Key Points
When donor and host coexpress ligands for sole donor inhibitory KIRs, NK-cell education is best after haplo-HSCT.
When donor and host coexpress ligands for all donor inhibitory KIRs, AML and MDS patients show the lowest relapse rates after haplo-HSCT.
Abstract
The rate and extent of natural killer (NK)–cell education after hematopoietic cell transplantation correlates with leukemia control. To study the effect of donor and host HLA on NK-cell reconstitution, single killer-cell immunoglobulin-like receptor (KIR)+ NK cells (exhibiting KIR2DL1, KIR2DL2/KIR2DL3, or KIR3DL1 as their sole receptor) were grouped into 4 groups based on the interaction between donor/host HLA and donor inhibitory KIR in 2 cohorts (n = 114 and n = 276, respectively). On days 90 to 180 after transplantation, the absolute number and responsiveness against K562 cells (CD107a or interferon-γ expression) of single-KIR+ NK cells were higher in pairs where donor and host HLA both expressed ligands for donor inhibitory KIRs than in pairs where 1 or both of the donor and recipient HLA lacked at least 1 KIR ligand. NK-cell responsiveness was tuned commensurate with the number of inhibitory receptors from the donor. When both donor and host expressed the 3 major KIR ligands (HLA-C1, HLA-C2, and HLA-Bw4), NK cells expressing 3 inhibitory receptors (KIR2DL1/2DL3/3DL1) reached the maximum responsiveness against K562 cells compared with those NK cells expressing only 1 or 2 inhibitory receptors. When donor and host HLA both expressed all ligands for donor inhibitory KIRs, patients with acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) showed the lowest recurrence rate after haploidentical hematopoietic stem cell transplantation (haplo-HSCT). In conclusion, this study demonstrates that when both donors and hosts present all the KIR ligands for donor KIRs, reconstituted NK cells achieve better functional education and contribute to least relapse among patients. This observation study was registered at www.clinicaltrials.gov as #NCT02978274.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the most potent antileukemic postremission treatment strategy for myeloid malignant disease. However, relapse remains the main cause of mortality after allo-HSCT.1 Natural killer (NK) cells, a major lymphocyte of the innate immune system, express surface receptors that mediate potent effector functions, including cytolytic activity and cytokine release, and play a central role in leukemia control.2-4 NK cells are the first cells to recover after transplant; their reconstitution occurs before that of T cells. The absolute number and functional reconstitution of NK cells correlates with leukemia control.5-9
Functional calibration of NK cells involves the surface inhibitory killer-cell immunoglobulin-like receptors (KIRs), which recognize self-HLA.10 In healthy donors, NK cells with inhibitory KIRs for self-HLA are hyperresponsive against targets lacking cognate HLAs. Cells capable of this “missing self” recognition are referred to as “licensed” or “educated” NK cells. NK cells with KIRs for non-self HLAs are hyporesponsive and referred to as “unlicensed” or “uneducated” NK cells.11 Educated NK cells prevent autoreactive behavior but also permit cytotoxicity against target cells that have downregulated HLA class I expression.10 When and how the process of education occurs have not been clearly discerned.
Raulet al12 and Lanier et al13 simultaneously demonstrated that when uneducated NK cells from HLA class I–deficient mice were transferred to hosts that expressed major histocompatibility complex (MHC) class I, they acquired responsiveness; however, when educated NK cells from wild-type mice were transferred to MHC-deficient mice, it reduced their responsiveness. This indicates that host MHC plays essential roles in NK responsiveness. Using a wild-type MHC-mismatched allo-HSCT mouse model with cytomegalovirus infection, Murphy et al14,15 found that depletion of only the educated and not the uneducated NK cells, as predicted by donor MHC haplotypes, resulted in significantly increased susceptibility to murine cytomegalovirus infection, indicating that donor MHC determines NK responsiveness. Using a humanized mouse model, Hsu and colleagues found that NK-cell responsiveness increased when MHC was acquired from neighboring cells but was maintained in educated cells where MHC was encoded, indicating that both donor and host HLA cooperate to maintain and adjust NK-cell responsiveness.16 Therefore, it seems that both the donor and host MHC could influence NK-cell education in mouse models.
What about the process of education in humans? In healthy persons, it would be hard to study the education process, but successful HLA-mismatched allo-HSCT provides a good platform to fully explore the education process.17,18 Whether determination of NK-cell education is dependent upon the interaction between donor KIRs and donor HLA, host HLA, or both has not yet been conclusively determined. Dulphy et al19 demonstrated that NK-cell education is shaped by donor HLA genotype. For example, in hosts whose donor was HLA-C group homozygous (C1C1 or C2C2), single-KIR+ NK cells expressing a receptor for donor HLA were more responsive than single-KIR+ NK cells expressing a receptor for host HLA and not donor HLA, but these kinds of pairings were too few and lack statistical analysis and dynamic analysis at different time points after transplantation. Therefore, the contribution of host HLA still could not be excluded. On the other hand, our previous study found NK-cell education was shaped by host HLA genotype, as hosts presenting HLA for all donor inhibitory KIRs were shown to be more capable of functional NK reconstitution.20,21 However, due to the lack of single-KIR+ NK cells, functional analysis limited the full evaluation of the donor/host HLA and donor inhibitory KIR’s interaction, so the contribution of donor HLA could not be excluded. The HLA-mismatched transplantation setting is highly complex, with different conditioning regimens with or without T-cell depletion in vitro. This heterogeneity could interfere with NK-cell reconstitution.6-9,22,23
To determine the relative contributions of donor and recipient HLA to NK-cell education, we prospectively enrolled a large enough sample size for group analysis and monitored the responsiveness of single-KIR+ NK cells at different time points after transplantation. We found that donor and host HLA cooperated to promote the functional reconstitution of NK cells. Meanwhile, in line with these results, donors and hosts coexpressing all KIR ligands for donor inhibitory KIRs displayed the lowest relapse rates and superior leukemia-free survival after haploidentical transplantation in another prospective cohort of myeloid leukemia patients. Collectively, our study sheds light on the interplay between functional reconstitution and the involvement of donor/host HLA interaction in NK-cell control of leukemic cells.
Materials and methods
Patients
Patients with hematological malignancies suitable for allo-HSCT but without HLA-identical related or unrelated donors were candidates for haploidentical stem cell transplantation (haplo-SCT). Two cohorts of patients were enrolled in this study. We first prospectively enrolled 114 patients undergoing haplo-SCT between May 2016 and April 2017 to explore NK-cell phenotypes and functional reconstitution. From June 2012 to April 2016, DNA from 900 haploidentical donors was prospectively collected. Among 900 haplo-SCTs, 400 patients were diagnosed with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS). There were 20 patients accepted with prophylactic donor lymphocyte infusion, and 104 patients lacked HLA-C allele typing. Thus, 276 AML/MDS patients who underwent haploidentical transplantation were enrolled in the second cohort to analyze the effect of donor-host KIR-HLA combinations on relapse after transplantation. All patients and donors provided written, informed consent, and the Institutional Review Board of the Peking University Institute of Hematology approved the study. This observation study was registered at www.clinicaltrials.gov as #NCT02978274.
Transplants
All patients received myeloablative regimens.24,25 Pretransplant conditioning was performed with cytarabine (4 g/m2 per day on days −10 to −9), busulfan (4 mg/kg per day, orally on days −8 to −6 before January 2008 and 3.2 mg/kg per day IV on days −8 to −6 thereafter), cyclophosphamide (1.8 g/m2 per day on days −5 to −4), semustine (250 mg/m2 on day −3), and rabbit anti-thymocyte globulin (Thymoglobulin; Imtix Sangstat, Lyon, France; 2.5 mg/kg per day IV on days −5 to −2). Graft-versus-host disease (GVHD) prophylaxis was with cyclosporine, mycophenolate mofetil, and short-course methotrexate, as described25,26 . Grafts were granulocyte-colony stimulating factor–mobilized bone marrow and blood cells, as described previously.25,26
HLA and KIR typing
Patient and donor DNA was prepared from peripheral blood mononuclear cells (PBMCs), used for HLA typing prior to transplantation, and stored at −40°C. Molecular HLA typing and KIR genotyping was performed according to the manufacturer’s instructions (One Lambda, Canoga Park, CA). Host HLA-C and HLA-B alleles were identified using high-resolution DNA-based HLA typing and segregated into the following epitope groups: HLA-C group 1 (HLA-CAsn80: HLA-Cw1, 3, 7, 8, 13, and 14 alleles, ligand of KIR2DL2/L3), HLA-C group 2 (HLA-CLys80: HLA-Cw2, 4, 5, 6, 12, 15, 17, and 18 alleles, ligand of KIR2DL1), and HLA-Bw4 (ligand of KIR3DL1).
Chimerism analyses were performed according to a report by Qin et al (Shanghai Tissuebank Diagnostics, Shanghai, China).27 Complete donor chimerism was defined as 100% donor short tandem repeat profiling.
Sample collection
For prospective studies, patient blood samples were collected following allo-HSCT at days 30, 90, and 180 after transplant. PBMCs of each sample were freshly isolated by density gradient centrifugation (Pancoll human, Pan-Biotech) and within 24 hours analyzed by flow cytometry (fluorescence-activated cell sorting). Samples of healthy controls were processed in exactly the same way as patient samples.
Flow cytometric analyses
The phenotype and function recovery of NK cells with different maturation expression levels of NKG2A, KIR, and CD57, as well as of activating receptors (NKp30, NKp46, NKG2D, and DNAM-1) and interferon-γ (IFN-γ), CD107a, Bcl-2, CD25, and CD122 on NK cells were monitored in patients after transplantation. The percentages of NKG2A+, KIR+, and CD57+ were calculated as the percentages of cells expressing each marker among single-KIR+ NK cells. The expression of DNAM-1, NKp46, NKp30, NKG2D, CD25, CD122, and BCl-2 was calculated as the mean fluorescence intensity (MFI) or percentage among single-KIR+ NK cells. The anti-human monoclonal antibodies used for flow cytometry are listed in supplemental Table 1.
Single-KIR+ NK subsets were analyzed as KIR2DL1 single+ (CD3−CD56dimNKG2A−KIR2DL2/L3−KIR3DL1−KIR2DL1+) NK cells, KIR2DL2/L3 single+ (CD3−CD56dimNKG2A−KIR2DL1−KIR3DL1−KIR2DL2/L3+) NK cells, and KIR3DL1 single+ (CD3−CD56dimNKG2A−KIR2DL1−KIR2DL2/L3−KIR3DL1+) NK cells. Representative raw flow plots for both gating and functional data of single-KIR+ NK subsets at days 30, 90, and 180 after transplantation are shown in supplemental Figure 1.
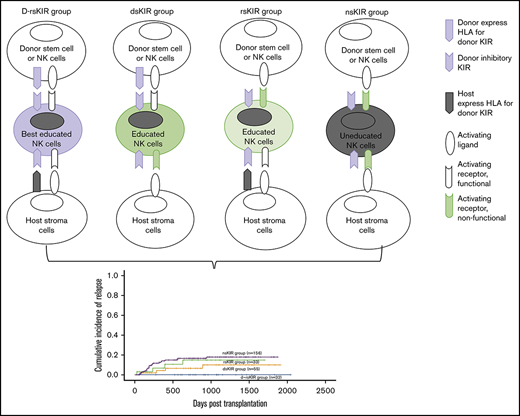
To study the effect of donor and host MHC class I on NK-cell education/licensing in the first cohort, single-KIR+ NK cells were grouped into the following groups (as described in supplemental Materials and methods): (A) non-self KIR (nsKIR), where both hosts and donors lacked HLA ligands for 1 donor KIR (ie, for donor KIR2DL1 [CD158a] derived from C1C1 donors and developed in C1C1 hosts); (B) donor-host “self” KIR (d-rsKIR), where donors and hosts encoded HLA ligands for donor KIRs (ie, for donor KIR2DL1 derived from C2Cx donors and developed in C2Cx hosts); (C) donor “self” KIR (dsKIR), where donors, but not hosts, encoded HLA ligands for donor KIRs (ie, for donor KIR2DL1 derived from C2Cx donors and developed in C1C1 hosts); and (D) host “self” KIR (rsKIR), where hosts, but not donors, encoded HLA ligands for donor KIR (ie, for donor KIR2DL1 derived from C1C1 donors and developed in C2Cx hosts).
In accordance with the grouping strategy in first cohort, to study the effect of donor and host MHC class I on clinical outcomes, patients were grouped into the following groups: (A) non-self KIR (nsKIR), where both hosts and donors lacked HLA ligands for all 3 donor KIRs (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and for host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); (B) donor-host “self” KIR (d-rsKIR), where donors and hosts encoded all HLA ligands for donor KIRs (ie, donor C1C2Bw4 to host C1C2Bw4); (C) donor “self” KIR (dsKIR), where donors, but not hosts, encoded all HLA ligands for donor KIR (ie, for donor C1C2Bw4 and for host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); and (D) host “self” KIR (rsKIR), where hosts, but not donors, encoded all HLA ligands for donor KIR (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and host C1C2Bw4).
Acquisition was performed on an LSR Fortessa instrument (BD Biosciences). Analysis was performed using FlowJo software (Tree Star) and visualized as heatmaps using the heatmap function in R.
Cytotoxicity assays
PBMCs were cultured in RPMI with 10% fetal calf serum with 1000 IU/mL interleukin-2 (Beijing Double-Crane Pharmaceutical) for 10 to 14 hours for use in both spontaneous and interleukin-2–stimulated (lymphokine-activated killer) NK cytotoxicity assays.20 PBMC samples were collected from patients at days 30 (n = 102), 100 (n = 75), and 180 (n = 93). The cytotoxicity and cytokine secretion of NK cells was determined using CD107a expression and IFN-γ production against the MHC class I–deficient human erythroleukemia K562 cell line as targets at an effector-to-target ratio of 5:1 for 4 hours. GolgiStop (0.7 µL/mL, BD Biosciences) was added after 1 hour.
Statistical analysis
Associations between patient characteristics and pretransplantation variables with posttransplantation outcomes were analyzed using the Kaplan-Meier method or calculated using cumulative incidence curves to accommodate competing risks. For transplant-related mortality (TRM), relapse was considered the event of interest and death prior to relapse the competitor. For relapse, death not caused by disease relapse was considered the event of interest and death caused by the malignancy the competitor. The R software program was used to calculate cumulative incidence, considering the presence of competing risk. Factors included in the models were host and donor ages, sex, sex mismatch, diagnosis (AML vs MDS), the donor and host’s 1 KIR/1 ligand mismatch/match, numbers of HLA-mismatched alleles, HLA-KIR, pretransplantation risk category, pretransplantation minimal residual disease (MRD), disease risk index, KIR haplotype (AA vs BX), and activating KIR genes (KIR2DS1, 2DS2, 2DS3, 2DS5, and 3DS1). The pretransplantation risk category included both low- and high-risk patients. High-risk patients were those with AML not first complete remission (CR1), MDS not refractory anemia (RA) or ringed sideroblasts (RARS), or refractory cytopenia with multilineage dysplasia (RCMD). Low-risk patients were those with AML CR1, CML-CP1, MDS RA, RARS, or RCMD.
The final multivariate models were built using a forward stepwise selection model. Patient characteristics between groups were compared using χ2 statistics for categorical variables and Mann-Whitney U for continuous variables. Instances with P < .05 were considered to represent statistically significant differences.
Results
Patient characteristics
Patient characteristics in the first and second cohort are described in Tables 1 and 2. All patients showed engraftment and complete donor chimerism following transplantation. In the first and second cohort, the median follow-up time for those patients who survived was 775 days (range, 611-1332 days) and 1626 days (range, 983-2510 days), respectively.
Patient characteristics in cohort 1
| . | d-rsKIR . | dsKIR . | rsKIR . | nsKIR . | P . |
|---|---|---|---|---|---|
| n | 17 | 20 | 15 | 62 | |
| Sex of host, male, n (%) | 8 (47.1) | 11 (55) | 10 (66.7) | 40 (64.5) | NS |
| Age of host, median (range), y | 29 (18-53) | 42.5 (26-63) | 28 (15-43) | 31.5 (15-58) | .003 |
| Age of donor, median (range), y | 45 (14-64) | 27 (8-65) | 40 (13-54) | 29 (12-60) | NS |
| Donor and host sex group, n (%) | |||||
| Male-male | 8 (47.1) | 8 (40) | 10 (66.7) | 28 (45.2) | |
| Male-female | 8 (47.1) | 4 (20) | 3 (20) | 15 (24.2) | |
| Female-male | 0 (0) | 5 (25) | 0 (0) | 12 (19.4) | |
| Female-female | 1 (5.9) | 3 (15) | 2 (13.3) | 7 (11.3) | |
| Donor and host relationship, n (%) | |||||
| Child-parent | 4 (23.5) | 12 (60) | 3 (20) | 22 (35.5) | |
| Sibling | 3 (17.6) | 3 (15) | 5 (33.3) | 22 (35.5) | |
| Parent-child | 10 (58.8) | 5 (25) | 7 (46.7) | 18 (29) | |
| Disease category, n (%) | <.01 | ||||
| AML | 10 (58.8) | 7 (35) | 4 (26.7) | 37 (50.9) | |
| ALL | 2 (11.8) | 3 (15) | 8 (53.3) | 21 (33.9) | |
| CML | 2 (11.8) | 0 (0) | 1 (6.7) | 2 (3.2) | |
| MDS | 3 (17.6) | 10 (50) | 2 (13.3) | 2 (3.2) | |
| Disease risk, n (%) | |||||
| High | 6 (35.3) | 10 (50) | 4 (26.7) | 9 (14.5) | .011 |
| HCT-CI score, n (%) | NS | ||||
| 0 | 15 (88.2) | 18 (90) | 11 (73.3) | 56 (90.3) | |
| ≥1 | 2 (11.8) | 2 (10) | 4 (26.7) | 6 (9.7) | |
| MRD status before transplantation, n (%) | NS | ||||
| Negative | 10 (58.8) | 8 (40) | 6 (40) | 29 (46.8) | |
| FCM and molecular positive | 5 (29.4) | 8 (40) | 4 (26.7) | 11 (17.7) | |
| Only FCM positive | 1 (5.9) | 3 (15) | 2 (13.3) | 6 (9.7) | |
| Only molecular positive | 1 (5.9) | 1 (5.0) | 3 (20) | 16 (25.8) | |
| Loci of HLA-A,B,DR mismatch, n (%) | NS | ||||
| 1 | 1 (5.9) | 0 (0) | 0 (0) | 3 (4.8) | |
| 2 | 4 (23.5) | 1 (5.0) | 0 (0) | 10 (16.1) | |
| 3 | 12 (70.6) | 19 (95) | 15 (100) | 49 (79) | |
| Missing self (donor ligand view), n (%) | 0 (0) | 0 (0) | 15 (100) | 62 (100) | <.01 |
| Missing self (host ligand view), n (%) | 0 (0) | 20 (100) | 0 (0) | 62 (100) | <.01 |
| Ligand-ligand GVH mismatch, n (%) | 0 (0) | 20 (100) | 0 (0) | 11 (28.2) | <.01 |
| Donor KIR haplotype, n (%) | NS | ||||
| AA | 9 (52.9) | 9 (45) | 12 (80) | 33 (55) | |
| BX | 8 (47.1) | 11 (55) | 3 (20) | 27 (45) | |
| KIR2DL1 single+group, n (%) | <.01 | ||||
| nsKIR(donor C1C1 to host C1C1) | 0 (0) | 0 (0) | 0 (0) | 43 (69.4) | |
| d-rsKIR(donor C2Cx to host C2Cx) | 17 (100) | 6 (30) | 7 (46.7) | 9 (14.5) | |
| dsKIR(donor C2CX to host C1C1) | 0 (0) | 14 (70) | 0 (0) | 9 (14.5) | |
| rsKIR(donor C1C1 to host C2Cx) | 0 (0) | 0 (0) | 8 (53.3) | 1 (1.6) | |
| KIR2DL2/L3 single+group, n (%) | <.01 | ||||
| nsKIR (donor C2C2 to host C2C2) | 0 (0) | 0 (0) | 0 (0) | 1 (1.6) | |
| d-rsKIR (donor C1Cx to host C1Cx) | 17 (100) | 16 (80) | 9 (60) | 54 (88.5) | |
| dsKIR(donor C1CX to host C2C2) | 0 (0) | 4 (20) | 0 (0) | 3 (4.8) | |
| rsKIR(donor C2C2 to host C1Cx) | 0 (0) | 0 (0) | 6 (40) | 4 (6.5) | |
| KIR3DL1 single+group, n (%) | <.01 | ||||
| nsKIR(donor Bw6Bw6 to host Bw6Bw6) | 0 (0) | 0 (0) | 0 (0) | 25 (40.3) | |
| d-rsKIR(donor Bw4Bwx to host Bw4Bwx) | 17 (100) | 15 (75) | 11 (73.3) | 12 (19.4) | |
| dsKIR(donor Bw4Bwx to host Bw6Bw6) | 0 (0) | 5 (25) | 0 (0) | 13 (21) | |
| rsKIR(donor Bw6Bw6 to host Bw4Bwx) | 0 (0) | 0 (0) | 4 (26.7) | 12 (19.4) | |
| Acute GVHD, n (%) | NS | ||||
| I | 6 (35.3) | 4 (20) | 4 (26.7) | 17 (27.4) | |
| II | 2 (11.8) | 2 (10) | 3 (20) | 12 (19.4) | |
| III | 0 (0) | 1 (5) | 2 (13.3) | 5 (8.1) | |
| IV | 1 (5.9) | 1 (5.0) | 0 (0) | 3 (4.8) | |
| Relapse | 2 (11.8) | 1 (5.0) | 2 (13.3) | 4 (6.5) | |
| Disease-free survival | 13 (76.5) | 16 (80) | 11 (73.3) | 51 (82.3) | |
| OS | 14 (82.4) | 16 (80) | 11 (73.3) | 53 (85.5) |
| . | d-rsKIR . | dsKIR . | rsKIR . | nsKIR . | P . |
|---|---|---|---|---|---|
| n | 17 | 20 | 15 | 62 | |
| Sex of host, male, n (%) | 8 (47.1) | 11 (55) | 10 (66.7) | 40 (64.5) | NS |
| Age of host, median (range), y | 29 (18-53) | 42.5 (26-63) | 28 (15-43) | 31.5 (15-58) | .003 |
| Age of donor, median (range), y | 45 (14-64) | 27 (8-65) | 40 (13-54) | 29 (12-60) | NS |
| Donor and host sex group, n (%) | |||||
| Male-male | 8 (47.1) | 8 (40) | 10 (66.7) | 28 (45.2) | |
| Male-female | 8 (47.1) | 4 (20) | 3 (20) | 15 (24.2) | |
| Female-male | 0 (0) | 5 (25) | 0 (0) | 12 (19.4) | |
| Female-female | 1 (5.9) | 3 (15) | 2 (13.3) | 7 (11.3) | |
| Donor and host relationship, n (%) | |||||
| Child-parent | 4 (23.5) | 12 (60) | 3 (20) | 22 (35.5) | |
| Sibling | 3 (17.6) | 3 (15) | 5 (33.3) | 22 (35.5) | |
| Parent-child | 10 (58.8) | 5 (25) | 7 (46.7) | 18 (29) | |
| Disease category, n (%) | <.01 | ||||
| AML | 10 (58.8) | 7 (35) | 4 (26.7) | 37 (50.9) | |
| ALL | 2 (11.8) | 3 (15) | 8 (53.3) | 21 (33.9) | |
| CML | 2 (11.8) | 0 (0) | 1 (6.7) | 2 (3.2) | |
| MDS | 3 (17.6) | 10 (50) | 2 (13.3) | 2 (3.2) | |
| Disease risk, n (%) | |||||
| High | 6 (35.3) | 10 (50) | 4 (26.7) | 9 (14.5) | .011 |
| HCT-CI score, n (%) | NS | ||||
| 0 | 15 (88.2) | 18 (90) | 11 (73.3) | 56 (90.3) | |
| ≥1 | 2 (11.8) | 2 (10) | 4 (26.7) | 6 (9.7) | |
| MRD status before transplantation, n (%) | NS | ||||
| Negative | 10 (58.8) | 8 (40) | 6 (40) | 29 (46.8) | |
| FCM and molecular positive | 5 (29.4) | 8 (40) | 4 (26.7) | 11 (17.7) | |
| Only FCM positive | 1 (5.9) | 3 (15) | 2 (13.3) | 6 (9.7) | |
| Only molecular positive | 1 (5.9) | 1 (5.0) | 3 (20) | 16 (25.8) | |
| Loci of HLA-A,B,DR mismatch, n (%) | NS | ||||
| 1 | 1 (5.9) | 0 (0) | 0 (0) | 3 (4.8) | |
| 2 | 4 (23.5) | 1 (5.0) | 0 (0) | 10 (16.1) | |
| 3 | 12 (70.6) | 19 (95) | 15 (100) | 49 (79) | |
| Missing self (donor ligand view), n (%) | 0 (0) | 0 (0) | 15 (100) | 62 (100) | <.01 |
| Missing self (host ligand view), n (%) | 0 (0) | 20 (100) | 0 (0) | 62 (100) | <.01 |
| Ligand-ligand GVH mismatch, n (%) | 0 (0) | 20 (100) | 0 (0) | 11 (28.2) | <.01 |
| Donor KIR haplotype, n (%) | NS | ||||
| AA | 9 (52.9) | 9 (45) | 12 (80) | 33 (55) | |
| BX | 8 (47.1) | 11 (55) | 3 (20) | 27 (45) | |
| KIR2DL1 single+group, n (%) | <.01 | ||||
| nsKIR(donor C1C1 to host C1C1) | 0 (0) | 0 (0) | 0 (0) | 43 (69.4) | |
| d-rsKIR(donor C2Cx to host C2Cx) | 17 (100) | 6 (30) | 7 (46.7) | 9 (14.5) | |
| dsKIR(donor C2CX to host C1C1) | 0 (0) | 14 (70) | 0 (0) | 9 (14.5) | |
| rsKIR(donor C1C1 to host C2Cx) | 0 (0) | 0 (0) | 8 (53.3) | 1 (1.6) | |
| KIR2DL2/L3 single+group, n (%) | <.01 | ||||
| nsKIR (donor C2C2 to host C2C2) | 0 (0) | 0 (0) | 0 (0) | 1 (1.6) | |
| d-rsKIR (donor C1Cx to host C1Cx) | 17 (100) | 16 (80) | 9 (60) | 54 (88.5) | |
| dsKIR(donor C1CX to host C2C2) | 0 (0) | 4 (20) | 0 (0) | 3 (4.8) | |
| rsKIR(donor C2C2 to host C1Cx) | 0 (0) | 0 (0) | 6 (40) | 4 (6.5) | |
| KIR3DL1 single+group, n (%) | <.01 | ||||
| nsKIR(donor Bw6Bw6 to host Bw6Bw6) | 0 (0) | 0 (0) | 0 (0) | 25 (40.3) | |
| d-rsKIR(donor Bw4Bwx to host Bw4Bwx) | 17 (100) | 15 (75) | 11 (73.3) | 12 (19.4) | |
| dsKIR(donor Bw4Bwx to host Bw6Bw6) | 0 (0) | 5 (25) | 0 (0) | 13 (21) | |
| rsKIR(donor Bw6Bw6 to host Bw4Bwx) | 0 (0) | 0 (0) | 4 (26.7) | 12 (19.4) | |
| Acute GVHD, n (%) | NS | ||||
| I | 6 (35.3) | 4 (20) | 4 (26.7) | 17 (27.4) | |
| II | 2 (11.8) | 2 (10) | 3 (20) | 12 (19.4) | |
| III | 0 (0) | 1 (5) | 2 (13.3) | 5 (8.1) | |
| IV | 1 (5.9) | 1 (5.0) | 0 (0) | 3 (4.8) | |
| Relapse | 2 (11.8) | 1 (5.0) | 2 (13.3) | 4 (6.5) | |
| Disease-free survival | 13 (76.5) | 16 (80) | 11 (73.3) | 51 (82.3) | |
| OS | 14 (82.4) | 16 (80) | 11 (73.3) | 53 (85.5) |
High-risk patients were those with AML not CR1, MDS not RARS, or RCMD.
ALL, acute lymphoid leukemia; BW4, HLA-BW4; C1, HLA-C group 1; C2, HLA-C group 2; CML chronic myeloid leukemia; FCM, flow cytometry; MDS, myelodysplastic syndrome; NS, not significant.
Patient characteristics in cohort 2
| . | d-rsKIR . | dsKIR . | rsKIR . | nsKIR . | P . |
|---|---|---|---|---|---|
| n | 32 | 55 | 33 | 156 | |
| Age of host, median (range), y | 37 (2-57) | 31 (5-64) | 31 (2-55) | 31 (2-61) | |
| Donor and host sex group, n (%) | NS | ||||
| Male-male | 12 (37.5) | 22 (40) | 14 (42.4) | 68 (43.6) | |
| Male-female | 5 (15.6) | 18 (32.7) | 9 (27.3) | 43 (27.6) | |
| Female-male | 11 (34.4) | 8 (14.5) | 7 (21.2) | 24 (15.4) | |
| Female-female | 4 (12.5) | 7 (12.7) | 3 (9.1) | 21 (13.5) | |
| Donor and host relationship, n (%) | NS | ||||
| Child-parent | 11 (34.4) | 14 (25.5) | 7 (21.2) | 32 (20.5) | |
| Sibling | 9 (28.1) | 16 (29.1) | 12 (36.4) | 52 (33.3) | |
| Parent-child | 12 (37.5) | 25 (45.5) | 14 (42.4) | 68 (43.6) | |
| Collateral relative | 0 (0) | 0 (0) | 0 (0) | 4 (2.6) | |
| Disease category, n (%) | NS | ||||
| AML | 23 (71.9) | 40 (72.7) | 25 (75.8) | 115 (73.7) | |
| MDS | 9 (28.1) | 15 (27.3) | 8 (24.2) | 41 (26.3) | |
| Disease risk, n (%) | NS | ||||
| High | 12 (37.5) | 19 (34.5) | 10 (30.3) | 58 (37.2) | |
| Disease risk index, n (%) | NS | ||||
| Low | 2 (6.5) | 5 (9.1) | 2 (6.1) | 25 (16) | |
| Intermediate | 23 (74.2) | 43 (78.2) | 24 (72.7) | 97 (62.2) | |
| High or very high | 6 (19.4) | 7 (12.7) | 7 (21.2) | 34 (21.8) | |
| MRD status before transplantation, n (%) | NS | ||||
| Negative | 11 (36.7) | 27 (49.1) | 14 (42.4) | 46 (29.7) | |
| FCM and molecular positive | 8 (26.7) | 15 (27.3) | 7 (21.2) | 52 (33.5) | |
| Only FCM positive | 2 (6.7) | 2 (3.6) | 3 (9.1) | 10 (6.5) | |
| Only molecular positive | 9 (30) | 11 (20) | 9 (27.3) | 47 (30.3) | |
| Loci of HLA-A,B,DR mismatch, n (%) | .027 | ||||
| 0 | 1 (3.1) | 0 (0) | 1 (3.0) | 0 (0) | |
| 1 | 2 (6.3) | 1 (1.8) | 0 (0) | 7 (4.5) | |
| 2 | 10 (31.3) | 5 (9.1) | 5 (15.2) | 38 (24.4) | |
| 3 | 19 (59.4) | 49 (89.1) | 27 (81.8) | 111 (71.2) | |
| Missing self (donor ligand view), n (%) | 0 (0) | 0 (0) | 33 (100) | 156 (100) | <.01 |
| Missing self (host ligand view), n (%) | 0 (0) | 55 (100) | 0 (0) | 156 (100) | <.01 |
| Ligand-ligand GVH mismatch, n (%) | 0 (0) | 55 (100) | 0 (0) | 41 (26.3) | <.01 |
| Donor KIR haplotype, n (%) | NS | ||||
| AA | 22 (71) | 29 (54.7) | 18 (56.3) | 77 (50.3) | |
| BX | 9 (29) | 24 (45.3) | 14 (43.8) | 76 (49.7) | |
| KIR2DL1 single+group, n (%) | <.01 | ||||
| nsKIR (donor C1C1 to host C1C1) | 0 (0) | 0 (0) | 0 (0) | 110 (70.5) | |
| d-rsKIR (donor C2Cx to host C2Cx) | 32 (100) | 14 (25.5) | 17 (51.5) | 21 (13.5) | |
| dsKIR (donor C2CX to host C1C1) | 0 (0) | 41 (74.5) | 0 (0) | 18 (11.5) | |
| rsKIR (donor C1C1 to host C2Cx) | 0 (0) | 0 (0) | 16 (48.5) | 7 (4.5) | |
| KIR2DL2/L3 single+group, n (%) | <.01 | ||||
| nsKIR (donor C2C2 to host C2C2) | 0 (0) | 0 (0) | 0 (0) | 1 (0.6) | |
| d-rsKIR (donor C1Cx to host C1Cx) | 32 (100) | 46 (83.6) | 23 (69.7) | 143 (91.7) | |
| dsKIR (donor C1CX to host C2C2) | 0 (0) | 9 (16.4) | 0 (0) | 2 (1.3) | |
| rsKIR (donor C2C2 to host C1Cx) | 0 (0) | 0 (0) | 10 (30.3) | 10 (6.4) | |
| KIR3DL1 single+group, n (%) | <.01 | ||||
| nsKIR (donor Bw6Bw6 to host Bw6Bw6) | 0 (0) | 0 (0) | 0 (0) | 73 (46.8) | |
| d-rsKIR (donor Bw4Bwx to host Bw4Bwx) | 32 (100) | 34 (61.8) | 22 (66.7) | 38 (24.4) | |
| dsKIR (donor Bw4Bwx to host Bw6Bw6) | 0 (0) | 21 (38.2) | 0 (0) | 19 (12.2) | |
| rsKIR (donor Bw6Bw6 to host Bw4Bwx) | 0 (0) | 0 (0) | 11 (33.3) | 26 (16.7) | |
| Acute GVHD, n (%) | NS | ||||
| I | 9 (28.1) | 17 (30.9) | 9 (27.3) | 52 (33.3) | |
| II | 6 (18.8) | 7 (12.7) | 7 (21.2) | 24 (15.4) | |
| III | 2 (6.3) | 2 (3.6) | 3 (9.1) | 10 (6.4) | |
| IV | 2 (6.3) | 2 (3.6) | 3 (9.1) | 2 (1.3) | |
| Relapse | 0 (0) | 4 (7.3) | 4 (12.1) | 23 (14.7) | .025 |
| TRM | 4 (12.5) | 10 (18.2) | 7 (21.2) | 27 (17.3) | NS |
| . | d-rsKIR . | dsKIR . | rsKIR . | nsKIR . | P . |
|---|---|---|---|---|---|
| n | 32 | 55 | 33 | 156 | |
| Age of host, median (range), y | 37 (2-57) | 31 (5-64) | 31 (2-55) | 31 (2-61) | |
| Donor and host sex group, n (%) | NS | ||||
| Male-male | 12 (37.5) | 22 (40) | 14 (42.4) | 68 (43.6) | |
| Male-female | 5 (15.6) | 18 (32.7) | 9 (27.3) | 43 (27.6) | |
| Female-male | 11 (34.4) | 8 (14.5) | 7 (21.2) | 24 (15.4) | |
| Female-female | 4 (12.5) | 7 (12.7) | 3 (9.1) | 21 (13.5) | |
| Donor and host relationship, n (%) | NS | ||||
| Child-parent | 11 (34.4) | 14 (25.5) | 7 (21.2) | 32 (20.5) | |
| Sibling | 9 (28.1) | 16 (29.1) | 12 (36.4) | 52 (33.3) | |
| Parent-child | 12 (37.5) | 25 (45.5) | 14 (42.4) | 68 (43.6) | |
| Collateral relative | 0 (0) | 0 (0) | 0 (0) | 4 (2.6) | |
| Disease category, n (%) | NS | ||||
| AML | 23 (71.9) | 40 (72.7) | 25 (75.8) | 115 (73.7) | |
| MDS | 9 (28.1) | 15 (27.3) | 8 (24.2) | 41 (26.3) | |
| Disease risk, n (%) | NS | ||||
| High | 12 (37.5) | 19 (34.5) | 10 (30.3) | 58 (37.2) | |
| Disease risk index, n (%) | NS | ||||
| Low | 2 (6.5) | 5 (9.1) | 2 (6.1) | 25 (16) | |
| Intermediate | 23 (74.2) | 43 (78.2) | 24 (72.7) | 97 (62.2) | |
| High or very high | 6 (19.4) | 7 (12.7) | 7 (21.2) | 34 (21.8) | |
| MRD status before transplantation, n (%) | NS | ||||
| Negative | 11 (36.7) | 27 (49.1) | 14 (42.4) | 46 (29.7) | |
| FCM and molecular positive | 8 (26.7) | 15 (27.3) | 7 (21.2) | 52 (33.5) | |
| Only FCM positive | 2 (6.7) | 2 (3.6) | 3 (9.1) | 10 (6.5) | |
| Only molecular positive | 9 (30) | 11 (20) | 9 (27.3) | 47 (30.3) | |
| Loci of HLA-A,B,DR mismatch, n (%) | .027 | ||||
| 0 | 1 (3.1) | 0 (0) | 1 (3.0) | 0 (0) | |
| 1 | 2 (6.3) | 1 (1.8) | 0 (0) | 7 (4.5) | |
| 2 | 10 (31.3) | 5 (9.1) | 5 (15.2) | 38 (24.4) | |
| 3 | 19 (59.4) | 49 (89.1) | 27 (81.8) | 111 (71.2) | |
| Missing self (donor ligand view), n (%) | 0 (0) | 0 (0) | 33 (100) | 156 (100) | <.01 |
| Missing self (host ligand view), n (%) | 0 (0) | 55 (100) | 0 (0) | 156 (100) | <.01 |
| Ligand-ligand GVH mismatch, n (%) | 0 (0) | 55 (100) | 0 (0) | 41 (26.3) | <.01 |
| Donor KIR haplotype, n (%) | NS | ||||
| AA | 22 (71) | 29 (54.7) | 18 (56.3) | 77 (50.3) | |
| BX | 9 (29) | 24 (45.3) | 14 (43.8) | 76 (49.7) | |
| KIR2DL1 single+group, n (%) | <.01 | ||||
| nsKIR (donor C1C1 to host C1C1) | 0 (0) | 0 (0) | 0 (0) | 110 (70.5) | |
| d-rsKIR (donor C2Cx to host C2Cx) | 32 (100) | 14 (25.5) | 17 (51.5) | 21 (13.5) | |
| dsKIR (donor C2CX to host C1C1) | 0 (0) | 41 (74.5) | 0 (0) | 18 (11.5) | |
| rsKIR (donor C1C1 to host C2Cx) | 0 (0) | 0 (0) | 16 (48.5) | 7 (4.5) | |
| KIR2DL2/L3 single+group, n (%) | <.01 | ||||
| nsKIR (donor C2C2 to host C2C2) | 0 (0) | 0 (0) | 0 (0) | 1 (0.6) | |
| d-rsKIR (donor C1Cx to host C1Cx) | 32 (100) | 46 (83.6) | 23 (69.7) | 143 (91.7) | |
| dsKIR (donor C1CX to host C2C2) | 0 (0) | 9 (16.4) | 0 (0) | 2 (1.3) | |
| rsKIR (donor C2C2 to host C1Cx) | 0 (0) | 0 (0) | 10 (30.3) | 10 (6.4) | |
| KIR3DL1 single+group, n (%) | <.01 | ||||
| nsKIR (donor Bw6Bw6 to host Bw6Bw6) | 0 (0) | 0 (0) | 0 (0) | 73 (46.8) | |
| d-rsKIR (donor Bw4Bwx to host Bw4Bwx) | 32 (100) | 34 (61.8) | 22 (66.7) | 38 (24.4) | |
| dsKIR (donor Bw4Bwx to host Bw6Bw6) | 0 (0) | 21 (38.2) | 0 (0) | 19 (12.2) | |
| rsKIR (donor Bw6Bw6 to host Bw4Bwx) | 0 (0) | 0 (0) | 11 (33.3) | 26 (16.7) | |
| Acute GVHD, n (%) | NS | ||||
| I | 9 (28.1) | 17 (30.9) | 9 (27.3) | 52 (33.3) | |
| II | 6 (18.8) | 7 (12.7) | 7 (21.2) | 24 (15.4) | |
| III | 2 (6.3) | 2 (3.6) | 3 (9.1) | 10 (6.4) | |
| IV | 2 (6.3) | 2 (3.6) | 3 (9.1) | 2 (1.3) | |
| Relapse | 0 (0) | 4 (7.3) | 4 (12.1) | 23 (14.7) | .025 |
| TRM | 4 (12.5) | 10 (18.2) | 7 (21.2) | 27 (17.3) | NS |
Donor KIR ligated by both donor and host HLA is associated with better single-KIR+ NK-cell education among the same patients
In order to exclude the bias caused by different individuals on single-KIR+ NK function, we compared different single-KIR+ NK-cell functions, identified by donor-host HLA and donor inhibitory KIR interactions, within the same individual.
KIR2DL2/L3 single+ NK cells exhibited higher reactivity than KIR2DL1 single+ NK cells in pairs of donor C1C1 or C1C2 and host C1C1
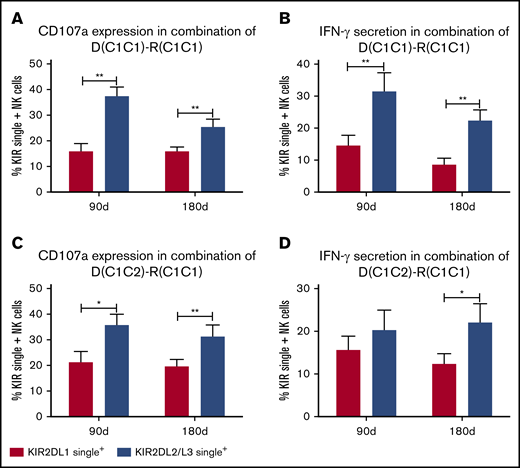
In the pairs of donor C1C1 and host C1C1 (n = 14), NK cells singly expressing KIR2DL2/L3 NK cells exhibited higher reactivity than KIR2DL1 single+ NK cells at days 90 and 180 regardless of the expression of CD107a or IFN-γ against K562 cells (Figure 1A-B), suggesting that when both donor and host express HLA-C1, KIR2DL2/L3+ NK cells exhibit the greatest function.
KIR2DL2/L3 single+NK cells show higher reactivity than KIR2DL1 single+NK cells in pairs of donor C1C1 or C1C2 and host C1C1. Expression of CD107a and IFN-γ against K562 by KIR2DL2/L3 single+ NK cells and KIR2DL1 single+ NK cells in pairs of donor C1C1 and host C1C1 (n = 14; A-B) and pairs of donor C1C2 and host C1C1 (n = 6; C-D) at days 90 and 180 after transplantation. **P < .01; *P < .05.
KIR2DL2/L3 single+NK cells show higher reactivity than KIR2DL1 single+NK cells in pairs of donor C1C1 or C1C2 and host C1C1. Expression of CD107a and IFN-γ against K562 by KIR2DL2/L3 single+ NK cells and KIR2DL1 single+ NK cells in pairs of donor C1C1 and host C1C1 (n = 14; A-B) and pairs of donor C1C2 and host C1C1 (n = 6; C-D) at days 90 and 180 after transplantation. **P < .01; *P < .05.
At the same time, in the pairs of donor C1C2 and host C1C1 (n = 6), CD107a expression of KIR2DL2/L3 single+ NK cells against K562 cells was higher compared with KIR2DL1 single+ NK cells at days 90 and 180 (Figure 1C), and IFN-γ expression of KIR2DL2/L3 single+ NK cells against K562 cells was also higher at day 180 compared with KIR2DL1 single+ NK cells (Figure 1D), further suggesting that both donors and hosts coexpressing HLA-C1 are good for KIR2DL2/L3 single+ NK-cell functional recovery, but only donors exhibiting HLA-C2 were not enough for KIR2DL1 single+ NK-cell functional recovery.
KIR2DL2/L3 single+ NK cells exhibited higher reactivity than KIR3DL1 single+ NK cells in pairs of donors Bw4C1Cx and host C1Cx
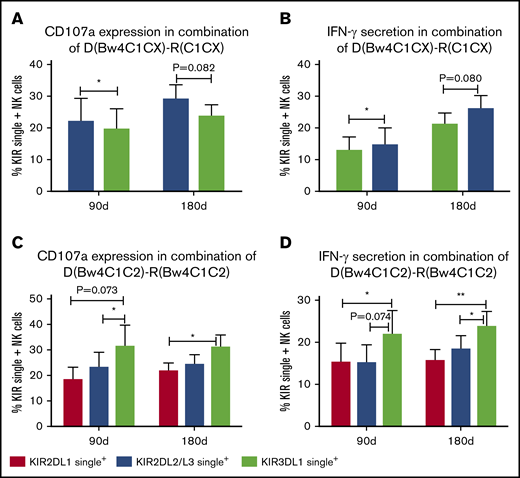
In the pairs of donor Bw4C1C1 and host C1C1 (n = 11), CD107a expression of KIR2DL2/L3 single+ NK cells was higher compared with KIR3DL1 single+ NK cells at days 90 and 180 (Figure 2A), and IFN-γ expression of KIR2DL2/L3 single+ NK cells against K562 cells was higher at day 180 compared with KIR3DL1 single+ NK cells (Figure 2B), suggesting that both donor and host coexisting HLA-C1 is good for KIR2DL2/L3 single+ NK-cell functional recovery, but only donor existing HLA-Bw4 is not enough for KIR3DL1 single+ NK-cell functional recovery.
KIR2DL2/L3 single+NK cells show higher reactivity than KIR3DL1 single+NK cells in pairs of donor Bw4C1C1 and host C1C1. Expression of CD107a and IFN-γ against K562 cells by KIR2DL2/L3 single+ NK cells and KIR3DL1 single+ NK cells in pairs of donor Bw4C1C1 and host C1C1 (n = 11; A-B) at days 90 and 180 after transplantation. Meanwhile, the expression of CD107a and IFN-γ against K562 by KIR2DL1 single+, KIR2DL2/L3 single+, and KIR3DL1 single+ NK cells in pairs of donor Bw4C1C2 and host Bw4C1C2 (n = 18; C-D) at days 90 and 180 after transplantation. **P < .01; *P < .05.
KIR2DL2/L3 single+NK cells show higher reactivity than KIR3DL1 single+NK cells in pairs of donor Bw4C1C1 and host C1C1. Expression of CD107a and IFN-γ against K562 cells by KIR2DL2/L3 single+ NK cells and KIR3DL1 single+ NK cells in pairs of donor Bw4C1C1 and host C1C1 (n = 11; A-B) at days 90 and 180 after transplantation. Meanwhile, the expression of CD107a and IFN-γ against K562 by KIR2DL1 single+, KIR2DL2/L3 single+, and KIR3DL1 single+ NK cells in pairs of donor Bw4C1C2 and host Bw4C1C2 (n = 18; C-D) at days 90 and 180 after transplantation. **P < .01; *P < .05.
KIR2DL1 single+ NK cells and KIR2DL2/L3 single+ NK cells exhibited comparable reactivity in pairs of donor Bw4C1C2 and host Bw4C1C2
In the pairs of donor Bw4C1C2 and host Bw4C1C2 (n = 18), CD107a and IFN-γ expression of KIR3DL1 single+ NK cells against K562 cells was higher compared with KIR2DL2/L3 single+ NK cell and KIR2DL1 single+ NK cells at day 90 and 180, but the reactivity of KIR2DL2/2DL3 single+ NK cells and KIR2DL1 single+ NK cells was comparable, suggesting that when both donor and host coexpress C1C2 for donor KIR2DL2/L3 and KIR2DL1, the comprehensiveness of education of KIR2DL2/L3 single+ NK cells and KIR2DL1 single+ NK cells is comparable. However, when both donor and host coexpress C1C2Bw4 for donor KIR2DL2/L3, KIR2DL1, and KIR3DL1, why the function of KIR3DL1 single+ NK cells is higher than that of KIR2DL2/L3 single+ and KIR2DL1 single+ NK cells cannot be explained by the coexistence of donor and host MHC alone (Figure 2C-D).
Donor KIR ligated by both donor and host HLA contributes to better single-KIR+ NK-cell education among the same single-KIR+ NK cells
The reconstitution kinetics of absolute number, phenotype, and reactivity of single KIR+ NK cells after transplantation can be found in supplemental Materials and supplemental Figures 2-7. To further exclude the potential bias caused by different single KIR+ NK cells, we compared the effects of different donor-host HLA combinations on the function of single KIR+ NK cells within the same single KIR+ NK cells.
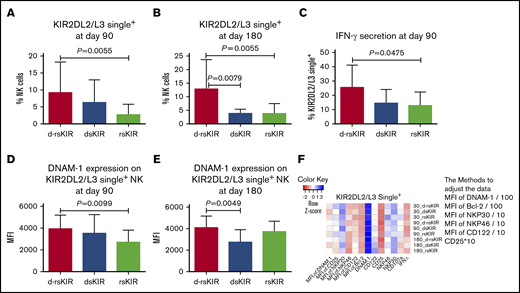
KIR2DL2/L3 single+ NK cells in the d-rsKIR group (C1Cx-C1Cx) exhibit higher reactivity compared with other groups (dsKIR, C1Cx-C2C2, and rsKIR C2C2-C1Cx)
Because only 1 patient carried nsKIR (C2C2-C2C2), no further comparisons were considered in this group. Among the d-rsKIR (n = 96), dsKIR (n = 7), and rsKIR (n = 10) groups, we analyzed differences in the expression of CD57, NKG2C, NKp46, NKp30, NKG2D, DNAM-1, CD25, CD122, BCl-2, CD107a, and IFN-γ on single KIR2DL2/L3+ NK cells.
On days 90 and 180, the percentage of KIR2DL2/L3 single+ NK cells was higher in the d-rsKIR group than the rsKIR or dsKIR groups (Figure 3A-B). At day 90, the secretion of IFN-γ on KIR2DL2/L3 single+ NK cells against K562 cells was higher in the d-rsKIR group than the rsKIR group (Figure 3C), and the MFI of DNAM-1 expression on single KIR2DL2/L3+ NK cells was higher in the d-rsKIR group than the rsKIR group at day 90 and the dsKIR group at day 180 (Figure 3D-E), suggesting further that both donor and host coexisting HLA-C1 is good for KIR2DL2/L3 single+ NK-cell education, but having only donor or host with HLA-C1 is not enough. No differences were found in the expression of CD107a on KIR2DL2/L3 single+ NK cells after stimulation with K562 cells among the d-rsKIR, dsKIR, and rsKIR groups (Figure 3F).
KIR2DL2/L3 single+NK cells in the d-rsKIR group (C1Cx-C1Cx) show higher reactivity compared with other groups (dsKIR and rsKIR). The percentage of KIR2DL2/L3 single+ NK cells at day 90 (A) and day 180 (B) after transplantation in the d-rsKIR group (pairs of donor C1Cx and host C1Cx, n = 96), dsKIR group (pairs of donor C1Cx and host C2C2, n = 7), and rsKIR group (pairs of donor C2C2 and host C1Cx, n = 10). Secretion of IFN-γ (C) of KIR2DL2/L3 single+ NK cells against K562 cells at day 90 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 on KIR2DL2/L3 single+ NK cells at day 90 (D) and 180 (E) after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR2DL2/L3+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel F. In order to adjust all the data to the same order of magnitude, we process the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100, the MFI of NKP30, NKP46, and CD122 was divided by 10, and the expression of CD25 was multiplied by 10.
KIR2DL2/L3 single+NK cells in the d-rsKIR group (C1Cx-C1Cx) show higher reactivity compared with other groups (dsKIR and rsKIR). The percentage of KIR2DL2/L3 single+ NK cells at day 90 (A) and day 180 (B) after transplantation in the d-rsKIR group (pairs of donor C1Cx and host C1Cx, n = 96), dsKIR group (pairs of donor C1Cx and host C2C2, n = 7), and rsKIR group (pairs of donor C2C2 and host C1Cx, n = 10). Secretion of IFN-γ (C) of KIR2DL2/L3 single+ NK cells against K562 cells at day 90 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 on KIR2DL2/L3 single+ NK cells at day 90 (D) and 180 (E) after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR2DL2/L3+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel F. In order to adjust all the data to the same order of magnitude, we process the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100, the MFI of NKP30, NKP46, and CD122 was divided by 10, and the expression of CD25 was multiplied by 10.
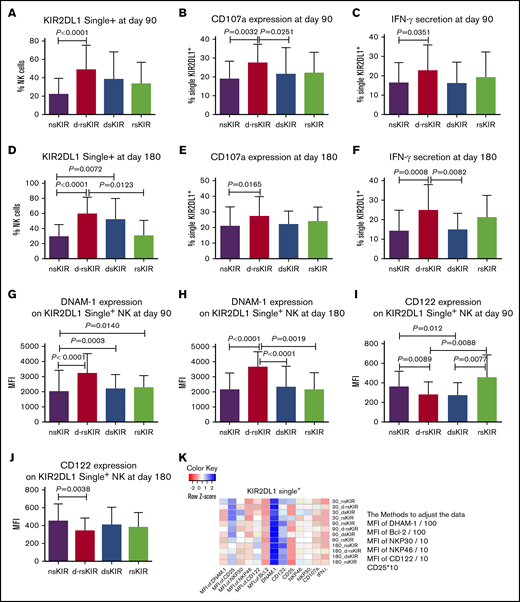
KIR2DL1 single+ NK cells in the d-rsKIR group (C2Cx-C2Cx) exhibit higher reactivity compared with other groups (nsKIR [C1C1-C1C1], dsKIR [C2Cx-C1C1], and rsKIR [C1C1-C2Cx])
In accordance with the above KIR2DL2/L3 single+ NK analysis, the percentage of KIR2DL1 single+ NK cells was higher in the d-rsKIR group (n = 39) than the nsKIR group (n = 43) or rsKIR group (n = 9) at days 90 and 180 (Figure 4A,D). The expression of CD107a as well as the secretion of IFN-γ on KIR2DL1 single+ NK cells against K562 cells was higher in the d-rsKIR group than the nsKIR or dsKIR groups (n = 23) group at days 90 and 180 (Figure 4B-C,E-F). These results further confirmed that coexistence of both donor and host HLA-C2 is good for KIR2DL1 single+ NK-cell functional recovery, but solely one or the other (donor or host) exhibiting HLA-C2 is not enough. The MFI expression of DNAM-1 on KIR2DL1 single+ NK cells was higher in the d-rsKIR group than the nsKIR, dsKIR, or rsKIR groups (Figure 4G-H) at days 90 and 180. Inversely, the MFI of CD122 expression on single KIR2DL1+ NK cells was actually higher in the nsKIR group than the d-rsKIR group at days 90 and 180 (Figure 4I, J). This suggests DNAM-1 and CD122 are inversely correlated with the education process. No differences were found in the expression of NKp30, NKp46, CD25, or Bcl-2 on KIR2DL1 single+ NK cells among the 4 groups (Figure 4K).
KIR2DL1 single+NK cells in the d-rsKIR group (C2Cx-C2Cx) showed higher reactivity compared with other groups (nsKIR, dsKIR, and rsKIR). The percentage of KIR2DL1 single+ NK cells at day 90 (A) and day 180 (D) after transplantation in the d-rsKIR (pairs of donor C2Cx and host C2Cx, n = 39), dsKIR (pairs of donor C2Cx and host C1C1, n = 23), rsKIR (pairs of donor C1C1 and host C2Cx, n = 9), and nsKIR groups (pairs of donor C1C1 and host C1C1, n = 43). The expression of CD107a (B,E) and IFN-γ (C,F) of KIR2DL1 single+ NK cells against K562 cells at days 90 and 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 (G-H) and CD122 (I-J) on KIR2DL1 single+ NK cells at days 90 and 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR2DL1+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel K. In order to adjust all the data to the same order of magnitude, we processed the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100; the MFI of NKP30, NKP46, and CD122 was divided by 10; and the expression of CD25 was multiplied by 10.
KIR2DL1 single+NK cells in the d-rsKIR group (C2Cx-C2Cx) showed higher reactivity compared with other groups (nsKIR, dsKIR, and rsKIR). The percentage of KIR2DL1 single+ NK cells at day 90 (A) and day 180 (D) after transplantation in the d-rsKIR (pairs of donor C2Cx and host C2Cx, n = 39), dsKIR (pairs of donor C2Cx and host C1C1, n = 23), rsKIR (pairs of donor C1C1 and host C2Cx, n = 9), and nsKIR groups (pairs of donor C1C1 and host C1C1, n = 43). The expression of CD107a (B,E) and IFN-γ (C,F) of KIR2DL1 single+ NK cells against K562 cells at days 90 and 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 (G-H) and CD122 (I-J) on KIR2DL1 single+ NK cells at days 90 and 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR2DL1+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel K. In order to adjust all the data to the same order of magnitude, we processed the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100; the MFI of NKP30, NKP46, and CD122 was divided by 10; and the expression of CD25 was multiplied by 10.
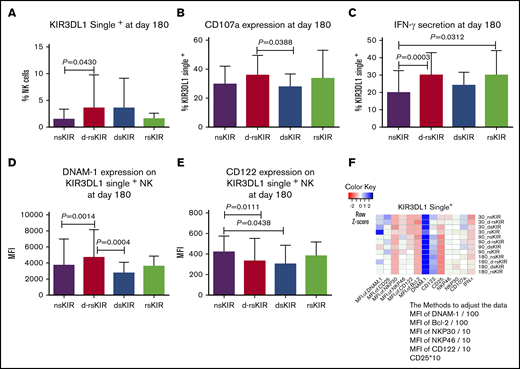
KIR3DL1 single+ NK cells in the d-rsKIR group (Bw4Bwx-Bw4Bwx) exhibit higher reactivity compared with other groups (nsKIR [Bw6Bw6-Bw6Bw6], dsKIR [Bw4Bwx-Bw6Bw6], and rsKIR [Bw6Bw6-Bw4Bwx])
In accordance with the above KIR2DL1 single+ and KIR2DL2/L3 single+ NK cells, the percentage of KIR3DL1 single+ NK cells was higher in the d-rsKIR group (n = 55) than the nsKIR group at day 180 (n = 25; Figure 5A). The expression of CD107a on KIR3DL1 single+ NK cells against K562 cells was higher in the d-rsKIR group than the dsKIR group (n = 18; Figure 5B). The secretion of IFN-γ on KIR3DL1 single+ NK cells against K562 cells was higher in the d-rsKIR group than the nsKIR group (Figure 5C). The MFI expression of DNAM-1 on KIR3DL1 single+ NK cells was higher in the d-rsKIR group than the nsKIR and dsKIR groups (Figure 5D). However, the MFI expression of CD122 on KIR3DL1 single+ NK cells was higher in the nsKIR group than the d-rsKIR and dsKIR groups (Figure 5E). No differences were found in the expression of NKp30, NKp46, CD25, and Bcl-2 on KIR3DL1 single+ NK cells among the 4 groups (Figure 5F).
KIR3DL1 single+NK cells in the d-rsKIR group (Bw4Bwx-Bw4Bwx) show higher reactivity compared with other groups (nsKIR, dsKIR, and rsKIR). The percentage of KIR3DL1 single+ NK cells at day 180 (A) after transplantation in the d-rsKIR (pairs of donor Bw4Bwx and host Bw4Bwx, n = 55), dsKIR (pairs of donor Bw4Bwx and host Bw6Bw6, n = 18), rsKIR (pairs of donor Bw6Bw6 and host Bw4Bwx, n = 16), and nsKIR groups (pairs of donor Bw6Bw6 and host Bw6Bw6, n = 25). The expression of CD107a (B) and IFN-γ (C) of KIR3DL1 single+ NK cells against K562 cells at day 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 (D) and CD122 (E) on KIR3DL1 single+ NK cells at day 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR3DL1+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel F. In order to adjust all the data to the same order of magnitude, we processed the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100; the MFI of NKP30, NKP46, and CD122 was divided by 10; and the expression of CD25 was multiplied by 10.
KIR3DL1 single+NK cells in the d-rsKIR group (Bw4Bwx-Bw4Bwx) show higher reactivity compared with other groups (nsKIR, dsKIR, and rsKIR). The percentage of KIR3DL1 single+ NK cells at day 180 (A) after transplantation in the d-rsKIR (pairs of donor Bw4Bwx and host Bw4Bwx, n = 55), dsKIR (pairs of donor Bw4Bwx and host Bw6Bw6, n = 18), rsKIR (pairs of donor Bw6Bw6 and host Bw4Bwx, n = 16), and nsKIR groups (pairs of donor Bw6Bw6 and host Bw6Bw6, n = 25). The expression of CD107a (B) and IFN-γ (C) of KIR3DL1 single+ NK cells against K562 cells at day 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. MFI expression of DNAM-1 (D) and CD122 (E) on KIR3DL1 single+ NK cells at day 180 after transplantation in the d-rsKIR, dsKIR, and rsKIR groups. Heatmap of average NK-cell marker expression on single KIR3DL1+ NK cells from each group at different time points. Sample origin color-coding according to the color key in panel F. In order to adjust all the data to the same order of magnitude, we processed the data as follows: the MFI of DNAM-1 and Bcl-2 was divided by 100; the MFI of NKP30, NKP46, and CD122 was divided by 10; and the expression of CD25 was multiplied by 10.
No matter NK cells stimulated with or without IL2, singleKIR+ NK cells in group of d-rsKIR exhibit higher reactivity compared to other groups
Considering cytokine activation can overcome hypo-function of the noneducated NK cells, the peripheral blood samples of patients at 1 year post transplantation were also selected from each group looking at the functional analysis without and with overnight IL-2 activation. As shown in supplemental Figure 11, without IL2 stimulation, there was higher IFN-γ expression in the d-rsKIR group compared to the nsKIR group, no matter whether we used KIR2DL1 single+ NK cells, KIR2DL2/L3 single+ NK cells, or KIR3DL1 single+ NK cells, and higher CD107a expression in the d-rsKIR group compared to the nsKIR group in KIR2DL1 single+ NK cells. NK cells stimulated with IL2 overnight show similar trends to those differences in samples without IL-2 activation.
Both donor and host HLA must coexist for maximum education of NK cells given donor 3 inhibitory KIRs
Considering the dose effect of KIRs for self-HLA on effector capacity in healthy donors, we further explored whether the dose effect of KIRs still exists during the recovery of NK cells after transplantation.28 To allow for the evaluation of inhibitory KIRs and minimize interference from potential class I–recognizing activating KIRs and non-self HLA, we analyzed NK responsiveness from 9 donor-patient pairs, both of donors and hosts presenting all HLAs (Bw4C1C2) for haplotype A donor KIRs presenting KIR3DL1, KIR2DL3, KIR2DL1 at the same time. We showed that there was a clear hierarchy of responses among NK populations. NK cells with 2 inhibitory KIRs for self-HLA exhibit higher NK responsiveness (CD107a and IFN-γ) than single KIR+ NK cells. NK cells with 3 inhibitory KIRs for self-HLA exhibit maximum responsiveness, indicating that presentation of all HLAs by both donor and host maximizes the NK-cell functional responsiveness after transplantation (Figure 6A-B).
Both donor and host coexist all HLAs for donor 3 inhibitory KIRs associated with maximum responsiveness recovery of NK cells and least relapse following haplo-SCT. To allow for the evaluation of inhibitory KIRs and minimize interference from potential class I–recognizing activating KIRs and non-self HLA, we analyzed NK responsiveness from 9 donor-patients pairs of both of donor and host presenting all HLAs (Bw4C1C2) for haplotype A donor KIR presenting KIR3DL1, KIR2DL3, KIR2DL1 at the same time. The hierarchy of expression of CD107a (A) and IFN-γ (B) against K562 cells of NK subsets with expression of 3 (NKG2A−KIR2DL1+KIR2DL3+KIR3DL1+ NK), 2 (NKG2A−KIR2DL1+KIR2DL2/L3+ NK, NKG2A−KIR2DL1+KIR3DL1+ NK, or NKG2A−KIR2DL3+KIR3DL1+ NK), 1 (NKG2A−KIR2DL1+ NK, NKG2A-KIR2DL3+ NK, or NKG2A−KIR3DL1+ NK), or 0 (NKG2A−KIR−) inhibitory KIRs for self-HLA. To evaluate the predictive roles of the interaction of donor and host coexisting HLAs for donor inhibitory KIRs and clinical outcomes, in the second cohort, 276 AML or MDS patients who underwent haplo-SCT were subgrouped into the nsKIR group (n = 156), where both hosts and donors lacked HLA ligands for all 3 donor KIRs (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); the d-rsKIR group (n = 31), where donors and hosts encoded all HLA ligands for donor KIRs (ie, donor C1C2Bw4 to host C1C2Bw4); the dsKIR group (n = 33), where donors, but not hosts, encoded all HLA ligands for donor KIR (ie, for donor C1C2Bw4 and host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); and the rsKIR group (n = 55), where hosts, but not donors, encoded all HLA ligands for donor KIR (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and host C1C2Bw4). The lowest relapse rate (C) was found in the d-rsKIR group (0%) compared with the rsKIR group (10.0% ± 4.9%, P = .115), dsKIR group (14.9% ± 7.0%, P = .039), or nsKIR group (18% ± 3.5%, P = .022). Therefore, higher LFS (D) and OS (E) were found in the d-rsKIR group compared with the nsKIR group (87.5% ± 5.8% vs 67.2% ± 3.8%, P = .046 and 87.5% ± 5.8% vs 68.1% ± 4.0%, P = .066).
Both donor and host coexist all HLAs for donor 3 inhibitory KIRs associated with maximum responsiveness recovery of NK cells and least relapse following haplo-SCT. To allow for the evaluation of inhibitory KIRs and minimize interference from potential class I–recognizing activating KIRs and non-self HLA, we analyzed NK responsiveness from 9 donor-patients pairs of both of donor and host presenting all HLAs (Bw4C1C2) for haplotype A donor KIR presenting KIR3DL1, KIR2DL3, KIR2DL1 at the same time. The hierarchy of expression of CD107a (A) and IFN-γ (B) against K562 cells of NK subsets with expression of 3 (NKG2A−KIR2DL1+KIR2DL3+KIR3DL1+ NK), 2 (NKG2A−KIR2DL1+KIR2DL2/L3+ NK, NKG2A−KIR2DL1+KIR3DL1+ NK, or NKG2A−KIR2DL3+KIR3DL1+ NK), 1 (NKG2A−KIR2DL1+ NK, NKG2A-KIR2DL3+ NK, or NKG2A−KIR3DL1+ NK), or 0 (NKG2A−KIR−) inhibitory KIRs for self-HLA. To evaluate the predictive roles of the interaction of donor and host coexisting HLAs for donor inhibitory KIRs and clinical outcomes, in the second cohort, 276 AML or MDS patients who underwent haplo-SCT were subgrouped into the nsKIR group (n = 156), where both hosts and donors lacked HLA ligands for all 3 donor KIRs (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); the d-rsKIR group (n = 31), where donors and hosts encoded all HLA ligands for donor KIRs (ie, donor C1C2Bw4 to host C1C2Bw4); the dsKIR group (n = 33), where donors, but not hosts, encoded all HLA ligands for donor KIR (ie, for donor C1C2Bw4 and host C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4); and the rsKIR group (n = 55), where hosts, but not donors, encoded all HLA ligands for donor KIR (ie, for donor C1C1, C2C2, C1C2, C1C1Bw4, or C2C2Bw4 and host C1C2Bw4). The lowest relapse rate (C) was found in the d-rsKIR group (0%) compared with the rsKIR group (10.0% ± 4.9%, P = .115), dsKIR group (14.9% ± 7.0%, P = .039), or nsKIR group (18% ± 3.5%, P = .022). Therefore, higher LFS (D) and OS (E) were found in the d-rsKIR group compared with the nsKIR group (87.5% ± 5.8% vs 67.2% ± 3.8%, P = .046 and 87.5% ± 5.8% vs 68.1% ± 4.0%, P = .066).
Both donor and host exhibiting all HLAs (Bw4C1C2) for donor 3 inhibitory KIRs contributes to the lowest relapse rates following haplo-HSCT
The superior responsiveness of single KIR+ NK and multiple KIR+ NK cells in the context of donor and host coexisting KIR ligand for relevant inhibitory KIRs motivated us to investigate whether the clinical outcome in AML was affected by the donor and host coexpressing ligands for all inhibitory KIRs.
A longitudinal analysis in the first cohort of 80 myeloid leukemia patients showed that there was no hematologic relapse when both the donor and host expressed ligands for donor 3 inhibitory KIRs. In contrast, 2 patients relapsed in other situations.
Because there were few relapsed patients in the first cohort of myeloid leukemia patients, we enrolled the second cohort. The lowest relapse rate was found in the d-rsKIR group (n = 31, 0%) compared with the rsKIR (n = 55, 0% vs 10.0 ± 4.9%, P = .115), dsKIR (n = 33, 0% vs 14.9% ± 7.0%, P = .039), or nsKIR groups (n = 156, 0% vs 18% ± 3.5%, P = .022; Figure 6C). Therefore, higher leukemia-free survival (LFS) (Figure 6D) and higher overall survival (OS) (Figure 6E) were found in the d-rsKIR group compared with nsKIR group (87.5% ± 5.8% vs 67.2% ± 3.8%, P = .046 and 87.5% ± 5.8% vs 68.1% ± 4.0%, P = .066).
No effect of KIR haplotype, KIR2DS2, KIR2DS3, KIR2DS5, or KIR3DS1 on relapse, TRM, OS, and LFS was found in the second cohort. As shown in Table 3, multivariate analysis demonstrated in the second haplo-SCT cohort setting that donor KIRs not ligated by donor or host MHC class I ligand were associated with a higher relapse rate and donor KIR2DS1 positivity was associated with a lower relapse rate.
Multivariate analysis: factors affecting transplantation outcome
| Outcome and factors . | HR (95% CI) . | P . |
|---|---|---|
| LFS | ||
| Disease risk index | 1.603 (1.082-2.376) | .019 |
| MRD status before transplantation (positive vs negative) | 2.039 (1.209-3.441) | .008 |
| OS | ||
| Disease risk index | 1.662 (1.110-2.487) | .014 |
| MRD status before transplantation (positive vs negative) | 1.968 (1.157-3.348) | .012 |
| Relapse | ||
| Disease risk index | 1.961 (1.045-3.680) | .036 |
| d-rsKIR vs dsKIR vs rsKIR vs nsKIR | 1.899 (1.167-3.090) | .090 |
| KIR2DS1 (positive vs negative) | 0.365 (0.148-0.899) | .028 |
| Outcome and factors . | HR (95% CI) . | P . |
|---|---|---|
| LFS | ||
| Disease risk index | 1.603 (1.082-2.376) | .019 |
| MRD status before transplantation (positive vs negative) | 2.039 (1.209-3.441) | .008 |
| OS | ||
| Disease risk index | 1.662 (1.110-2.487) | .014 |
| MRD status before transplantation (positive vs negative) | 1.968 (1.157-3.348) | .012 |
| Relapse | ||
| Disease risk index | 1.961 (1.045-3.680) | .036 |
| d-rsKIR vs dsKIR vs rsKIR vs nsKIR | 1.899 (1.167-3.090) | .090 |
| KIR2DS1 (positive vs negative) | 0.365 (0.148-0.899) | .028 |
CI, confidence interval; HR, hazard ratio.
Discussion
Through the first cohort, at serial time points after transplantation, we found that NK-cell reconstitution and restoration of functional education were best when both the donor and host HLA presented the ligands for donor inhibitory KIRs. We found that NK-cell responsiveness was tuned upward with increasing doses of the ligands for inhibitory receptors from donor and host. When both donors and hosts expressed the 3 KIR ligands (HLA-C1, HLA-C2, and HLA-Bw4), NK cells expressing 3 inhibitory receptors (KIR2DL1/2DL3/3DL1) reached maximum functional education compared with those NK cells expressing 2 or 1 inhibitory receptor. The second cohort confirmed that when both the donor and host coexpressed all 3 KIR ligands (HLA-C1, HLA-C2, and HLA-Bw4), and the donor expressed 3 inhibitory KIRs genotype, the host showed the lowest recurrence rate, confirming that this configuration where NK cells were highly educated was associated with the lowest relapse following bone marrow transplantation. This suggests a functional significance of these NK cells.
The presence of HLA ligands for inhibitory KIR in both the donor and host was associated with the highest proportions of educated NK cells in the repopulating NK-cell repertoire and the strongest education of donor NK cells for missing-self responsiveness against K562 cells. This finding is in agreement with a recent report that demonstrated that NK-cell education was similarly conveyed by both host and donor HLA composition using a humanized mouse model and posttransplant peripheral blood samples collected from patients receiving HLA-disparate umbilical cord blood transplantations.29 The education of the NK cells could be tuned upward depending on the MHC environment, as the acquisition of cognate HLA from trans sources is associated with increased responsiveness among human NK cells bearing cognate KIR receptors. However, this previous report29 only analyzed the NK cells derived in patients 200 to 365 days after cord blood HLA-mismatched HCT, lacking the reconstitution kinetics analysis and the comparison at different time points posttransplantation. Meanwhile, besides function reconstitution, the absolute number recovery of single-KIR+ NK cells also increased when both the donor and host HLA coexisted for donor inhibitory KIR, suggesting that the presence of HLA class I molecules in trans and in cis supports enhanced survival of NK cells.30,31 However, when we further analyzed the Ki67 data of individual KIR2DL1 single+, KIR2DL2/L3 single+, and KIR3DL1 single+ NK cells at day 30 posttransplantation, the expression of Ki67 on individual single-KIR+ NK cells seemed to be higher when their corresponding ligand was absent (ns-KIR group) than when it was present (dsKIR and d-rsKIR groups). The expression of BCL-2 on individual single-KIR+ cells was similar regardless of whether the corresponding ligand was present. The mechanism regarding why there was better reconstitution of individual KIR+ NK cells when their corresponding ligand was present needs to be explored further.
Recognizing the crucial role of education for establishing NK responsiveness and its characteristic phenotypic markers would help NK biologists estimate education and regulate the process of education. However, through phenotype or profiling, previous reports have failed to find the unique markers identifying the mechanism underlying education. In humans, allo-HSCT provides an opportunity to study NK development and the acquisition of tolerance to self-MHC. Through monitoring the phenotype of single KIR+ NK cells at different time points after haploidentical transplantation, we found that the expression of NKp30, NKp46, CD25, CD57, and NKG2D was detectable on the surface of NK cells harvested from the blood and did not differ at indicated KIR single+ NK cells among the nsKIR, d-rsKIR, dsKIR and rsKIR groups. However, DNAM-1 was higher at single KIR2DL1+ NK, single KIR2DL2/L3+ NK, or single KIR3DL1+ NK cells in the d-rsKIR group compared with those cells in the nsKIR group, suggesting that DNAM-1 would be a good marker to distinguish education.32 Meanwhile, compared with NKG2A−CD57+NKG2C+ NK cells (mature NK cells), the expression levels of DNAM-1 were lower in NKG2A+ NK cells (immature NK cells), also indicating that high expression levels of DNAM-1 correlate with more educated NK cells. In mice, although the activating receptor DNAM-1 is not required for the acquisition or maintenance of MHC class I–mediated NK education, its expression is significantly correlated with the educated state.33 Human educated NK cells may also be distinguished from their uneducated counterparts by expression of DNAM-1.32 However, in humans, whether DNAM-1 is associated with or determines the education process is not very clear.34 In the future, single-cell transcriptional analysis could help to identify the molecular mechanisms by which DNAM-1 modifies the education process after transplantation in the context of the donor and host MHC environment.
Besides the interaction of single KIR and single HLA, the class and quantity of inhibitory KIR receptors for HLA-B and HLA-C ligands also influence NK-cell reactivity. In a mouse model, Brodin, Joncker, et al12,35-37 reported a relationship between the extent of inhibitory signals for self-MHC during education and functional responsiveness. In healthy humans, Yu et al28 also demonstrated that there was a dose effect of KIR for self-MHC on effector capacity; in NK cells expressing 3 inhibitory KIRs, NK cells to both HLA-B and HLA-C ligands achieved significantly higher effector capacity. However, in the allogeneic transplantation setting, no previous study had considered how the number and type of MHC class I alleles quantitatively tune the responsiveness of individual KIR subsets. In this study, we demonstrated that NK cells expressing multiple inhibitory KIRs achieve the highest effector capacity when donor and host pairs exhibit all 3 KIR ligands, confirming the effect of class and quantity of donor and/or host HLA on the responsiveness of an individual KIR subset in the allogeneic transplantation setting. This indicated that when discussing the effects of NK cells on leukemia recurrence after allo-HSCT, we should consider not only missing self or missing ligand but also the number of KIR ligands simultaneously expressed by the donor and host.
Valiante et al38 demonstrated that the first donor possesses 3 class I ligands for KIR, and a majority of NK cells use KIR as their inhibitory receptor; the second donor possesses only a single ligand for KIR, and a majority of NK cells use the more broadly reactive NKG2A as their inhibitory receptor. Because of these differences, the first donor has subpopulations of NK cells that kill cells of the second donor, whereas the NK cells of the second donor are universally tolerant of cells from the first donor. Considering HLA in the setting of a tumor or leukemia, the ligand of KIR was selectively decreased on leukemia cells.39 Therefore, the pairs possessing 3 class I ligands for KIR might have more of an advantage in killing leukemia. Through the second cohort, we firstly confirmed that when NK cells reconstituted in the setting of both donor and host possessing 3 class I ligands for KIR, the patient outcome was associated with decreased leukemia relapse and therefore better OS and LFS. From the point of NK education and the rheostat model, these results explained the conflicting roles of alloreactive NK cells on leukemia relapse in different haplo-SCT settings.7,20-23,40,41
In conclusion, in this study, we found that NK-cell functional reconstitution after haploidentical transplantation reflects how the education of NK cells could be tuned to adjust the cognate MHC environment. When both the donor and host present all of the KIR ligands for donor KIRs, reconstituted NK cells achieve a better functional reconstitution and contribute to lower relapse rates among patients.
Acknowledgments
The authors thank Dong Z (School of Medicine and Institute for Immunology, Beijing Key Laboratory for Immunological Research on Chronic Diseases, Tsinghua University, Beijing, China) and Jeanette E. Boudreau (Department of Microbiology and Immunology, Dalhousie University, Halifax, NS, Canada) for their help scientifically editing this manuscript. The authors thank all of the core facilities at the Peking University Institute of Hematology for sample collection.
This study was supported by grants from the National Natural Science Foundation of China (81670166, 81870140, and 81530046), the Innovative Research Groups of the National Natural Science Foundation of China (81621001), the National Key Research and Development Program of China (2017YFA0104500), the Beijing Municipal Science & Technology Commission (Z171100001017098), the project of health collaborative innovation of Guangzhou City (201704020214), the Scientific Research Foundation for Capital Medicine Development (2018-2-4084), the Peking University Clinical Scientist Program (BMU2019LCKXJ003), and the Milstein Medical Asian American Partnership Foundation Research Project Award in Hematology.
Authorship
Contribution: X.-Y.Z. and X.-J.H. conceived and designed the study; X.-X.Y. and X.-H.C. conducted the in vitro experiments; Z.-L.X., Y.-J.C., X.-S.Z., Y.W., L.-P.X., K.-Y.L., X.-H.Z., and M.-R.H. performed the clinical examination; X.-Y.Z. and X.-J.H. analyzed and interpreted the data; X.-J.H. and X.-Y.Z. wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiao-Jun Huang, Peking University People’s Hospital, Peking University Institute of Hematology, National Clinical Research Center for Hematologic Disease, Peking-Tsinghua Center for Life Sciences, and Beijing Engineering Laboratory for Cellular Therapy, 11 Xizhimen South St, Beijing 100044, China; e-mail: huangxiaojun@bjmu.edu.cn.
References
Author notes
For original data, please e-mail the corresponding author, Xiao-Jun Huang (huangxiaojun@bjmu.edu.cn).
The full-text version of this article contains a data supplement.