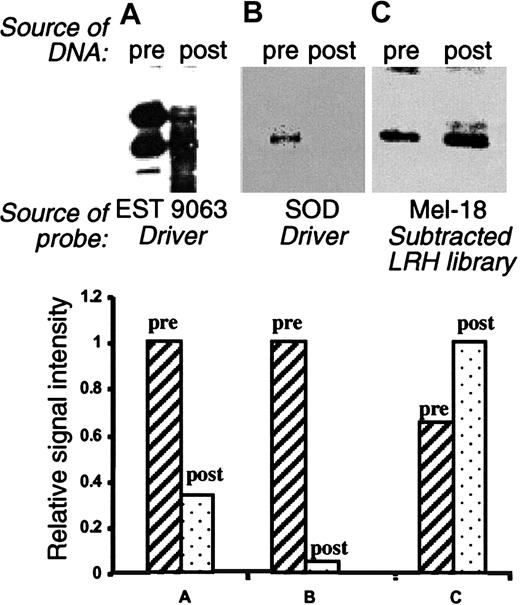
To assess the efficacy of the subtraction process, we performed Southern blot analysis of library-derived cDNA populations derived before and after normalization. As hybridization probes, we used 3 different sequences known to be present in both the driver and the tracer populations. The results (Figure2) clearly indicate that the subtraction was effective in greatly reducing the abundance of these clones, but the degree of reduction for these genes was variable. For example, clone ID9063 (IMAGE: 421622) was reduced around 3-fold with subtraction, but superoxide dismutase precursor was reduced more than 20-fold (Figure 2). We also tested for enrichment of genes present only in the tracer population, not in the driver. In this instance, some low-copy genes were enriched more than 1.5- to 5-fold through hybridization (eg, Mel-18; Figure 2C).
Southern blot analysis of subtracted LRH cDNA library.
Pooled plasmids from the presubtracted (pre) and postsubtracted (post) libraries were double-digested with BamHI/EcoR1 and analyzed by Southern blot. The probes were as follows: EST 9063, derived from the driver pool (panel A); superoxide dismutase (SOD), derived from the driver pool (panel B); and Mel-18, derived from subtracted LRH cDNA library (panel C). The bar graphs present quantitation of the data, revealing the effective elimination of the representative driver sequences and enrichment of the selected clones not present in the driver.
Southern blot analysis of subtracted LRH cDNA library.
Pooled plasmids from the presubtracted (pre) and postsubtracted (post) libraries were double-digested with BamHI/EcoR1 and analyzed by Southern blot. The probes were as follows: EST 9063, derived from the driver pool (panel A); superoxide dismutase (SOD), derived from the driver pool (panel B); and Mel-18, derived from subtracted LRH cDNA library (panel C). The bar graphs present quantitation of the data, revealing the effective elimination of the representative driver sequences and enrichment of the selected clones not present in the driver.
The subtracted LRH cDNA library contains a high percentage of novel sequences
To determine the identity of the cDNA clones derived from the library subtraction, we subjected 2304 LRH clones to partial sequence determination and analysis (Table2). Of the clones, 54% (1255) were nonredundant cDNAs, representing protein-encoding mRNAs. Of these, 247 (20%) were novel sequences; 386 (31%) ESTs; and 622 (50%) known genes. Of the LRH sequences, 46% were not useful, the majority because of being redundant, ribosomal, or empty vector. Sequence data for all of the novel genes have been submitted to GenBank (dbEST).
To facilitate the analysis, retrieval, and further accrual of information concerning these genes, we created a database that is accessible via the World Wide Web (http://yale130132115135.med.yale.edu/).
Examples of the known genes derived from the subtracted cDNA library are shown in Table 3, categorized by the functional criteria. Of interest is the presence of 3 members of theC/ebp family of transcription factors, as well as Cbfb, Klf9, Lrf, Sox4, Tal1, and Xbp1,each of which is an important regulator of cell differentiation of blood cell lineages and/or other organs.26 It is also notable that while genes for growth factors (eg, Hdgf, Hegfl, Efnb1) and growth factor receptors (eg,Fgfr1, Tnfrsf1b, Oprs1) are present, none of the classical hematopoietic-specific cytokines or their receptors is present in our subtracted library. Components of apoptotic pathways are represented byTnfrsf1b, Traf1, Traf6,Prg2, Tax1bp1, Bnip3l, andCasp6. Calcium signaling transducers are included, eg, Calm, Calr, Cmkk2, and Itpr3. Also present are regulators of the cell cycle, including Ccnd1, Hus1, andLats.
Protein structure analysis of novel genes
We identified 247 novel sequences among the subtracted LRH clones. These clones are considered novel because of our inability to find any matching sequence in available databases. For each of the potentially novel genes, we subjected clones to additional sequencing from both the 5′ and 3′ ends of the clones. After compiling the 5′ and 3′ sequence data, we derived potential open reading frames from these sequences and analyzed them for domains and/or functional motifs (Table 4). This revealed that our novel sequences contained 13 potential nucleic acid–binding proteins, including 4 transcription factors, 11 signal transducers including 2 with similarity to Jak3, 1 with homolgy to Flt3 ligand, 1 bearing resemblance to the insulin receptor, and 1 mapk/erk kinase kinase–like protein. Sixteen proteins with similarities to known enzymes or enzyme inhibitors were identified, including some potential drug targets (eg, farnesyltransferase, prenyltransferase, and adenylate cyclase). What is notable is the relative paucity of more structural proteins (Table 4). In Figure 3, we show detailed analyses of 9 potentially important novel genes and their homology to known proteins.
Alignments of important motifs for 9 proteins identified in hematopoietic stem cells.
(A) Clone 1317 with 3 closely related WEE1 kinase protein family members, which play an important role in mitosis. (B) Clone 1508 with 5 closely related members of the Zinc-finger protein CNBP DNA-binding family, which play a role in gene transcription. (C) Clone 2265 with 6 GLI protein family members, all of which are Zinc-finger DNA-binding proteins. (D) Clone 1405 with 2 C–ETA type protein kinases. (E) Clone 1797 with 5 closed relative insulin/IGF/relaxin family members, which play a role in insulin expression and distribution. (F) Clone 2131 with 6 GPR1/FUN34/yaaH family members. (G) Clone 2995 with a region of homology to MEKK1 proteins. (H) (I) Regions of similarity between 2 clones (3001 and 2858) and the Jak3 protein.
Alignments of important motifs for 9 proteins identified in hematopoietic stem cells.
(A) Clone 1317 with 3 closely related WEE1 kinase protein family members, which play an important role in mitosis. (B) Clone 1508 with 5 closely related members of the Zinc-finger protein CNBP DNA-binding family, which play a role in gene transcription. (C) Clone 2265 with 6 GLI protein family members, all of which are Zinc-finger DNA-binding proteins. (D) Clone 1405 with 2 C–ETA type protein kinases. (E) Clone 1797 with 5 closed relative insulin/IGF/relaxin family members, which play a role in insulin expression and distribution. (F) Clone 2131 with 6 GPR1/FUN34/yaaH family members. (G) Clone 2995 with a region of homology to MEKK1 proteins. (H) (I) Regions of similarity between 2 clones (3001 and 2858) and the Jak3 protein.
Development of a cDNA microarray for analysis of early hematopoiesis
A major goal of this endeavor was to create a cDNA microarray for evaluating gene expression changes during hematopoietic differentiation with specific interest in the myeloid lineage. Thus, the second prong of our approach was to supplement the subtracted library with genes known to be expressed in myeloid cells as well as genes encoding proteins that regulate cell cycle, apoptosis, differentiation, and cell signaling. Thus, we added 587 cDNAs for known genes from an IMAGE Consortium clone set, 310 genes from EML cells isolated by 2 separate subtractive cloning procedures,36 96 putative Evi-1 target genes,27 and 576 T-cell–expressed genes (B. Lu, S. Kim, and R. A. Flavell, unpublished data, 2001) (Table5). A significant number of cytokines, hematopoietic transcription factors, growth factors, and growth factor receptors are also on the array (Table6). A detailed description of these cDNAs and their sources can be found on the Web-accessible database mentioned earlier (http://yale130132115135.med.yale.edu/). Purified PCR-amplified cDNA inserts from this collection of plasmids were robotically spotted on polylysine-coated glass slides.
Myeloid cell differentiation is accompanied by abundant fluctuations in gene expression
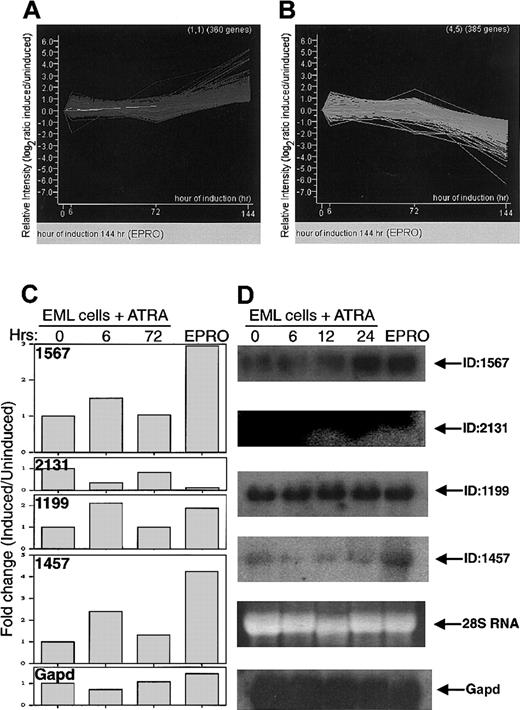
We tested the cDNA microarray by employing it to analyze patterns of gene expression during induced differentiation of EML cells, a myeloid progenitor cell line.20 We compared the spectrum of gene expression in uninduced EML cells to that of EML cells induced to differentiate for 6, 24, and 72 hours, as well as to that of EPRO cells, which represent a promyelocytelike stage derived from the EML cells.28 In each experiment, a competitive hybridization was performed between labeled cDNA from uninduced EML cell and from induced EML or EPRO cells, except for the 24-hour time point samples, for which some hybridizations were not competitive. Fluorescently labeled cDNA samples (with either cy-3 or cy-5) (“Materials and methods”) were hybridized. Following washing of the slide, the amount of hybridized probe was quantitated as pixel intensity of fluorescence; low-intensity signals were discarded; and the normalized data were expressed as a log2 of the ratio of signal from induced RNA to uninduced RNA cells. These values for EML cells induced for 6, 24, and 72 hours, and for EPRO cells were subjected to clustering by means of a self-organizing map algorithm. This yielded 20 different sets of genes, each of which contained genes that varied in expression level in a similar manner across the samples. Figure4A-B shows composite graphs for the sets containing genes that increased the most (Figure 4A) or decreased the most (Figure 4B) over the time points. Tables7 and 8list the named genes in each of these sets, respectively.
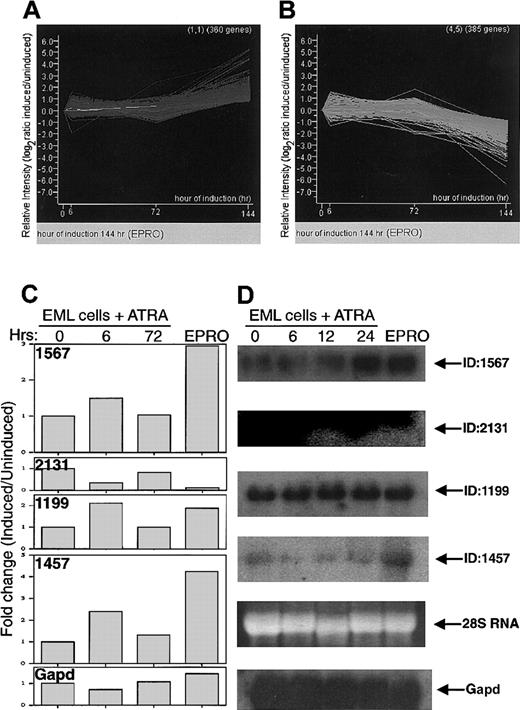
Graphical representation of the change in gene expression over time in EML cells treated with ATRA/IL-3.
(A) (B) Each line represents a different gene. The time points (x-axis) represent 0, 6, 24, and 72 hours. The last point on this axis represents EPRO cells, a stage in differentiation beyond 72 hours. The y-axis shows the relative change in gene expression level as log2 (induced/uninduced). Thus, a value of +1 represents a 2-fold increase, and −1 equals a decrease to 50% of the value at time zero. (C) Quantitation of fold change in gene expression (as a ratio of induced/uninduced), for 4 novel genes and (Gapd),derived from microarray data, for 0 hours, 6 hours, and 72 hours of induction, and for EPRO cells. (D) Northern blot to confirm microarray data obtained with RNA from EML cells. Presented are hybridizations with 4 novel clones from our subtracted myeloid cDNA library, as well as Gapd for normalization. Also shown is the ethidium bromide staining of 28S RNA. RNAs are from EML cells treated with ATRA plus IL-3 for 0, 6, 12, and 24 hours to induce myeloid differentiation as well as from EPRO cells.
Graphical representation of the change in gene expression over time in EML cells treated with ATRA/IL-3.
(A) (B) Each line represents a different gene. The time points (x-axis) represent 0, 6, 24, and 72 hours. The last point on this axis represents EPRO cells, a stage in differentiation beyond 72 hours. The y-axis shows the relative change in gene expression level as log2 (induced/uninduced). Thus, a value of +1 represents a 2-fold increase, and −1 equals a decrease to 50% of the value at time zero. (C) Quantitation of fold change in gene expression (as a ratio of induced/uninduced), for 4 novel genes and (Gapd),derived from microarray data, for 0 hours, 6 hours, and 72 hours of induction, and for EPRO cells. (D) Northern blot to confirm microarray data obtained with RNA from EML cells. Presented are hybridizations with 4 novel clones from our subtracted myeloid cDNA library, as well as Gapd for normalization. Also shown is the ethidium bromide staining of 28S RNA. RNAs are from EML cells treated with ATRA plus IL-3 for 0, 6, 12, and 24 hours to induce myeloid differentiation as well as from EPRO cells.
The major class of genes expressed at higher level in RA-induced EML cells and EPRO cells relative to EML cells was the class encoding ribosomal proteins (Table 7) and included proteins in both the large- and small-ribosomal subunit. These data, together with the increase observed for elongation factors 1α1 and Tu-binding and polyA–binding protein, are consistent with a generalized increase in protein synthesis. The calcium signaling pathway also appeared up-regulated: calmodulin, calreticulin, and annexin A1 were all higher. Iκbα, an inhibitor of nuclear factor–κB (NF-κB), was also induced, suggesting down-regulation of the NF-κB–signaling pathway. In additon, there were 74 uncharacterized genes (ESTs) and 37 novel genes that increased in expression during EML cell differentiation. The latter were derived from our library subtractions, and this demonstration that they are differentially expressed and thus likely to be of interest, shows the utility of this undertaking for investigating myelopoiesis.
Genes that were down-regulated in this differentiation pathway were more varied (Table 8), but also included 13 novel genes derived from our cloning effort, as well as 68 uncharacterized genes. Several key transcription factors were down during ATRA/IL-3–induced differentiation of EML cells (Table 8). Some of these, such asKlf1,29Hoxb4,30 andXbp1,31 have known regulatory roles in hematopoietic cells. These data provide an important starting point for further analyses aimed at understanding myelopoiesis at the molecular level, studies that are ongoing in the laboratory.
Northern blot analysis was performed as confirmation of the microarray results obtained for several novel genes (ID1567, ID2131, ID1199, and ID1457). For example, ID2131 (which contains a GTW motif of G-protein receptor [GPR1]/FUN34/yaaH family proteins) is dramatically down-regulated during differentiation, and ID1567 (containing a KGR motif) and ID1457 are up-regulated during EML cell differentiation (Figure 5). Novel gene ID1199 showed similar expression levels before and after induction of differentiation.
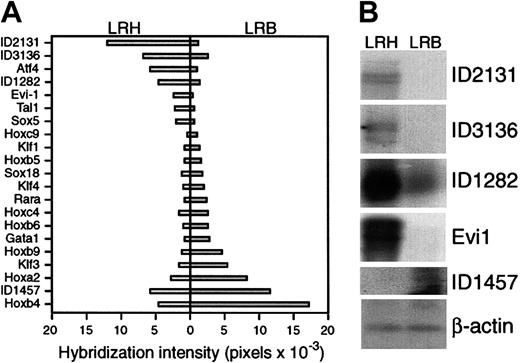
(A) Graphic representation of hybridization intensity in pixels of 21 genes to the cDNA microarray.
Values for LRH are depicted by bars to the left; for Lin−HoechstLowrhodamineBright(LRB), by bars to the right. Gene name or novel gene ID number is indicated to the left. (B) Southern blot of amplified cDNAs derived by reverse-transcription PCR of entire mRNA populations from 2 cell preparations from primary sorted mouse bone marrow cells. The samples in each lane are as indicated on the figure. The probes used for hybridization are listed to the right of each panel. Hybridization with β-actin serves as a control for loading.
(A) Graphic representation of hybridization intensity in pixels of 21 genes to the cDNA microarray.
Values for LRH are depicted by bars to the left; for Lin−HoechstLowrhodamineBright(LRB), by bars to the right. Gene name or novel gene ID number is indicated to the left. (B) Southern blot of amplified cDNAs derived by reverse-transcription PCR of entire mRNA populations from 2 cell preparations from primary sorted mouse bone marrow cells. The samples in each lane are as indicated on the figure. The probes used for hybridization are listed to the right of each panel. Hybridization with β-actin serves as a control for loading.
While the EML cell culture system has proved useful in identifying changes in gene expression during hematopoietic differentiation, it is nonetheless an immortalized cell line and thus may not accurately represent normal hematopoietic cells. To compare the changes in gene expression observed in EML cells with normal hematopoiesis, we analyzed RNA from sorted primary bone marrow cells. We obtained 2 pools of cDNAs from sorted primary mouse bone marrow cells: lin−HoechstLowrhodamineBright (LRB), representing late-stage progenitor cells; and LRH, representing more primitive progenitors.23These cDNAs were amplified by PCR with the use of primers specific to adaptor sequences and concomitantly labeled with fluorescent dyes. The LRH and LRB pools were competitively hybridized to the cDNA microarray. Hybridizations were performed in triplicate, and data were normalized to internal control (Gapd). This analysis revealed differences in expression between the LRH and LRB preparations of a number of key regulatory transcription factors, including Hox, Klf, and Sox family genes, Evi-1, Tal-1, GATA-1, and Rara (Figure 5A). In addition, a number of novel genes (designated by ID number) were differentially expressed, including ID2131 and ID1457, which were up-regulated and down-regulated, respectively, during EML cell differentiation (Figure 4). To confirm these microarray results, samples of amplified cDNA from LRH and LRB cells were fractionated by gel electrophoresis and then subjected to Southern blot analysis with specific cDNAs used as probes. These data (Figure 5B) support the microarray data in that they demonstrate differential expression between LRH and LRB preparations.
Discussion
We have described the creation of a resource for the in-depth study of gene expression in early hematopoietic cells that should be useful in the study of the molecular regulation of myeloid cell differentiation. Several features of this work are notable and essentially novel. First, the library that we exploited for the creation of the subtracted library represents an early stage of hematopoietic differentiation distinct from that used previously.32 Second, we undertook a library subtraction step that successfully removed most commonly expressed genes, leaving a residual that was relatively enriched in regulatory genes and novel genes. In aggregate, we netted 1255 different gene sequences from the subtraction effort. Given that more than 50% of the clones picked were nonredundant, it is likely that further sequencing of clones from this subtracted library will allow isolation of additional interesting genes.
Third, we have been successful in creating a glass slide–based microarray from the gene sequences we have isolated. To complement the clones from the subtracted library, we have added genes from a variety of other sources (Table 5). We have tested the utility of this array in 2 initial hybridization experiments. The first identifies genes that are up-regulated or down-regulated during ATRA/IL-3–induced differentiation of EML cells. The second documents transcriptional differences between sorted primary bone marrow cells. This investigation is continuing. However, some of our initial results with the microarray have been confirmed by Northern blot analysis (Figure4D) or by Southern blot analysis of cDNA populations (Figure 5B), which attests to the validity of the microarray-based quantitation of mRNA or cDNA copies.
Fourth, we have created a Web-accessible database for the genes on the microarray. Using this database, one can download a list of genes present on the array and can query to obtain information regarding specific genes. This Web site represents the starting point for a variety of features, including posting of downloadable microarray data and accrual of information on genes important to hematopoietic progenitor and myeloid cell biology.
A remarkable feature of our sequence analysis was the high number of novel gene sequences present in the subtracted library. This should prove to be an important resource for the isolation of genes that play regulatory roles in early hematopoiesis. Initial protein motif analysis reveals the presence of numerous interesting motifs (Table 4; Figure 3) within these genes. Also remarkable is the paucity of growth factor receptors or cytokines among the known genes in the subtracted library. This is likely to be due to their being present in the driver or to their lack of expression in the LRH library. Our finding of Ephrin-B1 in LRH is novel. Previous studies have shown that a related transmembrane ligand, Ephrin B2, is expressed in certain leukemias and lymphomas.33 It has also been shown that the receptor for Ephrin B2, EphB4 (hepatoma transmembrane kinase), is expressed on human erythroid progenitors cord blood cells and that it was regulated by SCF.34 35 However, no report of expression of EphB1, the receptor for Ephrin B1, in hematopoietic cells has been made. The role of this signaling system in hematopoietic cells is unknown. Interestingly, in the subtracted library, we also identified Nsp3, which encodes a protein that couples Eph receptors to Ras, further suggesting that this is an important pathway in early hematopoietic cells.
Our studies complement and extend data reported by Phillips et al,32 who reported on 2119 nonredundant gene products and the creation of a Stem Cell Database as a repository for these sequences. In aggregate, our effort, combined with theirs, provide an abundance of cloned sequences from early hematopoietic progenitors that allow for investigation into the molecular control of hematopoiesis.
We thank Troy Moore of Research Genetics for the contribution of the cDNA clone set used for the subtractive hybridization and arraying; Sherman Weissman for use of the LRH library; Nathan Lawson for construction of the EML and derivative libraries; Barbara Degar for contributing the LRH, LRB, and Lin+ cDNA; Bernard Forget for helpful comments and suggestions; and Janet Hager and the Yale Microarray Facility for the arraying of cDNAs.
Supported by NIH, NHLBI grant PO1 HL63357-02 (to N.B., D.S.K., and A.S.P) and NIDDK Microarray Biotechnology Center Grant (NIM5 U24 DK58776; PI: Kenneth Williams).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Archibald S. Perkins, Department of Pathology, Yale University School of Medicine, PO Box 208023, New Haven, CT 06520-8023; e-mail: archibald.perkins@yale.edu.