Abstract
Although DNA repair processes have been shown to considerably modulate the cytotoxic effects of alkylating agents, little information is available on the role of these mechanisms in chemotherapy-induced myelosuppression. Therefore, we have analyzed in detail the DNA repair capacity of primary human hematopoietic cells from cord blood (CB) or bone marrow (BM) by 2 functional assays, the immunocytologic assay (ICA) and single-cell gel electrophoresis (comet assay). Besides substantial interindividual differences, we consistently observed significantly lower repair capacity of CD34+ cells in comparison to CD34−, CD19+, or CD33+ cells of the same donor. After exposure to the alkylating agent ethylnitrosourea (EtNU), the comet assay displayed on average twice as many DNA single-strand breaks (SSBs) in CD34+ cells and a tripled half-life of these lesions in comparison to corresponding CD34− cells. Similarly, reduced SSB repair activity in CD34+ cells was detected following melphalan or cisplatin application. When specific antibodies were used to monitor DNA reaction products of these drugs, adduct levels were significantly higher and lesions persisted longer in the CD34+ fraction. To assess the contribution of individual pathways to overall DNA repair, modulators blocking defined steps in repair processes were coapplied with alkylating drugs. Similar “modulation pattern” in corresponding CD34+ and CD34− cell fractions indicated a generalized reduction in DNA repair capacity of CD34+ cells, rather than deficiencies in a specific pathway. Because CD34+ cells also displayed higher frequencies of apoptosis in response to melphalan or cisplatin, these findings may help to explain the myelosuppression after exposure to alkylating agents.
Introduction
Myelosuppression is one of the major side effects of cancer chemotherapy with cytotoxic agents. Following dose-intensified or high-dose therapy, hematotoxicity might become life-threatening and require treatment with hematopoietic growth factors or autologous stem cell support. In general, damage inflicted on primitive cells of the hematopoietic system, such as precursor cells, clonogenic progenitor cells, or stem cells, has been attributed to underlie this toxicity and the marked sensitivity of hematopoietic cells to chemotherapeutic agents has been related to the high turnover rate and the high S-phase percentage of these populations. However, this does not explain the considerable variations in myelosuppression induced by distinct antineoplastic agents nor does it explain the preferential toxicity of certain cancer drugs for defined subpopulations of hematopoietic cells. Thus, for anthracyclines or taxoids, a role of active cellular export mechanisms linked to high expression of the gene for the detoxifying efflux pump protein MDR-1 has been demonstrated in the relative resistance of primitive stem cells.1 Similarly, resistance of hematopoietic stem cells to the alkylating agent cyclophosphamide has been attributed to high activity levels of the detoxifying enzyme cytosolic aldehyde dehydrogenase.2
Little information is available on the molecular basis of the hematotoxicity induced by other DNA-reacting agents in clinical use, such as mitomycin C, melphalan, the chloroethylnitrosoureas, busulfan, temozolomide, or cisplatin and carboplatin. Although the principle molecular mechanisms underlying the cytotoxicity of these agents have been delineated and a number of primary lesions in the cellular DNA have been defined, little is known about the specific cellular factors that modulate the toxicity of these drugs in a specific target cell population, such as hematopoietic cells.
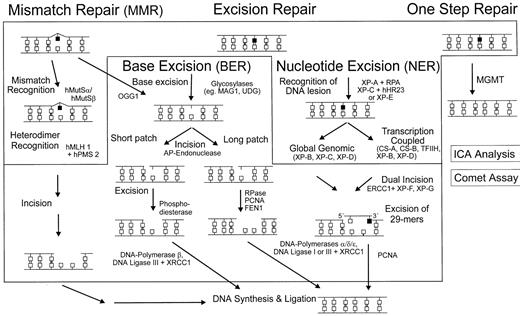
Mammalian cells are equipped with specific mechanisms to protect them from the mutagenic and cytotoxic effects of damaged DNA, and our knowledge about the DNA repair machinery has dramatically increased during recent years.3,4 A type of “repair network” is now emerging, which displays various main routes and a number of subpathways and crossroads (Figure 1). With the exception of the direct, one-step removal of adducts from the O6 position of guanine by the repair protein methylguanine-DNA methyltransferase (MGMT), all other routes are multistep pathways comprising the consecutive action of different proteins. In general, sites of structurally altered DNA are recognized by specific proteins followed by clipping off the damaged bases by glycosylases via base excision repair (BER)5 or by removing a longer patch around the lesion, for example, by nucleotide excision repair (NER)6 mechanisms. The repair-induced gaps are then filled in by DNA polymerases and finally closed by a ligation step. In addition, a number of accessory proteins are involved in these processes, for instance, for local unwinding of helical structures prior to excision or trimming and protecting the incised strands from degradation.
Multiple mechanisms of DNA repair in mammalian cells.
Removal of primary adducts is performed by several repair pathways, each depending on a couple of distinct proteins. These repair proteins eliminate specific DNA adducts either directly (eg, MGMT at the O6-position of guanine) or, after damage recognition, initiate a cascade of steps leading to excision reactions, resynthesis, and subsequently, religation of the repaired patches.
Multiple mechanisms of DNA repair in mammalian cells.
Removal of primary adducts is performed by several repair pathways, each depending on a couple of distinct proteins. These repair proteins eliminate specific DNA adducts either directly (eg, MGMT at the O6-position of guanine) or, after damage recognition, initiate a cascade of steps leading to excision reactions, resynthesis, and subsequently, religation of the repaired patches.
In general, the significance of DNA repair processes for the sensitivity of mammalian cells to DNA-reactive agents is well documented.7,8 Experiments with cell lines and, more convincingly, with primary cells of transgenic or “knock-out” mice, have shown that genetic alterations along specific DNA repair pathways not only resulted in alterations of repair capacity but also in pronounced modulation of cellular drug sensitivity.9 Due to the complexity of this machinery little is known about the time courses of complete repair processes from initial damage recognition to the resealing steps. Especially in primary hematopoietic cells the steps or components that are rate-limiting for the entire process of damage processing along distinct repair pathways are still poorly defined. Only recently was it recognized that DNA repair may exhibit a “Janus character” with respect to cellular drug sensitivity. Cell death programs can be triggered not only by persisting primary adducts in genomic DNA, but also by a delayed processing of repair intermediates like single-strand breaks (SSBs). The same holds true for “futile repair cycles” misdirected to the undamaged DNA strand as discussed, for example, for mismatch repair functions at sites of O6-methylated guanine residues.10 Cellular response to DNA damage, therefore, is not likely to be determined exclusively by the activity of a distinct repair mechanism, but is probably governed by the relative activity of different pathways to each other. For this reason, complete time courses of repair processing of primary and secondary lesions seems a superior parameter to be correlated to cellular drug sensitivity than expression or activity of single components of a specific repair pathway.
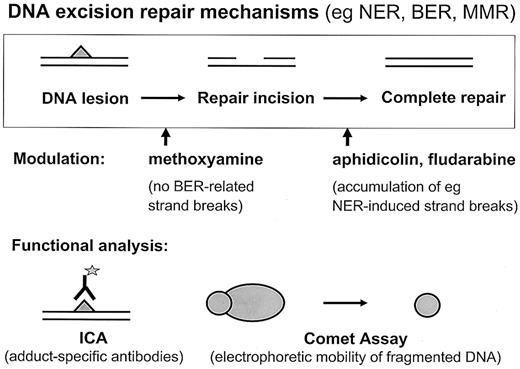
Thus, to study the processes triggered in primary hematopoietic cells by DNA-reactive cytostatic agents in more detail and to assess the role of DNA repair in myelosuppression following therapy with these drugs, we have systematically analyzed different subsets of chemotherapy-naive human hematopoietic cells, such as CD34+progenitor cells and more differentiated CD34−, CD19+, or CD33+ cells. The performance of the repair machinery was determined by using 2 functional assays, the immunocytologic assay (ICA) technology to measure formation and persistence of drug-induced adducts and single-cell gel electrophoresis (comet assay), which illuminates the occurrence and processing of repair intermediates in the nuclear DNA (Figure2).
Schematic depiction of functional assays to measure DNA repair at the single-cell level.
Formation and removal of drug-induced adducts in nuclear DNA is detected by the immunocytological assay (ICA). The amount of repair incisions is monitored by single-cell gel electrophoresis (comet assay).
Schematic depiction of functional assays to measure DNA repair at the single-cell level.
Formation and removal of drug-induced adducts in nuclear DNA is detected by the immunocytological assay (ICA). The amount of repair incisions is monitored by single-cell gel electrophoresis (comet assay).
Materials and methods
Chemicals
Ethylnitrosourea (EtNU; Serva, Heidelberg, Germany) was dissolved in dimethyl sulfoxide (DMSO) and stored as stock solution (100 mg/mL) at −20°C. Before use, the EtNU concentration was measured by UV spectroscopy (λ = 236 nm) in MES buffer (60 mM NaCl, 1mM 2-morpholino-ethane-sulfonacid, 0.5 mM EDTA; pH6). Melphalan (Sigma, Deisenhofen, Germany) was stored as a stock solution (1 mg/mL DMSO) at −20°C. Cisplatin (Platinex; Bristol Medicine, München, Germany) was used from a fresh infusion solution. DNA repair modulators, methoxyamine, aphidicolin, and fludarabine (Sigma), were prepared as stock solutions in sodium phosphate buffer (pH 7.2) and stored at −20°C.
Antibodies
Rat anti–cisplatin-(G-G) monoclonal antibody (mAb) “R-C18“11 was established in the Institute of Cell Biology (Essen, Germany), and rat anti–melphalan-DNA mAb “Amp4/32”12 was kindly provided by Dr M. J. Tilby (University of Newcastle, Newcastle, United Kingdom). Mouse anti–single-stranded DNA mAb “3299” was purchased from Chemicon International (Temecula, CA), secondary goat anti–rat IgG antibody labeled with fluorescein isothiocyanate (FITC; Dianova, Hamburg, Germany) and rabbit antifluorescein or goat anti–mouse IgM antibodies labeled with ALEXA FLUOR 488 from Molecular Probes (Leiden, Netherlands).
Preparation of cells
Leukocytes were isolated from samples of peripheral blood, cord blood (CB), or bone marrow (BM) by density centrifugation. Heparinized blood (10 mL) was layered onto 10-mL Ficoll-Hypaque (Lymphoprep; Nycomed, Birmingham, United Kingdom) and centrifuged for 25 minutes at 200g at room temperature. Cells from the interface were removed, washed twice with phosphate-buffered saline (PBS), and resuspended in prewarmed RPMI medium supplemented with 10% fetal calf serum (FCS).
Fractions of CD34+, CD34−, CD33+or CD19+ cells were isolated by immunomagnetic procedures from the leukocyte fraction of BM or CB samples. CD34+cells were purified using antibody-coated magneto beads and MS+-Separation Columns (miniMACS; Miltenyi, Bergisch-Gladbach, Germany) according to the manufacturer's instructions. After separation, purity of cell fractions, tested by flow cytometry, was more than 80%. CD34− cells were taken from the column wash. For control experiments specific subpopulations of CD33+ or CD19+ cells were separated by the same procedure using antigen-specific magneto beads. Cells were kept in supplemented RPMI medium (10% FCS) at 37°C in a humidified atmosphere containing 5% CO2.
Exposure to drug
Cells were exposed to melphalan (1 hour; 5 μg/mL) or to cisplatin (2 hours; 10 μg/mL) in serum-free RPMI medium or to EtNU (20 minutes; 100 μg/mL) in PBS/Hepes (25 mM; pH 7.25) at 37°C. After exposure cells were washed twice with PBS and resuspended in prewarmed RPMI supplemented with 1% FCS for further incubation. Cell aliquots were taken immediately and at several times after drug treatment. Where appropriate, cells were preincubated with RPMI medium containing methoxyamine (5 mM) and/or aphidicolin (1 mM) or fludarabine (20 ng/mL) 1 hour before exposure to the drug and throughout the experiment.
Comet assay
The comet assay was performed essentially as described.13 Briefly, after drug exposure, cells were washed twice with ice-cold PBS; aliquots of 5 × 104cells were suspended in low melting point (LMP) agarose (0.6%), and spread onto fully frosted microscope slides precoated with a thin layer of 0.7% Seakem LE agarose (Biozyme, Hameln, Germany). Cells were exposed to lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 10% DMSO, 1% Triton X-100, 1% sodium sarcosinate, pH 10) at 4°C overnight and DNA was unwound by alkaline treatment (300 mM NaOH, 1 mM EDTA, 10 mM Tris, pH 12.5) for 20 minutes (EtNU) or 60 minutes (melphalan, cisplatin) at 4°C. Slides were then subjected to alkaline electrophoresis in the same buffer (20 minutes; 4°C; 4 V/cm), neutralized, and stained with ethidium bromide (2 μg/mL).
lmmunofluorescence staining
lmmunofluorescence staining for adducts in the nuclear DNA of single cells was performed essentially as described14,15with the following modifications.11 Cells were suspended in PBS containing 25% “HAES-steril” solution (Fresenius, Bad Homburg, Germany) and spread onto microscopic slides precoated with 2% 3-aminopropyl-triethoxysilan. Cells were air-dried, fixed with methanol (−20°C; overnight) and rehydrated in PBS. After treatment with RNases,14 DNA was partly denatured by alkali treatment (NaOH, 70 mM in 140 mM NaCl, 40% methanol for 5 minutes, 0°C). Cells were treated with pepsin (200 μg/mL in 20 mM HCl; 37°C; 5 minutes) and proteinase K (2 μg/mL in 20 mM Tris/HCl, 2 mM CaCl2, pH 7.5; 37°C; 5 minutes), blocked with PBS/1% casein (30 minutes; room temperature), and immunostained with anti–cisplatin-DNA mAb “R-C18” (0.2 μg/mL) or with anti–melphalan-DNA mAb “Amp4/42” diluted 1:50, both in PBS/1% casein (12 hours; 4°C). A double-sandwich staining was performed using secondary goat anti–mouse Ig antibody labeled with FITC (6.5 μg/mL PBS/1% casein; 1 hour; room temperature) and rabbit anti–FITC labeled with ALEXA FLUOR 488 (5 μg/mL PBS/1% casein; 1 hour; 37°C). The nuclear DNA was counterstained with DAPI (3 × 10−7 M in PBS; 30 minutes), and slides were covered with an antifading solution.
Quantification of fluorescence signals and nuclear DNA of individual cells
A Zeiss fluorescence photomicroscope (Axioplan) equipped with an HBO 100 W mercury arc lamp and Zeiss standard filter combinations were used for the visualization of fluorescence signals from individual nuclei. Signals were amplified and recorded by a dual-mode CCD camera (Photonics, Hamamatsu City, Japan), and fed into a 4-parameter image analysis program (ACAS Cytometry Analysis System; Ahrens Electronics, Bargteheide, Germany).
For the ICA analysis, adduct levels in nuclear DNA were determined by normalizing antibody-derived signals for the corresponding DNA content of the same cell.18 For the comet assay, DNA fluorescence and integral area of nuclear DNA was determine as a measure for DNA strand breaks in single nuclei.13 Average values were computed from the fluorescence intensities of more than 100 individual nuclei.
To determine the amount of repair induced DNA strand breaks in single nuclei by the comet assay the intensity and the total area of stained nuclear DNA were measured by image analysis. Thresholds were defined to give homogeneously positive integrated areas throughout individual comets. The increase of these values in exposed cells compared to the corresponding untreated cells from the same donor in the same experiment was used to calculate the relative rates of strand breaks.13
Chemosensitivity of hematopoietic cells
The fraction of apoptotic cells after drug exposure was determined by immunofluorescence staining of single-stranded DNA in individual nuclei essentially as described elsewhere.16 In this assay patches of single-stranded DNA are induced by mild denaturation at sites of altered DNA-histone interaction, which occurs during apoptotic processes. Briefly, cells were exposed to different doses of melphalan or cisplatin in normal medium, kept at 37°C for 24 hours, washed with PBS, fixed by addition of 3 volumes of ice-cold ethanol (96%), centrifuged, and stored at −20°C. Cells were suspended in formamide (100 μL) and incubated for 10 minutes at 75°C. After adding 2 mL PBS/1% BSA cells were incubated (10 minutes; room temperature), immunostained with mouse anti–single-stranded DNA antibody “3299” (1 μg in 100 μL PBS/BSA; 15 minutes; room temperature), and with goat anti–mouse IgM secondary antibody labeled with ALEXA FLUOR 488 (2 μg/100 μL; 15 minutes; room temperature). After washing with PBS, cells were counterstained with DAPI (see above), transferred to microscopic slides, and fluorescence signals from individual nuclei were evaluated by image analysis (see above). The frequency of nuclei showing ALEXA FLUOR 488-related fluorescence signals was determined in more than 200 cells/sample.
Results
Strand break formation in CD34+ and CD34−cells following exposure to EtNU
Primary hematopoietic cells (BM or CB) expressing the CD34 antigen were compared with more differentiated CD34− cells from the same donor for their functional DNA repair capacity using single-cell gel electrophoresis (comet assay). This assay monitors the amount of repair-induced strand breaks in damaged DNA (Figure 2). After isolation, cells were challenged in vitro by a short treatment (20 minutes) with the monofunctional standard alkylator EtNU. Under physiologic conditions, EtNU decomposes extremely fast (half-life, 8 minutes) and induces a reproducible level and pattern of about a dozen specific DNA alkylation products, which then trigger a number of distinct repair pathways.13 17 By shifting the cells back to normal medium after exposure the repair kinetics can be analyzed without disturbance by ongoing adduct formation.
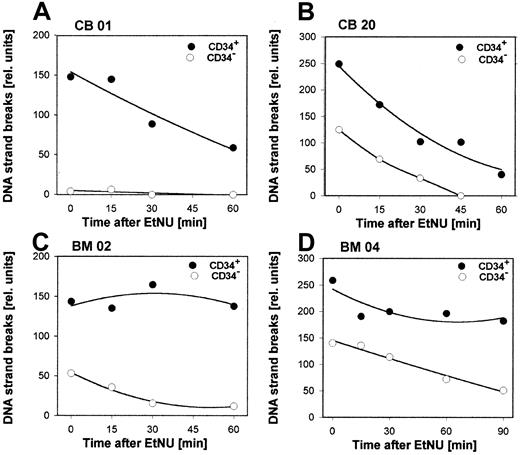
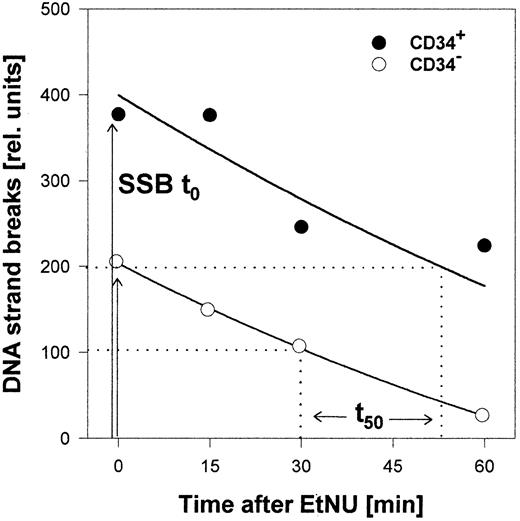
Both populations, CD34+ and CD34− cells, exhibited significant interindividual differences for the cellular repair capacity, as exemplified for 4 specimens in Figure3 (cumulative data for all samples in Table 1). Differences were observed for the initial amount of DNA strand breaks already induced during the incubation period with EtNU, as well as for the time course of the subsequent removal of these lesions. Interestingly, however, CD34+ cells reproducibly exhibited augmented numbers of breaks and delayed repair kinetics in comparison to CD34−cells derived from the same donor. To analyze this phenomenon in a quantitative manner, 2 characteristic parameters of the individual SSB processing were used: (1) the relative amount of repair incisions immediately after drug exposure (SSBt0) and (2) the time interval for repairing 50% of these lesions (t50) as shown in Figure 4. The respective values from repair kinetics of 18 independent specimens (6 CB; 12 BM) are summarized in Table 1. Following exposure to the same dose of EtNU, CD34+ cells, irrespective of the interindividual variation, accumulated more strand breaks at t0 (mean factor: 2.2;P < .0001) and exhibited longer repair half-times (mean factor, 3.1; P < .0001) in comparison to CD34− cells. Because both parameters, SSBt0and t50, reflect different aspects of the DNA repair activity, the product of SSBt0 and t50 was used to characterize overall repair activity by a single parameter. When the relative ratios of overall repair activity for CD34− and CD34+ cells were calculated, this parameter indicated that the DNA repair capacity in CD34+ cells was about 6 times lower than in the CD34− population (mean, 6.2;P < .0001). No significant differences in the overall repair capacity were observed between cell fractions isolated either from BM (n = 12; factor 6.5) or from CB (n = 6; factor 5.9).
Repair kinetics for secondary DNA lesions in hematopoietic cells.
CD34+ or CD34− cells were isolated from samples of CB (A,B) or BM (C,D) of 4 individual donors and exposed ex vivo to a short treatment (20 min) with EtNU (100 μg/mL). Repair-induced DNA strand breaks were determined by the comet assay in cell aliquots at different time points after exposure.
Repair kinetics for secondary DNA lesions in hematopoietic cells.
CD34+ or CD34− cells were isolated from samples of CB (A,B) or BM (C,D) of 4 individual donors and exposed ex vivo to a short treatment (20 min) with EtNU (100 μg/mL). Repair-induced DNA strand breaks were determined by the comet assay in cell aliquots at different time points after exposure.
DNA strand break processing in CD34+ and CD34− cells isolated from BM or CB after pulse exposure to EtNU
| Sample no. . | Source . | Repair-induced SSBt0 . | Time of 50% repair (t50), min . | Relative repair ratio (X × Y) . | ||||
|---|---|---|---|---|---|---|---|---|
| CD34+ . | CD34− . | Ratio CD34+/CD34−(X) . | CD34+ . | CD34− . | Ratio CD34+/CD34− (Y) . | |||
| 2 | BM | 143.3 | 53 | 2.7 | 105 | 20 | 5.3 | 14.2 |
| 3 | BM | 133.2 | 116.9 | 1.1 | 115 | 30 | 3.8 | 4.4 |
| 4 | BM | 258.6 | 140.2 | 1.8 | 130 | 70 | 1.9 | 3.4 |
| 5 | BM | 320.8 | 244.6 | 1.3 | 135 | 45 | 3 | 3.9 |
| 6 | BM | 105.2 | 14.4 | 7.3 | 50 | 25 | 2 | 14.6 |
| 8 | BM | 179.7 | 112 | 1.6 | 95 | 25 | 3.8 | 6.1 |
| 10 | BM | 196 | 73 | 2.7 | 25 | 15 | 1.6 | 4.3 |
| 11 | BM | 134.8 | 71.5 | 1.9 | 70 | 22 | 3 | 6 |
| 12 | BM | 207 | 125.9 | 1.6 | 45 | 30 | 1.5 | 2.5 |
| 13 | BM | 258.9 | 110.2 | 2.3 | 25 | 55 | 0.5 | 1.2 |
| 14 | BM | 182.3 | 93.9 | 1.9 | 60 | 30 | 2 | 3.9 |
| 17 | BM | 285.2 | 112.2 | 2.5 | 60 | 15 | 4 | 10.2 |
| Mean BM | 2.4 | 2.7 | 6.5 | |||||
| 15 | CB | 142 | 55 | 2.6 | 40 | 18 | 2.1 | 5.4 |
| 16 | CB | 377.4 | 203.3 | 1.9 | 70 | 31 | 2.1 | 3.9 |
| 18 | CB | 142 | 126.7 | 1.1 | 40 | 12.5 | 3.2 | 3.6 |
| 19 | CB | 119.4 | 67.3 | 1.8 | 120 | 15 | 8 | 14.2 |
| 20 | CB | 248.7 | 124.5 | 2 | 30 | 20 | 1.5 | 3 |
| 1 | CB | 89.9 | 4.3 | 20.9* | 30 | 20 | 1.5 | 31.3* |
| Mean CB | 1.9 | 3.1 | 5.9 | |||||
| Total mean | 2.2 | 2.8 | 6.2 | |||||
| Sample no. . | Source . | Repair-induced SSBt0 . | Time of 50% repair (t50), min . | Relative repair ratio (X × Y) . | ||||
|---|---|---|---|---|---|---|---|---|
| CD34+ . | CD34− . | Ratio CD34+/CD34−(X) . | CD34+ . | CD34− . | Ratio CD34+/CD34− (Y) . | |||
| 2 | BM | 143.3 | 53 | 2.7 | 105 | 20 | 5.3 | 14.2 |
| 3 | BM | 133.2 | 116.9 | 1.1 | 115 | 30 | 3.8 | 4.4 |
| 4 | BM | 258.6 | 140.2 | 1.8 | 130 | 70 | 1.9 | 3.4 |
| 5 | BM | 320.8 | 244.6 | 1.3 | 135 | 45 | 3 | 3.9 |
| 6 | BM | 105.2 | 14.4 | 7.3 | 50 | 25 | 2 | 14.6 |
| 8 | BM | 179.7 | 112 | 1.6 | 95 | 25 | 3.8 | 6.1 |
| 10 | BM | 196 | 73 | 2.7 | 25 | 15 | 1.6 | 4.3 |
| 11 | BM | 134.8 | 71.5 | 1.9 | 70 | 22 | 3 | 6 |
| 12 | BM | 207 | 125.9 | 1.6 | 45 | 30 | 1.5 | 2.5 |
| 13 | BM | 258.9 | 110.2 | 2.3 | 25 | 55 | 0.5 | 1.2 |
| 14 | BM | 182.3 | 93.9 | 1.9 | 60 | 30 | 2 | 3.9 |
| 17 | BM | 285.2 | 112.2 | 2.5 | 60 | 15 | 4 | 10.2 |
| Mean BM | 2.4 | 2.7 | 6.5 | |||||
| 15 | CB | 142 | 55 | 2.6 | 40 | 18 | 2.1 | 5.4 |
| 16 | CB | 377.4 | 203.3 | 1.9 | 70 | 31 | 2.1 | 3.9 |
| 18 | CB | 142 | 126.7 | 1.1 | 40 | 12.5 | 3.2 | 3.6 |
| 19 | CB | 119.4 | 67.3 | 1.8 | 120 | 15 | 8 | 14.2 |
| 20 | CB | 248.7 | 124.5 | 2 | 30 | 20 | 1.5 | 3 |
| 1 | CB | 89.9 | 4.3 | 20.9* | 30 | 20 | 1.5 | 31.3* |
| Mean CB | 1.9 | 3.1 | 5.9 | |||||
| Total mean | 2.2 | 2.8 | 6.2 | |||||
Cells were exposed to EtNU (100 μg/mL) for 20 minutes.
Values were excluded from mean.
Quantitative evaluation of the DNA repair capacity.
CD34+ and CD34− cells were isolated from a sample of CB, exposed to EtNU and measured by comet analysis as in Figure 3. The relative amount of “initial” strand breaks (SSBt0) directly after treatment and the time for repairing 50% of these lesions (t50) were determined as parameters for the cellular DNA repair capacity.
Quantitative evaluation of the DNA repair capacity.
CD34+ and CD34− cells were isolated from a sample of CB, exposed to EtNU and measured by comet analysis as in Figure 3. The relative amount of “initial” strand breaks (SSBt0) directly after treatment and the time for repairing 50% of these lesions (t50) were determined as parameters for the cellular DNA repair capacity.
To exclude the possibility that the reduced repair capacity measured in CD34+ cells was influenced by cell handling during the magnetic separation step, we used the same technique to isolate 2 subfractions of CD34− cells, CD19+ B cells and CD33+ myeloid cells, from 4 samples of CB or BM. When these subgroups were exposed to EtNU and analyzed in parallel to the CD34+ and CD34− fractions from the same donor, the kinetics of strand-break processing in CD19+ or CD33+ cells in all cases reflected the data observed for unfractionated CD34− cells (data not shown).
DNA repair following exposure of hematopoietic cells to melphalan or cisplatin
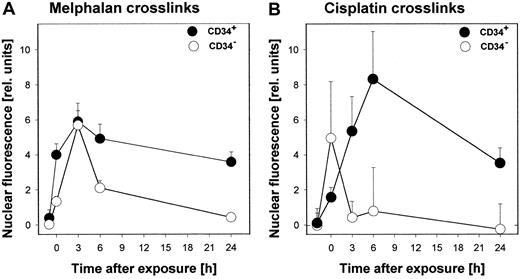
In addition to studying DNA repair after treatment with the standard alkylator EtNU, we have exposed hematopoietic cells to the DNA reactive agents melphalan and cisplatin, used in the treatment of both solid and hematologic malignancies. To measure the repair of primary adducts induced by these drugs, an immunohistochemical procedure (ICA) was used employing mAbs against specific DNA cross-links (melphalan-DNA adducts; cisplatin-(G-G) intrastrand adducts). With this assay, a direct determination of adduct formation and elimination can be performed. Again, CD34− cells turned out to be much more repair efficient than their CD34+ counterparts (Figure5). Although in the first hours after exposure only a weak tendency of higher adduct levels in CD34+ cells was observed, data generated 6 hours after incubation and thereafter demonstrated significantly elevated adduct levels corresponding to reduced elimination capacities for DNA cross-links in CD34+ cells. This phenomenon was observed in all samples analyzed so far (melphalan, n = 5; cisplatin, n = 8;P < .0002).
Formation and repair of specific DNA adducts in CD34+ and CD34− cells after exposure to melphalan or cisplatin ex vivo.
Cells were isolated from CB and exposed to melphalan (5 μg/mL) for one hour or cisplatin (10 μg/mL) for 2 hours and then replaced to fresh medium. Cell aliquots were taken at different time points after treatment and specific DNA adducts were measured by ICA analysis using anti–melphalan-DNA or anti–cisplatin (G-G) antibodies as described in “Materials and methods.”
Formation and repair of specific DNA adducts in CD34+ and CD34− cells after exposure to melphalan or cisplatin ex vivo.
Cells were isolated from CB and exposed to melphalan (5 μg/mL) for one hour or cisplatin (10 μg/mL) for 2 hours and then replaced to fresh medium. Cell aliquots were taken at different time points after treatment and specific DNA adducts were measured by ICA analysis using anti–melphalan-DNA or anti–cisplatin (G-G) antibodies as described in “Materials and methods.”
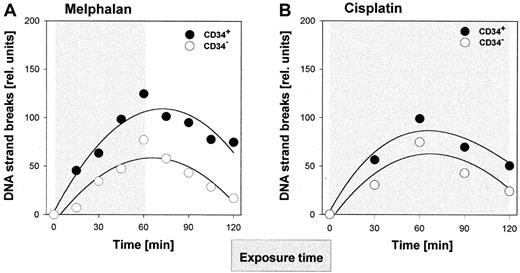
In parallel, the comet assay was applied to monitor the kinetics of strand break formation and processing during and after the exposure to melphalan (exposure time, 1 hour) or cisplatin (exposure time, 2 hours). Data from representative samples are given in Figure6. After exposure to each drug, repair intermediates in the CD34+ progenitor fraction not only accumulated to higher levels but also persisted longer than in the corresponding CD34− cells. Similar differences in strand break processing were observed for all samples analyzed (melphalan, n = 5; cisplatin, n = 6).
Repair-induced DNA strand breaks in hematopoietic cells after treatment with melphalan or cisplatin.
CD34+ and CD34- cells from CB were exposed ex vivo to melphalan (A) or cisplatin (B) as in Figure 5. Cell aliquots were taken at different time points during or after treatment and comet analysis was performed as described in Figure 3 and in “Materials and methods.”
Repair-induced DNA strand breaks in hematopoietic cells after treatment with melphalan or cisplatin.
CD34+ and CD34- cells from CB were exposed ex vivo to melphalan (A) or cisplatin (B) as in Figure 5. Cell aliquots were taken at different time points during or after treatment and comet analysis was performed as described in Figure 3 and in “Materials and methods.”
Contribution of individual pathways to the overall repair result
Most alkylating agents are capable of inducing a set of structurally different DNA adducts, which can be repaired in mammalian cells in parallel via different pathways (Figure 1). Therefore, a lower overall repair capacity, such as observed in CD34+ in comparison to CD34− cells, might be due to loss of function either preferentially in one pathway or reduced activity in a number of pathways. To address this question, we used specific modulators of DNA repair, which block key steps within the repair network13 15 and allow assessment of the relative contribution of individual pathways to the overall processing of drug-induced DNA damage. Examples of such modifiers are methoxyamine, aphidicolin, and fludarabine. Methoxyamine is able to react with abasic sites in DNA and by this prevents the incision step(s) of BER, leading to reduced numbers (the fraction resulting from BER activity) of total SSBs in the comet assay. On the other hand, aphidicolin and fludarabine are able to inhibit late, resealing steps (DNA polymerases and DNA ligases) of the NER, “long-patch” BER, or mismatch repair (MMR) mechanisms, which should result in SSB accumulation when adducts are processed via these pathways (Figures 1 and 2).
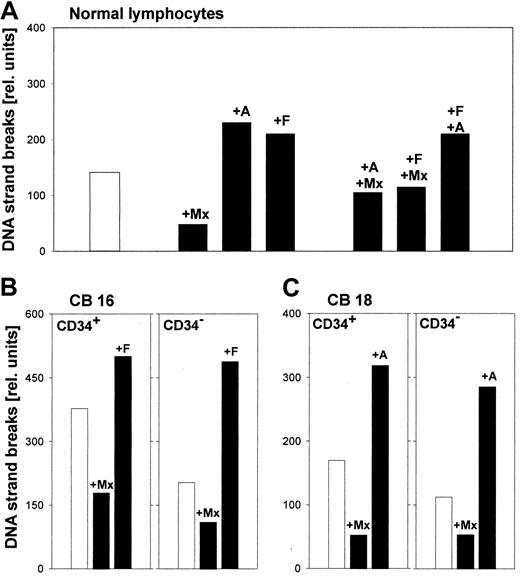
To establish the effects of these compounds on the rate of repair incisions, a pilot experiment was performed with peripheral blood lymphocytes exposed to EtNU ex vivo (Figure7A). Coapplication of methoxyamine reduced the number of SSBs in the comet assay by 65% indicating a significant contribution of BER to the removal of EtNU-induced ethyl adducts in these cells. The remaining 35% of SSBs indicated the contribution of other pathways. On the other hand, aphidicolin and fludarabine augmented the number of persisting strand breaks by approximately 60%, reflecting the involvement of NER, MMR, or long-patch BER mechanisms, either alone or in combination with one another. The combined treatment with the inhibitors aphidicolin and fludarabine did not further increase the rate of SSBs, indicating that both compounds execute their activity within the same pathway(s). Thus, the modulation pattern obtained in this pilot experiment established the contribution of different mechanisms to the repair of EtNU-damaged DNA in primary lymphocytes.
Modulation of DNA strand break formation by repair modifiers in peripheral lymphocytes and hematopoietic cells after exposure to EtNU.
Cells were isolated from peripheral blood (A) or from CB (B,C) and exposed to EtNU (50 μg/mL; 20 minutes; white column) or were pretreated with different repair modifiers for 30 minutes prior to and throughout EtNU exposure (black columns). Modifiers were applied alone or in combinations: methoxyamine (Mx; 5 mM); aphidicolin (A; 1 mM); and fludarabine (F; 20 nM). To monitor the contribution of different DNA repair pathways, cells were analyzed directly after EtNU exposure by comet analysis (see Figure 3).
Modulation of DNA strand break formation by repair modifiers in peripheral lymphocytes and hematopoietic cells after exposure to EtNU.
Cells were isolated from peripheral blood (A) or from CB (B,C) and exposed to EtNU (50 μg/mL; 20 minutes; white column) or were pretreated with different repair modifiers for 30 minutes prior to and throughout EtNU exposure (black columns). Modifiers were applied alone or in combinations: methoxyamine (Mx; 5 mM); aphidicolin (A; 1 mM); and fludarabine (F; 20 nM). To monitor the contribution of different DNA repair pathways, cells were analyzed directly after EtNU exposure by comet analysis (see Figure 3).
We next adopted this experimental strategy to analyze the relative contribution of individual repair pathways in CD34+ and CD34− cells (Figure 7B,C). Again, the reduced repair capacity of CD34+ cells was obvious, leading to an increased accumulation of strand breaks in these cells in the absence of specific modulators. The “modulation pattern” (relative reduction or augmentation of SSBs induced by individual modulators), however, was rather similar for CD34+ progenitor and more differentiated CD34− cells and reflected the pattern found in the pilot experiment with primary lymphocytes: significantly decreased rates of SSBs after pretreatment with methoxyamine and higher accumulation after aphidicolin or fludarabine. These results, which were confirmed for BM- as well as CB-derived cells, seem to imply that in both, more differentiated and CD34+ hematopoietic cells, the major DNA repair mechanisms are operative. The relative velocity of damage processing, however, is much slower in CD34+ cells leading to a prolonged persistence of adducts (Figure 5) and of secondary lesions (Figure 3).
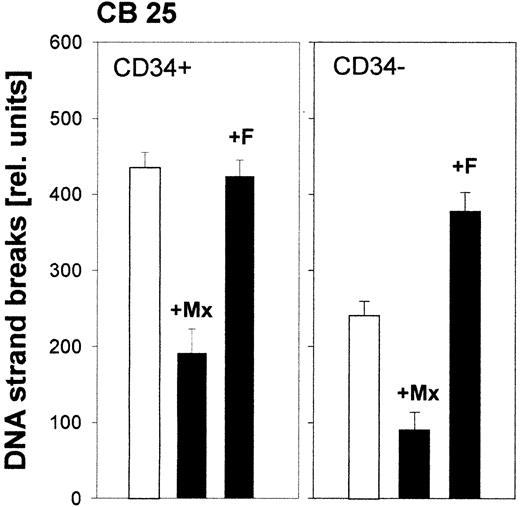
Contribution of individual repair pathways to removal of melphalan- or cisplatin-induced damage in hematopoietic cells
Up to now, there are no clear-cut results available about the contribution of specific repair mechanisms to the removal of melphalan- or cisplatin-induced adducts from DNA of hematopoietic cells. Therefore, we have applied the repair modulators also in combination with those drugs. When cells were pretreated with methoxyamine, aphidicolin, or fludarabine prior to melphalan exposure, the modulation pattern observed hardly differed from the pattern seen after EtNU exposure. A reduction of SSBs by methoxyamine and a higher accumulation after blocking repair synthesis steps by aphidicolin or fludarabine in both cell fractions (Figure8). This result suggests that BER, as well as at least one of the other repair pathways, most likely NER, is involved in parallel in the processing of melphalan-induced adducts.
Contribution of different repair pathways in hematopoietic cells to the processing of melphalan-induced DNA damage.
CD34+ and CD34- cells isolated from CB were exposed to melphalan alone (1 h; white columns) or were pretreated with DNA repair modulators (30 minutes), and throughout melphalan exposure (black columns) were analyzed for DNA strand breaks (see Figure 7for details).
Contribution of different repair pathways in hematopoietic cells to the processing of melphalan-induced DNA damage.
CD34+ and CD34- cells isolated from CB were exposed to melphalan alone (1 h; white columns) or were pretreated with DNA repair modulators (30 minutes), and throughout melphalan exposure (black columns) were analyzed for DNA strand breaks (see Figure 7for details).
In contrast to melphalan, the coexposure of CD34+ or CD34− cells to cisplatin and methoxyamine did not result in a significant reduction of repair-induced SSBs in comparison to cisplatin alone (data not shown). From this observation we can exclude a major contribution of BER mechanisms to the removal of DNA platination products in both cell fractions.
Sensitivity of hematopoietic cells to the cytotoxic effects of melphalan and cisplatin
Finally, we have determined whether the reduced repair capacity of CD34+ progenitor cells translates into augmented drug sensitivity. As an indicator for the onset of apoptotic processes, we have determined the amount of single-stranded DNA patches in individual nuclei by an immunoanalytical procedure. In 4 independent samples analyzed, the frequency of apoptotic cell death after exposure to melphalan or cisplatin was higher by factors of 1.2 to 3.4 in CD34+ cells when compared to their CD34−counterparts (Table 2). This difference was observed both at low and high doses (melphalan, 10 and 20 μg/mL; cisplatin, 25 and 50 μg/mL). Not only the percentage of apoptotic cells was regularly higher in the fraction of progenitor cells but also the mean intensity of DNA fragmentation in positively stained cells as measured by quantitative image analysis.
Apoptotic response of CD34+ and CD34− cells on exposure to melphalan or cisplatin ex vivo*
| Cell sample . | Melphalan . | Cisplatin . | |||
|---|---|---|---|---|---|
| 10 μg/mL . | 20 μg/mL . | 25 μg/mL . | 50 μg/mL . | ||
| CB 30 | CD34− | 27 | 33 | ||
| CD34+ | 34 | 40 | |||
| CB 41 | CD34 | 65 | 74 | ||
| CD34+ | 77 | 89 | |||
| CB 42 | CD34− | 38 | 51 | ||
| CD34+ | 47 | 73 | |||
| BM 40 | CD34− | 14 | 16 | ||
| CD34+ | 48 | 52 | |||
| Cell sample . | Melphalan . | Cisplatin . | |||
|---|---|---|---|---|---|
| 10 μg/mL . | 20 μg/mL . | 25 μg/mL . | 50 μg/mL . | ||
| CB 30 | CD34− | 27 | 33 | ||
| CD34+ | 34 | 40 | |||
| CB 41 | CD34 | 65 | 74 | ||
| CD34+ | 77 | 89 | |||
| CB 42 | CD34− | 38 | 51 | ||
| CD34+ | 47 | 73 | |||
| BM 40 | CD34− | 14 | 16 | ||
| CD34+ | 48 | 52 | |||
Apoptotic cells (%) 24 h after exposure. Mean values of more than 200 cells measured (see “Materials and methods”).
Discussion
DNA repair mechanisms have been convincingly demonstrated to modulate the cytotoxicity of DNA-damaging agents, both by eliminating disturbing adducts from the genome and by creating strand incisions, which, by themselves, can initiate cell death programs. However, little information is available on the role of DNA repair processes in primary hematopoietic cells. By application of functional assays covering the majority of cellular DNA repair mechanisms, we have found in this study significant interindividual differences in the repair capacity of hematopoietic cells isolated from human BM or CB as observed earlier for mature peripheral lymphocytes.13 18 Besides this individual “repair phenotype,” however, CD34+progenitor cells in all cases exhibited a lower efficiency in removing DNA lesions compared to their more differentiated CD34−, CD19+, or CD33+ counterparts, irrespective whether the cells were derived from BM or CB.
Previously, low repair activity has been reported for unfractionated cells from BM or spleen. Thus, in comparison to other tissues, such as liver or lung, murine as well as human BM cells have been shown to express 3- to 10-fold reduced levels of the detoxifying repair protein MGMT, which eliminates highly toxic O6-guanine alkylations from the DNA.19 These characteristics have been advocated to explain the pronounced hematotoxic potential of cytostatic agents alkylating the O6-position of guanines, such as the chloroethylnitrosourea-type drugs ACNU, BCNU, and CCNU or the triazene derivatives dacarbazine or temozolomide.20 Low expression in the murine spleen has also been described for proteins involved in early steps of BER, such as the 8-oxoguanine DNA glycosylase (OGG-1) protein, which catalyzes the repair of imidazole ring opening (formamidopyrimidine [Fapy]) lesions.21
The idea of DNA repair processes as critical modulators of hematopoietic toxicity is further supported by studies aiming to reduce this toxicity by overexpression of DNA repair proteins in the hematopoietic system via retroviral gene transfer. This strategy has been successfully applied in murine models using transgenic expression of MGMT to reduce myelosuppression by drugs alkylating the O6-position of guanine residues.22,23 More recently, proteins catalyzing early steps in BER repair such as the bacterial Fapy-DNA glycosylase (FPG) or the murine OGG-1 protein have been used to reduce the hematotoxicity following the application of triethylenethiophosphoramide (thiotepa), an aziridine-type alkylating agent leading to amino-ethyl adducts of guanine and Fapy lesions.24
Only a few studies have addressed the question of DNA repair capacity in subfractions of hematopoietic cells. In one of these studies, substantial interindividual differences similar to the alterations described here have been reported for the one-step mechanism driven by the alkyltransferase MGMT.25 In extracts of BM-derived CD34− cells the repair activity of this protein varied by a factor of 5 among 3 individual specimens. Interestingly, MGMT activities were not significantly different between CD34+and CD34− cells of the same donor. This finding supports our notion that the approximately 6-fold less efficient strand break processing in CD34+ cells after EtNU treatment (Table 1) is related to genuine differences in excision repair pathways like BER, NER, or MMR rather than being provoked by a reduced removal of O6-ethylguanines via the MGMT mechanism. In another study, CD34+ cells were reported to exhibit higher efficiency in NER than more maturated CD34− cells by measuring repair kinetics of SSBs in UV-irradiated human BM cells.26 This conclusion is clearly controversial with the findings in the present study and may be due to comparing cell fractions isolated from different donors. With regard to the pronounced interindividual variation in repair capacity (see above), results of this study may be biased by accidental selection.
The advantage of using EtNU as a standard alkylator for measuring cellular repair capacities is the fast reactive decomposition of this compound under physiologic conditions (half-life, 8 minutes) and the exact knowledge about the pattern of its reaction products with nuclear DNA.13 After 20 minutes of exposure, more than 80% of all possible adducts have already been formed. During the postincubation time, therefore, the comet assay merely reflects the kinetics of cellular repair reactions undisturbed by ongoing adduct formation.
In the case of melphalan and cisplatin the situation is more complicated. Both drugs react comparatively slowly with DNA by initially forming monoadducts predominantly with N7 atoms of guanine and, subsequently, within hours, interstrand and intrastrand cross-links.27 28 Both types of lesions, monoadducts and cross-links, are most likely substrates/targets for initiating repair reactions via different excision pathways detectable in the comet analysis. The chemical formation of interstrand cross-links, however, partly masks the persistence of repair gaps in this assay by reducing the electrophoretic mobility of DNA fragments in the nuclei. Therefore, the number of SSBs is likely to be underestimated at later time points of repair kinetics using bifunctional drugs. This may be causal for the less pronounced differences between CD34+ and CD34− cells for their strand break kinetics after melphalan or cisplatin exposure in contrast to the processing of repair-induced lesions after monofunctional EtNU. Nevertheless, SSB rates were significantly higher in the CD34+ progenitor fraction at all time points (Figure 5).
Although, in general, decreased activity of DNA repair pathways is associated with cellular sensitivity, the opposite phenotype has been described with MMR activities, that is, increased resistance with impairment of the pathway in various cell lines.29,30Additionally, experiments with mice deficient in more than one pathway revealed that in primary hematopoietic cells the relative activity of different pathways is important for the cellular phenotype.31 Thus, it is interesting to note that the reduced repair capacity of CD34+ cells observed in our studies most likely is not due to the low expression or activity of a single component of the complex DNA repair network (Figure 1). Because these cells are slow in removing drug-induced adducts (by melphalan or cisplatin; Figure 5) as well as in processing secondary lesions like SSBs (after EtNU, melphalan, or cisplatin), both early and late steps along repair pathways are probably working less effectively than in CD34− cells. Reduced DNA repair activity in more than one pathway is also suggested by our data generated with specific DNA repair modifiers (Figure 6), showing that both major pathways, BER and NER, are still operative in CD34+ cells.
Clinical application of most cytotoxic agents is followed by a myelosuppressive episode of 3 to 4 weeks duration, with nadirs usually occurring around days 10 to 14.32 This time course fits well with the notion that the damage inflicted by these agents on the hematopoietic system is most severe on the level of a CD34+progenitor cell population. Thus, the deficiencies in DNA repair, which we observed for this population, may very well contribute to the toxicity. Several of the alkylating agents, however, produce a more protracted pattern of myelosuppression. Besides the chloroethylnitrosourea- and triazene-type compounds, this has been described for mitomycin C, busulfan, and melphalan, and in murine models this delayed toxicity has been linked to extensive damage to the stem cell compartment.33-35 In view of these data, DNA repair analysis in a more primitive “stem cell” subfraction of the hematopoietic department, such as CD34+/CD38−cells, seems warranted. This type of analysis also appears interesting to clarify the role of DNA repair in drug-induced leukemias. Low levels of MGMT expression have been correlated with the development of methylnitrosourea-induced thymic lymphoma/leukemia, and it has been speculated that the low MGMT expression measured in primary human myeloid leukemic cells may be an etiologic factor in the induction of secondary leukemias following chemotherapy with O6-alkylating agents.36,37 Our data suggest that insufficient DNA repair may be a more widespread etiologic factor in human leukemias and may also be operative in the development of secondary leukemias after therapy with alkylating drugs such as nitrogen mustard, cyclophosphamide, chlorambucil, platinum derivatives, or the epipodophyllotoxins etoposide and teniposide.38-40
To our knowledge, we have shown here for the first time that human CD34+ progenitor cells have a grossly reduced overall repair capacity for a variety of drug-induced DNA lesions. This observation coincides with a higher risk of these cells in comparison to their CD34− counterparts to undergo apoptosis after exposure to melphalan or cisplatin ex vivo. From these observations we conclude that the pronounced sensitivity of the hematopoietic system to the toxic side effects of cancer chemotherapy with DNA-reactive drugs is, at least partly, due to its repair deficiency. On the other hand, our data indicate that the individual risk of suffering from myelosuppression may be very different according to the individual “repair phenotype.” If peripheral lymphocytes reflect the repair capacity of hematopoietic cells of BM, as shown here, functional repair assays in these cells prior to treatment might give valuable hints for severe side effects, a hypothesis which we will test in future studies.
The authors are indebted to Dr M. J. Tilby (University of Newcastle) for providing monoclonal antibodies against DNA adducts of melphalan. In addition, the authors would like to thank M. Stuhl, B. Karow, and B. Meurer for expert technical assistance, and C. Wartchow for critical reading of the manuscript.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2002-01-0022.
Supported in part by Wilhelm Sander-Stiftung (grant 1999.082.1) and Deutsche Forschungsgemeinschaft (grant FOR 236).
Each laboratory contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jürgen Thomale, Institute of Cell Biology, University of Essen Medical School, Virchowstr 173, D-45122 Essen, Germany; e-mail: juergen.thomale@uni-essen.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal