Key Points
Expression of VNN2 identifies B-cell precursor ALL cases with increased resistance to chemotherapy.
VNN2 positivity occurs across different genomic ALL subtypes and is associated with mutations in epigenetic regulators.
Abstract
Most relapses of acute lymphoblastic leukemia (ALL) occur in patients with a medium risk (MR) for relapse on the Associazione Italiana di Ematologia e Oncologia Pediatrica and Berlin-Frankfurt-Münster (AIEOP-BFM) ALL protocol, based on persistence of minimal residual disease (MRD). New insights into biological features that are associated with MRD are needed. Here, we identify the glycosylphosphatidylinositol-anchored cell surface protein vanin-2 (VNN2; GPI-80) by charting the cell surface proteome of MRD very high-risk (HR) B-cell precursor (BCP) ALL using a chemoproteomics strategy. The correlation between VNN2 transcript and surface protein expression enabled a retrospective analysis (ALL-BFM 2000; N = 770 cases) using quantitative polymerase chain reaction to confirm the association of VNN2 with MRD and independent prediction of worse outcome. Using flow cytometry, we detected VNN2 expression in 2 waves, in human adult bone marrow stem and progenitor cells and in the mature myeloid compartment, in line with proposed roles for fetal hematopoietic stem cells and inflammation. Prospective validation by flow cytometry in the ongoing clinical trial (AIEOP-BFM 2009) identified 10% (103/1069) of VNN2+ BCP ALL patients at first diagnosis, primarily in the MRD MR (48/103, 47%) and HR (37/103, 36%) groups, across various cytogenetic subtypes. We also detected frequent mutations in epigenetic regulators in VNN2+ ALLs, including histone H3 methyltransferases MLL2, SETD2, and EZH2 and demethylase KDM6A. Inactivation of the VNN2 gene did not impair leukemia repopulation capacity in xenografts. Taken together, VNN2 marks a cellular state of increased resistance to chemotherapy that warrants further investigations. Therefore, this marker should be included in diagnostic flow cytometry panels.
Introduction
Despite the remarkable improvements in molecular classification,1 our understanding of the factors associated with treatment resistance remains scarce. Thus far, assessment of minimal residual disease (MRD) remains 1 of the most important parameters for risk stratification in acute lymphoblastic leukemia (ALL).2 More recently, improved classifiers were proposed based on the combination of cytogenetic information and MRD. Such a score can help to classify patients with a very low risk for relapse,3,4 providing a rationale to decrease treatment intensity. Markers to identify patients at very high risk (VHR) for relapse have been defined for rare subgroups, such as TCF3-HLF+ ALL,5 hypodiploid ALL,6 or KMT2A (MLL) rearrangements.7 On the Associazione Italiana di Ematologia e Oncologia Pediatrica and Berlin-Frankfurt-Münster (AIEOP-BFM) ALL 2009 treatment protocol,2 the majority of relapses occur in the group of patients who are at medium risk (MR) by MRD. Recently, a subgroup with very poor outcome has been identified with deletions in IKAROS, together with other mutations (IKZF1plus).8 The significance of BCR-ABL–like mutations in tyrosine kinase genes9 for VHR for relapse classification remains to be clarified, because precise information from next-generation sequencing is emerging.1 Clearly, more efforts are needed to improve our understanding and molecular classification of resistant disease.
Mapping the leukemic cell surface more precisely will be relevant in the era of antigen receptor–directed immunotherapy. Using the chemoproteomics cell surface capturing (CSC) technology to phenotype cells without antibodies, a bird’s eye view of the repertoire of expressed cell surface proteins and their relative abundance (“surfaceome”) can be determined in development10 and disease.11,12 Here, we report about surfaceome proteins that are differentially present on leukemia cells with increased resistance to conventional treatment in ALL compared with good-risk ALL and identify an association between the glycosylphosphatidylinositol (GPI)-anchored surface protein vanin-2 (VNN2; GPI-80) and MRD in the AIEOP-BFM ALL 2009 study. The human vanin gene family consists of VNN1, VNN2, and VNN3, whereas in mice, only Vnn1 and Vnn3 orthologs have been identified.13 Human VNN1 and VNN2 are membrane-associated proteins, whereas VNN3 lacks GPI anchoring and is secreted.14 The function of the vanin family remains to be explored. VNN1 has pantetheinase activity to generate pantothenic acid and cysteamine, activating stress pathways and inflammation by inhibition of gluthatione.14,15 Additionally, VNN1 is involved in the thymus homing of immature T lymphocytes in mice.16 VNN2 is mainly expressed in the hematopoietic system on myeloid cells and, to a lesser extent, on lymphoid cells, contributing to leukocyte adhesion and migration to inflammatory sites.17 Recently, VNN2 has been implicated as a human fetal liver hematopoietic stem cell (FLHSC) marker, identifying a subset of FLHSCs in xenotransplantation assays,18 as well as a marker of CD34− human cord blood–derived primitive hematopoietic stem cells (HSCs).19 Our study establishes the association between VNN2 and drug resistance across different molecular leukemia subtypes, providing a new marker for phenotypic exploration.
Materials and methods
Patient material
Patient specimens were included for the retrospective analysis of patients who were enrolled on the ALL-FM 2000 trial (NCT00430118) between June 1999 and December 2004, according to availability in the central biological specimen bank (supplemental Figure 1A). The characteristics of this cohort are provided in Table 1.20 The prospective analysis was performed by the reference laboratories of the AIEOP-BFM ALL 2009 (NCT01117441) for patients enrolled between 2009 and 2018. Relapse samples were available from patients included in the BFM relapse trial 2002 (ALL-REZ BFM 2002; NCT00114348). Informed consent, in accordance with the Declaration of Helsinki, was obtained for all patients. Patients were classified as standard risk (SR), MR, or high risk (HR) for relapse, according to the clinical criteria used in the ALL-BFM 2000 trial.21,22 Briefly, HR patients are those presenting with 1 of the following features: poor response to prednisone, no complete remission at the end of induction on day 33, MRD ≥1 × 10−3 after consolidation at time point 2 (day 78), or KMT2A-AFF1 positivity. SR patients are MRD negative at the end of induction (day 33) and after consolidation (day 78), with ≥2 markers with a sensitivity of 10−4 and no HR criteria. All other patients were classified as MR.
Clinical features of VNN2+ patients
| Classifier . | Retrospective cohort (PCR) . | Retrospective cohort (PCR) . | Prospective cohort (FACS) . | Retrospective cohort (n = 770) . | ALL-BFM-2000 (n = 4152) . |
|---|---|---|---|---|---|
| VNN2 | Negative | Positive | Positive | ||
| 615 (79.8) | 155 (20.1) | 72 (—) | |||
| Age at diagnosis, y | |||||
| 1-10 | 456 (74.1) | 101 (65.1) | 41 (57.9) | 557 (72.3) | 3099 (74.6) |
| ≥10 | 159 (25.8) | 54 (34.8) | 31 (43.0) | 213 (27.6) | 1053 (25.4) |
| WBC count, ×109/L | |||||
| <50 | 458 (74.4) | 89 (57.4) | 52 (72.2) | 547 (71.0) | 3351 (80.7) |
| 50-100 | 74 (12.0) | 28 (16.1) | 5 (6.9) | 102 (13.2) | 383 (9.2) |
| ≥100 | 83 (13.4) | 38 (24.5) | 3 (4.1) | 121 (15.7) | 403 (9.7) |
| CNS status | |||||
| Negative (1) | 518 (84.2) | 134 (86.4) | 53 (73.6) | 712 (92.5) | 3757 (90.7) |
| Positive (2, 3) | 22 (3.5) | 10 (6.4) | 12 (16.6) | 32 (4.1) | 142 (3.4) |
| Risk (MRD) | |||||
| SR | 209 (33.9) | 32 (20.6) | 18 (25.0) | 241 (31.2) | 1341 (32.3) |
| MR | 268 (43.5) | 78 (50.3) | 4 (66.6) | 346 (44.9) | 1779 (42.8) |
| HR | 30 (4.9) | 14 (9.0) | 37 (51.4) | 44 (5.7) | 146 (5.9) |
| TEL-AML1 | |||||
| Negative | 434 (70.6) | 135 (87.7) | 69 (95.8) | 569 (73.8) | 3019 (74) |
| Positive | 160 (26.0) | 16 (10.3) | 3 (4.1) | 176 (22.8) | 868 (20.9) |
| KMT2A | |||||
| Negative | 612 (99.5) | 152 (99.5) | 71 (99) | 764 (99.2) | 4130 (99.5) |
| Positive | 3 (0.2) | 3 (0.2) | 1 (1.3) | 3 (0.5) | 6 (0.8) |
| CRLF2 aberrations | |||||
| Immunoglobulin H fusion | 4 (0.6) | — (—) | 1 (1.3) | 4 (0.7) | 4 (0.5) |
| P2RY8 fusion | 16 (2.6) | 3.2 (—) | — (—) | 21 (2.7) | 21 (2.7) |
| 3X CRLF2 | 20 (3.2) | 4 (2.5) | — (—) | 24 (3.1) | 24 (3.1) |
| Classifier . | Retrospective cohort (PCR) . | Retrospective cohort (PCR) . | Prospective cohort (FACS) . | Retrospective cohort (n = 770) . | ALL-BFM-2000 (n = 4152) . |
|---|---|---|---|---|---|
| VNN2 | Negative | Positive | Positive | ||
| 615 (79.8) | 155 (20.1) | 72 (—) | |||
| Age at diagnosis, y | |||||
| 1-10 | 456 (74.1) | 101 (65.1) | 41 (57.9) | 557 (72.3) | 3099 (74.6) |
| ≥10 | 159 (25.8) | 54 (34.8) | 31 (43.0) | 213 (27.6) | 1053 (25.4) |
| WBC count, ×109/L | |||||
| <50 | 458 (74.4) | 89 (57.4) | 52 (72.2) | 547 (71.0) | 3351 (80.7) |
| 50-100 | 74 (12.0) | 28 (16.1) | 5 (6.9) | 102 (13.2) | 383 (9.2) |
| ≥100 | 83 (13.4) | 38 (24.5) | 3 (4.1) | 121 (15.7) | 403 (9.7) |
| CNS status | |||||
| Negative (1) | 518 (84.2) | 134 (86.4) | 53 (73.6) | 712 (92.5) | 3757 (90.7) |
| Positive (2, 3) | 22 (3.5) | 10 (6.4) | 12 (16.6) | 32 (4.1) | 142 (3.4) |
| Risk (MRD) | |||||
| SR | 209 (33.9) | 32 (20.6) | 18 (25.0) | 241 (31.2) | 1341 (32.3) |
| MR | 268 (43.5) | 78 (50.3) | 4 (66.6) | 346 (44.9) | 1779 (42.8) |
| HR | 30 (4.9) | 14 (9.0) | 37 (51.4) | 44 (5.7) | 146 (5.9) |
| TEL-AML1 | |||||
| Negative | 434 (70.6) | 135 (87.7) | 69 (95.8) | 569 (73.8) | 3019 (74) |
| Positive | 160 (26.0) | 16 (10.3) | 3 (4.1) | 176 (22.8) | 868 (20.9) |
| KMT2A | |||||
| Negative | 612 (99.5) | 152 (99.5) | 71 (99) | 764 (99.2) | 4130 (99.5) |
| Positive | 3 (0.2) | 3 (0.2) | 1 (1.3) | 3 (0.5) | 6 (0.8) |
| CRLF2 aberrations | |||||
| Immunoglobulin H fusion | 4 (0.6) | — (—) | 1 (1.3) | 4 (0.7) | 4 (0.5) |
| P2RY8 fusion | 16 (2.6) | 3.2 (—) | — (—) | 21 (2.7) | 21 (2.7) |
| 3X CRLF2 | 20 (3.2) | 4 (2.5) | — (—) | 24 (3.1) | 24 (3.1) |
All data are n (%).
—, no sample of the corresponding genetic group observed; WBC, white blood cell.
Identification of proteins that are preferentially expressed in HR ALL
To select proteins preferentially identified in HR ALL, we used a proteomics dataset that was obtained using CSC technology, as previously reported.12 To identify the ALL surfaceome proteotype, cell surface residing glycoproteins were chemically tagged on living cells and subsequently analyzed by data-dependent acquisition mass spectrometry.12 A comparable number of peptides that were matched to proteins were identified in VHR and SR patients, with an average 5481 peptides in SR ALL and 5714 peptides in VHR ALL. Data from Mirkowska et al12 are included in supplemental Table 1. To identify candidates of the cell surface proteome that are expressed more often at the surface of 8 HR ALL samples compared with the 11 SR ALL samples in that dataset, we filtered for proteins that are identified in ≥50% of all HR cases (≥4/8 samples) and not more than 30% of SR cases (≤3 samples).
Immunophenotyping
Immunophenotyping was done in the corresponding BFM centers, and data were generated using the stringent diagnostic standards developed and approved by the AIEOP-BFM ALL FLOW-SG,23 which are based on 2016 World Health Organization criteria, the EGIL classification, and the Bethesda 2006 recommendations.
Validation of VNN2 antibodies by flow cytometry and definition of VNN2 positivity
We tested 2 commercial monoclonal VNN2 antibodies: 1 that was generated with recombinant human VNN2 (clone 04) and another that was generated with extracts from neutrophils (clone 3H9). We stained 3 human leukemia samples and observed a stronger signal with the 3H9 clone (supplemental Figure 1B). We further validated this clone by staining peripheral blood (PB) cells from a healthy adult donor (supplemental Figure 1C) and by generating lentiviral VNN2-knockout patient-derived xenograft (PDX) cells and VNN2-overexpressing cells (supplemental Figure 1D).
To define positivity in patient samples, the internal cross-lineage CD45bright lymphocyte population for each sample was used to set the VNN2 gate. The frequency of VNN2 positivity was evaluated based on the standard operating procedure of AIEOP-BFM ALL Immunophenotyping Consensus Guidelines.23 A frequency of VNN2+ cells ≥10% of control was considered positive for VNN2. To define positivity in xenograft samples, we used a fluorescence-minus-one control, which consisted of a sample stained with all antibodies except VNN2. VNN2 staining always detected a homogeneous population with increased fluorescence compared with in-sample control lymphocyte populations.
Quantitative polymerase chain reaction
Total RNA was isolated with TRIzol reagent and subsequently extracted with an RNeasy Mini Kit (cat. no, 74106; Qiagen), according to standardized protocols. The amount and quality of the extracted RNA were measured with a spectrophotometer (Nanodrop ND-1000) and a Bioanalyzer.
TaqMan Gene Expression Assays were used to measure messenger RNA (mRNA) abundance of VNN2, with SDHA as control gene. TaqMan Gene Expression Master Mix (cat. no. 4369016; Applied Biosystems, Foster City, CA), Assay-On-Demand (TaqMan Gene Expression Assays, Hs01546812_m1 VNN1, Hs00190581_m1 VNN2, Hs00417200_m1 SDHA; Applied Biosystems), and water were mixed (5:3:2), and complementary DNA (1:3 dilution) was added. The same was done using a SYBR Green assay for VNN2 and SDHA as control gene (QuantiTect SYBR Green PCR Kit, Power SYBR Green PCR Master Mix, cat. no. 4367659, Applied Biosystems; Quantitect Primer Assay, Hs_VNN2_1_SG, cat. no. QT00034902 and Hs_SDHA_1_SG, cat. no. QT00059486, Qiagen).
A 7900HT Fast Real-Time PCR System (Applied Biosystems) was used for both methods. We used the following cycles: 2 minutes 50°C, 10 minutes 95°C, 50× (10 seconds 95°C/1 minute 60°C). Analysis was performed with SDS 2.2 software using the 2-ΔCT value.
Statistics
VNN2 threshold was defined based on SYBR Green PCR values, which divided a group of 770 patients from the ALL-BFM 2000 study into a training set (one third of the patients) and a test set (two thirds of the patients). The VNN2 threshold associated with a higher risk for relapse was defined based on the lowest P value in a univariate Cox model in the training set. This threshold was used for dichotomizing the patients in the test set. Samples with 2-ΔΔCT > 1.13 (cutoff VNN2 transcript level) were tested by flow cytometry (FCM) based on sample availability. Event-free survival was defined as the time from diagnosis to the date of last follow-up or first event. Events were nonresponse, relapse, secondary neoplasm, or death from any cause. Failure to achieve remission due to death before complete remission or nonresponse was considered as event at time 0. The Kaplan-Meier method was used to estimate survival rates; differences between groups were compared with the log-rank test. Cumulative incidence functions for competing events were constructed using the method of Kalbfleisch and Prentice and were compared with Gray’s test.24 Statistical analysis was done using GraphPad Prism version 8.0 (GraphPad Software). The prognostic value of VNN2 was analyzed with a Cox regression model including covariates that are known to be risk factors in the ALL-BFM 2000 study and used as covariates in another study8 : ETV6-RUNX1 positivity, poor prednisone response, white blood cell count ≥100 × 103/μL, MRD low risk, MRD HR, and MRD slow early response. Statistical analysis was done using GraphPad Prism 8 (GraphPad software) and SAS 9.4 (SAS Institute, Cary, NC).
Results
A set of proteins implicated in cell adhesion and immune response is preferentially present on the surface of MRD+ leukemia
To identify cell surface markers that are characteristic of resistant disease, we interrogated the cell surface proteomes of 19 B-cell precursor (BCP) ALL PDXs previously acquired using CSC technology.12 This dataset included specimens from 8 patients with VHR ALL, as defined by persistence of MRD (>5 × 10−4) after induction chemotherapy, consolidation therapy, and 3 highly intensive postconsolidation blocks according to the ALL-BFM 2000 protocol,21,25,26 and 11 patients with an excellent clinical outcome (SR ALL). We identified 13 proteins that were detected more frequently in VHR ALL (Table 2), including several surface proteins that are involved in cell adhesion and migration: the GPI-anchored vanin family of proteins (VNN1 and VNN2 [GPI-80]), with VNN2 being exclusively detected on the cell surface of VHR ALL cases, the immunoglobulin-like CEA family member CEAM1, the C-type lectin receptor CD302, and the sialic acid binding immunoglobulin-like lectin CD33, as well as ITGAM (CD11b), which is a subunit of the complement receptor 3 and has been linked to chemotherapy resistance in ALL.27 Furthermore, the receptors for activated complement CR1 and CR2, as well as components of prosurvival signaling pathways (TNFR1B, IGF1R), were detected in VHR ALL cases.
Proteins identified in MRD VHR ALL by CSC
| . | SR . | VHR . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 . | 02 . | 03 . | 04 . | 05 . | 06 . | 07 . | 14 . | 15 . | 16 . | 17 . | 08 . | 09 . | 10 . | 11 . | 12 . | 13 . | 18 . | 19 . | |
| Adhesion and migration | |||||||||||||||||||
| VNN2 | + | + | + | + | |||||||||||||||
| VNN1 | + | + | + | + | + | + | + | + | |||||||||||
| ITGAM | + | + | + | + | + | + | |||||||||||||
| CD33 | + | + | + | + | + | ||||||||||||||
| CD302 | + | + | + | + | + | ||||||||||||||
| CEAM1 | + | + | + | + | |||||||||||||||
| Signaling | |||||||||||||||||||
| TNR1B | + | + | + | + | + | ||||||||||||||
| IGF1R | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| Immune response | |||||||||||||||||||
| CR1 | + | + | + | + | + | ||||||||||||||
| CR2 | + | + | + | + | + | + | + | ||||||||||||
| Others | |||||||||||||||||||
| SORL | + | + | + | + | + | ||||||||||||||
| 2B19 | + | + | + | + | + | + | + | ||||||||||||
| GI24 | + | + | + | + | + | + | + | + | + | ||||||||||
| . | SR . | VHR . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 . | 02 . | 03 . | 04 . | 05 . | 06 . | 07 . | 14 . | 15 . | 16 . | 17 . | 08 . | 09 . | 10 . | 11 . | 12 . | 13 . | 18 . | 19 . | |
| Adhesion and migration | |||||||||||||||||||
| VNN2 | + | + | + | + | |||||||||||||||
| VNN1 | + | + | + | + | + | + | + | + | |||||||||||
| ITGAM | + | + | + | + | + | + | |||||||||||||
| CD33 | + | + | + | + | + | ||||||||||||||
| CD302 | + | + | + | + | + | ||||||||||||||
| CEAM1 | + | + | + | + | |||||||||||||||
| Signaling | |||||||||||||||||||
| TNR1B | + | + | + | + | + | ||||||||||||||
| IGF1R | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| Immune response | |||||||||||||||||||
| CR1 | + | + | + | + | + | ||||||||||||||
| CR2 | + | + | + | + | + | + | + | ||||||||||||
| Others | |||||||||||||||||||
| SORL | + | + | + | + | + | ||||||||||||||
| 2B19 | + | + | + | + | + | + | + | ||||||||||||
| GI24 | + | + | + | + | + | + | + | + | + | ||||||||||
+, detected by CSC as described in Mirkowska et al.12
The GPI-anchored surface protein VNN2 is associated with MRD and a higher risk for relapse
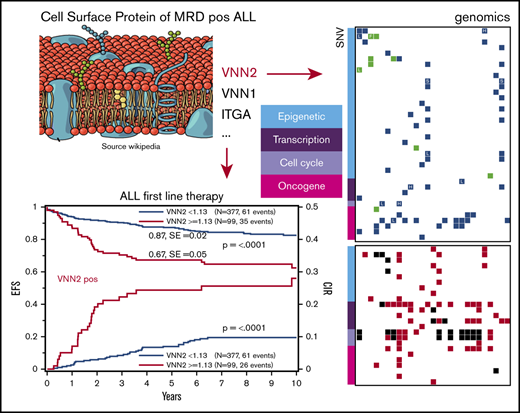
Given that VNN2 is implicated in early immature compartments and that we identified 2 members of the same vanin protein family in our experiment, we next focused on the validation of VNN2 in ALL. To explore retrospective banked samples, we first established a correlation between VNN2 RNA and protein expression in 34 PDXs derived from HR, MR, and SR patients based on their MRD after induction chemotherapy (Figure 1A). Detection of VNN2 by FCM correlated with detection of VNN2 mRNA by quantitative PCR (q-PCR) (Figure 1A). VNN2 was detected preferentially on the surface of leukemia cells from patients with MR ALL and HR ALL. We compared VNN2 expression by FCM on matched samples from 11 patients from diagnosis, relapse, and PDX to verify that VNN2 expression is maintained during disease evolution and in PDXs (supplemental Table 2). Thus PDXs constitute an acceptable model to study this phenotype. We then performed a retrospective quantitative RT-PCR analysis on banked RNA from 770 pediatric ALL patients treated on the ALL-BFM-2000 protocol (Table 1). Using 294 (38%) of 770 patients as a training set, we established a cutoff (q-PCR value ≥1.13) to determine the association between risk and VNN2+ samples (see Materials and methods) and explored the correlation between VNN2 positivity and outcome in a test set of the remaining 476 patients with BCP ALL (Figure 1B-E; supplemental Figure 1A). In total, 155 of 770 patients had VNN2 expression ≥ 1.13. The 5-year probability of event-free survival (pEFS) was inferior in VNN2+ patients (0.87; standard error, 0.02 for VNN2 <1.13 and 0.67; standard error, 0.05 for VNN2 ≥1.13; P < .0001) (Figure 1B). We detected a significant increase in relapses, predominantly in the MR ALL subgroup (P = .0003) (Figure 1D). In the HR group, the increased incidence of relapse for VNN2+ patients was not significant, which may be explained by the comparable state of drug resistance in other subtypes of the HR cohort or the fact that some VNN2+ patients may benefit from HR treatment intensification, including stem cell transplantation (Figure 1E). Thus, further exploration of this phenotype may be most relevant for the MR group, for which new markers to improve risk stratification are warranted. Importantly, using a Cox regression analysis, VNN2 was found to be an independent prognostic factor compared with MRD and other clinical factors (Table 3). We also noticed a higher proportion of central nervous system (CNS) involvement in VNN2+ patients (6.4% in the retrospective cohort, 16% in the prospective cohort) compared with the frequency of CNS+ patients in the ALL-BFM-2000 cohort (3.4% CNS+; Tables 2 and 3).
VNN2 expression is associated with an increased risk for relapse in BCP ALL. (A) VNN2 mRNA expression by q-PCR correlates with detection of VNN2 at the cell surface by FCM. PDXs from patients included in the ALL-BFM-2000 study were used for correlation. The percentage of VNN2+ cells by FCM with respect to the fluorescence-minus-one control is plotted on the y-axis against the mRNA levels by q-PCR as logarithmic value of 2-ΔCT (ΔCT = CTVNN2 − CTSDHA) on the x-axis. MRD-based risk stratification in ALL-BFM-2000 is indicated as SR, eMR, and HR. Patients with the translocation t(17;19) leading to TCF3-HLF are highlighted as TCF3-HLF+. (B) Five-year pEFS (upper part of the panel) and probability of cumulative incidence of relapse (pCIR; lower part of the panel) of 476 patients (test set) treated on the ALL-BFM 2000 study stratified according to VNN2 positivity by q-PCR. The cutoff for positivity was set to 1.13 ΔCT, as described in Materials and methods. (C-E) Five-year pEFS (upper part of the panel) and pCIR (lower part of the panel) of the same 476 patients in a subgroup analysis based on their risk stratification [SR (C), MR (D), and HR (E) for relapse] in the ALL-BFM-2000 study. A statistically significant difference was observed for pEFS and pCIR in VNN2+ vs VNN2− patients in the MR group (P < .01). (F) Retrospective analysis of VNN2 expression by FCM on available archived samples from VNN2+ ALL patients based on q-PCR. Sample selection is described in supplemental Figure 1. Twenty-two of 40 positive samples by q-PCR were confirmed VNN2+ by FCM. Cutoff for VNN2 positivity by FCM > 10% (Materials and methods). pEFS of 22 VNN2+ patients based on FCM is plotted in comparison with pEFS of 615 VNN2− patients based on q-PCR from the total retrospective cohort. P < .0001, log-rank test.
VNN2 expression is associated with an increased risk for relapse in BCP ALL. (A) VNN2 mRNA expression by q-PCR correlates with detection of VNN2 at the cell surface by FCM. PDXs from patients included in the ALL-BFM-2000 study were used for correlation. The percentage of VNN2+ cells by FCM with respect to the fluorescence-minus-one control is plotted on the y-axis against the mRNA levels by q-PCR as logarithmic value of 2-ΔCT (ΔCT = CTVNN2 − CTSDHA) on the x-axis. MRD-based risk stratification in ALL-BFM-2000 is indicated as SR, eMR, and HR. Patients with the translocation t(17;19) leading to TCF3-HLF are highlighted as TCF3-HLF+. (B) Five-year pEFS (upper part of the panel) and probability of cumulative incidence of relapse (pCIR; lower part of the panel) of 476 patients (test set) treated on the ALL-BFM 2000 study stratified according to VNN2 positivity by q-PCR. The cutoff for positivity was set to 1.13 ΔCT, as described in Materials and methods. (C-E) Five-year pEFS (upper part of the panel) and pCIR (lower part of the panel) of the same 476 patients in a subgroup analysis based on their risk stratification [SR (C), MR (D), and HR (E) for relapse] in the ALL-BFM-2000 study. A statistically significant difference was observed for pEFS and pCIR in VNN2+ vs VNN2− patients in the MR group (P < .01). (F) Retrospective analysis of VNN2 expression by FCM on available archived samples from VNN2+ ALL patients based on q-PCR. Sample selection is described in supplemental Figure 1. Twenty-two of 40 positive samples by q-PCR were confirmed VNN2+ by FCM. Cutoff for VNN2 positivity by FCM > 10% (Materials and methods). pEFS of 22 VNN2+ patients based on FCM is plotted in comparison with pEFS of 615 VNN2− patients based on q-PCR from the total retrospective cohort. P < .0001, log-rank test.
Prognostic significance of VNN2 expression in a multivariable setting (n = 476)
| Group . | HR . | LL . | UL . | P (χ2) . |
|---|---|---|---|---|
| Cox EFS | ||||
| MRD HR | 5.16 | 2.58 | 10.32 | <.001 |
| VNN2 | 2.14 | 1.28 | 3.55 | .003 |
| SET | 2.67 | 1.19 | 6.00 | .017 |
| WBC ≥100 000 | 2.01 | 1.08 | 3.73 | .027 |
| MRD SR | 0.74 | 0.43 | 1.27 | .275 |
| Pred. resp. | 0.71 | 0.33 | 1.52 | .379 |
| ETV6-RUNX1 | 0.93 | 0.51 | 1.70 | .812 |
| Cox RFS | ||||
| VNN2 | 1.97 | 1.13 | 3.44 | .016 |
| MRD HR | 2.85 | 1.20 | 6.80 | .018 |
| WBC ≥100 000 | 2.17 | 1.12 | 4.21 | .021 |
| SER | 2.41 | 1.01 | 5.74 | .046 |
| MRD SR | 0.67 | 0.37 | 1.21 | .182 |
| ETV6-RUNX1 | 0.74 | 0.38 | 1.45 | .379 |
| Pred. resp. | 0.81 | 0.35 | 1.86 | .623 |
| Group . | HR . | LL . | UL . | P (χ2) . |
|---|---|---|---|---|
| Cox EFS | ||||
| MRD HR | 5.16 | 2.58 | 10.32 | <.001 |
| VNN2 | 2.14 | 1.28 | 3.55 | .003 |
| SET | 2.67 | 1.19 | 6.00 | .017 |
| WBC ≥100 000 | 2.01 | 1.08 | 3.73 | .027 |
| MRD SR | 0.74 | 0.43 | 1.27 | .275 |
| Pred. resp. | 0.71 | 0.33 | 1.52 | .379 |
| ETV6-RUNX1 | 0.93 | 0.51 | 1.70 | .812 |
| Cox RFS | ||||
| VNN2 | 1.97 | 1.13 | 3.44 | .016 |
| MRD HR | 2.85 | 1.20 | 6.80 | .018 |
| WBC ≥100 000 | 2.17 | 1.12 | 4.21 | .021 |
| SER | 2.41 | 1.01 | 5.74 | .046 |
| MRD SR | 0.67 | 0.37 | 1.21 | .182 |
| ETV6-RUNX1 | 0.74 | 0.38 | 1.45 | .379 |
| Pred. resp. | 0.81 | 0.35 | 1.86 | .623 |
Analysis was performed on 476 samples from the test set from ALL-BFM 2000.
EFS, event-free survival; HR, hazard ratio; LL, lower limit of 95% confidence interval; Pred. resp, prednisone good responders; RFS, relapse-free survival; SER, slow early responders, a HR criterium; UL, upper limit of 95% confidence interval; WBC, white blood cell.
For these retrospective analyses, biobanked material with bone marrow (BM) involvement >80% was selected. Because VNN2 is expressed in the normal myeloid compartment, residual myeloid cells may express higher levels of VNN2 and, thus, generate a background signal in the entire cohort. Given the strong correlation between RNA and FCM data, we propose to use FCM to determine VNN2 expression at the single-cell level in prospective studies. To gather pilot information, we used 40 of 155 VNN2+ samples for which viably frozen specimens were available from the biobanks and confirmed VNN2 by FCM in 22 of 40 (supplemental Figure 1); this indicates that FCM may define VNN2 positivity more precisely. Eleven (50%) of these 22 VNN2+ patients eventually relapsed, which again indicates that VNN2 expression is an indicator for resistant disease (Figure 1F). Taken together, pediatric BCP ALL leukemias with VNN2 expression are associated with a lower event-free survival and a higher MRD-based risk for relapse.
VNN2 is detected predominantly in MRD HR and MR patients by prospective FCM
To validate VNN2 in pediatric BCP ALL prospectively, we included VNN2 in the diagnostic FCM panel of the AIEOP-BFM ALL 2009 study. Positivity was defined as ≥10% of cells being positive compared with the internal cross-lineage, CD45bright control (supplemental Figure 2). In total, 1069 patients were evaluated, of whom 103 were VNN2+, which translates to 10% VNN2 positivity in BCP ALL (Figure 2A). We confirmed that VNN2 is detected predominantly in MRD HR and MR ALL patients and at lower frequency in MRD SR ALL (17%) compared with what has been reported28 (Table 1). VNN2 is detected across different genetic subtypes (Figure 2B). It was associated with detection of CD11a, an integrin implicated in cell adhesion, but not with bilineage acute leukemia or mixed phenotype acute leukemia status (Figure 2B; supplemental Table 3). Of note, we detected VNN2 in all leukemias bearing the translocation t(17;19)(q22;p13) (Figure 1A), which results in the oncogenic TCF3-HLF transcription factor fusion and defines a rare, but generally incurable, ALL subtype (<1% of pediatric ALL).29,30 Having established an international consortium to study the genomic landscape of this rare subtype,5 we evaluated 18 TCF3-HLF+ ALL samples by FCM and confirmed consistent positivity for VNN2 in patient samples and in PDXs (Figures 1A, 2C). VNN2 positivity remained stable when comparing samples taken at diagnosis and at day 15 from 9 patients with ALL (Figure 2D). Matched samples from diagnosis and first relapse were available for 22 cases, selected from a cohort of isolated and combined BM (involvement of extramedullary compartment, ALL-REZ BFM 2002 study) based on sample availability. Two samples were VNN2+ at both diagnosis and relapse, whereas 5 cases were positive at first relapse (Figure 2E). Relapses in VNN2+ patients occurred very early/early compared with VNN2− patients (86% vs 26.6%; P = .02). Taking advantage of our ex vivo drug response profiling platform,31 we screened 10 VNN2+ ALL samples for sensitivity to standard chemotherapeutic agents of induction therapy. Comparison with 8 samples from ALL samples from the SR group revealed lower sensitivity of VNN2 ALL to dexamethasone, bortezomib, and anthracyclines (Figure 2E), further supporting the notion that VNN2+ ALL is associated with chemoresistance.
Prospective validation of VNN2 by FCM confirms its association with MRD. (A) FCM evaluation of VNN2 in diagnostic samples from 1069 B-cell ALL patients enrolled in the AIEOP-BFM 2009 study. (B) Classification of 72 BFM 2009 VNN2+ patients based on cytogenetic information and FCM data. Cytogenetic data were available for 72 of 103 patients identified by FCM. (C) VNN2 expression by FCM identifies TCF3-HLF–rearranged samples. Green dots indicate TCF3-HLF–rearranged samples (patient and PDX), and pink dots indicate TCF3-PBX1–rearranged PDX samples. Arrows indicate matched patient-PDX sample or matched xenograft samples at diagnosis-relapse. Asterisks (*) indicate matched diagnosis-relapsed samples. TCF3-HLF rearranged samples scored positive for VNN2 by FCM (>10%). (D) Analysis of VNN2 in matched samples from 9 patients taken at diagnosis of ALL (day 0) and at day 15 by FCM. (E) Comparison of matched diagnosis (black circles) and relapse (gray rhombus) pairs indicated conservation of VNN2 positivity; in some cases, an increase in VNN2 from diagnosis to relapse could be observed. FCM was used to evaluate VNN2 of matched diagnosis and first relapse of 22 patients with ALL. (F) Ex vivo drug response profiling of 10 VNN2+ ALL samples (red circles) compared with 8 VNN2− SR cases (blue circles). Patient samples were tested for dexamethasone (DEX), mitoxantrone (MITOX), bortezomib (BORTEZ), vincristine (VINCR), doxorubicin (DOX), and idarubicin (IDA). **P < .05. BAL, biphenotypic acute leukemia according to EGIL classification; B-ALL, B-cell ALL; dx, diagnosis; FMO, fluorescence-minus-one; IC50, 50% inhibitory concentration; MPAL, mixed phenotype ALL, according to World Health Organization classification23 ; rz, relapse; SE, standard error.
Prospective validation of VNN2 by FCM confirms its association with MRD. (A) FCM evaluation of VNN2 in diagnostic samples from 1069 B-cell ALL patients enrolled in the AIEOP-BFM 2009 study. (B) Classification of 72 BFM 2009 VNN2+ patients based on cytogenetic information and FCM data. Cytogenetic data were available for 72 of 103 patients identified by FCM. (C) VNN2 expression by FCM identifies TCF3-HLF–rearranged samples. Green dots indicate TCF3-HLF–rearranged samples (patient and PDX), and pink dots indicate TCF3-PBX1–rearranged PDX samples. Arrows indicate matched patient-PDX sample or matched xenograft samples at diagnosis-relapse. Asterisks (*) indicate matched diagnosis-relapsed samples. TCF3-HLF rearranged samples scored positive for VNN2 by FCM (>10%). (D) Analysis of VNN2 in matched samples from 9 patients taken at diagnosis of ALL (day 0) and at day 15 by FCM. (E) Comparison of matched diagnosis (black circles) and relapse (gray rhombus) pairs indicated conservation of VNN2 positivity; in some cases, an increase in VNN2 from diagnosis to relapse could be observed. FCM was used to evaluate VNN2 of matched diagnosis and first relapse of 22 patients with ALL. (F) Ex vivo drug response profiling of 10 VNN2+ ALL samples (red circles) compared with 8 VNN2− SR cases (blue circles). Patient samples were tested for dexamethasone (DEX), mitoxantrone (MITOX), bortezomib (BORTEZ), vincristine (VINCR), doxorubicin (DOX), and idarubicin (IDA). **P < .05. BAL, biphenotypic acute leukemia according to EGIL classification; B-ALL, B-cell ALL; dx, diagnosis; FMO, fluorescence-minus-one; IC50, 50% inhibitory concentration; MPAL, mixed phenotype ALL, according to World Health Organization classification23 ; rz, relapse; SE, standard error.
Because VNN2 defines a human FLHSC compartment18 and marks human cord blood–derived primitive CD34− HSCs,19 we evaluated BM and PB samples from healthy adults and children. VNN2 was readily detectable in HSCs from normal BM, including HSC subpopulations with very high VNN2 surface expression (supplemental Figure 3). Intriguingly, VNN2 was not expressed in mobilized PB HSCs. As expected, VNN2 was highly expressed on mature granulocytes and monocytes and weakly expressed on other lymphocytes. These results support the use of FCM for VNN2 detection in clinical studies. The prognostic value of VNN2 will require further maturation of the clinical data.
The gene expression signature of VNN2+ ALL reveals immune and cell adhesion features, as well as myeloid characteristics
To obtain a gene expression signature associated with VNN2 expression, we performed a transcriptome analysis of 50 B-cell ALL (B-ALL) leukemia samples with low to high VNN2 RNA expression with TCF3-HLF–rearranged, TCF3-PBX1–rearranged, IKZF1plus, IKZF1deleted, hyperdiploid, hypodiploid, and additional ALL without known genetic features (so-called B-others) (Figure 3A). After fitting a linear model using VNN2 as a predictor of gene expression, we identified a VNN2 signature with 1061 genes that strongly correlated with VNN2; 549 and 512 genes were positively and negatively correlated, respectively. This VNN2 signature includes some of the surface proteins that we detected in HR MRD ALL by proteomics (Figure 3A). We found an overrepresentation of features functionally related to cell adhesion, the immune response, and the JAK-STAT pathway (Figure 3B). By gene-set enrichment analysis (Figure 3C) and using a different method based on text mining annotations (Figure 3D; supplemental Table 4), we found an association between the VNN2 signature and myeloid features and detected a significant reduction in PAX5 expression, which is in line with the genomic features of VNN2+TCF3-HLF ALL.5 However, VNN2 appears to be dispensable for leukemia maintenance and propagation, because we could generate and serially passage 2 VNN2-knockout samples using clustered regularly interspaced short palindromic repeats/Cas9 in NSG mice (supplemental Figure 1D). More studies will be required to carefully evaluate the impact of VNN2 on sites of leukemia involvement in this PDX model.
The signature of VNN2+ALL is enriched with myeloid features and functional annotations related to immune response, interferon signaling, cell adhesion, and the JAK-STAT pathway. (A) Heat map with 1061 differentially expressed genes showing the transcriptome signature of 50 B-ALL samples ordered by increasing VNN2 RNA expression (left panel). Using edgeR,48 a linear model was fitted across the dataset of 50 ALL samples using VNN2 expression as predictor of gene expression. Genes that showed a significant linear relationship with VNN2 expression were selected (false discovery rate [FDR] ≤0.01). Genes were additionally filtered using Spearman correlation (FDR ≤0.04). A total of 549 genes and 512 genes appeared to be positively or negatively correlated with VNN2, respectively. Genes positively correlated with VNN2 include 3 genes that were identified by proteomics in an independent cohort using CSC technology (right panels). (B) Positively correlated genes were grouped by pathways using the online tool and database ConsensusPathDB49 (FDR ≤0.01). The most significantly overrepresented pathways (according to P value) included the immune system, interferon signaling, β-2 integrin–mediated interactions, and the JAK-STAT signaling pathway. Node size indicates the number of genes contained and the color refers to significance (P value). Two nodes are connected by a line if they share members. The line width reflects the relative overlap between the nodes, whereas the line color represents the number of shared members that are also found in the input. (C) Enrichment plot for the myeloid signature obtained after performing gene set enrichment analysis.50 (D) Components of the VNN2+ ALL signature generated using the Genomatix genome analyzer reveal functional annotations related to myeloid cells and their cellular location (adjusted P ≤ .05; P ≤ .01). Lines between genes indicate a functional association. The line type indicates the evidence level. Dashed lines indicate a functional association based on cocitation or experimental validation. Solid lines indicate a functional association based on expert curation. Diamonds indicate that a gene carries a binding site of the associated transcription factor. Arrows indicate that a gene is altered by the associated gene. Curved lines indicate that a gene alters itself on a transcriptional or protein level. The line type indicates the evidence level. Myeloid_DN, myeloid gene expression signature; NES, normalized enrichment score.
The signature of VNN2+ALL is enriched with myeloid features and functional annotations related to immune response, interferon signaling, cell adhesion, and the JAK-STAT pathway. (A) Heat map with 1061 differentially expressed genes showing the transcriptome signature of 50 B-ALL samples ordered by increasing VNN2 RNA expression (left panel). Using edgeR,48 a linear model was fitted across the dataset of 50 ALL samples using VNN2 expression as predictor of gene expression. Genes that showed a significant linear relationship with VNN2 expression were selected (false discovery rate [FDR] ≤0.01). Genes were additionally filtered using Spearman correlation (FDR ≤0.04). A total of 549 genes and 512 genes appeared to be positively or negatively correlated with VNN2, respectively. Genes positively correlated with VNN2 include 3 genes that were identified by proteomics in an independent cohort using CSC technology (right panels). (B) Positively correlated genes were grouped by pathways using the online tool and database ConsensusPathDB49 (FDR ≤0.01). The most significantly overrepresented pathways (according to P value) included the immune system, interferon signaling, β-2 integrin–mediated interactions, and the JAK-STAT signaling pathway. Node size indicates the number of genes contained and the color refers to significance (P value). Two nodes are connected by a line if they share members. The line width reflects the relative overlap between the nodes, whereas the line color represents the number of shared members that are also found in the input. (C) Enrichment plot for the myeloid signature obtained after performing gene set enrichment analysis.50 (D) Components of the VNN2+ ALL signature generated using the Genomatix genome analyzer reveal functional annotations related to myeloid cells and their cellular location (adjusted P ≤ .05; P ≤ .01). Lines between genes indicate a functional association. The line type indicates the evidence level. Dashed lines indicate a functional association based on cocitation or experimental validation. Solid lines indicate a functional association based on expert curation. Diamonds indicate that a gene carries a binding site of the associated transcription factor. Arrows indicate that a gene is altered by the associated gene. Curved lines indicate that a gene alters itself on a transcriptional or protein level. The line type indicates the evidence level. Myeloid_DN, myeloid gene expression signature; NES, normalized enrichment score.
VNN2+ ALL includes different B-ALL subtypes, combining inactivation of B-cell developmental genes, frequent homozygous CDKN2A/B deletions, and mutations affecting epigenetic regulators
To identify underlying genetic alterations, we applied next-generation sequencing analysis integrating short and large insert size paired-end whole-genome and whole-exome sequencing to a discovery cohort consisting of 22 VNN2+, TCF3-HLF− diagnostic samples (supplemental Figure 4). Genomic information from TCF3-HLF+ patients was available from our previous study.5
Of note, we detected frequent monoallelic deletions of PAX5, almost always combined with deletion of CDKN2A/B and sometimes combined with deletions of IKZF1 (Figure 4). In 1 sample we detected the PAX5 P80R mutation, in combination with biallelic deletion of CDKNA/B. We observed mutations affecting IKZF1 in 8 of 22 samples (36%), primarily hemizygous deletions (5/8), but also homozygous deletions (2/8) and 1 single-nucleotide variant. Five of the patients (23%) with IKZF1 deletions, all from non-SR subgroups, fit the definition of IKZF1plus cases, which represent a new subtype with very poor outcome on the AIEOP-BFM protocol.8 This frequency of IKZF1 deletions is expected in ALL,32 but IKZF1plus appears to be enriched compared with the 6% frequency reported previously.8 In addition to TCF3-HLF fusions, we identified rearrangements of TCF3 with ZNF384, a recurrent mutation in ALL,33 as well as with JAK2, which has not been described previously. We also found a JAK2-BCR fusion, previously described in BCR-ABL–like ALL.34 Thus, the VNN2 phenotype was detected across different leukemia-initiating lesions, including IKZF1plus, possibly B-ALLs corresponding to the recently defined PAX5alt subtype,1 TCF3-rearranged ALL, and, rarely, BCR-ABL–like subtypes.
Whole-exome and transcriptome sequencing identified frequent deletions in PAX5, IKZF1, CDKN2A/B and mutations in epigenetic regulators in VNN2+patients. Whole-exome sequencing was performed in 27 VNN2+ patient samples (22 diagnosis, 5 relapse), and transcriptome sequencing was performed in 16 VNN2+ patient samples (13 diagnosis, 3 relapse). Details about sample selection are described in supplemental Figure 4. Sample identifiers are shown at the bottom of the figure. «V_a» indicates samples at diagnosis, and «V_c» indicates samples at relapse. Samples in which transcriptome sequencing was performed can be identified by an additional «_dx» or «_rz» at the end of the identifier. dx, diagnosis; rz, relapse.
Whole-exome and transcriptome sequencing identified frequent deletions in PAX5, IKZF1, CDKN2A/B and mutations in epigenetic regulators in VNN2+patients. Whole-exome sequencing was performed in 27 VNN2+ patient samples (22 diagnosis, 5 relapse), and transcriptome sequencing was performed in 16 VNN2+ patient samples (13 diagnosis, 3 relapse). Details about sample selection are described in supplemental Figure 4. Sample identifiers are shown at the bottom of the figure. «V_a» indicates samples at diagnosis, and «V_c» indicates samples at relapse. Samples in which transcriptome sequencing was performed can be identified by an additional «_dx» or «_rz» at the end of the identifier. dx, diagnosis; rz, relapse.
As expected in these B-ALL subtypes, we detected frequent gain-of-function single-nucleotide variants in the Ras pathway, such as PTPN11, NRAS, and KRAS (Figure 4; supplemental Table 6). In addition, we noticed frequent mutations in epigenetic regulators in 13 of 22 (60%) VNN2+ patients at first diagnosis (Figure 4), suggesting a marked increase compared with frequencies reported previously for pediatric B-ALL (12/42 = 30%).35 Interestingly, we did not find these epigenetic mutations in TCF3-HLF cases.5 To confirm possible enrichment of epigenetic mutations in MRD MR ALL, we performed targeted sequencing of the histone-lysine methyltransferases EZH2, KMT2D, and SETD2, as well as the lysine demethylase KDM6A, on 185 samples from the retrospective ALL-BFM-2000 MRD-MR cohort in which we also determined VNN2 mRNA levels by q-PCR (supplemental Table 7). In this MRD MR cohort, 15 of 35 (43%) VNN2+ patients and 19 of 150 (13%) VNN2− patients displayed ≥1 mutation in these 4 genes. Mutations in this functional class have been reported for a subset of patients with PAX5alt and PAX5 P80R.1 Taken together, we identify a VNN2 signature that is clearly associated with resistance to chemotherapy but that is found in different molecular B-ALL subtypes, mostly with lesions in B-cell developmental genes and, frequently, in genes affecting epigenetic modification. These features should be included in upcoming prospective international trials to evaluate their contribution to more precise risk stratification in ALL.
Discussion
This study identifies a recurrent VNN2 signature that is associated with increased resistance to chemotherapy in 10% of BCP ALL cases across different ALL subtypes, primarily stratified in the MR and HR groups based on resistance to treatment, as assessed by MRD. Most of the relapses in ALL still occur in the MR group by MRD.8,36,37 Although patients with more resistant leukemia may need a different intervention, many patients in the MR group may benefit instead from a reduction in treatment intensity to minimize long-term toxicity.
Expression and cell surface display of the glycosylphosphatidylinositol-anchored surface protein VNN2/GPI-80 is associated with myeloid features that are not captured by current definitions of bilineage and mixed immunophenotypes by diagnostic FCM.23 VNN2 itself was recently identified as a marker of self-renewing human FLHSCs18 and cord blood–derived HSCs.19 In this study, we detected strong VNN2 expression in subsets of human adult BM HSCs from healthy individuals, confirming its implication in the very early stages of hematopoiesis. We also found recurrent surface markers associated with VNN2, including VNN1 and members of the integrin and selectin family. Functionally, these proteins are involved in cell adhesion and processes linked to inflammation. VNN2 has been implicated in inflammation, contributing to leukocyte adhesion and migration to inflammatory sites in association with cell surface integrins.17 It is conceivable that this signature reflects functional states that are important for interactions with the leukemia microenvironment and involvement at sanctuary sites. Given the strong correlation between mRNA and protein levels, VNN2 is regulated at the transcriptional level. Because of the marked expression of VNN2 in the normal myeloid compartment, FCM represents the methodology of choice to detect VNN2 positivity at the single-cell level on ALL cells.
From a genomics standpoint, we detected a reduction in PAX5 gene dosage in the majority of samples that we tested thus far, which is reminiscent of our observation in TCF3-HLF+ ALL.5 In our cohort, events leading to PAX5 loss of function were primarily monoallelic, which is in contrast with the majority of the samples for 2 recently described entities in ALL, PAX5alt1 and PAX5 P80R,38 in which both alleles of PAX5 are generally affected. Indeed, VNN2 expression was also detected in the IKZF1plus ALL subtype, which has been identified based on the combination of IKZF1 deletions that co-occurred with deletions in CDKN2A, CDKN2B, PAX5, or PAR1 in the absence of ERG deletions, marking a subtype with poor outcomes in patients with MRD MR ALL or MRD HR ALL.8
Of note, we detected genetic alterations affecting epigenetic modifiers in 13 of 22 VNN2+ ALLs. The methyltransferases EZH2, KMT2D, and SETD2, as well as the lysine demethylase KDM6A, were affected recurrently. Similar mutations have been reported in a subset of PAX5alt and PAX5 P80R B-ALL patients.1 Loss-of-function mutations in EZH2 have been reported in T-cell ALL and B-ALL at low frequencies,39-41 as well as in myeloid disorders.42 Moreover, loss-of-function mutations in EZH2 have very recently been associated with poor outcome in AML.43 MLL2 deficiency impedes B-cell differentiation and is a driver of lymphoma in vivo.44 Moreover, mutations in SETD2 and KDM6A have been reported to be enriched in relapsed pediatric B-ALL,45 which suggests that they could be relevant for chemotherapy resistance. Inactivation of KDM6A is crucial in B-cell lymphoma44 and T-cell ALL,46 contributing to tumorigenesis by deregulation of complexes that control histone methylation in important regulatory regions.47
Taken together, we have identified a new signature in ALL, whereby surface expression of VNN2 is associated with increased resistance to induction chemotherapy. Our results support a prospective evaluation of VNN2 by FCM in the ongoing AIEOP-BFM ALL trial. We propose to explore the prognostic significance of this marker, along with more complex genomic information, to improve the identification of patients who will eventually relapse, in particular those with IR ALL, the largest risk group, on the current treatment protocol.
For original data, please contact Jean-Pierre Bourquin (jean-pierre.bourquin@kispi.uzh.ch).
Acknowledgments
The authors thank Renate Siegenthaler for data management, Astrid Mecklenbräuker for assistance with FCM, and Yi-Chien Tsai and Silvia Jenni for technical help.
This work was supported by Kinderkrebsforschung Schweiz (J.-P.B.), Krebsliga Zurich (J.-P.B.), the Swiss National Science Foundation (310030-156407 and 310030-182269) (J.-P.B.), the Sassella Foundation (J.-P.B.), the Hanne Liebermann Foundation (J.-P.B.), Clinical Research Priority Program (CRPP) Human Hemato-Lymphatic Diseases (HHLD) (J.-P.B.), and Czech Ministry of Health NV18-03-00343 (E.M.).
Authorship
Contribution: V.R.M., M. Schütte, A.R., G.C., T.R., H.-J.W., N.S., P.M., A.H., M.T., C.M., A.S., M.R., E.M., B. Bornhauser, B. Buldini, P.S., O.M., E.V., B.M., M.J.B., T.L.S., M.Z., and B.W. performed experiments and analyzed data; Q.A.N. analyzed data; M.D., A.A., M.B., G.B., G.G., G.t.K., R.P.-G., M.H.-H., F.N., C.E., M. Schrappe, and M. Stanulla provided samples and clinical data; B. Bornhauser and J.-P.B. wrote the manuscript; and B. Bornhauser, M.-L.Y., and J.-P.B. conceived and supervised the study.
Conflict of interest disclosure: M. Schütte is an employee of Alacris Theranostics GmbH. The remaining authors declare no competing financial interests.
Correspondence: Jean-Pierre Bourquin, Department of Oncology, University Children’s Hospital Zurich and Children’s Research Center, Steinwiesstr 75, 8032 Zurich, Switzerland; e-mail: jean-pierre.bourquin@kispi.uzh.ch.
References
Author notes
B. Bornhauser, G.C., A.R., T.R., V.R.M., and M. Schütte contributed equally to this work.
The full-text version of this article contains a data supplement.

![VNN2 expression is associated with an increased risk for relapse in BCP ALL. (A) VNN2 mRNA expression by q-PCR correlates with detection of VNN2 at the cell surface by FCM. PDXs from patients included in the ALL-BFM-2000 study were used for correlation. The percentage of VNN2+ cells by FCM with respect to the fluorescence-minus-one control is plotted on the y-axis against the mRNA levels by q-PCR as logarithmic value of 2-ΔCT (ΔCT = CTVNN2 − CTSDHA) on the x-axis. MRD-based risk stratification in ALL-BFM-2000 is indicated as SR, eMR, and HR. Patients with the translocation t(17;19) leading to TCF3-HLF are highlighted as TCF3-HLF+. (B) Five-year pEFS (upper part of the panel) and probability of cumulative incidence of relapse (pCIR; lower part of the panel) of 476 patients (test set) treated on the ALL-BFM 2000 study stratified according to VNN2 positivity by q-PCR. The cutoff for positivity was set to 1.13 ΔCT, as described in Materials and methods. (C-E) Five-year pEFS (upper part of the panel) and pCIR (lower part of the panel) of the same 476 patients in a subgroup analysis based on their risk stratification [SR (C), MR (D), and HR (E) for relapse] in the ALL-BFM-2000 study. A statistically significant difference was observed for pEFS and pCIR in VNN2+ vs VNN2− patients in the MR group (P < .01). (F) Retrospective analysis of VNN2 expression by FCM on available archived samples from VNN2+ ALL patients based on q-PCR. Sample selection is described in supplemental Figure 1. Twenty-two of 40 positive samples by q-PCR were confirmed VNN2+ by FCM. Cutoff for VNN2 positivity by FCM > 10% (Materials and methods). pEFS of 22 VNN2+ patients based on FCM is plotted in comparison with pEFS of 615 VNN2− patients based on q-PCR from the total retrospective cohort. P < .0001, log-rank test.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/4/17/10.1182_bloodadvances.2019000938/8/m_advancesadv2019000938f1.png?Expires=1765900274&Signature=Dv7K5oGJCq94iswxe0LvFon2lmEags2qAVMtLCl36QZfmb1ItTQg3QQEI-GK-WX08CKyIWkKoFLKtPnfXxfIXUrHMv79UwG9uj~SzcjoZ555uQpM3Y6RQyxoz84Wnnjc0eVRwnYqjB4JIMTQVUDwMTx85LsrpfkPMwibgZv0mvShn5B3kuiDDZI4~X-ExXIuw~J5GFrRDusRtFSAUThwUPUqLAGB~cm8Tm89HjQ4SauJoyF0FOekuD0DnTSjGMtw0c8MG2Tr1jbEbRuZAGg1iWaK~xvYND9CLTUsiwTZoDI8hDIFCmyWlfARgV513v0jcGW3elZ7XG-kqoDudReNUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![The signature of VNN2+ALL is enriched with myeloid features and functional annotations related to immune response, interferon signaling, cell adhesion, and the JAK-STAT pathway. (A) Heat map with 1061 differentially expressed genes showing the transcriptome signature of 50 B-ALL samples ordered by increasing VNN2 RNA expression (left panel). Using edgeR,48 a linear model was fitted across the dataset of 50 ALL samples using VNN2 expression as predictor of gene expression. Genes that showed a significant linear relationship with VNN2 expression were selected (false discovery rate [FDR] ≤0.01). Genes were additionally filtered using Spearman correlation (FDR ≤0.04). A total of 549 genes and 512 genes appeared to be positively or negatively correlated with VNN2, respectively. Genes positively correlated with VNN2 include 3 genes that were identified by proteomics in an independent cohort using CSC technology (right panels). (B) Positively correlated genes were grouped by pathways using the online tool and database ConsensusPathDB49 (FDR ≤0.01). The most significantly overrepresented pathways (according to P value) included the immune system, interferon signaling, β-2 integrin–mediated interactions, and the JAK-STAT signaling pathway. Node size indicates the number of genes contained and the color refers to significance (P value). Two nodes are connected by a line if they share members. The line width reflects the relative overlap between the nodes, whereas the line color represents the number of shared members that are also found in the input. (C) Enrichment plot for the myeloid signature obtained after performing gene set enrichment analysis.50 (D) Components of the VNN2+ ALL signature generated using the Genomatix genome analyzer reveal functional annotations related to myeloid cells and their cellular location (adjusted P ≤ .05; P ≤ .01). Lines between genes indicate a functional association. The line type indicates the evidence level. Dashed lines indicate a functional association based on cocitation or experimental validation. Solid lines indicate a functional association based on expert curation. Diamonds indicate that a gene carries a binding site of the associated transcription factor. Arrows indicate that a gene is altered by the associated gene. Curved lines indicate that a gene alters itself on a transcriptional or protein level. The line type indicates the evidence level. Myeloid_DN, myeloid gene expression signature; NES, normalized enrichment score.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/4/17/10.1182_bloodadvances.2019000938/8/m_advancesadv2019000938f3.png?Expires=1765900274&Signature=R0xhpggofipoeNw6RpI82icRHi-9G7fMvy3WTEAb43sZQsOEmA-oJehWByUTsm2RuTH18cWO8I6HoaGsE-c3RDAjVZWWt00rPjWBBVwnR694SD0mIxTnRxzZpARg-~RX~~Aw4yDtG7cWk85EZ3WDfq2QsW4TkESFpDtjDlfixvHNE62g7p15dRmUvOULRs0wAch~2cbvvR1vtrmVkRKpGXv8ZT0n-mM-9AXSokuAQeYhu0poeKW1Iuric6AkEqkLj5ly3A-6ycOxtT19U2eQDSc8I07dCVRj-QQAIsnXQ0uWjd-m4kGw~3SJ6ve1IuIWQhvh93kLEnfR77APBc9-XQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
