Abstract
Patients with mantle cell lymphoma (MCL) may present with either nodal or leukemic disease. The molecular determinants underlying this different biologic behavior are not known. This study compared the pattern of genetic abnormalities in patients with nodal and leukemic phases of MCL using comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH) for specific gene loci. Although both leukemic and nodal MCL showed similar genomic patterns of losses (involving 6q, 11q22-q23, 13q14, and 17p13) and gains (affecting 3q and 8q), genomic loss of chromosome 8p occurred more frequently in patients with leukemic disease (79% versus 11%,P < .001). Subsequent CGH analysis confirmed the genomic loss of 8p21-p23 in 6 of 8 MCL cell lines. Interestingly,MYC gene amplification was restricted to cases with 8p deletion. These data indicate the presence of a novel tumor suppressor gene locus on 8p, whose deletion may be associated with leukemic dissemination and poor prognosis in patients with MCL.
Introduction
Mantle cell lymphoma (MCL) is characterized by the translocation t(11;14)(q13;q32) resulting in overexpression of cyclin D1.1-3 Patients with MCL present frequent extranodal disease at diagnosis, with peripheral blood (PB) involvement observed in one third of the cases. However, the natural history of MCL eventually includes involvement of the PB in almost all cases.1-4 Leukemic MCL has been associated with a worse prognosis than nodal MCL,4 although a small percentage of patients with leukemic disease may have an indolent course.1,5 Although many genetic aberrations in addition to the t(11;14)(q13;q32) have been correlated with specific features of the disease, whether these abnormalities are different in the leukemic and in the nodal forms of MCL remains unknown.5-11 To address this issue we have compared the pattern of secondary genetic abnormalities in patients with leukemic and nodal MCL, as well as in 8 MCL-derived cell lines.
Study design
Twenty-eight patients with MCL with t(11;14)(q13;q32) orBCL1-IGH gene rearrangement or both, fulfilling the World Health Organization criteria,3 were diagnosed between 1995 and 2000 among 400 consecutive patients newly diagnosed with B-cell lymphoproliferative disorders. Histologic, immunophenotypic, cytologic, and cytogenetic studies were performed in all cases. Screening forBCL1-IGH gene rearrangement by dual-color fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR) assays were performed in all patients with a suspected MCL.12,13Among 28 patients diagnosed with MCL, 19 presented with PB involvement at diagnosis (defined as > 10% CD5/CD19+ cells in the PB), and 9 patients were classified as having nodal MCL. One nodal and 6 leukemic cases were classified as blastoid variants of MCL (Table1). Six MCL-derived cell lines (Granta 519, NCEB-1, HBL2, SP-49, REC1, and Z-138), the JVM-2 cell line derived from a prolymphocytic leukemia, and the SKMM2 cell line derived from a multiple myeloma, both carrying t(11;14)(q13;q32), were also included in the study.14 References for the derivation of these cell lines may be obtained on request.
Cytogenetic, FISH, and molecular studies of patients with leukemic and nodal MCL
| Case . | Source . | Morphology . | Karyotype* . | FISH/PCRBCL1-IGH† . | FISH‡ . | CGH1-153 . |
|---|---|---|---|---|---|---|
| 1 | PB | Typical | 46,X,t(X;4)(p22;p12),ins(1)(?q22),t(11;14)(q13;q32)(2)/46,XX(19) | +/+ | −13q14 | Rev ish enh(3q13q27),dim(Xp22,Xq28,4p15,13q14q21) |
| 2 | PB | Large-cell1-155 | 46,XX,t(11;14)(q13;q32)(2)/45,idem,der(16)t(12;16)(q13;q23),−18(1)/48,idem,−X,−6,del(6)(p12),del(8)(p11),add(9)(p21),del(13)(q12q14),add(15)(q26),add(20)(q13.3),+1-4mar(1)/46,XX(15) | +/+ | −13q14,+8q24 | Rev ish enh(8q24),dim(Xp21p22,Xq28,8p21p23,10p14p15,12p13,13q14q21,14q31q32,19q13) |
| 3 | BM | Typical | 46,XY,t(11;14)(q13;q32)(17)/46,XY(3) | +/− | N | No imbalances |
| 4 | BM | Typical | 47,XX,+3,del(6)(q14q24),t(11;14)(q13;q32)(10)/47,idem,del(18)(q22q23)(20) | +/− | N | Rev ish enh(3,10p15),dim(6q13q27,8p21p23,17p13),amp(Xp22) |
| 5 | PB | Typical | 46,XX,del(8)(p12),add(14)(q32)(4)/46,XX(10) | +/+ | N | Rev ish dim(8p12p23) |
| 6 | PB | Large-cell | 44,X,Y,t(3;10)(p14;q22),+8,dic(8;9)(q10;q10),t(11;14)(q13;q32),del(12)(p11),del(17)(p11),−21,+mar(7)/44,idem,der(9)t(9;11)(q11;p11),−der+ (14)t(11;14)(q13;q32),der(14)t(11;14)(q13;q32)t(9;11)(q11;p11),−18, +2mar(5) | +/− | +8q24,−17p13 | Rev ish enh(8q13q24.3,15q12q26),dim(Yp11q11,3p13p25,8p11p23,9p12p24,12p11,12p13,17p11p13) |
| 7 | PB | Large-cell | 46,XY(20) | +/+ | +8q24,−17p13 | Rev ish enh(8q11q24.3),dim(4p15,8p12p23,16p13,17p11p13,20p13) |
| 8 | PB | Typical | 46,XY,−6,t(11;14)(q13;q32),+mar(5)/46,XY(15) | +/− | N | Rev ish enh(1q12q31,9p23),dim(X) |
| 9 | PB | Blastoid | 47-48,XY,+der(X)del(q25q27),t(11;14)(q13;q32)+mar(cp5) | +/− | −11q22,BCL1-IGHx2 | Rev ish enh(Xp11q25,16p13),dim(8p21p23,6q24q27,11q22q24,14q24q32,22q11q13),amp(3p25) |
| 10 | BM | Blastoid | 88,XX,−Y,−Y,−1,del(1)(p22p31.2),add(3)(q27)x2,del(6)(q14q24)x2,t(11;14) (q13;q32)x2,add(13)(q32)x2,add(15)(q25)x2,−16,−16,+mar(10)/46,XY(2) | +/+ | +8q24,−13q14,BCL1-IGHx2 | Rev ish enh(Xq22q26,3q24q27,8q21q24.3,11p11q13,14q32,15q21q23),dim(Xp21p22,Yq11, 1p13p36,1q31q44,6q21q27,8p21p23,13q31q34,15q24q26,16q21q23) |
| 11 | Spleen | Blastoid | 75-80,XY,+X,+Y,+2,+3,add(3)(?p13),+4,+4,+5,+6,+7,+8,+8,+9,+10,+11,t(11;14)(q13;q32),+12,+14,+15,+16,+17,+18,+18,+19,+20,+21,+22,+1-5mar(cp10) | +/− | +8q24,−13q14,−17p13 | Rev ish enh(Xp22q26,4p13q13,8q11q24.3,18q22q23),dim(Yq11q12,6q23q27,8p12p23, 10q24q26,13q14q34,17p11p13),amp(Xq25,18p11,19p13.3) |
| 12 | BM | Typical | 46,XX,t(11;14)(q13;q32)(2)/46,XX(8) | +/+ | −11q22,−13q14 | Rev ish enh(3q13q27,9q21q34,15q13q24),dim(Xp21p22,1p13p31,7q32q36,8p22p23,10p12p15,11q14q23,13q14q34,18p11) |
| 13 | PB | Blastoid | 46,XY(20) | +/+ | −11q22,−17p13,BCL1-IGHx2 | Rev ish enh(3q21q29,9q11q21,13q21q31),dim(Y,8p22p23,9q21q23,13q32q34,17p11p13, 18q22q23,20q13) |
| 14 | Lnode | Typical | ND | +/+ | −11q22 | Rev ish enh(5p13q31),dim(1p21p31,8p23,11p14p15,11q23q25,22q12q13) |
| 15 | Spleen | Typical | 46,XY,t(1;5)(q23;p15),t(11;14)(q13;q32)(3)/46,XY(15) | +/− | N | Rev ish enh(q25q26),dim(1p13p31,6q23q27,8p23) |
| 16 | Lnode | Blastoid | 47,XY,+7,dic(8;9)(p10;p10),+9,t(11;14)(q13;q32)(4) | +/+ | N | Rev ish enh(7p22p21,9p11q13),dim(8p23) |
| 17 | PB | Typical | 46,XY,del(6)(q14q25),t(11;14)(q13;q32)(7)/46,XY(3) | +/+ | −13q14 | Rev ish enh(Xp22q22,3q26,9p23p24),dim(6p25p12,6q21q27,8p12p23,11q22q24,12p12p13) |
| 18 | Lnode | Typical | ND | +/+ | −11q22 | Rev ish enh(12p12q21),dim(8q23q24,9p21p24,11q21q24) |
| 19 | PB | Blastoid | 46,XX,der(1)add(1)(p21)del(1)(q31),t(11;14)(q13;q32),dup(18)(q12q23)(inc 20) | +/− | +8q24,−11q22,+18q21 | Rev ish enh(1p34p36,2p12p25,8q23q24.3,9q34,11q13,14q32,16p13q13,17,22q11q13),dim (Xp22q26,1p13p31,1q23q44,2q24q35,4q11q33,8q11p23,9p13p24),amp(18q12q21.3,19) |
| 20 | Lnode | Typical | 46,XY,i(3)(q10),t(11;14)(q13;q32),del(13)(q14q22)(9) | +/− | −17p13 | Rev ish enh(3q11q29,9q33q34,15q21q26),dim(3p11p26,17p11p13) |
| 21 | Lnode | Typical | 45-47,X,−Y,+X,del(2)(p?15,del(3)(?q26),t(11;14)(q13;q32),−18,+mar(cp10) | +/+ | −13q14 | Rev ish enh(Xp21q22,5p11q32),dim(6q24q27,8p21p23,18p11) |
| 22 | Lnode | Typical | 48,XY,+3,t(11;14)(q13;q32),+mar(2) | +/− | N | Rev ish enh(3),dim(1p13p22,9p22p24,18p11) |
| 23 | Lnode | Typical | 46,XY,t(11;14)(q13;q32)(4)/46,XY(6) | +/+ | N | No imbalances |
| 24 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(1p32p36,15q21q25,16p13q12,17,19p13q13.2,22q11q13.2),dim(9p11p24,9q21q34,13q12q34) |
| 25 | Lnode | Blastoid | 89-90,XXYY,der(1)add(1)(q21)del(1)(?p21)x2,+der(3)del(3)(p21), t(11;14)(q13;q32)x2,add(?18)(q21)x2,−19,−22x2,+1-3mar(cp20) | +/− | −17p13,BCL1-IGHx2 | Rev ish enh(Xq12q23,3p13q26,4p13q32,5q11q23,6q11q21),dim(Xq26q27,1p32p36,6q24q26,10q25q26,17p11p13,19,22q11q13) |
| 26 | Lnode | Typical | 47,XY,+3,del(6)(q11),t(11;14)(q13;q32)(7) | +/+ | N | Rev ish enh(Xp22q21,3,15q21q25),dim(6q11q27) |
| 27 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(9q32q34),dim(Xp11p22,1p12p36,11q13q25,13,17q21) |
| 28 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(2q32q37,3q11q29,7),dim(6q21q27,9q32q34,11q22q24) |
| Case . | Source . | Morphology . | Karyotype* . | FISH/PCRBCL1-IGH† . | FISH‡ . | CGH1-153 . |
|---|---|---|---|---|---|---|
| 1 | PB | Typical | 46,X,t(X;4)(p22;p12),ins(1)(?q22),t(11;14)(q13;q32)(2)/46,XX(19) | +/+ | −13q14 | Rev ish enh(3q13q27),dim(Xp22,Xq28,4p15,13q14q21) |
| 2 | PB | Large-cell1-155 | 46,XX,t(11;14)(q13;q32)(2)/45,idem,der(16)t(12;16)(q13;q23),−18(1)/48,idem,−X,−6,del(6)(p12),del(8)(p11),add(9)(p21),del(13)(q12q14),add(15)(q26),add(20)(q13.3),+1-4mar(1)/46,XX(15) | +/+ | −13q14,+8q24 | Rev ish enh(8q24),dim(Xp21p22,Xq28,8p21p23,10p14p15,12p13,13q14q21,14q31q32,19q13) |
| 3 | BM | Typical | 46,XY,t(11;14)(q13;q32)(17)/46,XY(3) | +/− | N | No imbalances |
| 4 | BM | Typical | 47,XX,+3,del(6)(q14q24),t(11;14)(q13;q32)(10)/47,idem,del(18)(q22q23)(20) | +/− | N | Rev ish enh(3,10p15),dim(6q13q27,8p21p23,17p13),amp(Xp22) |
| 5 | PB | Typical | 46,XX,del(8)(p12),add(14)(q32)(4)/46,XX(10) | +/+ | N | Rev ish dim(8p12p23) |
| 6 | PB | Large-cell | 44,X,Y,t(3;10)(p14;q22),+8,dic(8;9)(q10;q10),t(11;14)(q13;q32),del(12)(p11),del(17)(p11),−21,+mar(7)/44,idem,der(9)t(9;11)(q11;p11),−der+ (14)t(11;14)(q13;q32),der(14)t(11;14)(q13;q32)t(9;11)(q11;p11),−18, +2mar(5) | +/− | +8q24,−17p13 | Rev ish enh(8q13q24.3,15q12q26),dim(Yp11q11,3p13p25,8p11p23,9p12p24,12p11,12p13,17p11p13) |
| 7 | PB | Large-cell | 46,XY(20) | +/+ | +8q24,−17p13 | Rev ish enh(8q11q24.3),dim(4p15,8p12p23,16p13,17p11p13,20p13) |
| 8 | PB | Typical | 46,XY,−6,t(11;14)(q13;q32),+mar(5)/46,XY(15) | +/− | N | Rev ish enh(1q12q31,9p23),dim(X) |
| 9 | PB | Blastoid | 47-48,XY,+der(X)del(q25q27),t(11;14)(q13;q32)+mar(cp5) | +/− | −11q22,BCL1-IGHx2 | Rev ish enh(Xp11q25,16p13),dim(8p21p23,6q24q27,11q22q24,14q24q32,22q11q13),amp(3p25) |
| 10 | BM | Blastoid | 88,XX,−Y,−Y,−1,del(1)(p22p31.2),add(3)(q27)x2,del(6)(q14q24)x2,t(11;14) (q13;q32)x2,add(13)(q32)x2,add(15)(q25)x2,−16,−16,+mar(10)/46,XY(2) | +/+ | +8q24,−13q14,BCL1-IGHx2 | Rev ish enh(Xq22q26,3q24q27,8q21q24.3,11p11q13,14q32,15q21q23),dim(Xp21p22,Yq11, 1p13p36,1q31q44,6q21q27,8p21p23,13q31q34,15q24q26,16q21q23) |
| 11 | Spleen | Blastoid | 75-80,XY,+X,+Y,+2,+3,add(3)(?p13),+4,+4,+5,+6,+7,+8,+8,+9,+10,+11,t(11;14)(q13;q32),+12,+14,+15,+16,+17,+18,+18,+19,+20,+21,+22,+1-5mar(cp10) | +/− | +8q24,−13q14,−17p13 | Rev ish enh(Xp22q26,4p13q13,8q11q24.3,18q22q23),dim(Yq11q12,6q23q27,8p12p23, 10q24q26,13q14q34,17p11p13),amp(Xq25,18p11,19p13.3) |
| 12 | BM | Typical | 46,XX,t(11;14)(q13;q32)(2)/46,XX(8) | +/+ | −11q22,−13q14 | Rev ish enh(3q13q27,9q21q34,15q13q24),dim(Xp21p22,1p13p31,7q32q36,8p22p23,10p12p15,11q14q23,13q14q34,18p11) |
| 13 | PB | Blastoid | 46,XY(20) | +/+ | −11q22,−17p13,BCL1-IGHx2 | Rev ish enh(3q21q29,9q11q21,13q21q31),dim(Y,8p22p23,9q21q23,13q32q34,17p11p13, 18q22q23,20q13) |
| 14 | Lnode | Typical | ND | +/+ | −11q22 | Rev ish enh(5p13q31),dim(1p21p31,8p23,11p14p15,11q23q25,22q12q13) |
| 15 | Spleen | Typical | 46,XY,t(1;5)(q23;p15),t(11;14)(q13;q32)(3)/46,XY(15) | +/− | N | Rev ish enh(q25q26),dim(1p13p31,6q23q27,8p23) |
| 16 | Lnode | Blastoid | 47,XY,+7,dic(8;9)(p10;p10),+9,t(11;14)(q13;q32)(4) | +/+ | N | Rev ish enh(7p22p21,9p11q13),dim(8p23) |
| 17 | PB | Typical | 46,XY,del(6)(q14q25),t(11;14)(q13;q32)(7)/46,XY(3) | +/+ | −13q14 | Rev ish enh(Xp22q22,3q26,9p23p24),dim(6p25p12,6q21q27,8p12p23,11q22q24,12p12p13) |
| 18 | Lnode | Typical | ND | +/+ | −11q22 | Rev ish enh(12p12q21),dim(8q23q24,9p21p24,11q21q24) |
| 19 | PB | Blastoid | 46,XX,der(1)add(1)(p21)del(1)(q31),t(11;14)(q13;q32),dup(18)(q12q23)(inc 20) | +/− | +8q24,−11q22,+18q21 | Rev ish enh(1p34p36,2p12p25,8q23q24.3,9q34,11q13,14q32,16p13q13,17,22q11q13),dim (Xp22q26,1p13p31,1q23q44,2q24q35,4q11q33,8q11p23,9p13p24),amp(18q12q21.3,19) |
| 20 | Lnode | Typical | 46,XY,i(3)(q10),t(11;14)(q13;q32),del(13)(q14q22)(9) | +/− | −17p13 | Rev ish enh(3q11q29,9q33q34,15q21q26),dim(3p11p26,17p11p13) |
| 21 | Lnode | Typical | 45-47,X,−Y,+X,del(2)(p?15,del(3)(?q26),t(11;14)(q13;q32),−18,+mar(cp10) | +/+ | −13q14 | Rev ish enh(Xp21q22,5p11q32),dim(6q24q27,8p21p23,18p11) |
| 22 | Lnode | Typical | 48,XY,+3,t(11;14)(q13;q32),+mar(2) | +/− | N | Rev ish enh(3),dim(1p13p22,9p22p24,18p11) |
| 23 | Lnode | Typical | 46,XY,t(11;14)(q13;q32)(4)/46,XY(6) | +/+ | N | No imbalances |
| 24 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(1p32p36,15q21q25,16p13q12,17,19p13q13.2,22q11q13.2),dim(9p11p24,9q21q34,13q12q34) |
| 25 | Lnode | Blastoid | 89-90,XXYY,der(1)add(1)(q21)del(1)(?p21)x2,+der(3)del(3)(p21), t(11;14)(q13;q32)x2,add(?18)(q21)x2,−19,−22x2,+1-3mar(cp20) | +/− | −17p13,BCL1-IGHx2 | Rev ish enh(Xq12q23,3p13q26,4p13q32,5q11q23,6q11q21),dim(Xq26q27,1p32p36,6q24q26,10q25q26,17p11p13,19,22q11q13) |
| 26 | Lnode | Typical | 47,XY,+3,del(6)(q11),t(11;14)(q13;q32)(7) | +/+ | N | Rev ish enh(Xp22q21,3,15q21q25),dim(6q11q27) |
| 27 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(9q32q34),dim(Xp11p22,1p12p36,11q13q25,13,17q21) |
| 28 | Lnode | Typical | ND | ND/+ | ND | Rev ish enh(2q32q37,3q11q29,7),dim(6q21q27,9q32q34,11q22q24) |
Leukemic (no. 1-19) and nodal MCL cases (no. 20-28).
Lnode indicates lymph node; ND, not done; and N, normal results.
Cytogenetic analysis was performed on 72-hour tetradecanoylphorbol-13-acetate (TPA)–stimulated cultures in the samples from PB and BM, and on nonstimulated 24-hour cultures in spleen and lymph node samples.
PCR analysis was performed in the same samples used for CGH analysis.
FISH was performed on fixed cell nuclei from cytogenetic analysis. BCL1-IGHx2: two fusions of BCL1 andIGH gene probes per cell.
All samples used for CGH had more than 50% of CD5/CD19+ cells.
Large-cell variant of MCL presenting transformed blastlike cells with prominent nucleoli (Zoldan and coworkers21).
Comparative genomic hybridization (CGH) and FISH for the presence of trisomy 12 and deletions of 13q14 (D13S25 locus) and of P53gene in 17p13 were performed in all patients at diagnosis; the cell lines were analyzed by CGH and cross-species color banding (RxFISH). The probes and methods used have been previously reported.13,15-17 In leukemic cases, studies were performed on samples from PB or bone marrow (BM) (n = 14), lymph node (n = 3), and spleen (n = 2), whereas all nodal cases were studied on lymph node samples. In selected cases and cell lines with genomic imbalances affecting specific gene loci, the number of copies ofMYC gene (using previously reported probes17), and ATM and BCL2 genes was determined by FISH using probes obtained from Dr M. Rocci (University of Bari, Italy,www.bioserver.biologia.uniba.it).
Results and discussion
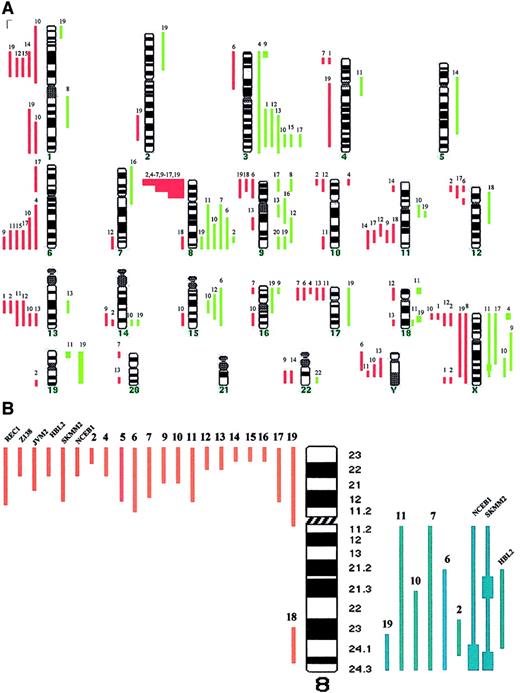
Table 1 shows the cytogenetic, FISH, and CGH studies in the patients with leukemic MCL (no. 1-19) and nodal MCL (no. 20-28). Those with leukemic disease showed genomic imbalances in 18 of 19 cases (95%), with losses being more frequent than gains (88 versus 50). The median number of chromosomal imbalances per case was 6 (range, 0-18). High-level amplifications were observed in 7 regions (Xp22, Xq25, 3p25, 18p11, 18q21, 19p, and 19q). The most frequent abnormalities included gains of 3q (37%), 8q affecting MYC gene (32%), 9q (26%), Xq (21%), and 15q (16%), and losses of 8p (79%), 13q encompassing D13S25 locus, 6q and Xp (32%), and 1p22, 11q involving ATMgene and 17p involving P53 gene (26%). Eight of the 9 patients with nodal MCL displayed chromosomal imbalances, showing a spectrum of abnormalities similar to that of leukemic MCL and consistent with previous reports.8-10 (Table 1 and Figure1A). Nevertheless, the genomic loss of 8p was associated with the leukemic forms because it was detected in 15 of 19 patients with leukemic MCL (79%) but in only 1 of 9 patients with nodal MCL (11%; P < .001). Comparison of cases with the deleted region allowed narrowing of the commonly deleted segment to 8p21-p23 (Figure 1B). Recent studies have reported this genetic abnormality in patients with human malignancies including T-cell prolymphocytic leukemia, and bladder, breast, head and neck, prostate, lung, and colorectal carcinoma.18 However, it has only been described in 13 of 99 reported cases of MCL studied by CGH,8-10 and very rarely in other B-cell lymphomas.11,15,18,19 It has been suggested that a tumor suppressor gene (TSG) may be located in the subtelomeric region of chromosome 8p, and associated with increased ability to metastasize in hepatocellular carcinoma.20 Our results suggest that the deletion of 8p may be a characteristic molecular marker of leukemic MCL and that this region may contain a novel TSG with a possible role in blood dissemination of MCL. Moreover, the loss of 8p was found in all 9 cases with aggressive variants of leukemic MCL (both blastoid and large-cell subtypes) but in 60% of cases of typical morphology, suggesting a correlation with aggressive tumors. Because of the location of the relevant segment at the subtelomeric region of 8p, the deletion was identified by cytogenetics only in 4 of the 16 patients with the genomic loss. This may probably explain why this abnormality has been identified only rarely in previous series of MCL.8-11,18,19 Additionally, most of our cases were studied on BM/PB samples, and this may have also contributed to these results. Interestingly, we found a high incidence of MYCamplification (6 of 19 cases, 32%), and all these abnormalities were detected in patients with deletion of 8p. All of them presented with aggressive MCL; 3 cases were blastoid variants and 3 were classified as large-cell variants of MCL showing circulating transformed blastlike cells.21 Confirming previous studies, a tetraploid karyotype with a double BCL1-IGH gene fusion, or a simultaneous genomic gain affecting to BCL1 andIGH loci at 11q13 and 14q32, respectively, was seen in 5 of the 7 blastoid variants, but not in any typical case.19
Chromosomal imbalances and genomic loss of 8p in MCL.
(A) Summary of chromosomal imbalances detected by CGH in patients with leukemic MCL. Cases 1 to 19 are as presented in Table 1. Red lines on the left of the ideogram indicate loss of chromosomal material, whereas the green lines to the right indicate gain of chromosomal material. Green squares represent high-level DNA amplification. (B) Schematic representation of chromosome 8 genomic abnormalities in patients with MCL and in cell lines with t(11;14)(q13;q32).
Chromosomal imbalances and genomic loss of 8p in MCL.
(A) Summary of chromosomal imbalances detected by CGH in patients with leukemic MCL. Cases 1 to 19 are as presented in Table 1. Red lines on the left of the ideogram indicate loss of chromosomal material, whereas the green lines to the right indicate gain of chromosomal material. Green squares represent high-level DNA amplification. (B) Schematic representation of chromosome 8 genomic abnormalities in patients with MCL and in cell lines with t(11;14)(q13;q32).
We subsequently expanded our study to 8 cell lines carrying t(11;14)(q13;q32). Deletion of 8p21-p23 was identified in 6 of them, including a genomic gain of 8q24 with MYC amplification in 3 (Figure 1B). In all of them the loss of 8p was caused by an unbalanced translocation involving 8p and varied partner chromosomes, or by a derivative isochromosome 8q. These results indicate that the genomic loss of 8p is a frequent event in MCL cell lines and that concomitantMYC amplification is seen in most cases. Overexpression and mutation of MYC has been identified in a subset of lymphomas including Burkitt lymphoma, where it plays a crucial pathogenic role.3,22 However, the clinical and biologic significance of MYC amplification in MCL is not well known.9 22 Based on our results, we suggest thatMYC amplification in MCL may be especially frequent in cases with a deletion of 8p. We may therefore hypothesize that an inactivated TSG at 8p cooperates with MYC in the pathogenesis of aggressive MCL.
In summary, our results show that genomic loss of 8p is a characteristic marker of leukemic MCL, suggesting the presence of a novel TSG locus related to blood dissemination of MCL. The deletion of 8p is frequently accompanied by MYC amplification and associated with an aggressive behavior of leukemic MCL.
We thank Dr A. Ferrandez (H. Clinico); Drs I. Navarro, R. Ferrer, J. Martinez (H. Gandia); M. Garcia, A. Carral (H. Sagunto); J. Marco, R. Garcia (H. Castellon); M. Montagut, M. D. Mirabet (H. Vinaroz); F. Ortuño (H. Murcia); and all the hematologists and pathologists from the “Club Citológico de la Comunidad Valenciana y Murcia” for providing samples and data from the patients; Dr Mariano Rocci (Bari, Italy) for ATM and BCL2 probes; and E. Cervello, M. Ordoñez, R. Marques, F. Domingo, and M. Botia, for excellent technical assistance.
Supported by grants from the Fondo de Investigación Sanitaria (FIS) FIS-98/0491 and FIS-01/0015, by the Deutsche Krebshilfe grants 10-1556-Schl4 and 10-1641-De1, and by the IZKF Kiel.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jose A. Martı́nez-Climent, Department of Hematology and Medical Oncology, Hospital Clı́nico, University of Valencia. Avda Blasco Ibañez, 17, 46010 Valencia, Spain; e-mail: martinez_jos@gva.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal