Abstract
It was previously shown that patients with chronic myeloid leukemia (CML) have a rare but consistently detectable population of quiescent (G0) leukemic (Philadelphia chromosome–positive and BCR-ABL–positive [BCR-ABL+]) CD34+ cells. In the study described here, most such cells expressed a primitive phenotype (CD38−, CD45RA−, CD71−, and HLA-DRlo) and cultures of these cells containing growth factors produced ultimately larger, but initially more slowly growing clones than do cultures of initially cycling CD34+ leukemic cells. Initially quiescent leukemic cells expressing BCR-ABLproliferated in single-cell cultures in the absence of added growth factors, thereby demonstrating their ability to spontaneously exit G0 and enter a continuously cycling state. Interestingly, on isolation, few of these quiescentBCR-ABL+ cells contained either interleukin-3 (IL-3) or granulocyte colony-stimulating factor (G-CSF) transcripts, whereas both were present in most cyclingBCR-ABL+ CD34+ cells. However, after 4 days of culture in the absence of added growth factors and in association with their entry into the cell cycle (as indicated by up-regulation of Ki-67 and cdc25 transcripts), IL-3 transcripts became detectable. These findings show that entry of leukemic (BCR-ABL–expressing) progenitors into a quiescent (G0) state in vivo is highest among the most primitive leukemic cell populations, associated with a down-regulation of IL-3 and G-CSF gene expression, and spontaneously reversible in association with up-regulation of IL-3 expression. These results highlight the potential physiologic relevance of quiescent CML progenitors, even in treated patients, in whom these cells would be predicted to have a proliferative advantage over their quiescent normal counterparts when cytokine concentrations are low.
Introduction
Chronic myeloid leukemia (CML) is a clonal multilineage myeloproliferative disorder1 in which there is a generalized expansion of many types of intermediate hematopoietic progenitors that leads to an excessive output of mature granulocytes.2 The strong association of this disease with the presence of a BCR-ABL fusion gene in the leukemic cells,3-5 coupled with independent experimental evidence that the protein it encodes has oncogenic properties,6-9suggests the primary involvement of BCR-ABL in the pathogenesis of early-stage CML. However, in spite of much information about how the BCR-ABL fusion-gene product can alter intracellular signaling,10-13 the relative importance of these molecular perturbations in the natural course of human CML is not clear.
Several studies have found that primitive CML cells isolated from patients can survive and proliferate in vitro in the absence of added growth factors, both with and without serum.14-16 We previously observed that such autonomous growth appears greatest in the most primitive, phenotypically defined subsets of CML cells. In these, it is consistently associated with and at least partly dependent on an abnormal activation of interleukin-3 (IL-3) and granulocyte colony-stimulating factor (G-CSF) production.17,18An autocrine mechanism involving IL-3 (and G-CSF) would provide an attractive explanation for both the increased proliferative activity2,19,20 and the decreased self-renewal capacity21,22 characteristic of primitive CML cells, because previous studies found that normal cells show both these responses when exposed to excess concentrations of IL-3 in vitro.23 However, more recent studies revealed a rare but consistently detectable population of primitive Philadelphia chromosome–positive (Ph+) andBCR-ABL–positive (BCR-ABL+) progenitor cells in CML patients that are quiescent.24 The current experiments were designed to investigate whether quiescent and cycling subsets of leukemic CML progenitors in the CD34+compartment differ in their potential for growth factor–independent proliferation in vitro and, if so, whether this activity correlates with constitutive expression of IL-3 and G-CSF. Our results support a model of clonal expansion of Ph+/BCR-ABL+ cells in which abnormal activation of IL-3 gene expression contributes to an autonomous proliferation of the most primitive neoplastic cells. In addition, our findings suggest that this may be dependent on but not exclusively regulated by the BCR-ABL oncoprotein.
Materials and methods
Cell samples
Heparin-treated blood and, in one case, bone marrow cells were obtained as part of the routine assessment of 9 untreated patients recently given a diagnosis of BCR-ABL+chronic-phase CML (Table 1). Samples of normal adult marrow were obtained from either harvests taken for allogeneic transplantation or cadavers (Northwest Tissue Center, Seattle, WA). Before use, all samples were enriched for CD34+ cells by means of negative immunomagnetic depletion of lineage-marker–positive cells (Stemsep; Stemcell Technologies, Vancouver, BC, Canada).25 All human cell samples were obtained after informed consent was given, and most samples were cryopreserved to allow repeated experimentation with the same samples.
Clinical data on patients with chronic myeloid leukemia from whom study samples were obtained
| Patient* . | WBC (109/L) . | Marrow metaphases† . | Cells used . | LTC-IC (Ph+/total)‡ . |
|---|---|---|---|---|
| 1 | 110 | 46XX, t(9;9), 25/25 | Blood | 0/61-153 |
| 2 | 540 | 46XY, t(9;22), 25/25 | Marrow | 0/7 |
| 3 | 52 | 46XX, t(9;22), 25/25 | Blood | 17/17 |
| 4 | 650 | 46XX, t(9;22), 25/25 | Blood | 33/33 |
| 5 | 180 | 46XY, t(9;22), 25/25 | Blood | 4/4 |
| 6 | 640 | 46XY, t(9;22), 25/25 | Blood | 2/2 |
| 7 | 520 | 46XY, t(9;22), 30/30 | Blood | 0/121-154 |
| 8 | 230 | 46XY, t(9;22), 30/30 | Blood | 0/31-154 |
| 9 | 400 | 46XY, t(9;22), 30/30 | Blood | 9/101-154 |
| Patient* . | WBC (109/L) . | Marrow metaphases† . | Cells used . | LTC-IC (Ph+/total)‡ . |
|---|---|---|---|---|
| 1 | 110 | 46XX, t(9;9), 25/25 | Blood | 0/61-153 |
| 2 | 540 | 46XY, t(9;22), 25/25 | Marrow | 0/7 |
| 3 | 52 | 46XX, t(9;22), 25/25 | Blood | 17/17 |
| 4 | 650 | 46XX, t(9;22), 25/25 | Blood | 33/33 |
| 5 | 180 | 46XY, t(9;22), 25/25 | Blood | 4/4 |
| 6 | 640 | 46XY, t(9;22), 25/25 | Blood | 2/2 |
| 7 | 520 | 46XY, t(9;22), 30/30 | Blood | 0/121-154 |
| 8 | 230 | 46XY, t(9;22), 30/30 | Blood | 0/31-154 |
| 9 | 400 | 46XY, t(9;22), 30/30 | Blood | 9/101-154 |
WBC indicates white blood cell count; LTC-IC, long-term-culture–initiating cells; Ph+, Philadelphia chromosome positive.
Patient samples 1 to 6 were described previously.24
Numbers after the karyotype indicate the number of metaphases with that karyotype of the total number analyzed at the time the sample was obtained.
Values are the number of LTC-IC–derived Ph+colony-forming cells (CFCs) detected and the total number of LTC-IC–derived CFCs for which 2 metaphases per colony were obtained. LTC-IC and CFCs were assayed as described previously.24
Based on the presence of t(9;9).
Positive for BCR-ABL on assessment with fluorescent in situ hybridization.
Routine isolation of CD34+ subpopulations and CD34− cells
Using previously described fluorescence-activated cell-sorter (FACS) methods,26 we isolated CD34+, CD34+CD38−, CD34+CD38+, and CD34− cells with greater than 99% purity from thawed lineage-marker–negative (lin−) cells. In a few experiments, FACS-purified CD34+ G0 and G1/S/G2/M cells were restained with antibodies to CD38, CD45RA, CD71, or HLA-DR as described previously26-28 and then reanalyzed. To obtain defined numbers of cells for reverse transcriptase–polymerase chain reaction (RT-PCR) analyses and for measuring the frequency of cells that generated clones in liquid cultures (ie, cloning efficiency), the automated cell-deposition unit of a FACStar Plus or FACSVantage device (Becton Dickinson, San Jose, CA) was used to obtain the required number of cells (1-1000) in individual wells of 96-well microtiter plates containing lysis buffer or serum-free medium (SFM).
Isolation of cells after Hoechst 33342 and pyronin Y staining
Cryopreserved lin− cells were thawed and, to allow reactivation of RNA synthesis, the cells were then incubated overnight in Iscoves medium (Stemcell Technologies) supplemented with a serum substitute (BIT; Stemcell Technologies), 40 μg/mL low-density lipoproteins (Sigma Chemical, St Louis, MO) and 10−4 M 2-mercaptoethanol (complete SFM) supplemented with 300 ng/mL each of recombinant human Flt3-ligand (FL; Immunex, Seattle, WA) and Steel factor (SF; Terry Fox Laboratory, Vancouver, BC, Canada), and 60 ng/mL each of recombinant human IL-3 (Novartis, Basel, Switzerland), IL-6 (Cangene, Mississauga, ON, Canada), and G-CSF (Stemcell Technologies). The next day, the cells were washed once and stained in Hanks balanced salt solution supplemented with 2% fetal-calf serum (HF/2), 10 μM Hoechst 33342 (Hst; Molecular Probes, Eugene, OR), 2.5 μg/mL pyronin Y (Py; Sigma Chemical) and anti-CD34–fluorescein isothiocyanate, conjugated (FITC; 8G12-FITC29; provided by P. Lansdorp, Terry Fox Laboratory), and 1 μg/mL propidium iodide (PI; Sigma Chemical). Cells were then sorted with gates set to collect HstloPylo (G0) and Hstlo/+Py+ (G1/S/G2/M) cells as separate fractions in the PI-negative (PI−) CD34+ populations.24 30
Isolation of cells labeled with carboxyfluorescein diacetate succinimidyl ester
Cells were incubated with carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) and cultured overnight in SFM supplemented with 50 ng/mL thrombopoietin (TPO; Genentech, San Francisco, CA) before being washed and labeled with anti-CD34–phycoerythrin (PE; Becton Dickinson) and 1 μg/mL PI as described previously.24,31 A relatively homogeneous subset of CFSE-positive PI− CD34+ cells (105 to 3 × 106, depending on the sample) was sorted by using a narrow fluorescence gate (36-40 channels wide with a 1024-channel log amplifier). These cells were then cultured for another 4 days in SFM containing 50 ng/mL TPO, both with and without 100 ng/mL colcemid (Gibco BRL, Burlington, ON, Canada). All the cells present (105 to 4 × 106) were then harvested, washed, and stained with PI (1 μg/mL). Cells cultured in the presence of colcemid were used to establish the range of fluorescence shown by cells that had not divided during the 4-day postlabeling incubation and, hence, their separation by FACS from cells that had divided (in the cultures to which no colcemid was added).24 31
Serum-free cultures
Single cells were cultured individually in 100-μL aliquots of SFM with or without added growth factors. Cultures with growth factors contained the same mixture of 5 growth factors described above for incubating cells overnight before Py and Hst staining. The number of viable (refractile) cells in each well was assessed after 4, 10, 20, and 35 days of incubation at 37°C. A clone was defined as the presence of at least 2 refractile cells. In this study, any cell capable of forming such a clone in liquid medium was described as “proliferating” and the proportion of any given population producing clones represented the cloning efficiency of that population. (This term should not be confused with the term “colony-forming cell” [CFC], which is commonly used to refer to cells that can proliferate and generate at least 20 mature myeloid or erythroid progeny when stimulated by appropriate growth factors in semisolid media.) Bulk populations of cells were cultured in 2.5-mL volumes of SFM with and without growth factors in 35-mm suspension dishes.
Isolation of viable cells from serum-free cultures for RT-PCR analyses
Cells were cultured in SFM for various periods, washed to remove any residual growth factors, and labeled with annexin-V–FITC (Pharmingen, San Diego, CA) and PI. The viable (annexin-V–negative [annexin-V−]/[PI−]) cells were then isolated by FACS and subjected to RT-PCR analyses.
RT-PCR analyses
Cells were sorted directly into a guanidinium isothiocyanate (GIT) lysis buffer (5 M GIT, 20 mM 1,4-diothioerythritol, 25 mM sodium citrate [pH 7.0], and 0.05% sarcosyl). A 2-step (nested) RT-PCR procedure using an initial oligo (dT)–based primer and poly (A) tailing strategy32,33 was then done. After electrophoresis of the amplified products, BCR-ABL–, actin-, IL-3–, and G-CSF–specific fragments were detected with Southern blotting using complementary DNA probes for BCR-ABL (provided by J. Griffin, Dana Farber Cancer Institute, Boston, MA) actin, IL-3, and G-CSF (provided by R. Kay and K. Humphries, Terry Fox Laboratory).17,32 33 Primers sets included ABL 1 and 2, 5′TTCAGCGGCCAGTAGCATCTGACTT3′ and 5′GGTACCAGGAGTGTTTCTCCAGACTG3′; BCR-ABL 1 and 2, 5′CAGGGTGCACAGCCGCAACGGCAA3′ and 5′GTCCAGCGAGAAGGTTTTCCTTGGA3′; actin 1 and 2, 5′GTGCGTGACATTAAGGAGAA3′ and 5′GGAGGGGCCGGACTCGTCA3′; IL-3 1 and 2, 5′GCTCCCATGACCCAGACAACGTCC3′ and 5′CAGATAGAACGTCAGTTTCCTCCG3′; G-CSF 1 and 2, 5′CTCTGGACAGTGCAGGAAGCCACC3′ and 5′GCTGGGCAAGGTGGCGTAGAACGC3′; CD34 1 and 2; 5′GGAATTCGAGGCCACAACAAACATCAC3′ and 5′GGAATTCGCAGATGCCCTGAGTCAATT3′; cdc25 1 and 2, 5′GGAAGTGCATTTAGCTGGGATGAA3′ and 5′CCACCTGCTTCAGTCTTGGCCTGT3′; Ki-67 1 and 2, 5′GCAAGAGGCAAATCATCCGAACCC3′ and 5′GAGAA- CCTTCGCACTCTTCTGCCC3′; p21 1 and 2, 5′CCTCACCTCCTCTAAGGTTGG3′ and 5′-CCCTTCCAGTCCATTGAGC3′; and cyclin D2 1 and 2, 5′CTGGCCATGAACTACCTGGA3′ and 5′CATGGCAA- ACTTAAAGTCGG3′.
Statistical analyses
Statistical analyses were done with the Studentt test.
Results
Various highly purified subpopulations of lin− cells were isolated from blood or marrow samples from 9 untreated patients with CML (Table 1). These populations included CD34−, CD34+CD38+, CD34+CD38−, CD34+ G0, CD34+ G1/S/G2/M, CD34+TPO-resistant, and CD34+ TPO-responsive cells (Figure1A-C). Four of the 9 CML samples studied were typical of those obtained from most initially presenting patients26 in that the most primitive hematopoietic cells (defined functionally as long-term culture-initiating cells [LTC-IC]) were predominantly normal in spite of a predominance of leukemic (Ph+/BCR-ABL+) cells in later compartments (Table 1). The other 5 samples were less typical in that the LTC-IC were predominantly Ph+. These were included to allow investigation of the behavior of very primitive leukemic cells.
FACS profiles of the cell populations tested.
(A) Dot plot of CD34-FITC versus CD38-PE–labeled PI−cells showing CD34+38−, CD34+CD38+, and CD34− populations. (B) Dot plot of Hst-stained versus Py-stained PI−CD34+ cells showing CD34+ G0 (R1) and CD34+ G1/S/G2/M (R2) populations. (C) Histogram of CFSE-stained PI−CD34+ cells showing TPO-resistant (M1) and TPO-responsive (M2) populations.
FACS profiles of the cell populations tested.
(A) Dot plot of CD34-FITC versus CD38-PE–labeled PI−cells showing CD34+38−, CD34+CD38+, and CD34− populations. (B) Dot plot of Hst-stained versus Py-stained PI−CD34+ cells showing CD34+ G0 (R1) and CD34+ G1/S/G2/M (R2) populations. (C) Histogram of CFSE-stained PI−CD34+ cells showing TPO-resistant (M1) and TPO-responsive (M2) populations.
Proliferating activity of CD34+CD38−, CD34+CD38+, and CD34−CML cells in single-cell serum-free liquid cultures
CD34+CD38− cells were isolated from the first 6 patients' samples and examined for their ability to proliferate (produce at least 2 progeny by day 10) when cultured as single cells in a serum-free liquid-suspension culture with or without added growth factors (FL, SF, IL-3, IL-6, and G-CSF). Preliminary time-course studies with cells from 3 of these samples showed that almost all single CD34+CD38− cells able to divide once did so within the first 4 days and that those able to divide at least 5 times did so within 10 days, whether or not growth factors were added (data not shown). As show in Table2, the frequency of proliferating CD34+CD38− cells (in the presence of growth factors) in the samples from all 6 patients with CML studied was high (range, 39%-90%). In fact, these frequencies were several times higher than the usual frequency of either CFCs (ie, cells able to proliferate in semisolid medium) or LTC-IC in the CD34+CD38− subpopulation of cells from patients with CML.22,26 This discrepancy between the frequency of either CFCs or LTC-IC and the frequency of CD34+CD38− cells able to proliferate in liquid medium was previously observed in normal marrow.34 Single CD34+CD38− cells from the same 6 samples from patients with CML also proliferated at high frequency in serum-free liquid cultures in the absence of added growth factors (range, 27%-68%; Table 2). Under the assumption that these growth factor–independent proliferating cells represented a subset of those that proliferate in the presence of growth factors, the frequency of growth factor–independent proliferating cells in each sample of CD34+CD38− cells was calculated. Values ranged from 54% to 96% (Table 2).
Cloning efficiency and growth factor independence in subpopulations of cells in samples from patients with chronic myeloid leukemia and from normal bone marrow, assessed in 10-day serum-free liquid cultures of single cells
| Patient . | CD34+CD38−cells . | CD34+CD3+cells . | CD34−cells . | CD34+ G0cells . | CD34+G1/S/G2/M cells . | CD34+ TPO-resistant† cells . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +GF (% Ph−*) . | −GF (% Ph−) . | % GFI . | +GF . | −GF . | % GFI . | +GF . | −GF . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | |
| 1 | 51 (68) | 38 (0) | 74 | 39 | 2 | 5 | 5 | 2 | 47 | 26 (39) | 20 (0) | 76 | 28 (0) | 10 (0) | 37 | 30 (64) | 23 (0) | 77 |
| 2 | 57 (0) | 46 (0) | 82 | 19 | 6 | 31 | 1 | 0 | 0 | 53 (33) | 50 (0) | 94 | 34 (0) | 22 (0) | 64 | 28 (13) | 27 (0) | 95 |
| 3 | 39 (0) | 38 (0) | 96 | — | — | — | — | — | — | — (0) | — (0) | — | 32 (—) | 18 (—) | 55 | 13 (—) | 9 (—) | 75 |
| 4 | 50 (—) | 27 (0) | 54 | — | — | — | — | — | — | 48 (0) | 24 (0) | 50 | 34 (—) | 9 (—) | 26 | 43 (0) | 18 (0) | 42 |
| 5 | 90 (0) | 68 (0) | 76 | 42 | 21 | 50 | 20 | 7 | 32 | 21 (0) | 3 (—) | 15 | 24 (—) | 1.5 (—) | 6 | 40 (0) | 22 (—) | 55 |
| 6 | 74 (0) | 48 (0) | 65 | 22 | 6 | 29 | 1 | 0 | 0 | — (0) | — (0) | — | — | — | — | 15 (0) | 4 (0) | 29 |
| Mean ± SE | 62 ± 9 | 46 ± 7 | 75 ± 6 | 28 ± 7 | 11 ± 5 | 37 ± 7 | 7 ± 6 | 2 ± 2 | 11 ± 11 | 41 ± 10 | 26 ± 14 | 53 ± 23 | 31 ± 2.4 | 13 ± 4.6 | 38 ± 13 | 28 ± 6 | 16 ± 4 | 59 ± 12 |
| NBM 1 | — | — | — | — | — | — | — | — | — | 46 | 0 | 0 | 48 | 0 | 0 | 65 | 0 | 0 |
| NBM 2 | — | — | — | — | — | — | — | — | — | 59 | 0 | 0 | 53 | 0 | 0 | 71 | 0 | 0 |
| Patient . | CD34+CD38−cells . | CD34+CD3+cells . | CD34−cells . | CD34+ G0cells . | CD34+G1/S/G2/M cells . | CD34+ TPO-resistant† cells . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +GF (% Ph−*) . | −GF (% Ph−) . | % GFI . | +GF . | −GF . | % GFI . | +GF . | −GF . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | +GF (% Ph−) . | −GF (% Ph−) . | % GFI . | |
| 1 | 51 (68) | 38 (0) | 74 | 39 | 2 | 5 | 5 | 2 | 47 | 26 (39) | 20 (0) | 76 | 28 (0) | 10 (0) | 37 | 30 (64) | 23 (0) | 77 |
| 2 | 57 (0) | 46 (0) | 82 | 19 | 6 | 31 | 1 | 0 | 0 | 53 (33) | 50 (0) | 94 | 34 (0) | 22 (0) | 64 | 28 (13) | 27 (0) | 95 |
| 3 | 39 (0) | 38 (0) | 96 | — | — | — | — | — | — | — (0) | — (0) | — | 32 (—) | 18 (—) | 55 | 13 (—) | 9 (—) | 75 |
| 4 | 50 (—) | 27 (0) | 54 | — | — | — | — | — | — | 48 (0) | 24 (0) | 50 | 34 (—) | 9 (—) | 26 | 43 (0) | 18 (0) | 42 |
| 5 | 90 (0) | 68 (0) | 76 | 42 | 21 | 50 | 20 | 7 | 32 | 21 (0) | 3 (—) | 15 | 24 (—) | 1.5 (—) | 6 | 40 (0) | 22 (—) | 55 |
| 6 | 74 (0) | 48 (0) | 65 | 22 | 6 | 29 | 1 | 0 | 0 | — (0) | — (0) | — | — | — | — | 15 (0) | 4 (0) | 29 |
| Mean ± SE | 62 ± 9 | 46 ± 7 | 75 ± 6 | 28 ± 7 | 11 ± 5 | 37 ± 7 | 7 ± 6 | 2 ± 2 | 11 ± 11 | 41 ± 10 | 26 ± 14 | 53 ± 23 | 31 ± 2.4 | 13 ± 4.6 | 38 ± 13 | 28 ± 6 | 16 ± 4 | 59 ± 12 |
| NBM 1 | — | — | — | — | — | — | — | — | — | 46 | 0 | 0 | 48 | 0 | 0 | 65 | 0 | 0 |
| NBM 2 | — | — | — | — | — | — | — | — | — | 59 | 0 | 0 | 53 | 0 | 0 | 71 | 0 | 0 |
Growth factor independence (GFI) was calculated as the cloning efficiency measured after 10 days in the absence of any added growth factors (GF) divided by the cloning efficiency obtained in the presence of 300 ng/mL Steel factor and Flt3-ligand plus 60 ng/mL interleukin 3, interleukin 6, and granulocyte colony-stimulating factor. Assays with and without growth factor (+GF and −GF, respectively) were set up with 1 to 3 96-well plates per condition. Differences between the CD34+CD38− cells and either the CD34+CD38+ cells or the CD34−cells, incubated either +GF or −GF, were significant (P = .02 and P < .001, respectively, for cultures +GF, and P = .002 and P < .001, respectively, for cultures −GF).
TPO indicates thrombopoietin; Ph−, Philadelphia chromosome negative; NBM, normal bone marrow; (—), analysis not done.
Cytogenetic or reverse transcriptase–polymerase chain reaction studies were performed on multiple individual clones (2-45 per condition) or, if clones were not available, samples from representative bulk cultures, to confirm Ph/BCR-ABL status.
TPO-resistant cells were those that did not divide during an initial 4-day culture in serum-free medium supplemented with TPO. These cells were isolated on the basis of their carboxyfluorescein diacetate succinimidyl ester fluorescence.
Because 2 of the 6 patients with CML were known to have predominantly normal LTC-IC (Table 1), it was important to determine the proportion of CD34+CD38− cells that could proliferate in liquid culture (in both the presence and absence of growth factors) that were Ph+/BCR-ABL+. Therefore, 118 growth factor–stimulated clones produced by single CD34+CD38− cells were individually genotyped. All but 17 were found to be Ph+ (Table 2). The 17 normal (Philadelphia chromosome–negative [Ph−]) clones were all from patient 1 (whose LTC-IC were predominantly Ph−; Table 1) and, in this case, represented more than 70% of clones obtained. Subsequently, Ph− cells able to proliferate in vitro were also detected in the quiescent (G0) or TPO-resistant subsets of this patient's CD34+ cells when these subsets were incubated with growth factors. In contrast, all of the 25 growth factor–stimulated clones from CD34+CD38− cells from patient 2 were Ph+, even though the LTC-IC from this patient were predominantly Ph− (Table 1) and Ph− clones were detected when quiescent G0 or TPO-resistant CD34+ cells from this patient were cultured with growth factors. Notably, all 62 of the clones that were obtained from CD34+CD38− CML cells in the absence of added growth factors and similarly genotyped were Ph+, confirming that the property of growth factor independence was restricted to members of the neoplastic clone.
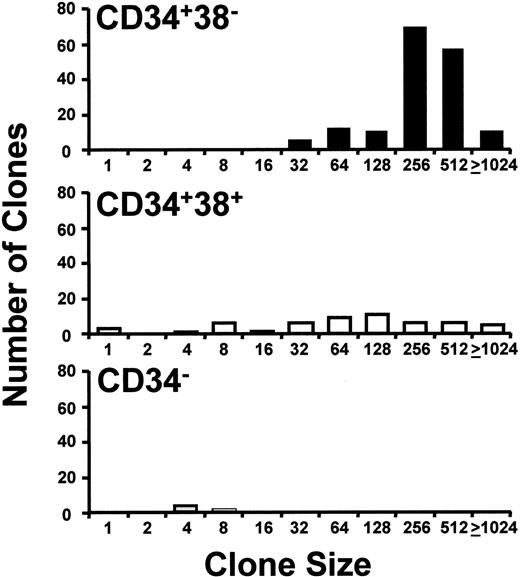
As expected,17 18 both the total frequency of cells that proliferated in single-cell cultures with growth factors (Table 2) and their proliferative potential, as indicated by the sizes of clones (number of cells) generated (Figure 2), were reduced in later compartments (CD34+CD38+and CD34− cells), relative to the CD34+CD38− population. Similarly, the proportion of CD34+CD38+ and CD34−cells that proliferated in the absence of added growth factors was also lower (Table 2).
Distribution of clone sizes generated by different phenotypes of single CML cells cultured in growth factor–supplemented SFM.
CD34+CD38− (▪), CD34+CD38+ (■), and CD34− (░) cells were cultured for 10 days in the presence of FL, SF, IL-3, IL-6, and G-CSF. The numbers on the abscissa indicate the minimum number of cells per clone for clones whose sizes ranged up to the next number shown. Results are from a representative experiment with cells from patient 2.
Distribution of clone sizes generated by different phenotypes of single CML cells cultured in growth factor–supplemented SFM.
CD34+CD38− (▪), CD34+CD38+ (■), and CD34− (░) cells were cultured for 10 days in the presence of FL, SF, IL-3, IL-6, and G-CSF. The numbers on the abscissa indicate the minimum number of cells per clone for clones whose sizes ranged up to the next number shown. Results are from a representative experiment with cells from patient 2.
Quiescent CD34+ CML cells have features of primitive cells
Because we previously identified a rare subset of quiescent CD34+ Ph+/BCR-ABL+ cells in CML patients that included some CFCs and LTC-IC,24 it was of interest to determine whether such cells could proliferate in serum-free single-cell cultures in the presence of added growth factors. Accordingly, quiescent CD34+ cells were isolated either on the basis of their low content of DNA and RNA (CD34+ G0 cells detected by Hst and Py staining, respectively; Figure 1B) or by using CFSE staining to isolate those that had not yet divided after being cultured for 4 days in SFM with TPO as the only added growth factor (ie, CD34+TPO-resistant cells; Figure 1C). Quiescent CD34+ cells isolated with use of either method represented a small fraction of the total CD34+ subset, in agreement with our previous findings.24 For the 9 CML patients studied, the CD34+ content of the samples ranged from 0.5% to 18% (mean ± SE, 6.4% ± 2.4%), but only between 0.8% and 31% (8.7% ± 3.4%) of the CD34+ cells were in G0;the remainder were in G1/S/G2/M. Likewise, the TPO-resistant CD34+ cells comprised only 2% to 6% of the entire CD34+ population and, by the end of the 4 days in culture with TPO, they constituted only 0.4% to 1.9% of those present at that time.
Single G0, G1/S/G2/M, and TPO-resistant cells were then cultured for 10 days in the presence or absence of FL, SL, IL-3, IL-6, and G-CSF. In addition, TPO-responsive cells were cultured under the same conditions in bulk cultures. In the presence of growth factors, the average frequency of proliferating G0, G1/S/G2/M, and TPO-resistant cells from the patients with CML ranged from 28% to 41%. These frequencies were somewhat lower than those measured by using analogous populations isolated from the marrow of normal persons (range of average values for normal cells, 51%-68%; Table 2). Genotyping analyses were again performed on the individual CML clones or bulk cultures. These analyses showed that all the cells derived from the cycling (G1/S/G2/M or TPO-responsive) CML cells were Ph+ (124/124). Most of the clones produced by quiescent (G0 or initially TPO-resistant) CML cells were also Ph+ (181/213), with the small number of normal (Ph−) proliferating cells detected in this population confined to the 2 patients' samples that contained predominantly normal LTC-IC (Table 1).
When the kinetics of growth factor–stimulated clone formation by quiescent cells (G0) was compared with that of cycling (G1/S/G2/M) CD34+ cells, more of the cycling cells (from both CML and normal marrow populations) were found to have completed at least 5 divisions by day 4. However, by day 20, the clones derived from the G0 cells (of either CML or normal marrow origin) were still expanding. In contrast, clones derived from cells that were already cycling initially had reached their maximum size by day 10. Moreover, by day 20, many of the cells in these clones were no longer viable (data not shown). In 2 experiments (with cells from patients 4 and 5), the growth factor–supplemented single-cell cultures were incubated for an additional 15 days (with the addition of fresh medium plus growth factors on days 20 and 27). By this time, about 10% of cultures initiated with G0 cells had produced large clones of more than 103 cells, all of which were Ph+ on cytogenetic analysis. In contrast, only about 1% of the G1/S/G2/M cells from the same samples produced clones of equivalent size. Thus, in the leukemic clone, as in normal marrow, quiescence appears to be a more common feature of CD34+ cells that have greater proliferative potential (measured in response to growth factor stimulation in vitro).
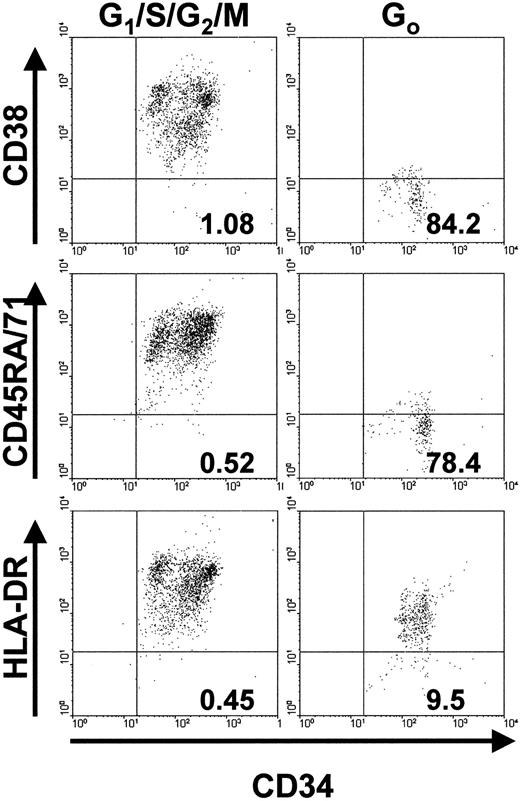
We next examined whether the greater proliferative potential of initially quiescent leukemic cells was reflected in their possession of a surface phenotype characteristic of very primitive hematopoietic cells.26-28 35 Accordingly, CD34+ G0 or G1/S/G2/M cells were isolated with use of FACS and then labeled with antibodies to CD45RA, CD71, HLA-DR, and CD38. Flow cytometric analyses of the final patterns of antigen expression on the cells from the 2 patients' samples examined in this way were similar (Figure3). A much higher proportion of CD34+ G0 cells than of the corresponding CD34+ G1/S/G2/M cells were negative for CD71, CD45RA, and CD38. In contrast, most of the G0cells and the G1/S/G2/M cells were positive for HLA-DR, although the average level of its expression on the G0 cells was approximately 10-fold lower.
Phenotype analyses of cycling (G1/S/G2/M) and quiescent (G0) subsets of CD34+ cell populations.
The numbers in the lower right quadrant of each plot indicate the percentage of PI− events of the corresponding phenotype (ie, positive for CD34 and negative for CD38, CD45RA, CD71, or HLA-DR). Results are from FACS analyses of cells from patient 1.
Phenotype analyses of cycling (G1/S/G2/M) and quiescent (G0) subsets of CD34+ cell populations.
The numbers in the lower right quadrant of each plot indicate the percentage of PI− events of the corresponding phenotype (ie, positive for CD34 and negative for CD38, CD45RA, CD71, or HLA-DR). Results are from FACS analyses of cells from patient 1.
Quiescent CD34+ CML cells are spontaneously activated in vitro
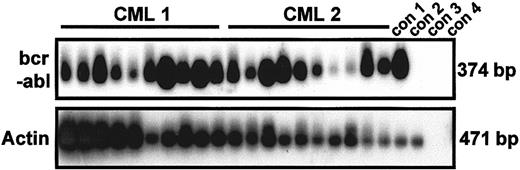
As expected, single-cycling CD34+ cells from all the samples from patients with CML studied produced growth factor–independent clones, whereas no normal cells (either initially quiescent or already cycling) were found to execute even a single division in SFM without growth factors (Table 2). Interestingly, some of the initially quiescent cells in all the CML samples also generated clones within 10 days in the absence of added growth factors, thereby demonstrating the spontaneous reversibility of their quiescent status in vitro. Indeed, for each patient studied, the frequency of growth factor–independent proliferating cells was almost 2-fold higher in the quiescent population than in the corresponding cycling population. The exclusively leukemic (Ph+/BCR-ABL+) origin of all the growth factor–independent clones obtained from CML patients' cells, regardless of their origin (initially quiescent or cycling cells), was again established by using genotyping studies of individual clones (55/55 and 110/110 clones, respectively; Table 2). Figure 4 shows representative RT-PCR data demonstrating the BCR-ABL positivity of 20 of 20 randomly selected clones generated in the absence of growth factors by single G0 cells obtained from patients 1 and 2. Because these samples also produced Ph− clones when growth factors were added, it is clear that culture in the absence of cytokines is selective for Ph+/BCR-ABL+ cells. Interestingly, the size attained by the leukemic clones after 10 days in the absence of added growth factors, though highly variable (up to 50 cells on day 10 and up to 100 cells on day 35), was, on average, consistently smaller than the size attained by cells cultured with growth factors (Figure 5).
Results showing that growth factor–independent G0 cells are exclusively
BCR-ABL+. RT-PCR analysis ofBCR-ABL and actin expression was done on 20 clones derived from single, initially quiescent CD34+ cells (10 each from patients 1 and 2) after culture for 10 days in the absence of added growth factors. PCR products were hybridized by using complementary DNA probes specific for BCR-ABL and actin, respectively. Con 1, 1000 CML cells; con 2, 1000 normal bone marrow cells; con 3, 1000 CML cells without reverse transcriptase; con 4, water.
Results showing that growth factor–independent G0 cells are exclusively
BCR-ABL+. RT-PCR analysis ofBCR-ABL and actin expression was done on 20 clones derived from single, initially quiescent CD34+ cells (10 each from patients 1 and 2) after culture for 10 days in the absence of added growth factors. PCR products were hybridized by using complementary DNA probes specific for BCR-ABL and actin, respectively. Con 1, 1000 CML cells; con 2, 1000 normal bone marrow cells; con 3, 1000 CML cells without reverse transcriptase; con 4, water.
Clone sizes obtained in 10-day SFM cultures of single, initially quiescent (G0) CD34+ cells.
Results from a representative experiment (patient 4) in the presence (▪) or absence (■) of added growth factors are shown. The numbers on the abscissa indicate the minimum number of cells per clone for clones whose sizes range up to the next number shown. The difference in average clone size for clones generated in the presence and absence of added growth factors was significant (P < .001).
Clone sizes obtained in 10-day SFM cultures of single, initially quiescent (G0) CD34+ cells.
Results from a representative experiment (patient 4) in the presence (▪) or absence (■) of added growth factors are shown. The numbers on the abscissa indicate the minimum number of cells per clone for clones whose sizes range up to the next number shown. The difference in average clone size for clones generated in the presence and absence of added growth factors was significant (P < .001).
Quiescent CML cells express BCR-ABL but not IL-3 or G-CSF
Using RT-PCR analyses of single cells, we previously found that more than 80% of all CD34+BCR-ABL+ cells from patients with CML contain transcripts for IL-3 and that more than 50% contain transcripts for G-CSF.17 Because only a rare subset of the CD34+ cells are quiescent,24 we wondered whether such cells also contain transcripts for these growth factor genes. To address this issue, G0 and G1/S/G2/M subpopulations of CD34+cells were isolated from 6 samples from patients with CML (patients 3-5 and 7-9) and RT-PCR analysis was done on separate aliquots of at least 10 cells from each fraction. In addition, single G0 cells from patients 3 to 5 and single G1/S/G2/M cells from patient 3 were analyzed individually. The results for the bulk populations are shown in Figure 6A; those for single cells are shown in Figure 6B and 6C. All bulk populations (consisting of at least 10 cells) but only a proportion of the single G0 cells (8/12, 6/12, and 6/12 from patients 3, 4, and 5, respectively) contained detectable BCR-ABLtranscripts, in spite of the fact that all previously examined CD34+ subpopulations from all 3 of these patients (ie, the growth factor–responsive CD34+CD38− cells, CFCs, and LTC-IC) were exclusively Ph+/BCR-ABL+. None of the CD34+ G0 cells that were BCR-ABLnegative (BCR-ABL−) contained detectable IL-3 or G-CSF transcripts (data not shown).
Results showing that quiescent CML cells rarely contain detectable levels of IL-3 or G-CSF transcripts.
(A) RT-PCR detection of transcripts for BCR-ABL, IL-3, G-CSF, and actin (samples 3-5) or c-ABL (samples 7-9) in extracts of at least 10 initially quiescent (G0) CD34+ cells or cycling (G1/S/G2/M [G1]) cells. (B) Single-cell RT-PCR analyses for IL-3 and G-CSF expression in initially quiescent (G0) CD34+BCR-ABL+ cells isolated from patients 3 to 5 and (C) for IL-3 expression in initially cycling (G1/S/G2/M) CD34+BCR-ABL+ cells isolated from patient 3 as a representative example confirming that more than 80% of such cells express IL-3.
Results showing that quiescent CML cells rarely contain detectable levels of IL-3 or G-CSF transcripts.
(A) RT-PCR detection of transcripts for BCR-ABL, IL-3, G-CSF, and actin (samples 3-5) or c-ABL (samples 7-9) in extracts of at least 10 initially quiescent (G0) CD34+ cells or cycling (G1/S/G2/M [G1]) cells. (B) Single-cell RT-PCR analyses for IL-3 and G-CSF expression in initially quiescent (G0) CD34+BCR-ABL+ cells isolated from patients 3 to 5 and (C) for IL-3 expression in initially cycling (G1/S/G2/M) CD34+BCR-ABL+ cells isolated from patient 3 as a representative example confirming that more than 80% of such cells express IL-3.
More recently, fluorescent in situ hybridization (FISH) was performed in parallel with RT-PCR on quiescent CD34+ cells purified from samples from patients with CML. These studies suggested that cells lacking BCR-ABL transcripts on RT-PCR are alsoBCR-ABL− on FISH; however, such data were not available for the patient samples included in the current study. In the single CD34+ G0 cells that wereBCR-ABL+, IL-3 and G-CSF transcripts were observed only occasionally (Figure 6B). In the examples shown in Figure 6B, 2 of 8, 3 of 6, and 1 of 6 single G0BCR-ABL+ cells were positive for IL-3 and none were positive for G-CSF. Assessment of the aliquots of more than 10 CD34+ G0BCR-ABL+ cells from 4 of 6 of the patients were negative for both IL-3 and G-CSF (Figure 6A). In the third case, IL-3 but not G-CSF was detected in the bulk population. Thus, up to 30% of the quiescent leukemic(BCR-ABL+) CD34+ cells showed evidence of constitutive expression of IL-3 or G-CSF. For the one example in which single CD34+G1/S/G2/M cells were analyzed for IL-3 expression, 9 of 10 individual cells that wereBCR-ABL+ coexpressed transcripts for IL-3 (Figure 6C).
Quiescent CML cells up-regulate expression of IL-3 on entry into the cell cycle
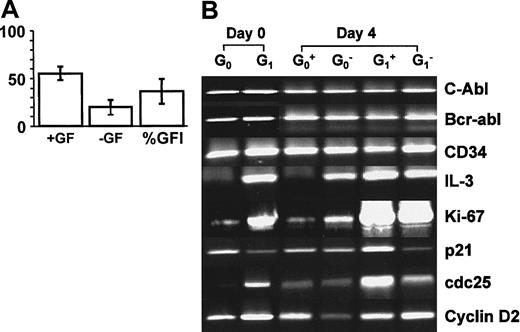
To determine whether cytokine gene expression becomes activated coincident with or before entry into the cell cycle, initially quiescent (G0) and cycling (G1/S/G2/M) CD34+ cells from 3 patients with CML (patients 7-9) were cultured in SFM with and without growth factors for up to 4 days. On days 1 and 4, single-cell cultures of CD34+ G0 cells were examined to determine the rate of entry of the cells into division. Gene-expression analysis using RT-PCR was done on the initially FACS-sorted cells and on viable (annexin V−/PI−) cells cultured in bulk for 4 days. As shown in Figure 7A, between day 0 and day 4, the quiescent cells became activated and divided in both the presence (cloning efficiency, 55% ± 7%) and absence of added growth factors (20% ± 8%). Initially (Figure 7B), the quiescent BCR-ABL+ cells expressed high levels of CD34, p21, and cyclin D2 and low levels of Ki-67 and cdc25 and IL-3 transcripts were almost undetectable. In comparison, initially cycling (G1/S/G2/M) BCR-ABL+cells expressed similar levels of CD34 and cyclin D2, less p21, and higher levels of Ki-67 and cdc25, and IL-3 transcripts were readily detectable. After 4 days of culture of initially quiescent cells in the absence of added growth factors, 20% of the cells had divided at least once and expression of Ki-67 and cdc25 was up-regulated. In addition, IL-3 transcripts were greatly increased (Figure 7B). Interestingly, in the presence of added growth factors, this was not the case: IL-3 expression remained almost undetectable under these conditions. However, for the initially cycling cells, IL-3 expression remained high regardless of whether growth factors were added. Thus, we found that in patients with CML, quiescence is accompanied by a lack of constitutive IL-3 expression in spite of the continued presence ofBCR-ABL and CD34 messenger RNA (mRNA) and that it is spontaneously reversible in the absence of growth factors, in association with an up-regulation of IL-3 expression.
Results showing that quiescent CML cells up-regulate IL-3 expression on entry into the cell cycle.
(A) The histograms show the level of proliferation by single, initially quiescent (G0) CD34+ CML cells cultured for 4 days in SFM in the presence or absence of added growth factors. The results shown are the mean level of proliferation for 288 single G0 cells analyzed from each of 3 similarly studied samples from patients with CML. (B) RT-PCR analyses of viable annexin-V−/PI− populations of G0or G1/S/G2/M CD34+ CML cells before (10 000 cells) and after (5000 cells) they were cultured for 4 days in either the absence (G0−, G1−) or presence (G0+, G1+) of added growth factors.
Results showing that quiescent CML cells up-regulate IL-3 expression on entry into the cell cycle.
(A) The histograms show the level of proliferation by single, initially quiescent (G0) CD34+ CML cells cultured for 4 days in SFM in the presence or absence of added growth factors. The results shown are the mean level of proliferation for 288 single G0 cells analyzed from each of 3 similarly studied samples from patients with CML. (B) RT-PCR analyses of viable annexin-V−/PI− populations of G0or G1/S/G2/M CD34+ CML cells before (10 000 cells) and after (5000 cells) they were cultured for 4 days in either the absence (G0−, G1−) or presence (G0+, G1+) of added growth factors.
Discussion
Human leukemia, like other malignancies, is likely to have a multistep pathogenesis in which several genes that normally regulate the proliferation, survival, and differentiation of normal hematopoietic cells are mutated.1 Autocrine mechanisms contribute to the uncontrolled proliferation of many malignant cell types, including several BCR-ABL+ cell lines.36-39 We previously showed that very primitive (CD34+CD45RA−CD71−)BCR-ABL+ chronic-phase CML cells differ from their normal counterparts in that they proliferate and differentiate in SFM in the absence of added growth factors.16-18 We also found that this activity is mediated by an autocrine mechanism involving IL-3 and G-CSF.17 The identification of a rare subset of leukemic CD34+ cells that are quiescent in vivo24 thus raised the question of whether such initially quiescent leukemic progenitors are capable of factor-independent growth in vitro and, if so, whether IL-3 and G-CSF gene expression occurs in these cells.
In this study, we examined the relation between growth factor independence and progenitor cell-cycle status, genotype, and expression of BCR-ABL, IL-3 and G-CSF. Factor-independent clones were obtained in serum-free single-cell cultures from 9 of 9 CML samples studied. Some of these factor-independent clones contained up to 100 cells and all were derived from BCR-ABL+ cells, even those in samples that, in serum-free single-cell cultures, also contained growth factor–responsive Ph− proliferating cells. Analysis of the subset of quiescent leukemic CD34+cells that were rare but consistently present in patients recently given a diagnosis of CML showed that the subset included cells subsequently able to generate clones in the absence of added growth factors in single-cell cultures. However, in marked contrast to the cycling cells, few of the quiescent CML cells expressed detectable levels of either growth factor transcript, in spite of the presence of the BCR-ABL mRNA (Figure 6). This last finding indicates that BCR-ABL gene expression alone is not sufficient to activate or maintain detectable levels of IL-3 and G-CSF mRNA when primitive leukemic cells become quiescent. However, it is possible that mechanisms associated with the initiation or maintenance of quiescence may influence BCR-ABL transcript levels in primitive CML cells that may, in turn, be dose limiting for some events initiated by the BCR-ABL oncoprotein, such as the activation of IL-3 and G-CSF expression.
It was previously suggested that BCR-ABL transcript levels may be lower in the CD34+ population of CML cells40 and when Ph+ cells terminally differentiate.41 We are confident that the quiescent leukemic cell populations isolated in this study included very primitive CD34+ cells, because we found previously that as few as 105 G0 CML cells isolated by using the same protocol consistently produced detectable numbers of leukemic (BCR-ABL+) progeny within 6 weeks in immunodeficient mice.24 In the current study, we also showed that quiescent CD34+ leukemic cells were markedly enriched in their content of CD38−, CD45RA−, and CD71− cells and expressed lower levels of HLA-DR than did cycling CD34+ leukemic cells. These finding are consistent with a prevalence of very primitive cell types in the quiescent population of CD34+ leukemic cells. In addition, quiescent leukemic CD34+ cells had a greater proliferative potential than did cycling CD34+ leukemic cells under conditions of optimal growth factor stimulation. Interestingly, the frequency of growth factor–independent cells in the quiescent population was also consistently higher. Thus, the observed lack of IL-3 and G-CSF transcripts in initially quiescent CD34+BCR-ABL+ cells was unlikely to be due to their contamination with more mature CML cells, because expression of these genes is greatly reduced in the CD34−compartment.17
The transient dissociation of BCR-ABL gene expression (present) and factor production (absent) in a small subset of freshly isolated primitive quiescent CML cells, as well as the loss of growth factor independence and IL-3 and G-CSF production during the differentiation of CML cells in vivo, raises many new questions about the precise relation between the 2 phenotypes and the underlying molecular mechanisms. The observation that some quiescent CML progenitor cells that did not initially express IL-3 or G-CSF subsequently reentered the cell cycle and proliferated in the absence of added growth factors provided an opportunity to determine whether IL-3 expression would be reactivated and, if so, to assess the conditions and timing of this change relative to the exit of the cells from G0 and other molecular events known to regulate this phase of cell-cycle control. Because G-CSF is known to be up-regulated in normal cells cultured in vitro, it was not included in the current analysis. The results of the time-course experiments confirmed the existence of a temporal relation between IL-3 transcription in primitive CML cells and quiescence compared with proliferation. However, in the absence of fully quantitative RT-PCR analyses, it is impossible to say whether a threshold level of BCR-ABL mRNA expression is required before IL-3 or G-CSF mRNA is induced and that it may not be reached by most quiescent leukemic cells until they enter the cell cycle.
Our findings confirm that the quiescent status of some CML progenitors is reversible and show that this can occur even in the absence of exogenous growth factors. However, it is not yet clear what causes these cells to enter a quiescent state in vivo. Some types of primitive CML CFCs are known to be unresponsive to the inhibitory activities that certain chemokines exert on their normal counterparts.42,43 However, these primitive CFCs also show a normal sensitivity to transforming growth factor β (TGF-β), which can force their entry into a reversible noncycling state in vitro.44,45 We therefore speculate that TGF-β (or other, still unknown factors that may mimic the effects of TFG-β) can partly regulate the proliferative activity of primitive CML cells in vivo by means of the same mechanisms shown to control their normal counterparts both in vitro43 and in vivo.46 Because quiescent (G0) cells are known to be resistant to most chemotherapeutic agents commonly used to treat CML, an improved understanding of the mechanisms that regulate the rate of primitive leukemic cell progression through G1 is likely to have clinical relevance. Similarly, it will be of interest to investigate whether interferon α or STI 57147 can affect the autocrine growth of primitive leukemic cells. The availability of the in vitro system described here, as well as in vivo xenotransplantation models of human CML48-50 and improved animal models for this disease,51-53 should help to answer these questions.
Acknowledgments
We thank our colleagues in the Division of Hematology of the University of British Columbia, the Stem Cell Assay Service of the BC Cancer Agency, and the Department of Haematology, Glasgow Royal Infirmary, for assistance in procuring and processing specimens; Gloria Shaw and Elaine Allan for cytogenetic and FISH analyses; Giovanna Cameron, Gayle Thornbury, and Charlie Pearson for FACS operation; Dianne Reid, Margaret Hale, and Linda Richmond for technical assistance, and Tara Palmater for typing the manuscript. We also thank K. Humphries, R. Kay, and P. Lansdorp (Terry Fox Laboratory); J. Griffin (Dana Farber Cancer Institute, Boston, MA); and Cangene, Genentech, Immunex, Novartis, and Stem Cell for generous gifts of reagents.
Supported by grants from the National Cancer Institute of Canada (NCIC) with funds from the Canadian Cancer Society and the Terry Fox Run. T.L.H. and H.G.J. are funded by the United Kingdom Leukaemia Research Fund and S.G. by the Glasgow Royal Infirmary Endowment Fund. C.J.E. is a Terry Fox Cancer Research Scientist of the NCIC.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tessa Holyoake, ATMU, Department of Medicine, Royal Infirmary, 10 Alexandra Parade, Glasgow G31 2ER, United Kingdom; e-mail: tlh1g@clinmed.gla.ac.uk.



![Fig. 6. Results showing that quiescent CML cells rarely contain detectable levels of IL-3 or G-CSF transcripts. / (A) RT-PCR detection of transcripts for BCR-ABL, IL-3, G-CSF, and actin (samples 3-5) or c-ABL (samples 7-9) in extracts of at least 10 initially quiescent (G0) CD34+ cells or cycling (G1/S/G2/M [G1]) cells. (B) Single-cell RT-PCR analyses for IL-3 and G-CSF expression in initially quiescent (G0) CD34+BCR-ABL+ cells isolated from patients 3 to 5 and (C) for IL-3 expression in initially cycling (G1/S/G2/M) CD34+BCR-ABL+ cells isolated from patient 3 as a representative example confirming that more than 80% of such cells express IL-3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/3/10.1182_blood.v97.3.720/6/m_h80310639006.jpeg?Expires=1765109610&Signature=g8oIcimKaa~0H8e7hMGtOEXwM-djSozB1~Q~dlSLnwIT9fcWjZmNA~lpxyAFjiqAXqmmUjlVrB54FWXzp6Z635T1DQ4QMUJkMa4Tn6ofnAigOqjHy9IMA68WLJteJL1plYP8E1pXX0olURr-s-emgugPa2CS6ZIiqhHgLUo42jITHJg7BJHouMYJx-qRA1Z9lslYcKOmRsJzIuklhgTwoeeki7uPP60HQrO~5yuxbuNfL2TO6GC7BpNVPPPUbUBXfzdbSvewm19zbHQJbmPaAinLTKE5qxBz1xTFzfSs3UdI9WI~630DrfQaxPvn05eGftFf6hTUSsu0mHwpdoObRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal