Key Points
Five easily accessible clinical variables allow accurate prediction of survival in systemic mastocytosis.
The clinical risk model for systemic mastocytosis is further enhanced by integration of mutation information.
Abstract
Systemic mastocytosis (SM) is a clinically heterogeneous disease with prognosis chiefly assigned based on World Health Organization (WHO) morphologic subclassification. We assessed the feasibility of developing contemporary risk models for SM based on clinical and integrated clinical-genetics information. Diagnosis of SM was per WHO criteria, and karyotype and next-generation sequencing data were available in a subset of the total 580 patients (median age, 55 years; range, 18-88 years) seen at the Mayo Clinic between 1968 and 2015. Morphologic subcategories were indolent/smoldering in 291 (50%) and “advanced” in 289 (50%): SM with an associated hematological neoplasm in 199, aggressive SM in 85, and mast cell leukemia in 5. Multivariable analysis of clinical variables identified age >60 years, advanced SM, thrombocytopenia <150 × 109/L, anemia below sex-adjusted normal, and increased alkaline phosphatase (ALP) as independent risk factors for survival; respective hazard ratios (HRs) 95% confidence intervals (95% CIs) were 2.5 (1.9-3.4), 2.7 (1.8-4.0), 2.5 (1.9-3.4), 2.2 (1.6-3.1), and 2.1 (1.5-3.0). In addition, ASXL1 (HR, 4.5; 95% CI, 2.6-7.6), RUNX1 (HR, 4.3; 95% CI, 1.3-10.8), and NRAS (HR, 5.0, 95% CI, 1.5-13.2) mutations were independently associated with inferior survival. Combined clinical, cytogenetic, and molecular risk factor analysis confirmed the independent prognostic contribution of adverse mutations (2.6, 1.6-4.4), advanced SM (4.0, 1.8-10.0), thrombocytopenia (2.8, 1.7-4.5), increased ALP (2.1, 1.2-4.0), and age >60 years (2.2, 1.3-3.6). These data were subsequently used to develop clinical and hybrid clinical-molecular risk models. The current study advances 2 complementary risk models for SM and highlights the independent prognostic contribution of mutations.
Introduction
The 2016 World Health Organization (WHO) system lists 5 morphological subcategories of systemic mastocytosis (SM): indolent (ISM), smoldering (SSM), SM with an associated hematological neoplasm (SM-AHN), aggressive (ASM), and mast cell leukemia (MCL).1 For practical purposes, SM variants other than ISM and SSM are jointly referred to as “advanced” SM. In 2009, we published the then-largest study in SM and highlighted the prognostic relevance of advanced SM vs ISM/SSM2 ; the study included 342 adult patients with SM seen at the Mayo Clinic between 1976 and 2007 and included 159 (46%) patients with ISM/SSM, 138 (40%) with SM-AHN, 41 (12%) with ASM, and 4 (1%) with MCL. The particular study suggested that life expectancy in patients with ISM/SSM may not be significantly different from that of the age- and sex-matched US population, but superior to that seen in patients with advanced SM.2 The study also identified advanced age, weight loss, anemia, thrombocytopenia, hypoalbuminemia, and excess bone marrow blasts as independent adverse prognostic factors for survival.
In a more recent study of next-generation sequencing (NGS) in 150 patients with SM,3 the most frequently mutated genes included TET2 (29%), ASXL1 (17%), and CBL (11%), with significantly higher mutation frequency in SM-AHN. In 1 study, ASXL1 and RUNX1 mutations predicted inferior survival in advanced SM3 ; also, the prognostic contribution of ASXL1 mutations was shown to be independent of other factors that were significant in multivariable analysis: advanced age, hemoglobin <10 g/dL, platelet count <150 × 109/L, and serum albumin <3.5 g/dL. Another study allowed the construction of a 3-tiered, mutation-augmented prognostic scoring system for advanced SM, with median survivals of 5 months for high-, 21 months for intermediate-, and 86 months for low-risk disease.3 In a subsequent study of advanced SM,4 inferior survival was also associated with CBL mutations.
Most recently, we examined the prognostic relevance of cytogenetic abnormalities in 348 informative patients with SM and reported an incidence figure of 15%, including 6% with ISM/SSM, 26% with SM-AHN, and 8% with ASM.5 . That particular study did not find a correlation between abnormal karyotype and adverse mutations; furthermore, although abnormal karyotype was associated with inferior survival, in univariate analysis, the significance was not sustained during multivariable analysis.5
In the current study, we capitalized on the large number of accumulated cases of SM seen at the Mayo Clinic between 1968 and 2015 (n = 580) to confirm our previous observations and devise prognostic models that are based on either clinically derived variables or a combination of both clinical and molecular information. In so doing, we took a broad approach involving all patients with WHO-defined SM and subsequently applied our prognostic models in ISM/SSM and advanced SM separately. In addition, we used an external cohort of 65 molecularly annotated cases of SM seen at the University of Florence, Florence, Italy, to validate the prognostic relevance of adverse mutations.
Methods
After approval from the institutional review board, study patients were recruited from the Mayo Clinic, Rochester, MN, with data updated in July 2018; the study period was 1968 to 2015. Diagnoses of SM and its morphological subcategories were confirmed by both clinical and bone marrow examinations, in line with the 2016 WHO criteria.1 For the purposes of the current paper, ASM, SM-AHN, and MCL were grouped together as “advanced” SM, whereas ISM and SSM were analyzed together. Previously described methods were used for NGS,3 which was performed in a subset of the study population. Targeted capture assays were carried out on bone marrow or whole blood DNA for the following genes: TET2, DNMT3A, IDH1, IDH2, ASXL1, EZH2, SUZ12, SRSF2, SF3B1, ZRSR2, U2AF1, PTPN11, TP53, SH2B3, RUNX1, CBL, NRAS, JAK2, CSF3R, FLT3, KIT, CALR, MPL, NPM1, CEBPA, IKZF1, and SETBP1. NGS-detected coding region variants were filtered through the Exome Aggregation Consortium database and annotated by the Catalogue of Somatic Mutations in Cancer database as mutants or variants of uncertain significance. Statistical analyses considered clinical and laboratory data collected at the time of initial diagnosis at the Mayo Clinic. Conventional statistics was used for calculation of overall survival and determination of risk factors. Receiver operating characteristic curves were used to determine the prognostically most discriminative platelet threshold. JMP Pro 13.0.0 software from SAS Institute, Cary, NC, was used for all calculations.
Results
A total of 580 patients with SM (median age, 55 years; range, 18-88 years; 52% males) were seen at our institution between 1968 and 2015; WHO morphologic subcategories were ISM (n = 274) or SSM (n = 17) in 291 (50%), ASM in 85 (15%), SM-AHN in 199 (34%), and MCL in 5 (1%). Table 1 lists clinical and laboratory characteristics at presentation; anemia defined by hemoglobin level below the lower limit of the sex-adjusted reference range, was present in 41% of the patients, hemoglobin <10 g/dL in 16%, red cell transfusion-dependency in 9%, platelet count below the receiver operating characteristic–determined limit of 150 × 109/L in 26%, serum albumin below the lower normal limit of the reference range in 22%, serum alkaline phosphatase (ALP) above the upper normal limit of the reference range in 54%, urticaria pigmentosa in 41%, mast cell mediator symptoms in 46%, palpable hepatomegaly in 21%, and palpable splenomegaly in 31%. As expected, there were significant differences in phenotype between ISM/SSM and advanced SM and, to a lesser degree, between ASM and SM-AHN, as outlined in Table 1.
Clinical and laboratory characteristics of 580 patients with SM seen at the Mayo Clinic between 1968 and 2015
| Variables . | All patients (N = 580) . | ISM/SSM (n = 291) . | Advanced SM (n = 289) . | P . | ASM (n = 85) . | SM-AHN (n = 199) . | MCL (n = 5) . | P . |
|---|---|---|---|---|---|---|---|---|
| Median age (range), y | 55 (18-88) | 48 (19-87) | 64 (18-88) | <.001 | 61 (32-86) | 65 (18-88) | 57 (45-74) | .08 |
| Age >60 y, n (%) | 235 (41) | 65 (22) | 170 (59) | <.001 | 44 (52) | 124 (62) | 2 (40) | .2 |
| Males, n (%) | 301 (52) | 127 (44) | 174 (60) | <.001 | 38 (45) | 132 (66) | 4 (80) | .002 |
| Hemoglobin, median (range), g/dL | 13.1 (5.1-17.4) | 13.9 (8.1-17.2) | 11.1 (5.1-17.4) | <.001 | 12.3 (5.1-17) | 10.8 (5.2-17.4) | 10.9 (9.5-11.5) | .03 |
| Anemia sex adjusted, n (%) | 236 (41) | 41 (14) | 195 (67) | <.001 | 44 (52) | 146 (73) | 5 (100) | <.001 |
| n eval = 574 | n eval = 285 | |||||||
| Leukocyte count, median (range), ×109/L | 7.1 (0.7-87.2) | 6.5 (1.6-22.2) | 8.3 (0.7-87.2) | <.001 | 7.3 (1.7-37.1) | 9 (0.7-87.2) | 4.5 (3.8-7) | .004 |
| n eval = 573 | n eval = 284 | |||||||
| Platelet count, median (range), ×109/L | 229 (2-1625) | 257.5 (39-563) | 157 (2-1625) | <.001 | 218.5 (19-570) | 134.5 (2-1625) | 145 (54-241) | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | n eval = 84 | n eval = 198 | ||||
| Platelet count <150 × 109/L, n (%) | 149 (26) | 13 (5) | 136 (47) | <.001 | 23 (27) | 110 (56) | 3 (60) | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | n eval = 84 | n eval = 198 | ||||
| Urticaria pigmentosa, n (%) | 234 (41) | 162 (56) | 72 (25) | <.001 | 32 (38) | 40 (20) | 0 (0) | .003 |
| n eval= 577 | n eval = 288 | |||||||
| Mast cell mediator symptoms, n (%) | 162 (46) | 106 (69) | 56 (29) | <.001 | 14 (27) | 40 (29) | 2 (50) | .6 |
| n eval = 349 | n eval = 153 | n eval = 196 | n eval = 52 | n eval = 140 | n eval = 4 | |||
| Serum tryptase <20; n (%) | 87 (22) | 62 (27) | 25 (14) | 5 (8) | 19 (17) | 1 (33) | ||
| Serum tryptase 20-200; n (%) | 266 (66) | 156 (68) | 110 (64) | 40 (68) | 69 (62) | 1 (33) | ||
| Serum tryptase >200; n (%) | 49 (12) | 11 (5) | 38 (22) | 14 (24) | 23 (21) | 1 (33) | ||
| n eval = 402 | n eval = 229 | n eval = 173 | n eval = 59 | n eval = 111 | n eval = 3 | |||
| BM mast cell <5%, n (%) | 31 (7) | 26 (12) | 5 (2) | 2 (3) | 3 (2) | 0 (0) | ||
| BM mast cell 5%-10%, n (%) | 211 (49) | 124 (56) | 87 (41) | 26 (38) | 61 (44) | 0 (0) | ||
| BM mast cell 11%-50%, n (%) | 144 (33) | 58 (26) | 86 (40) | 23 (34) | 63 (45) | 0 (0) | ||
| BM mast cell >50%, n (%) | 46 (11) | 12 (5) | 34 (16) | 17 (25) | 13 (9) | 4 (100) | ||
| n evaluable = 432 | n eval = 220 | n eval = 212 | n eval = 68 | n eval = 140 | n eval = 4 | |||
| Palpable hepatomegaly, n (%) | 119 (21) | 25 (9) | 94 (33) | <.001 | 28 (33) | 65 (33) | 1 (20) | .8 |
| n eval = 579 | n eval = 288 | n eval = 198 | ||||||
| Palpable splenomegaly, n (%) | 179 (31) | 33 (11) | 146 (51) | <.001 | 36 (42) | 107 (54) | 3 (60) | .2 |
| n eval = 578 | n eval = 290 | n eval = 288 | n eval = 198 | |||||
| Serum albumin, median (range), g/dL | 3.9 (2-5.1) | 4 (2.9-5.1) | 3.8 (2-4.9) | <.001 | 3.8 (2-4.9) | 3.8 (2-4.8) | 3.85 (3.1-4.4) | .6 |
| n eval = 389 | n eval = 157 | n eval = 232 | n eval = 71 | n eval = 157 | n eval = 4 | |||
| Serum albumin <3.5 g/dL, n (%) | 87 (22) | 18 (11) | 69 (30) | <.0001 | 18 (25) | 50 (32) | 1 (25) | .6 |
| n eval = 389 | n eval = 157 | n eval = 232 | n eval = 71 | n eval = 157 | n eval = 4 | |||
| Serum ALP, median (range), U/L | 124 (19-3680) | 92 (30-1957) | 178.5 (19-3680) | <.001 | 195 (33-2004) | 170 (19-3680) | 240 (139-1423) | .4 |
| n eval = 547 | n eval = 269 | n eval = 278 | n eval = 82 | n eval = 191 | ||||
| Serum ALP > UNL, n (%) | 296 (54) | 101 (38) | 195 (70) | <.001 | 58 (71) | 132 (68) | 5 (100) | .3 |
| n eval = 547 | n eval = 269 | n eval = 278 | n eval = 82 | n eval = 191 | ||||
| KITD816V, n (%) | 279 (78) | 141 (82) | 138 (75) | .09 | 41 (87) | 97 (71) | 0 (0) | .01 |
| n eval = 357 | n eval = 172 | n eval = 185 | n eval = 47 | n eval = 137 | n eval = 1 | |||
| ASXL1 mutated, n (%) | 25 (17) | 0 (0) | 25 (23) | <.001 | 4 (15) | 21 (26) | 0 (0) | .4 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| RUNX1 mutated, n (%) | 5 (3) | 0 (0) | 5 (5) | .15 | 0 (0) | 5 (6) | 0 (0) | .4 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| NRAS mutated, n (%) | 4 (3) | 0 (0) | 4 (4) | .19 | 0 (0) | 3 (4) | 1 (100) | <.001 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| Adverse mutations, n (%) | 31 (21) | 0 (0) | 31 (29) | <.001 | 4 (15) | 26 (33) | 1 (100) | .07 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| Abnormal karyotype, n (%) | 53 (15) | 8 (7) | 45 (22) | <.001 | 4 (8) | 40 (26) | 1 (50) | .02 |
| n eval = 348 | n eval = 142 | n eval = 206 | n eval = 2 | |||||
| Median follow-up (range), mo | 34 (0-496) | 51 (0-357) | 23 (0-496) | <.001 | 35 (0-496) | 20 (0-291) | 2 (1-30) | .06 |
| Deaths, n (%) | 239 (41) | 44 (15) | 195 (67) | <.001 | 42 (49) | 150 (75) | 3 (60) | <.001 |
| Leukemic transformations, n (%) | 9 (2) | 0 (0) | 9 (3) | <.001 | 1 (1) | 8 (4) | 0 (0) | .4 |
| Variables . | All patients (N = 580) . | ISM/SSM (n = 291) . | Advanced SM (n = 289) . | P . | ASM (n = 85) . | SM-AHN (n = 199) . | MCL (n = 5) . | P . |
|---|---|---|---|---|---|---|---|---|
| Median age (range), y | 55 (18-88) | 48 (19-87) | 64 (18-88) | <.001 | 61 (32-86) | 65 (18-88) | 57 (45-74) | .08 |
| Age >60 y, n (%) | 235 (41) | 65 (22) | 170 (59) | <.001 | 44 (52) | 124 (62) | 2 (40) | .2 |
| Males, n (%) | 301 (52) | 127 (44) | 174 (60) | <.001 | 38 (45) | 132 (66) | 4 (80) | .002 |
| Hemoglobin, median (range), g/dL | 13.1 (5.1-17.4) | 13.9 (8.1-17.2) | 11.1 (5.1-17.4) | <.001 | 12.3 (5.1-17) | 10.8 (5.2-17.4) | 10.9 (9.5-11.5) | .03 |
| Anemia sex adjusted, n (%) | 236 (41) | 41 (14) | 195 (67) | <.001 | 44 (52) | 146 (73) | 5 (100) | <.001 |
| n eval = 574 | n eval = 285 | |||||||
| Leukocyte count, median (range), ×109/L | 7.1 (0.7-87.2) | 6.5 (1.6-22.2) | 8.3 (0.7-87.2) | <.001 | 7.3 (1.7-37.1) | 9 (0.7-87.2) | 4.5 (3.8-7) | .004 |
| n eval = 573 | n eval = 284 | |||||||
| Platelet count, median (range), ×109/L | 229 (2-1625) | 257.5 (39-563) | 157 (2-1625) | <.001 | 218.5 (19-570) | 134.5 (2-1625) | 145 (54-241) | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | n eval = 84 | n eval = 198 | ||||
| Platelet count <150 × 109/L, n (%) | 149 (26) | 13 (5) | 136 (47) | <.001 | 23 (27) | 110 (56) | 3 (60) | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | n eval = 84 | n eval = 198 | ||||
| Urticaria pigmentosa, n (%) | 234 (41) | 162 (56) | 72 (25) | <.001 | 32 (38) | 40 (20) | 0 (0) | .003 |
| n eval= 577 | n eval = 288 | |||||||
| Mast cell mediator symptoms, n (%) | 162 (46) | 106 (69) | 56 (29) | <.001 | 14 (27) | 40 (29) | 2 (50) | .6 |
| n eval = 349 | n eval = 153 | n eval = 196 | n eval = 52 | n eval = 140 | n eval = 4 | |||
| Serum tryptase <20; n (%) | 87 (22) | 62 (27) | 25 (14) | 5 (8) | 19 (17) | 1 (33) | ||
| Serum tryptase 20-200; n (%) | 266 (66) | 156 (68) | 110 (64) | 40 (68) | 69 (62) | 1 (33) | ||
| Serum tryptase >200; n (%) | 49 (12) | 11 (5) | 38 (22) | 14 (24) | 23 (21) | 1 (33) | ||
| n eval = 402 | n eval = 229 | n eval = 173 | n eval = 59 | n eval = 111 | n eval = 3 | |||
| BM mast cell <5%, n (%) | 31 (7) | 26 (12) | 5 (2) | 2 (3) | 3 (2) | 0 (0) | ||
| BM mast cell 5%-10%, n (%) | 211 (49) | 124 (56) | 87 (41) | 26 (38) | 61 (44) | 0 (0) | ||
| BM mast cell 11%-50%, n (%) | 144 (33) | 58 (26) | 86 (40) | 23 (34) | 63 (45) | 0 (0) | ||
| BM mast cell >50%, n (%) | 46 (11) | 12 (5) | 34 (16) | 17 (25) | 13 (9) | 4 (100) | ||
| n evaluable = 432 | n eval = 220 | n eval = 212 | n eval = 68 | n eval = 140 | n eval = 4 | |||
| Palpable hepatomegaly, n (%) | 119 (21) | 25 (9) | 94 (33) | <.001 | 28 (33) | 65 (33) | 1 (20) | .8 |
| n eval = 579 | n eval = 288 | n eval = 198 | ||||||
| Palpable splenomegaly, n (%) | 179 (31) | 33 (11) | 146 (51) | <.001 | 36 (42) | 107 (54) | 3 (60) | .2 |
| n eval = 578 | n eval = 290 | n eval = 288 | n eval = 198 | |||||
| Serum albumin, median (range), g/dL | 3.9 (2-5.1) | 4 (2.9-5.1) | 3.8 (2-4.9) | <.001 | 3.8 (2-4.9) | 3.8 (2-4.8) | 3.85 (3.1-4.4) | .6 |
| n eval = 389 | n eval = 157 | n eval = 232 | n eval = 71 | n eval = 157 | n eval = 4 | |||
| Serum albumin <3.5 g/dL, n (%) | 87 (22) | 18 (11) | 69 (30) | <.0001 | 18 (25) | 50 (32) | 1 (25) | .6 |
| n eval = 389 | n eval = 157 | n eval = 232 | n eval = 71 | n eval = 157 | n eval = 4 | |||
| Serum ALP, median (range), U/L | 124 (19-3680) | 92 (30-1957) | 178.5 (19-3680) | <.001 | 195 (33-2004) | 170 (19-3680) | 240 (139-1423) | .4 |
| n eval = 547 | n eval = 269 | n eval = 278 | n eval = 82 | n eval = 191 | ||||
| Serum ALP > UNL, n (%) | 296 (54) | 101 (38) | 195 (70) | <.001 | 58 (71) | 132 (68) | 5 (100) | .3 |
| n eval = 547 | n eval = 269 | n eval = 278 | n eval = 82 | n eval = 191 | ||||
| KITD816V, n (%) | 279 (78) | 141 (82) | 138 (75) | .09 | 41 (87) | 97 (71) | 0 (0) | .01 |
| n eval = 357 | n eval = 172 | n eval = 185 | n eval = 47 | n eval = 137 | n eval = 1 | |||
| ASXL1 mutated, n (%) | 25 (17) | 0 (0) | 25 (23) | <.001 | 4 (15) | 21 (26) | 0 (0) | .4 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| RUNX1 mutated, n (%) | 5 (3) | 0 (0) | 5 (5) | .15 | 0 (0) | 5 (6) | 0 (0) | .4 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| NRAS mutated, n (%) | 4 (3) | 0 (0) | 4 (4) | .19 | 0 (0) | 3 (4) | 1 (100) | <.001 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| Adverse mutations, n (%) | 31 (21) | 0 (0) | 31 (29) | <.001 | 4 (15) | 26 (33) | 1 (100) | .07 |
| n eval = 150 | n eval = 43 | n eval = 107 | n eval = 26 | n eval = 80 | n eval = 1 | |||
| Abnormal karyotype, n (%) | 53 (15) | 8 (7) | 45 (22) | <.001 | 4 (8) | 40 (26) | 1 (50) | .02 |
| n eval = 348 | n eval = 142 | n eval = 206 | n eval = 2 | |||||
| Median follow-up (range), mo | 34 (0-496) | 51 (0-357) | 23 (0-496) | <.001 | 35 (0-496) | 20 (0-291) | 2 (1-30) | .06 |
| Deaths, n (%) | 239 (41) | 44 (15) | 195 (67) | <.001 | 42 (49) | 150 (75) | 3 (60) | <.001 |
| Leukemic transformations, n (%) | 9 (2) | 0 (0) | 9 (3) | <.001 | 1 (1) | 8 (4) | 0 (0) | .4 |
Bold indicates statistical significance (P < .05).
BM, bone marrow; n eval, number evaluable; UNL, upper normal limit.
Cytogenetic information was available in 342 cases, including 51 (15%) with abnormal karyotype. NGS-derived mutation information was available in 150 cases; the most frequent mutations were KIT (75%), TET2 (29%), ASXL1 (17%), CBL (11%), SF3B1 (6%), DNMT3A (6%), JAK2V617F (6%), U2AF1 (4%), RUNX1 (3%), NRAS (3%), SETBP1 (3%), and IDH1/2 (3%). Non-KIT mutations were limited to TET2 in ISM/SSM only (7% incidence), whereas the majority of other mutations clustered with SM-AHN. Adverse mutations were generally absent in patients with ISM/SSM (0 of 43 tested vs 29% incidence in advanced SM); ISM/SSM also expressed a low frequency of abnormal karyotype (7% vs 22% in advanced SM).
Risk factor analysis and development of clinical and clinical-molecular risk models
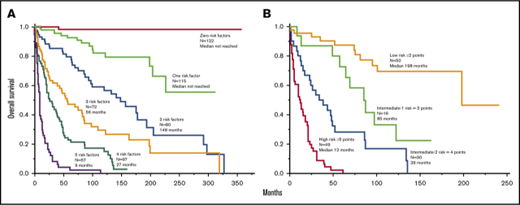
Median follow-up was 34 months, with 239 (41%) deaths and 9 (1.5%) leukemic transformations documented. Table 2 outlines clinical and laboratory parameters found to adversely affect overall survival in univariate analysis. Multivariable analysis limited to clinical variables identified 5 interindependent risk factors for overall survival: hazard ratio (HR) 95% confidence interval (95% CI) were 2.5 (1.9-3.4) for age >60 years, 2.7 (1.8-4.0) for advanced SM vs ISM/SM, 2.5 (1.9-3.4) for thrombocytopenia <150 × 109/L, 2.2 (1.6-3.1) for anemia below sex-adjusted normal, and 2.1 (1.5-3.0) for increased ALP; a total of 543 patients were informative for all 5 clinically derived risk factors. Considering the similarity in HR, survival data were prepared based simply on the number of risk factors (Figure 1A); median survival was not reached in the presence of ≤1 risk factor and declined progressively based on the number of risk factors ranging in median <3 years for patients with ≥4 risk factors to 5 to 11 years in the presence of 2 or 3 risk factors (Figure 1A). A similar approach in advanced SM (Figure 1B) and ISM/SSM (Figure 1C) produced similar results. In other words, thrombocytopenia <150 × 109/L, anemia below sex-adjusted normal, and increased ALP are the 3 age- and morphologic category-independent risk factors that determine survival in both ISM/SSM and advanced SM.
Univariate analysis of risk factors for overall survival among 580 patients with SM
| Variables . | All patients univariate P (n = 580) . | ISM univariate P (n = 291) . | Advanced SM univariate P (n = 289) . |
|---|---|---|---|
| Median age | <.001 | <.001 | <.001 |
| Age >60 y | <.001 | <.001 | <.001 |
| Males | <.001 | .4 | .17 |
| Anemia sex adjusted | <.001 | <.001 | <.001 |
| n eval = 574 | n eval = 285 | ||
| Leukocyte count | <.001 | .3 | .03 |
| n eval = 573 | n eval = 284 | ||
| Platelet count | <.001 | .4 | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | |
| Platelet count <150 × 109/L | <.001 | .08 | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | |
| Urticaria pigmentosa | <.001 | .7 | <.001 |
| n eval = 577 | n eval = 288 | ||
| Mast cell mediator symptoms | <.001 | .009 | .68 |
| n eval = 349 | n eval = 153 | n eval = 196 | |
| Palpable hepatomegaly | <.001 | .03 | .01 |
| n eval = 579 | n eval = 288 | ||
| Palpable splenomegaly | <.001 | .006 | <.001 |
| n eval = 578 | n eval = 290 | n eval = 288 | |
| Serum albumin | <.001 | .4 | <.001 |
| n eval = 389 | n eval = 157 | n eval = 232 | |
| Serum albumin <3.5 g/dL | <.001 | .2 | .01 |
| n eval = 389 | n eval = 157 | n eval = 232 | |
| Serum ALP | <.001 | <.001 | <.001 |
| n eval = 547 | n eval = 269 | n eval = 278 | |
| Serum ALP > UNL | <.001 | .001 | <.001 |
| n eval = 547 | n eval = 269 | n eval = 278 | |
| KITD816V | .4 | .1 | .16 |
| n eval = 357 | n eval = 172 | n eval = 185 | |
| ASXL1 mutated | <.001 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| RUNX1 mutated | .03 | No adverse mutations | .05 |
| n eval = 150 | n eval = 107 | ||
| NRAS mutated | .002 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| Adverse mutations | <.001 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| Abnormal karyotype | <.001 | .3 | <.001 |
| n eval = 53 | n eval = 8 | n eval = 45 |
| Variables . | All patients univariate P (n = 580) . | ISM univariate P (n = 291) . | Advanced SM univariate P (n = 289) . |
|---|---|---|---|
| Median age | <.001 | <.001 | <.001 |
| Age >60 y | <.001 | <.001 | <.001 |
| Males | <.001 | .4 | .17 |
| Anemia sex adjusted | <.001 | <.001 | <.001 |
| n eval = 574 | n eval = 285 | ||
| Leukocyte count | <.001 | .3 | .03 |
| n eval = 573 | n eval = 284 | ||
| Platelet count | <.001 | .4 | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | |
| Platelet count <150 × 109/L | <.001 | .08 | <.001 |
| n eval = 567 | n eval = 280 | n eval = 287 | |
| Urticaria pigmentosa | <.001 | .7 | <.001 |
| n eval = 577 | n eval = 288 | ||
| Mast cell mediator symptoms | <.001 | .009 | .68 |
| n eval = 349 | n eval = 153 | n eval = 196 | |
| Palpable hepatomegaly | <.001 | .03 | .01 |
| n eval = 579 | n eval = 288 | ||
| Palpable splenomegaly | <.001 | .006 | <.001 |
| n eval = 578 | n eval = 290 | n eval = 288 | |
| Serum albumin | <.001 | .4 | <.001 |
| n eval = 389 | n eval = 157 | n eval = 232 | |
| Serum albumin <3.5 g/dL | <.001 | .2 | .01 |
| n eval = 389 | n eval = 157 | n eval = 232 | |
| Serum ALP | <.001 | <.001 | <.001 |
| n eval = 547 | n eval = 269 | n eval = 278 | |
| Serum ALP > UNL | <.001 | .001 | <.001 |
| n eval = 547 | n eval = 269 | n eval = 278 | |
| KITD816V | .4 | .1 | .16 |
| n eval = 357 | n eval = 172 | n eval = 185 | |
| ASXL1 mutated | <.001 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| RUNX1 mutated | .03 | No adverse mutations | .05 |
| n eval = 150 | n eval = 107 | ||
| NRAS mutated | .002 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| Adverse mutations | <.001 | No adverse mutations | <.001 |
| n eval = 150 | n eval = 107 | ||
| Abnormal karyotype | <.001 | .3 | <.001 |
| n eval = 53 | n eval = 8 | n eval = 45 |
Bold indicates statistical significance (P < .05).
Clinical risk model. (A) “Clinical” risk model for SM (n = 543) based on number of risk factors: (1) advanced SM vs ISM/SSM (HR, 2.7); (2) age >60 years (HR, 2.5); (3) platelets <150 × 109/L (HR, 2.5); (4) anemia below sex-adjusted normal (HR, 2.2); and (5) serum ALP above normal range (HR, 2.1). HR (95% CI) values listed are calculated against the next lower risk level. (B-C) Application of the clinical risk model in advanced SM (n = 277) vs ISM/SSM (n = 266), respectively.
Clinical risk model. (A) “Clinical” risk model for SM (n = 543) based on number of risk factors: (1) advanced SM vs ISM/SSM (HR, 2.7); (2) age >60 years (HR, 2.5); (3) platelets <150 × 109/L (HR, 2.5); (4) anemia below sex-adjusted normal (HR, 2.2); and (5) serum ALP above normal range (HR, 2.1). HR (95% CI) values listed are calculated against the next lower risk level. (B-C) Application of the clinical risk model in advanced SM (n = 277) vs ISM/SSM (n = 266), respectively.
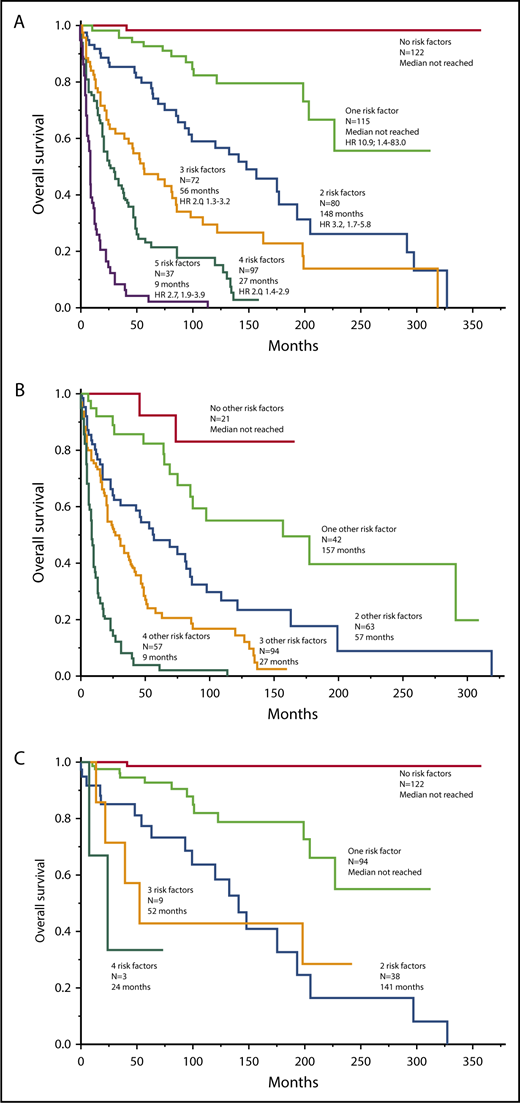
Mutations-wise, ASXL1 (HR, 4.5; 95% CI, 2.6-7.6), RUNX1 (HR, 4.3; 95% CI, 1.3-10.8), and NRAS (HR, 5.0; 95% CI, 1.5-13.2) were independently associated with inferior survival; accordingly, these 3 mutations were labeled as being “adverse.” Figure 2A-B illustrates the effect of adverse mutations on survival of all 150 molecularly annotated SM cases (Figure 2A) and in the 107 cases with advanced SM (Figure 2B). The prognostic contribution of adverse mutations was externally validated in 65 molecularly annotated patients with SM seen at the University of Florence (Figure 2C); in the Florence cohort, adverse mutations included ASXL1, RUNX1, and SRSF2. In both the Mayo and Florence patient cohorts, adverse mutations were not seen in ISM/SM, whereas their frequencies in advanced SM were 29% in the Mayo cohort and 50% in the Florence cohort.
Survival data. (A) Survival data in 150 molecularly annotated Mayo Clinic patients with SM (107 advanced, 43 indolent/smoldering) stratified by the presence or absence of adverse mutations. (B) Survival data in 107 molecularly annotated Mayo Clinic patients with advanced SM, stratified by the presence or absence of adverse mutations. (C) Survival data in 65 molecularly annotated patients with SM (10 advanced, 55 indolent/smoldering) seen at the University of Florence, Italy, stratified by the presence or absence of adverse mutations.
Survival data. (A) Survival data in 150 molecularly annotated Mayo Clinic patients with SM (107 advanced, 43 indolent/smoldering) stratified by the presence or absence of adverse mutations. (B) Survival data in 107 molecularly annotated Mayo Clinic patients with advanced SM, stratified by the presence or absence of adverse mutations. (C) Survival data in 65 molecularly annotated patients with SM (10 advanced, 55 indolent/smoldering) seen at the University of Florence, Italy, stratified by the presence or absence of adverse mutations.
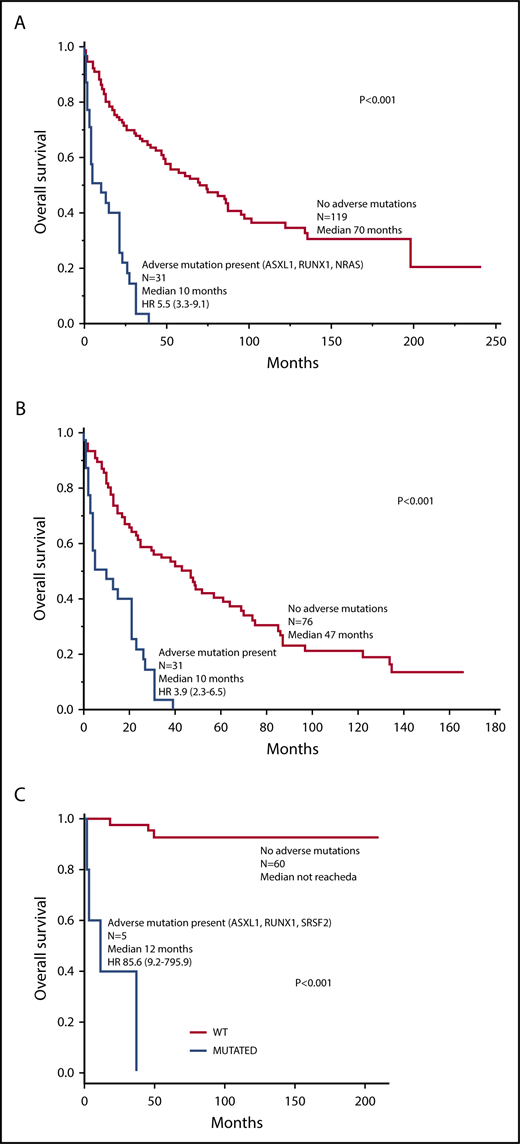
Combined clinical, cytogenetic (normal vs abnormal), and molecular risk factor analysis involving Mayo Clinic patients confirmed the independent prognostic contribution of adverse mutations (2.6, 1.6-4.4), advanced SM (4.0, 1.8-10.0), thrombocytopenia <150 × 109/L (2.8, 1.7-4.5), increased ALP (2.1, 1.2-4.0), and age >60 years (2.2, 1.3-3.6). In other words, the hybrid clinical-molecular model retained 4 of the 5 risk factors used in the previously elaborated clinical model and replaced anemia with adverse mutations as the fifth risk factor. Unlike the case with our clinical model, the HR value for advanced SM was notably higher than the HR values for the other risk factors in the clinical-molecular model, thus warranting assignment of 2 risk points for advanced SM and 1 risk point each for the other 4 risk factors. Figure 3A illustrates HR-weighted risk categories with significantly different survival data; the model was equally useful when patients with advanced SM were analyzed separately (Figure 3B). Because adverse mutations were not seen in ISM/SM, the clinical-molecular model was not applied to such patients. The small sample size from Florence did not allow similar all-inclusive multivariable analysis.
Clinical-molecular risk model. (A) Risk model for systemic mastocytosis based on HR-weighted risk points: (1) advanced SM vs ISM/SSM (HR, 4.0; 2 points); (2) age >60 years (HR, 2.2; 1 point); (3) platelets <150 × 109/L (HR, 2.8; 1 point); (4) serum ALP above normal range (HR, 2.1; 1 point); and (5) adverse mutations (HR, 2.6; 1 point). HR (95% CI) values listed are calculated against the next lower risk level. (B) Application of the clinical-molecular risk model in advanced SM. HR (95% CI) values listed are calculated against the next lower risk level.
Clinical-molecular risk model. (A) Risk model for systemic mastocytosis based on HR-weighted risk points: (1) advanced SM vs ISM/SSM (HR, 4.0; 2 points); (2) age >60 years (HR, 2.2; 1 point); (3) platelets <150 × 109/L (HR, 2.8; 1 point); (4) serum ALP above normal range (HR, 2.1; 1 point); and (5) adverse mutations (HR, 2.6; 1 point). HR (95% CI) values listed are calculated against the next lower risk level. (B) Application of the clinical-molecular risk model in advanced SM. HR (95% CI) values listed are calculated against the next lower risk level.
Discussion
The main objective for the current study was to devise a clinical prognostic model for SM that is widely applicable by internists, hematologists, oncologists, allergists, and other practitioners. We were able to accomplish this task by defining 5 easily accessible clinical risk factors: age >60 years, WHO-defined advanced SM vs ISM/SSM, thrombocytopenia <150 × 109/L, anemia defined as hemoglobin level below the sex-adjusted normal reference range, and increased serum ALP. As illustrated in Figure 1A, survival was directly and proportionally correlated with the number of risk factors, with an outstanding prognosis for patients with ≤1 risk factor (median survival not reached) and poor outcome for patients with 4 or 5 risk factors (median survival, 9-27 months). Figure 1B-C demonstrates that our clinical model was equally effective when applied to either advanced SM or ISM/SSM separately.
The current study also provides a hybrid clinical-molecular prognostic model that integrates mutation data (Figure 3). In this regard, it is important to underscore the powerful prognostic contribution of adverse mutations (ie, ASXL1, RUNX1, and NRAS), which was independent of the previously listed clinical variables as well as karyotype. The clinical-molecular model was also based on 5 risk factors, including age >60 years, advanced vs ISM/SM, thrombocytopenia <150 × 109/L, increased serum ALP, and adverse mutations; the inclusion of mutation data during multivariable analysis overrode prognostic contributions from anemia and abnormal karyotype. In regard to the latter, abnormal karyotype in SM clusters with SM-AHN and specifically with SM-AHN-myeloid5 ; this would suggest that its prognostic relevance, if any, might be more apparent in the context of the myeloid AHN component, rather than the associated SM. As was the case with the previously elaborated clinical model, the clinical-molecular model was successfully applied in patients with advanced SM (Figure 2B). Adverse mutations were not seen in patients with ISM/SSM in both the Mayo Clinic and Florence patient cohorts; therefore, the clinical-molecular model is primarily applicable in advanced SM.
It is important to recognize ongoing efforts by other investigators to decipher the prognostic role of karyotype6 and mutations in SM.7,8 Cytogenetic findings in SM were recently examined in 109 patients, including 26 ISM and 83 advanced cases.6 The authors of that particular study cited an abnormal karyotype incidence of 15% in their patients, and all abnormal cases occurred in patients with SM-AHN6 ; a possible association between abnormal karyotype and leukemic transformation and shorter survival was preliminarily suggested and found to be independent of somatic mutations. In another study of 62 patients with SM-AHN, non-KIT mutations included TET2 in 27%, ASXL1 in 14%, and CBL in 11%7 ; in multivariable analysis that included other clinical risk factors, only ASXL1 mutations carried an independent prognostic relevance for survival. In a more comprehensive and larger study of 70 patients with advanced SM,8 multivariable analysis identified ASXL1 and SRSF2 mutations as risk factors for overall survival; the study also suggested additional prognostic relevance of the number of adverse mutations.8 Unlike the case with the previously mentioned somatic mutations, the typical KIT mutations seen in >80% of patients with SM9 have not been shown, either qualitatively or quantitatively, to independently affect survival.
In conclusion, the current study advances 2 separate but complementary risk models for SM, illustrates the additional prognostic contribution of mutations, and underlines the importance of thrombocytopenia and serum ALP levels in both risk models. Furthermore, we show effective application of the clinical risk model, not only without a priori distinction of advanced SM vs ISM/SSM, but also in the context of 1 or the other. Whether these risk models will perform as well in the context of new targeted therapy, including the multi-kinase inhibitor midostaurin10 or the specific inhibitor of KIT activation loop mutants BLU-285 (avapritinib),11 remains to be seen. In this regard, in 1 study of patients treated with midostaurin,12 the drug did not appear to overcome the adverse effect of high-risk mutations. Regardless, additional studies with larger number of patients and inclusion of those treated with BLU-285 are required before making any conclusions.
Acknowledgments
This study was supported in part by research funding from the Henry J. Predolin Foundation grant (Madison, WI) and AIRC 5×1000 (“Metastatic disease: the key unmet need in oncology”) to the MYNERVA project (#21267; Myeloid Neoplasms Research Venture, AIRC). A detailed description of the MYNERVA project is available at http://www.progettoagimm.it.
Authorship
Contribution: A.P. and A.T. designed the study, conducted the analysis, and wrote the first manuscript draft; S.S. and Y.C.E. extracted data from patient charts; M.M.P. and N.G. contributed patients to the study; T.L.L. conducted molecular/mutation testing; R.P.K. analyzed the cytogenetics/karyotype data; K.K.R. and C.A.H. analyzed bone marrow pathology; F.M., P.G., and A.M.V. provided clinical and molecular data for patients from the Italian site; and all authors reviewed the manuscript and provided approval for the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ayalew Tefferi, Division of Hematology, Department of Medicine, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: tefferi.ayalew@mayo.edu.