Key Points
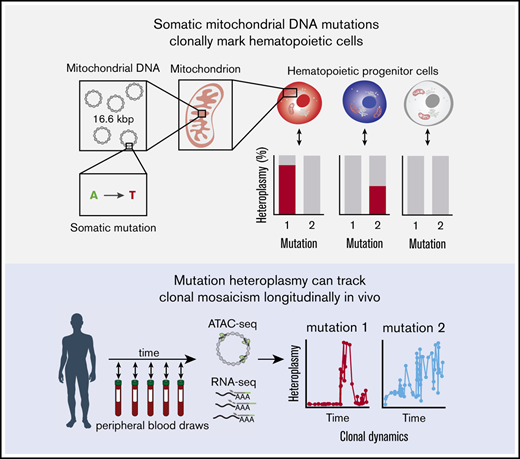
Tracking of somatic mtDNA mutations in the peripheral blood enables the longitudinal assessment of clonal dynamics.
This approach could enable clonal inference in vivo without reliance on genetic labeling.
Abstract
Our ability to track cellular dynamics in humans over time in vivo has been limited. Here, we demonstrate how somatic mutations in mitochondrial DNA (mtDNA) can be used to longitudinally track the dynamic output of hematopoietic stem and progenitor cells in humans. Over the course of 3 years of blood sampling in a single individual, our analyses reveal somatic mtDNA sequence variation and evolution reminiscent of models of hematopoiesis established by genetic labeling approaches. Furthermore, we observe fluctuations in mutation heteroplasmy, coinciding with specific clinical events, such as infections, and further identify lineage-specific somatic mtDNA mutations in longitudinally sampled circulating blood cell subsets in individuals with leukemia. Collectively, these observations indicate the significant potential of using tracking of somatic mtDNA sequence variation as a broadly applicable approach to systematically assess hematopoietic clonal dynamics in human health and disease.
Introduction
Recent studies have described the application of lineage tracing in model organisms1,2 and genetically modified cells in humans undergoing gene therapy.3,4 These studies have provided insights into clonal dynamics in complex tissues. In the hematopoietic system, such inferences have provided previously unappreciated knowledge about the contributions of hematopoietic stem and progenitor cells (HSPCs) to blood cell production.5 However, because most methods rely on the introduction of exogenous genetic labels (eg, lentiviral- and transposon-based barcoding or Cre-loxP based recombination), these techniques are not readily amenable to the broad study of physiologic and pathologic processes in humans. Assessing the dynamics of, and outputs from, HSPCs in an unperturbed setting in humans represents a methodological challenge, leaving open questions about their frequency, functionality, and longevity.6 This raises the important question of how we can effectively and longitudinally study clonal dynamics in humans.
Although somatic mutations in the nuclear genome have been leveraged to perform clonal lineage tracing in humans, these approaches are expensive and often prone to error in single cells, limiting broader or routine applications.7,8 Recently, we and other investigators have demonstrated the utility of somatic mitochondrial DNA (mtDNA) mutations as natural genetic barcodes that may be stably propagated across cell divisions.9,10 Importantly, common genomic techniques, including the assay for transposase-accessible chromatin sequencing (ATAC-seq) and RNA sequencing (RNA-seq), provide the means to concomitantly assess cell type and state with mtDNA genotypes. Because our previous work demonstrated substantial somatic mtDNA mutational diversity within HSPCs, we reasoned that tracking these mutations would enable assessment of clonal contributions to blood production. Specifically, because progenitor cell–specific mutations would be propagated to differentiated circulating blood cells, we hypothesized that fluctuations in mtDNA mutations should be reflective of the clonal output of progenitor cells over time. However, the utility of this approach to evaluate longitudinal clonal dynamics remains unexplored.
Methods
Raw sequencing reads were downloaded from Gene Expression Omnibus accession numbers GSE33029, GSE85853, GSE111015, and GSE111405. Alignment to the hg19 reference genome was performed using appropriate tools for RNA-seq (STAR11 ), ATAC-seq (bowtie212 ), and whole-genome bisulfite sequencing (bismark13 ). Reads aligning to the mtDNA genome were extracted using SAMtools,14 and polymerase chain reaction–duplicated reads were removed using picard tools. Per-sample and per-mutation heteroplasmy abundances were estimated using our previously reported pipeline.9 All depicted mutations were selected on the basis of supervised analyses. Mutations in RNA-seq were specifically filtered against a set of purported RNA-editing events as we have previously described.9 All meta-data (eg, sample, time point) were curated from the Gene Expression Omnibus accessions that contained the raw high-throughput sequencing data.
Results and discussion
We reasoned that assessment of somatic mtDNA mutations in data from recent studies that have longitudinally profiled human peripheral blood using genomic approaches could enable clonal inferences in circulating blood and immune cells (Figure 1A; supplemental Figure 1A).15,16 Because nearly the entirety of human mtDNA is transcribed, we reasoned that we could examine patterns of somatic mutation dynamics from bulk RNA-seq data. To these ends, we processed 57 RNA-seq datasets that had been serially sampled over the course of 161 weeks from a single individual. Using our previously reported pipeline, we were able to identify numerous high-confidence mtDNA mutations9 and illustrate their dynamics over nearly 3 years of peripheral blood sampling (Figure 1B). These mutations were selected because they did not show evidence of RNA editing or other known biases.9 For example, although the 10000A>G allele was gradually lost over the course of 3 years, the 295C>T allele increased in heteroplasmy during this time. In contrast, the 13636T>C allele appeared to be stably propagated over the full 3 years in vivo. Other mutations, such as 829A>G and 10278A>C, became more prominent in discrete windows spanning several months. Collectively, these observations support distinct models of hematopoiesis, including those involving clonal succession (progressive recruitment of distinct clones, marked by specific mtDNA mutations) and others involving stability of specific clones over periods of time.6,17 Considering all available alternative allele frequencies, we observed a decay in the Spearman correlation of mutation frequencies comparing baseline with subsequent time points (Figure 1C; supplemental Figure 1B), further reflecting the dynamic evolution of mitochondrial mutations in the sampled circulating blood and immune cells.
Evidence of clonal mosaicism from mtDNA mutations over 3 years in vivo. (A) Schematic diagram of somatic mtDNA mutations in human cells. Each cell contains multiple mitochondria, which, in turn, contain multiple copies of mtDNA. (B) Examples of variable mutations in vivo across 3 years of observation that reflect clonal mosaicism in 1 donor. (C) Spearman correlation of 57 time points (ordered by relative time of sampling) across time points sampled. Correlation value is measured with the baseline sample. (D) Heteroplasmy of 2394T>A allele, which is associated with human RSV detection in this donor; inset shows heteroplasmy levels for 2394T>A for ∼23 days after the initial detection of RSV. (E) Corroboration of 2394T>A allele at the time of RSV infection using whole-genome bisulfite sequencing data at the 6 time points (on specific days) that correspond to infection. (F) Heteroplasmic mutations specific to the day of detection for adenovirus (ADV; 1575A>G) and human rhinovirus (HRV; 10310T>G) infections.
Evidence of clonal mosaicism from mtDNA mutations over 3 years in vivo. (A) Schematic diagram of somatic mtDNA mutations in human cells. Each cell contains multiple mitochondria, which, in turn, contain multiple copies of mtDNA. (B) Examples of variable mutations in vivo across 3 years of observation that reflect clonal mosaicism in 1 donor. (C) Spearman correlation of 57 time points (ordered by relative time of sampling) across time points sampled. Correlation value is measured with the baseline sample. (D) Heteroplasmy of 2394T>A allele, which is associated with human RSV detection in this donor; inset shows heteroplasmy levels for 2394T>A for ∼23 days after the initial detection of RSV. (E) Corroboration of 2394T>A allele at the time of RSV infection using whole-genome bisulfite sequencing data at the 6 time points (on specific days) that correspond to infection. (F) Heteroplasmic mutations specific to the day of detection for adenovirus (ADV; 1575A>G) and human rhinovirus (HRV; 10310T>G) infections.
Because we previously observed highly heteroplasmic mtDNA mutations in clonal lymphocytes (defined by T-cell receptor rearrangements), we hypothesized that a subset of mutations may reflect clonal expansion of lymphocytes in response to foreign pathogens (supplemental Figure 1C). Indeed, we observed a rare mutation (2394T>A) emerge specifically when the donor was exposed to human respiratory syncytial virus (RSV; Figure 1D), noting the heteroplasmy was 0% at the previous time point (34 days prior). We confirmed the occurrence of this specific mutation in matched whole-genome bisulfite sequencing data comparing time points at which viral infections were detected (Figure 1E). These results, paired with our previous observations, suggest that a subset of lymphocytes carrying the 2394T>A allele clonally expanded upon RSV infection and persisted at detectable frequencies in peripheral blood for ≥10 days. Furthermore, we note the recurrence of 2 mutations (1575A>G and 10310T>G) at times of clinically documented infection with adenovirus and human rhinovirus (Figure 1F), respectively, suggesting virus-specific proliferation of distinct clonal lymphocyte populations in response to these infections. Together, the association of heteroplasmic variation with these clinical infections indicate that heteroplasmy can enable the assessment of clonal dynamics and would be of particular value in settings in which other clonal markers (eg, lymphocyte receptor sequences) are unavailable.
Because HSPCs can give rise to multiple lineages, an extension of our results from bulk peripheral blood measurements would be to examine the relative contributions of HSPCs to specific blood cell lineages, marked by the presence of distinct somatic mtDNA mutations that are absent in other lineages (Figure 2A). To explore this concept, we reanalyzed 188 ATAC-seq profiles from surface phenotype-sorted circulating blood cell populations from a cohort of 8 patients with chronic lymphocytic leukemia (CLL) that were collected up to 40 weeks following the initiation of ibrutinib treatment.15 Importantly, because mtDNA is nucleosome-free and, therefore, is highly susceptible to transposon insertion, ATAC-seq provides a facile approach for capturing somatic mutations in mtDNA. Strikingly, we observed many instances of recurrently detected lineage-specific mutations across the sampled time points, suggesting the presence of these somatic mtDNA mutations in a lineage-biased progenitor, including 1496T>C in CD4+ T lymphocytes (Donor CLL7), 10685G>A in CD8+ T lymphocytes (Donor CLL5), and 822G>A in natural killer cells (Donor CLL1) (Figure 2B). Alternatively, some of these may represent mtDNA mutations in clonally expanded and long-lived T lymphocytes. The persistence of these 3 mutations over the course of sampling is distinguished from 6453T>C, a CD19+CD5− B-lymphocyte–specific mutation that declined over >20 weeks of sampling (Figure 2C). Furthermore, we identified mutations that were shared among multiple lineages, indicating that these mtDNA mutations may exist in multipotent progenitor populations (Figure 2D). The incidence of these mutations in CD19+CD5+ leukemic cells and in CD19+CD5− B lymphocytes further supports the notion that mtDNA mutations could be informative to trace subclonal structure in response to targeted therapies, such as ibrutinib.9 Indeed, we observed instances of mutations (2885T>C and 7496T>C) decreasing in frequency with treatment, suggesting that particular subclones carrying these alleles are sensitive to the administered therapy (Figure 2E). To further verify the utility of our approach in potentially tracking clonal evolution in response to treatment, we processed an additional 81 bulk ATAC-seq samples from patients with cutaneous T-cell lymphoma treated with histone deacetylase inhibitors.18 Reanalysis of these longitudinally collected samples confirmed the detection of mtDNA sequence variation, further highlighting the utility of these mutations to track clonal dynamics in response to therapies, including putative treatment–sensitive and -resistant clones (supplemental Figure 2). Although our analyses largely elucidated specific examples of heteroplasmic mutations and their dynamics, the bulk nature and relative rarity of mtDNA transcriptome/genome coverage of these data (RNA-seq, Figure 1; single-end sequencing ATAC-seq, Figure 2) limit confident detection of low-frequency variants/clones that would enable more comprehensive analyses. We suggest that complementary bulk and single-cell genotyping assays optimized for mtDNA sequence capture are used in future studies, because we have previously shown that these can increase the resolution of inferences for clonal HSPC population dynamics.9
Inference of putative multilineage contributions from HSPCs. (A) Schematic diagram of HSPCs (unobserved) and 6 populations of cells that were sorted using fluorescence activated cell sorting. (B) Examples of cell type–specific mutations in vivo across up to 240 days of evaluation. Donor is indicated at the top of the panel (eg, CLL7). (C) Heteroplasmy of 6453T>C allele in donor CLL5; this is a CD19+CD5− B-cell–specific mutation that decreases in frequency over 150 days of observation. (D) Mutations in 2 donors that are present in both CD19+CD5+ (CLL) cells and CD19+CD5− B cells. Arrow highlights 1 observed CD19+CD5− sample. (E) Examples of shared CLL and B-cell mutations that decay at different rates for 2 donors.
Inference of putative multilineage contributions from HSPCs. (A) Schematic diagram of HSPCs (unobserved) and 6 populations of cells that were sorted using fluorescence activated cell sorting. (B) Examples of cell type–specific mutations in vivo across up to 240 days of evaluation. Donor is indicated at the top of the panel (eg, CLL7). (C) Heteroplasmy of 6453T>C allele in donor CLL5; this is a CD19+CD5− B-cell–specific mutation that decreases in frequency over 150 days of observation. (D) Mutations in 2 donors that are present in both CD19+CD5+ (CLL) cells and CD19+CD5− B cells. Arrow highlights 1 observed CD19+CD5− sample. (E) Examples of shared CLL and B-cell mutations that decay at different rates for 2 donors.
Overall, our results illustrate the potential to leverage somatic mtDNA mutations to longitudinally study clonal dynamics and somatic mosaicism in human hematopoiesis in vivo, and we hope that this further stimulates the design of such prospective studies in this poorly charted area of biomedical research. For example, such studies could enable assessments of cellular dynamics and responses to stressors, such as infections or acute blood loss, or complement existing strategies to track subclonal evolution in leukemia via bulk and single-cell analyses. Although these results reflect a multitude of scenarios in which bulk heteroplasmy changes could reflect clonal mosaicism, we note that mtDNA heteroplasmy has been described to drift over time.19 However, our previous work has shown that mtDNA mutations, depending on heteroplasmy, may be stably propagated to daughter cells over many cellular generations.9 In this respect, we emphasize the need for systematic longitudinal studies with single-cell technologies and computational tools to comprehensively model and reliably infer clonal dynamics for future analyses. Taken together, our analyses illustrate a broadly applicable strategy to facilitate our understanding of clonal dynamics in human health and disease.
Acknowledgments
The authors thank members of the Sankaran laboratory for valuable discussions.
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant R01 DK103794 and National Institutes of Health, National Heart, Lung, and Blood Institute grant R33 HL120791, as well as the New York Stem Cell Foundation (V.G.S.). C.A.L. is supported by National Institutes of Health, National Cancer Institute grant F31 CA232670. V.G.S. is a New York Stem Cell Foundation–Robertson Investigator.
Authorship
Contribution: C.A.L., L.S.L., and V.G.S. conceived and designed the study and wrote the manuscript; C.A.L. performed analyses; and V.G.S. supervised all aspects of this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vijay G. Sankaran, Boston Children’s Hospital/Broad Institute, 1 Blackfan Cir, Karp 7211, Boston, MA 02115; e-mail: sankaran@broadinstitute.org.
References
Author notes
C.A.L. and L.S.L. contributed equally to this work.
The full-text version of this article contains a data supplement.
Data sharing requests should be sent to Vijay G. Sankaran (sankaran@broadinstitute.org).