TO THE EDITOR:
The Wilms tumor 1 (WT1) gene, located at locus 11p13, is a tumor suppressor gene that encodes a zinc-finger transcription factor.1,2 The presence of WT1 gene mutation (mWT1) among adult patients with acute myeloid leukemia (AML) has been associated with poor prognoses.1,3 Outcomes of allogeneic stem cell transplantation (alloSCT) in patients with mWT1 AML have been explored in only a few studies limited by small sample size and inclusion of other gene mutations.4-7 The goal of this study was to determine factors affecting post-alloSCT survival in patients with mWT1 myeloid neoplasms.
We retrospectively reviewed patients with mWT1 myeloid neoplasms, as defined by the World Health Organization.8 The study was approved by the Mayo Clinic institutional review board and conducted in accordance with the Declaration of Helsinki. Patients with ≥2 WT1 mutations were deemed to have multihit WT1 (mhWT1) mutations. Given the limited sample size, we decided a priori upon variables deemed important from previous transplantation studies9-14 to be included in stepwise regression analysis and arrived at the final multivariate model. Please refer to supplemental Methods for a detailed description of methods.
A total of 6887 patients were tested, and 75 (1.1%) patients were found to harbor a WT1 mutation. Fifty-six (74.7%) had AML, 7 (9.3%) had myelodysplastic syndrome (MDS), 6 (8%) had mixed phenotypic acute leukemia (MPAL), and 6 (8%) patients had other myeloid neoplasms. Median age at diagnosis was 60 years (interquartile range [IQR], 42-67 years). Among patients with MDS, 4 (57.1%) had MDS with increased blasts (MDS-IB): 3 (42.9%) had MDS-IB2 and 1 (14.3%) had MDS-IB1. Median overall survival (OS) for the entire cohort was 1.9 years (95% confidence interval [CI], 1.45-2.42).
A total of 33 (44%) patients (21 [63.6%] males) underwent alloSCT at a median age of 44 years (IQR, 33-62 years). Twenty-five (75.8%) patients had AML, 3 (9.1%) had MDS, 2 (6.1%) had MPAL, and 3 (9.1%) had other myeloid neoplasms (supplemental Table 1). The median time to alloSCT after diagnosis was 6 months (IQR, 4-16 months). Twenty-seven (81.8%) patients were in complete remission (CR)/CR with incomplete count recovery at the time of alloSCT, 7 (21.2%) of whom were in second CR or beyond, whereas 5 (18.2%) patients, including 2 with AML and 1 with MPAL, had active disease. Post-alloSCT maintenance therapy was used in 13 (39.4%) patients, 8 (61.5%) of whom had a FLT3 mutation (supplemental Table 2; supplemental Figure 1).
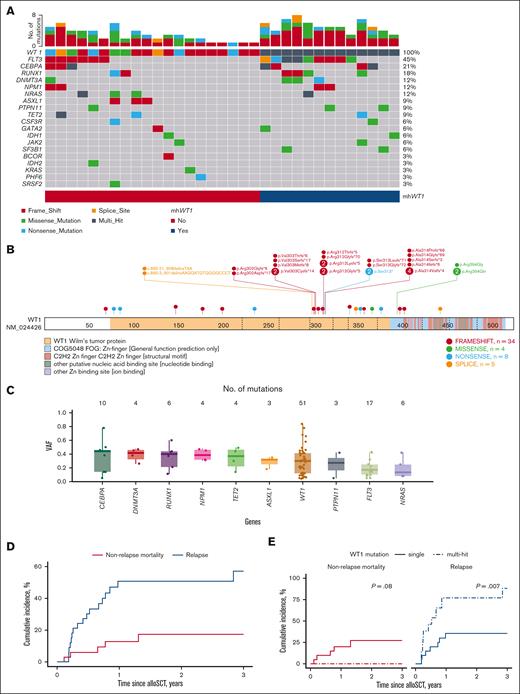
Five (15.2%) patients only had a WT1 mutation, whereas 28 (84.8%) had at least 1 other comutated gene (Figure 1A). Twenty-one (63.6%) patients were found to have WT1 at initial diagnosis, whereas 12 (36.4%) of the patients were found to have a WT1 mutation at first relapse or later (supplemental Table 3). The median number of comutations in the 28 patients was 2 (range, 1-5). The most frequently comutated gene was FLT3 (n = 15, 45.4%): 12 (80%) had FLT3–internal tandem duplication and 3 (20%) had FLT3–tyrosine kinase domain mutations. Several studies have shown that mWT1 AML is associated with FLT3 mutations.15-17 High-risk comutations such as BCOR or TP53 were rare (1 [3.3%] patient each). Thirteen (39.4%) patients had mhWT1 (Figure 1A; Table 1). Two hot-spot regions were found: codons 301 to 303 (7 [21.2%] patients), and codons 312 to 314 (13 [42.4%] patients; Figure 1B). Median WT1 variant allele frequency for patients undergoing alloSCT was 30% (IQR, 12%-42%; Figure 1C).
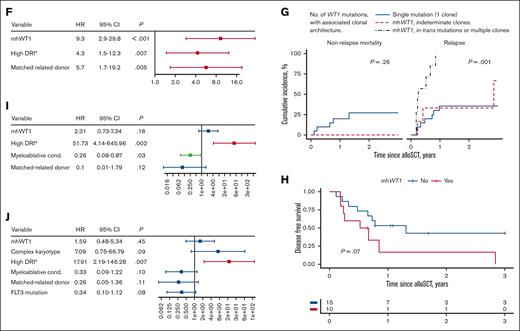
Genetic landscape and clinical outcomes of patients with WT1-mutated myeloid neoplasms undergoing alloSCT. (A) Oncoplot depicting comutations associated with WT1. Thirteen of 33 patients had mhWT1. (B) Lollipop plot showing position of mutations among 33 patients. Two hot spots were observed: codons 301-303 and codons 312-314. (C) Variant allele frequency (VAF) of top 10 mutated genes. (D) Post-alloSCT non-relapse mortality (NRM) and RI of the entire cohort. (E) Post-alloSCT NRM and RI stratified by single or mhWT1 mutations. (F) Multivariate competing risk regression analysis for relapse among patients undergoing alloSCT. (G) Cumulative incidence of NRM and relapse stratified by WT1 mutations and associated clonal architecture. (H) DFS after alloSCT, stratified by mhWT1. (I) Multivariate Cox proportional hazard analysis for 3-year DFS after transplantation in patients with mWT1 AML. (J) Multivariate Cox proportional hazard analysis for 3-year OS after transplantation. ∗None of the patients had a very high DRI.
Genetic landscape and clinical outcomes of patients with WT1-mutated myeloid neoplasms undergoing alloSCT. (A) Oncoplot depicting comutations associated with WT1. Thirteen of 33 patients had mhWT1. (B) Lollipop plot showing position of mutations among 33 patients. Two hot spots were observed: codons 301-303 and codons 312-314. (C) Variant allele frequency (VAF) of top 10 mutated genes. (D) Post-alloSCT non-relapse mortality (NRM) and RI of the entire cohort. (E) Post-alloSCT NRM and RI stratified by single or mhWT1 mutations. (F) Multivariate competing risk regression analysis for relapse among patients undergoing alloSCT. (G) Cumulative incidence of NRM and relapse stratified by WT1 mutations and associated clonal architecture. (H) DFS after alloSCT, stratified by mhWT1. (I) Multivariate Cox proportional hazard analysis for 3-year DFS after transplantation in patients with mWT1 AML. (J) Multivariate Cox proportional hazard analysis for 3-year OS after transplantation. ∗None of the patients had a very high DRI.
Characteristics and outcomes of patients with mWT1 stratified by single WT1 or mhWT1
| Variable . | mhWT1 . | P value . | |
|---|---|---|---|
| No . | Yes . | ||
| (n = 20) . | (n = 13) . | ||
| Age at diagnosis, y | |||
| Median (min, max) | 46.8 (18.4, 67.5) | 42.7 (26.7, 71.8) | .85 |
| Age at alloSCT, y | |||
| Median (min, max) | 48.0 (18.7, 68.0) | 44.1 (27.5, 72.2) | .8 |
| Sex | |||
| Female | 9 (45.0%) | 3 (23.1%) | .36 |
| Male | 11 (55.0%) | 10 (76.9%) | |
| Ethnicity | |||
| Caucasian | 17 (85.0%) | 11 (84.6%) | 1 |
| Other | 3 (15.0%) | 2 (15.4%) | |
| WT1 first detected | |||
| Initial diagnosis | 14 (70.0%) | 7 (53.8%) | .57 |
| Relapse | 6 (30.0%) | 6 (46.2%) | |
| Disease characteristics | |||
| Disease | |||
| AML | 15 (75%) | 10 (76.9%) | .39 |
| MDS | 1 (5%) | 2 (15.4%) | |
| MPAL | 1 (5%) | 1 (7.7%) | |
| Others | 3 (15%) | 0 (0%) | |
| Hemoglobin ≥ 10 g/dL at diagnosis | |||
| No | 17 (85.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 1 (7.7%) | |
| Missing | 1 (5.0%) | 1 (7.7%) | |
| Platelets ≥ 100 × 103/μL at diagnosis | |||
| No | 12 (60.0%) | 10 (76.9%) | .42 |
| Yes | 7 (35.0%) | 2 (15.4%) | |
| Missing | 1 (5.0%) | 1 (7.7%) | |
| Abnormal karyotype at diagnosis | |||
| No | 8 (40.0%) | 8 (61.5%) | .47 |
| Yes | 11 (55.0%) | 5 (38.5%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Monosomy 7 at diagnosis | |||
| No | 18 (90.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Complex karyotype at diagnosis | |||
| No | 16 (80.0%) | 13 (100%) | .38 |
| Yes | 3 (15.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Monosomal karyotype at diagnosis | |||
| No | 18 (90.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| maxWT1 VAF | |||
| Median [min, max] | 34.5 [4.00, 84.0] | 33.0 [12.0, 49.0] | .66 |
| Isolated WT1 (no comutation) | |||
| No | 15 (75.0%) | 13 (100%) | .14 |
| Yes | 5 (25.0%) | 0 (0%) | |
| mWT1 in codons 301-303 | |||
| No | 19 (95.0%) | 7 (53.8%) | .02 |
| Yes | 1 (5.0%) | 6 (46.2%) | |
| mWT1 in codons 312-314 | |||
| No | 14 (70.0%) | 5 (38.5%) | .15 |
| Yes | 6 (30.0%) | 8 (61.5%) | |
| mWT1 in zinc-finger motif | |||
| No | 17 (85.0%) | 11 (84.6%) | 1 |
| Yes | 3 (15.0%) | 2 (15.4%) | |
| CR/CRi at alloSCT | |||
| No | 2 (10.0%) | 3 (23.1%) | .64 |
| Yes | 17 (85.0%) | 10 (76.9%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| DRI | |||
| Low | 3 (15%) | 1 (7.7%) | .83 |
| Intermediate | 13 (65%) | 9 (69.2%) | |
| High | 3 (15%) | 2 (15.4%) | |
| NA | 1 (5%) | 1 (7.7%) | |
| Comutations | |||
| ASXL1 | |||
| No | 17 (85.0%) | 13 (100%) | .40 |
| Yes | 3 (15.0%) | 0 (0%) | |
| BCOR | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| CEBPA | |||
| No | 17 (85.0%) | 9 (69.2%) | .52 |
| Yes | 3 (15.0%) | 4 (30.8%) | |
| CSF3R | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| DNMT3A | |||
| No | 19 (95.0%) | 10 (76.9%) | .31 |
| Yes | 1 (5.0%) | 3 (23.1%) | |
| FLT3 | |||
| No | 14 (70.0%) | 4 (30.8%) | .06 |
| Yes | 6 (30.0%) | 9 (69.2%) | |
| IDH1 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| IDH2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| GATA2 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| JAK2 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| KRAS | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| NPM1 | |||
| No | 18 (90.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 2 (15.4%) | |
| NRAS | |||
| No | 17 (85.0%) | 12 (92.3%) | .93 |
| Yes | 3 (15.0%) | 1 (7.7%) | |
| PHF6 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| PTPN11 | |||
| No | 19 (95.0%) | 11 (84.6%) | .69 |
| Yes | 1 (5.0%) | 2 (15.4%) | |
| RUNX1 | |||
| No | 18 (90.0%) | 9 (69.2%) | .29 |
| Yes | 2 (10.0%) | 4 (30.8%) | |
| SF3B1 | |||
| No | 20 (100%) | 11 (84.6%) | .29 |
| Yes | 0 (0%) | 2 (15.4%) | |
| SRSF2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| TET2 | |||
| No | 18 (90.0%) | 12 (92.3%) | 1 |
| Yes | 2 (10.0%) | 1 (7.7%) | |
| TP53 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| ZRSR2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Transplantation characteristics and outcomes | |||
| HCT-CI ≥ 3 | |||
| No | 11 (55.0%) | 9 (69.2%) | .65 |
| Yes | 9 (45.0%) | 4 (30.8%) | |
| Conditioning intensity | |||
| Myeloablative | 12 (60.0%) | 8 (61.5%) | 1 |
| Reduced intensity | 7 (35.0%) | 5 (38.5%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Conditioning | |||
| Bu/Cy | 4 (20.0%) | 1 (7.7%) | NA |
| Bu/Flu | 4 (20.0%) | 3 (23.1%) | |
| Bu/Flu/ATG | 1 (5.0%) | 0 (0%) | |
| Cy/Flu/thiotepa/TLI | 2 (10.0%) | 0 (0%) | |
| Cy/TBI | 2 (10.0%) | 2 (15.4%) | |
| Flu/Mel | 4 (20.0%) | 4 (30.8%) | |
| Flu/TBI | 2 (10.0%) | 1 (7.7%) | |
| Bu/Cy/ATG | 0 (0%) | 1 (7.7%) | |
| Flu/BCNU/Mel | 1 (5%) | 1 (7.7%) | |
| Graft source | |||
| Peripheral blood | 18 (90.0%) | 13 (100%) | 1 |
| Bone marrow | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Donor type | |||
| MRD | 5 (25.0%) | 1 (7.7%) | .36 |
| MUD | 11 (55.0%) | 9 (69.2%) | |
| MMUD | 0 (0%) | 1 (7.7%) | |
| Haploidentical | 4 (20.0%) | 2 (15.4%) | |
| Major/bidirectional ABO mismatch | |||
| No | 15 (75.0%) | 10 (76.9%) | 1 |
| Yes | 4 (20.0%) | 3 (23.1%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| GVHD prophylaxis | |||
| CD34 selection | 1 (5.0%) | 0 (0%) | .16 |
| Tacrolimus + methotrexate (± ATG) | 13 (65%) | 9 (69.3%) | |
| Cyclosporine + methotrexate | 2 (10.0%) | 1 (7.7%) | |
| Tacrolimus + mycophenolate | 3 (15.0%) | 0 (0%) | |
| PT-Cy based | 0 (0%) | 3 (23.1%) | |
| None | 1 (5.0%) | 0 (0%) | |
| Grade 2-4 acute GVHD | |||
| No | 17 (85.0%) | 10 (76.9%) | .9 |
| Yes | 3 (15.0%) | 3 (23.1%) | |
| Grade 3-4 acute GVHD | |||
| No | 13 (65.0%) | 12 (92.3%) | .17 |
| Yes | 7 (35.0%) | 1 (7.7%) | |
| Moderate/severe chronic GVHD | |||
| No | 18 (90.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 2 (15.4%) | |
| Variable . | mhWT1 . | P value . | |
|---|---|---|---|
| No . | Yes . | ||
| (n = 20) . | (n = 13) . | ||
| Age at diagnosis, y | |||
| Median (min, max) | 46.8 (18.4, 67.5) | 42.7 (26.7, 71.8) | .85 |
| Age at alloSCT, y | |||
| Median (min, max) | 48.0 (18.7, 68.0) | 44.1 (27.5, 72.2) | .8 |
| Sex | |||
| Female | 9 (45.0%) | 3 (23.1%) | .36 |
| Male | 11 (55.0%) | 10 (76.9%) | |
| Ethnicity | |||
| Caucasian | 17 (85.0%) | 11 (84.6%) | 1 |
| Other | 3 (15.0%) | 2 (15.4%) | |
| WT1 first detected | |||
| Initial diagnosis | 14 (70.0%) | 7 (53.8%) | .57 |
| Relapse | 6 (30.0%) | 6 (46.2%) | |
| Disease characteristics | |||
| Disease | |||
| AML | 15 (75%) | 10 (76.9%) | .39 |
| MDS | 1 (5%) | 2 (15.4%) | |
| MPAL | 1 (5%) | 1 (7.7%) | |
| Others | 3 (15%) | 0 (0%) | |
| Hemoglobin ≥ 10 g/dL at diagnosis | |||
| No | 17 (85.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 1 (7.7%) | |
| Missing | 1 (5.0%) | 1 (7.7%) | |
| Platelets ≥ 100 × 103/μL at diagnosis | |||
| No | 12 (60.0%) | 10 (76.9%) | .42 |
| Yes | 7 (35.0%) | 2 (15.4%) | |
| Missing | 1 (5.0%) | 1 (7.7%) | |
| Abnormal karyotype at diagnosis | |||
| No | 8 (40.0%) | 8 (61.5%) | .47 |
| Yes | 11 (55.0%) | 5 (38.5%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Monosomy 7 at diagnosis | |||
| No | 18 (90.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Complex karyotype at diagnosis | |||
| No | 16 (80.0%) | 13 (100%) | .38 |
| Yes | 3 (15.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Monosomal karyotype at diagnosis | |||
| No | 18 (90.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| maxWT1 VAF | |||
| Median [min, max] | 34.5 [4.00, 84.0] | 33.0 [12.0, 49.0] | .66 |
| Isolated WT1 (no comutation) | |||
| No | 15 (75.0%) | 13 (100%) | .14 |
| Yes | 5 (25.0%) | 0 (0%) | |
| mWT1 in codons 301-303 | |||
| No | 19 (95.0%) | 7 (53.8%) | .02 |
| Yes | 1 (5.0%) | 6 (46.2%) | |
| mWT1 in codons 312-314 | |||
| No | 14 (70.0%) | 5 (38.5%) | .15 |
| Yes | 6 (30.0%) | 8 (61.5%) | |
| mWT1 in zinc-finger motif | |||
| No | 17 (85.0%) | 11 (84.6%) | 1 |
| Yes | 3 (15.0%) | 2 (15.4%) | |
| CR/CRi at alloSCT | |||
| No | 2 (10.0%) | 3 (23.1%) | .64 |
| Yes | 17 (85.0%) | 10 (76.9%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| DRI | |||
| Low | 3 (15%) | 1 (7.7%) | .83 |
| Intermediate | 13 (65%) | 9 (69.2%) | |
| High | 3 (15%) | 2 (15.4%) | |
| NA | 1 (5%) | 1 (7.7%) | |
| Comutations | |||
| ASXL1 | |||
| No | 17 (85.0%) | 13 (100%) | .40 |
| Yes | 3 (15.0%) | 0 (0%) | |
| BCOR | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| CEBPA | |||
| No | 17 (85.0%) | 9 (69.2%) | .52 |
| Yes | 3 (15.0%) | 4 (30.8%) | |
| CSF3R | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| DNMT3A | |||
| No | 19 (95.0%) | 10 (76.9%) | .31 |
| Yes | 1 (5.0%) | 3 (23.1%) | |
| FLT3 | |||
| No | 14 (70.0%) | 4 (30.8%) | .06 |
| Yes | 6 (30.0%) | 9 (69.2%) | |
| IDH1 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| IDH2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| GATA2 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| JAK2 | |||
| No | 19 (95.0%) | 12 (92.3%) | 1 |
| Yes | 1 (5.0%) | 1 (7.7%) | |
| KRAS | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| NPM1 | |||
| No | 18 (90.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 2 (15.4%) | |
| NRAS | |||
| No | 17 (85.0%) | 12 (92.3%) | .93 |
| Yes | 3 (15.0%) | 1 (7.7%) | |
| PHF6 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| PTPN11 | |||
| No | 19 (95.0%) | 11 (84.6%) | .69 |
| Yes | 1 (5.0%) | 2 (15.4%) | |
| RUNX1 | |||
| No | 18 (90.0%) | 9 (69.2%) | .29 |
| Yes | 2 (10.0%) | 4 (30.8%) | |
| SF3B1 | |||
| No | 20 (100%) | 11 (84.6%) | .29 |
| Yes | 0 (0%) | 2 (15.4%) | |
| SRSF2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| TET2 | |||
| No | 18 (90.0%) | 12 (92.3%) | 1 |
| Yes | 2 (10.0%) | 1 (7.7%) | |
| TP53 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| ZRSR2 | |||
| No | 19 (95.0%) | 13 (100%) | 1 |
| Yes | 1 (5.0%) | 0 (0%) | |
| Transplantation characteristics and outcomes | |||
| HCT-CI ≥ 3 | |||
| No | 11 (55.0%) | 9 (69.2%) | .65 |
| Yes | 9 (45.0%) | 4 (30.8%) | |
| Conditioning intensity | |||
| Myeloablative | 12 (60.0%) | 8 (61.5%) | 1 |
| Reduced intensity | 7 (35.0%) | 5 (38.5%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Conditioning | |||
| Bu/Cy | 4 (20.0%) | 1 (7.7%) | NA |
| Bu/Flu | 4 (20.0%) | 3 (23.1%) | |
| Bu/Flu/ATG | 1 (5.0%) | 0 (0%) | |
| Cy/Flu/thiotepa/TLI | 2 (10.0%) | 0 (0%) | |
| Cy/TBI | 2 (10.0%) | 2 (15.4%) | |
| Flu/Mel | 4 (20.0%) | 4 (30.8%) | |
| Flu/TBI | 2 (10.0%) | 1 (7.7%) | |
| Bu/Cy/ATG | 0 (0%) | 1 (7.7%) | |
| Flu/BCNU/Mel | 1 (5%) | 1 (7.7%) | |
| Graft source | |||
| Peripheral blood | 18 (90.0%) | 13 (100%) | 1 |
| Bone marrow | 1 (5.0%) | 0 (0%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| Donor type | |||
| MRD | 5 (25.0%) | 1 (7.7%) | .36 |
| MUD | 11 (55.0%) | 9 (69.2%) | |
| MMUD | 0 (0%) | 1 (7.7%) | |
| Haploidentical | 4 (20.0%) | 2 (15.4%) | |
| Major/bidirectional ABO mismatch | |||
| No | 15 (75.0%) | 10 (76.9%) | 1 |
| Yes | 4 (20.0%) | 3 (23.1%) | |
| Missing | 1 (5.0%) | 0 (0%) | |
| GVHD prophylaxis | |||
| CD34 selection | 1 (5.0%) | 0 (0%) | .16 |
| Tacrolimus + methotrexate (± ATG) | 13 (65%) | 9 (69.3%) | |
| Cyclosporine + methotrexate | 2 (10.0%) | 1 (7.7%) | |
| Tacrolimus + mycophenolate | 3 (15.0%) | 0 (0%) | |
| PT-Cy based | 0 (0%) | 3 (23.1%) | |
| None | 1 (5.0%) | 0 (0%) | |
| Grade 2-4 acute GVHD | |||
| No | 17 (85.0%) | 10 (76.9%) | .9 |
| Yes | 3 (15.0%) | 3 (23.1%) | |
| Grade 3-4 acute GVHD | |||
| No | 13 (65.0%) | 12 (92.3%) | .17 |
| Yes | 7 (35.0%) | 1 (7.7%) | |
| Moderate/severe chronic GVHD | |||
| No | 18 (90.0%) | 11 (84.6%) | 1 |
| Yes | 2 (10.0%) | 2 (15.4%) | |
ATG, anti-thymocyte globulin; BCNU, carmustine; Bu, busulfan; CRi, complete remission with incomplete count recovery; Cy, cyclophosphamide; Flu, fludarabine; GVHD, graft-versus-host disease; HCT-CI, hematopoietic stem cell transplant comorbidity index; max, maximum; Mel, melphalan; min, minimum; MRD, matched related donor; MMUD, mismatched unrelated donor; MUD, matched unrelated donor; PT, posttransplant; TBI, total body irradiation; TLI, total lymphoid irradiation; VAF, variant allele frequency.
The cumulative relapse incidence (RI) was 21.1% at 100 days, 50.7% at 1 year, and 57.1% at 3 years after alloSCT (Figure 1D). Patients with mhWT1 had higher post-alloSCT RI at 100 days (38.5% vs 10%), 1 year (76.9% vs 35.6%), and 3 years (88.5% vs 35.6%; P = .007; Figure 1E).
In multivariate analysis, mhWT1 (hazard ratio [HR], 9.3; 95% CI, 2.9-29.8; P < .001), matched-related donor, and high disease risk index (DRI) were associated with an increased risk of posttransplant relapse (Figure 1F). We then studied the impact of mhWT1 mutations in cis vs in trans/multiple clones on relapse. Of the 13 patients with mhWT1, 7 (53.8%) had WT1 mutations either in trans or had multiple WT1 clones, whereas the clonal architecture could not be determined in 6 (46.2%) patients with mhWT1. Patients with mutations in trans/multiple clones had a significantly higher RI than patients with mhWT1 with indeterminate clonal architecture, or a single WT1 mutation (1-year RI, 100% vs 33.3% vs 35.6%, P = .001; and 3-year RI, 100% vs 66.7% vs 35.6%, P = .001; supplemental Figure 1G).
Among patients with AML, those with mhWT1 had an inferior 3-year disease-free survival (DFS; 3-year DFS, 0% vs 42.7%; P = .07; Figure 1H). Data on multiparameter flow cytometry–based minimal residual disease (MRD) testing were available in 14 (56%) patients with AML, 8 (57.1%) of whom were MRD negative (supplemental Table 4). Patients with MRD positive or unknown status before transplantation had a significantly higher relapse rate than patients who were MRD negative at 1 year (66.7% vs 63.6% vs 0.00%; P = .03) and 3 years (not applicable [NA] vs 72.7% vs 0.00%; P = .02) after alloSCT (supplemental Figure 2). Multivariate analysis showed that a high DRI was associated with an inferior DFS (HR, 51.73; 95% CI, 4.14-645.96; P = .002). Although mhWT1 was associated with an inferior DFS, it was not statistically significant (HR, 2.31; 95% CI, 0.73-7.34; P = .16; Figure 1I).
Median follow-up after alloSCT was 3.6 years (95% CI, 1.69-NA). Median OS for the entire cohort was 1.31 years (95% CI, 1.12-NA) and was similar to that of patients with mWT1 AML undergoing alloSCT (median OS, 1.45 years; 95% CI, 1.12-NA; P = .81; supplemental Figure 3). Post-alloSCT 3-year OS was comparable among patients with AML vs those with non-AML disease (median, 1.45 vs 1.2 years; P = .57). Multivariate analysis confirmed that a high DRI was associated with a worse 3-year OS (HR, 17.91; 95% CI, 2.19-146.28; P = .007), whereas mhWT1 was not associated with OS (HR, 1.59; 95% CI, 0.48-5.34; P = .45; Figure 1J).
Our study shows that the majority of patients with mWT1 myeloid malignancies have either acute leukemia or MDS-IB. These findings have been reported previously.18,19 We found a high post-alloSCT RI in this subset of patients. Luskin et al evaluated 112 patients with AML, 8 of whom had mWT1, and found that mWT1 was associated with an increased post-alloSCT relapse (HR, 2.07; P = .07).6 Similarly, Quek et al also reported that mWT1 was associated with an increased risk of relapse (HR, 4.81; P = .018).7 Most importantly, we found that mhWT1 was associated with a high relapse rate, particularly among those with mutations in trans or with multiple WT1 clones. Further studies involving single-cell sequencing will help confirm this finding.
Although mhWT1 was not associated with an inferior post-alloSCT OS, there was a trend toward poor DFS. Likely, the early relapse detection through MRD and molecular-based studies and improved postrelapse treatment strategies influenced postrelapse survival. Post-alloSCT maintenance therapy was associated with a numerically superior 3-year post-alloSCT OS (median, 21 vs 11 months; P = .16) and DFS (median, 13 vs 7 months; P = .39; supplemental Figure 4). We did not find any specific comutation confounding the effect of mhWT1 on post-alloSCT relapse (supplemental Table 5). Post-alloSCT survival was similar among patients with WT1 mutation detected at initial diagnosis vs at relapse (median OS, 1.31 vs 1.23 years; P = .61).
Some of the limitations of our study include its retrospective nature and the small sample size.
In conclusion, this is, to our knowledge, the first study evaluating posttransplant outcomes in patients with WT1-associated myeloid malignancies. Our study shows that patients with WT1 mutation are at a high risk of posttransplant relapse, particularly patients with mhWT1 and, most importantly, those with mhWT1 mutations in trans or multiple WT1 clones. Post-alloSCT survival is poor, predominantly driven by relapse. Larger studies are needed to validate these findings.
Acknowledgment:Figure 1B was created using ProteinPaint (https://proteinpaint.stjude.org/).
Contribution: H.B.A. conceptualized the study; A.B. and H.B.A. contributed to study design, analyzed data, and wrote the manuscript; R.B. contributed to data acquisition and reviewed the manuscript; R.H., D.V., and P.G. provided cytogenetics and molecular data; H.S.M., J.F., J.P., W.J.H., M.R.L., M.H., A.M., M.V.S., and A.A.-K. recruited patients; and all authors reviewed the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hassan B. Alkhateeb, Mayo Clinic, Division of Hematology, 200 1st St SW, Rochester, MN 55905; email: alkhateeb.hassan@mayo.edu.
References
Author notes
The data sets generated and analyzed in this study may be obtained upon reasonable request from the corresponding author, Hassan B. Alkhateeb (alkhateeb.hassan@mayo.edu).
The full-text version of this article contains a data supplement.