IN HUMANS AND OTHER mammals, decreased oxygen tension triggers specific and tightly regulated cellular, vascular, and erythropoietic responses. An association between polycythemia and people living at high altitudes was first reported in 1863.1 Erythropoietin (Epo), a 34.4-kD glycoprotein hormone, was subsequently identified as the humoral regulator of red blood cell production. Decreased tissue oxygen tension modulates Epo levels by increasing expression of the Epo gene. Since the cloning of the Epo gene in 1985,2 3 considerable progress has been made in understanding the molecular mechanisms by which the Epo gene is regulated by environmental, tissue-specific, and developmental cues.
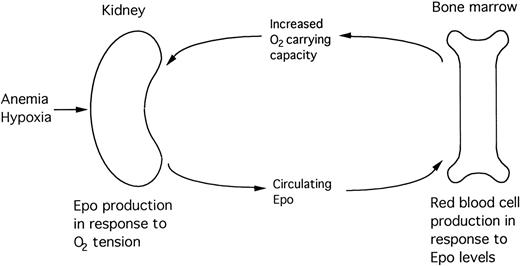
Erythropoiesis, which normally proceeds at a low basal level to replace aged red blood cells, is highly induced by loss of red blood cells, decreased ambient oxygen tension, increased oxygen affinity for hemoglobin, and other stimuli that decrease delivery of oxygen to the tissues. In states of severe hypoxia, production of Epo is increased up to 1,000-fold. The secreted hormone circulates in the blood and binds to receptors expressed specifically on erythroid progenitor cells, thereby promoting the viability, proliferation, and terminal differentiation of erythroid precursors, resulting in an increase in red blood cell mass. The oxygen carrying capacity of the blood is thus enhanced, increasing tissue oxygen tension, thereby completing the negative feedback loop (Fig 1).4 5
Research on the regulation of the Epo gene has shown a general system of oxygen-sensing, signaling, and transcriptional regulation of a broad range of physiologically relevant genes, including those encoding angiogenic growth factors, glucose transporters, and enzymes involved in adaptation to hypoxia.
TISSUE SPECIFICITY
Temporal and tissue-specific signals limit expression of the Epogene primarily to specific cells in the fetal liver and the adult kidney.
Localization of Epo production to the kidneys was first demonstrated by Jacobsen et al,6 who showed that, after bilateral nephrectomy, rats and rabbits do not respond to hemorrhage with an appropriate increase in plasma Epo levels. Erslev et al7have proposed that the peritubular region of the renal cortex is the ideal location for Epo production. Oxygen consumption in the kidney is determined largely by sodium reabsorption, which in turn depends on the filtered load. Because the glomerular filtration rate is roughly proportional to renal blood flow, renal oxygen consumption is linked to renal blood flow. Therefore, oxygen tension at the site of the Epo-producing cells is relatively independent of changes in renal blood flow. At high hematocrit levels, viscosity increases to the point that blood flow to tissues is compromised. If the primary site of Epo production were in an organ other than the kidney, the resultant decrease in tissue oxygen tension would lead to a vicious cycle of increasing erythropoiesis causing worsening hypoxia.
When kidney cells were separated into glomerular and tubular fractions,Epo mRNA was found only in the tubular fraction that included the peritubular interstitium.8 Consistent with these findings, in situ hybridization studies with 35S-labeledEpo RNA9,10 or DNA11 probes on kidney tissue from anemic mice showed Epo mRNA in peritubular interstitial cells. The number of these cells expressing EpomRNA increased with a decreasing hematocrit level.12 In contrast to these studies, other investigators have demonstrated Epo production by renal tubular cells using in situ hybridization forEpo mRNA,13 immunohistochemistry with Epo-specific antibodies,13 and detection of β-galactosidase in transgenic mice bearing a 7-kb fragment of the Epo gene linked to the lacZ gene.14 Human renal tumor cells of tubular origin can express Epo.15Immunohistochemistry using Epo-specific antibodies is confounded by the reabsorption of circulating Epo by renal tubular cells.
Two studies have demonstrated the colocalization of Epo-producing cells and immunoreactivity to 5′-ectonucleotidase, suggesting that the cells are likely to be fibroblasts.16,17 Maxwell et al17 prepared transgenic mice using regulatory sequences from the mouse Epo gene flanking the SV40 T antigen as a marker gene. In one line of these transgenic mice, the transgene was fortuitously integrated into the endogenous Epo locus by homologous recombination. This provided a model in which the marker gene, inserted into the Epo locus, is subject to all of the same tissue-specific controls as the endogenous Epo gene. Consequently, expression of the SV40 T antigen permitted immunohistochemical identification of the Epo-producing cells to be the fibroblast-like type I interstitial cells.17 When all of the somewhat contradictory studies cited above are carefully weighed, the bulk of convincing evidence favors a peritubular interstitial cell as the primary site of regulated Epo production in the kidney.
Before birth, Epo is primarily produced in the liver. The primary site of Epo production switches from liver to kidney shortly after birth,18,19 but the signals governing this change are poorly understood. In the liver, an oxygen gradient is established as oxygen-rich blood from the portal triads becomes depleted of oxygen as it flows towards the central vein. Consequently, in transgenic mice, both the Epo transgene and the endogenous Epo gene are preferentially expressed near the central vein where oxygen tension is lowest.10
Two Epo-producing cell types were identified in the liver: hepatocytes and a nonparenchymal cell type.10,20 The identity of the nonparenchymal cells was established using transgenic mice bearing the SV40 T antigen homologously recombined into the Epo locus. In these mice, SV40 T antigen expression was observed in a subset of nonparenchymal cells, identified as Ito cells, as well as in a subset of hepatocytes.21
Highly sensitive assays have shown low levels of Epo mRNA in the kidneys and livers of unstimulated mice and rats,9,22-24 consistent with low basal levels of Epo in serum. Low levels of Epo expression have also been detected in the lung, spleen, brain, and testis of rats.23 24
Expression and production of both Epo and Epo-receptors has been demonstrated in the brain.23,25-27 Oxygen-regulated expression of Epo has been observed in astrocytes both in vitro in cultured astrocytes25,27 and in vivo.23,27 The presence of Epo receptors, the inability of Epo to cross the blood-brain barrier, and the regulated expression of Epo in the brain have led researchers to propose a paracrine function for Epo in neural tissue. Recent evidence demonstrates that Epo can protect neurons from ischemic damage in vivo.28
The discovery that both Epo mRNA and Epo protein are expressed in erythroid progenitors29 30 has raised the intriguing possibility that tonic low-level erythropoiesis may be supported by autocrine stimulation, whereas circulating (hormonal) Epo provides a more robust stimulus to erythropoiesis during hypoxic stress.
For decades, a tissue culture model eluded investigators studying the regulation of Epo. Some cells, such as rat kidney mesangial cells,31 the renal cell line RC-1,32 and hepatic carcinomas,33 produced Epo at very low levels with minimal induction by hypoxia. Cell lines that produce significant amounts of Epo in a regulated fashion were discovered by screening a range of renal and hepatic cells in culture.34 Two human hepatoma cell lines, Hep3B and HepG2, were shown to produce significant amounts of Epo constitutively, with marked induction by hypoxia. The magnitude and time course of the induction of Epo mRNA paralleled Epo protein production. The discovery of a tissue culture model demonstrated that individual cells contain the apparatus necessary for oxygen sensing and the consequent regulation of gene expression. Hep3B and HepG2 cells have proved to be an invaluable tool in exploring the molecular basis of Epo gene regulation.
REGULATION OF Epo GENE EXPRESSION
In addition to tissue-specific and developmental signals, Epogene expression is modulated by a number of physiological and pharmacological agents (Table 1). Regulation of Epo by hypoxia and other stimuli occurs at the mRNA level. In the kidneys of mice made anemic by blood loss,Epo mRNA was increased approximately 200-fold over the level in the kidneys of normal control mice.35Epo mRNA was induced in the liver as well, but at a lower level of expression. The increase in Epo mRNA reached a maximum at 4 to 8 hours after induction. The magnitude of induction was proportional to the degree of anemia. Similarly, injection of cobalt chloride into rats inducedEpo expression in the kidney and, to a lesser degree, in the liver.36 The time course and level of induction of EpomRNA paralleled induction of Epo in serum measured by radioimmunoassay.36
Agents That Affect Epo Gene Expression
| Induction in vivo and in vitro |
| Hypoxia |
| Transition metals: Co2+, Ni2+, Mn2+ 118,192 |
| Iron chelators: desferrioxamine120 |
| Abrogation of induction in Hep3B and HepG2 cells |
| Carbon monoxide91,118 193 |
| Nitric oxide, nitroprusside91 |
| Hydrogen peroxide86 194 |
| Inflammatory cytokines: TNF-α, IL-1181 182 |
| Phorbol esters: Forskolin193 195-197 |
| Protein synthesis inhibitors: cycloheximide83 118 |
| Induction in vivo and in vitro |
| Hypoxia |
| Transition metals: Co2+, Ni2+, Mn2+ 118,192 |
| Iron chelators: desferrioxamine120 |
| Abrogation of induction in Hep3B and HepG2 cells |
| Carbon monoxide91,118 193 |
| Nitric oxide, nitroprusside91 |
| Hydrogen peroxide86 194 |
| Inflammatory cytokines: TNF-α, IL-1181 182 |
| Phorbol esters: Forskolin193 195-197 |
| Protein synthesis inhibitors: cycloheximide83 118 |
This list is not comprehensive. Other agents have been shown either to activate or suppress HIF-1 and therefore would be expected to affect Epo gene expression.
Nuclear run-on assays using nuclei prepared from the kidneys of rats that had been made hypoxic or treated with cobalt showed an increase in transcription of the Epo gene.8 Similar results were obtained with Hep3B cells.37 Transcriptionally active nuclear extracts from hypoxic Hep3B cells were compared with extracts from cells cultured in normoxia.38 The hypoxic nuclear extracts supported a higher rate of Epo transcription in vitro, demonstrating the presence of hypoxically inducible trans-acting factors capable of interacting with cis-acting sequences from theEpo gene.
REGULATORY DOMAINS IN THE Epo GENE
Comparison of the human and murine Epo genes provided clues to the location of key regulatory domains in the Epogene.39-41 Three noncoding segments of the Epogene are highly conserved between human and mouse sequences: the promoter, the first intron, and a 120-bp region 100 bp 3′ to the polyadenylation site.
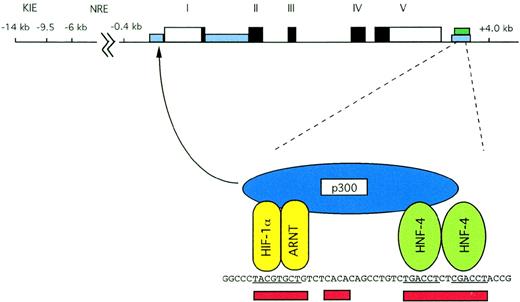
Both transient transfection experiments and studies with transgenic mice have been used to identify functionally important cis-acting elements. Experiments with transgenic mice mapped broad regions of theEpo gene that regulate Epo expression in response to tissue-specific and developmental signals. Transient transfections of cultured cells have been used to characterize cis-acting sequences that are critical for the response to hypoxia. Conserved sequences both 5′ and 3′ of the Epo gene proved to be important for regulation of the Epo gene, but deletion of the conserved sequences in the first intron did not influence hypoxic induction.42 Regulatory elements in the Epo gene are portrayed in Fig 2.
Structure of the human Epo gene. Exons are indicated by solid black boxes; 5′ and 3′ untranslated regions are indicated by open rectangles. Areas of homology between human and murine noncoding sequences are shown with blue rectangles, and the region of liver specific DNase I hypersensitivity is shown with a green rectangle. The 3′ enhancer is expanded for greater detail. Sites that are functionally critical for hypoxic induction are underscored in red. Binding of HIF-1, HNF-4, and p300 is illustrated. As indicated by the arrow, p300 is capable of interacting with the basal transcriptional machinery in the promoter.
Structure of the human Epo gene. Exons are indicated by solid black boxes; 5′ and 3′ untranslated regions are indicated by open rectangles. Areas of homology between human and murine noncoding sequences are shown with blue rectangles, and the region of liver specific DNase I hypersensitivity is shown with a green rectangle. The 3′ enhancer is expanded for greater detail. Sites that are functionally critical for hypoxic induction are underscored in red. Binding of HIF-1, HNF-4, and p300 is illustrated. As indicated by the arrow, p300 is capable of interacting with the basal transcriptional machinery in the promoter.
Transgenic experiments.
Transgenic mice were initially produced containing a 4-kb fragment that included the human Epo gene, 400 bp of 5′-flanking sequence, and 700 bp of 3′-flanking sequence. The transgene was widely expressed, causing the mice to become polycythemic, indicating that additional cis-acting sequences, required for tissue-specific and developmental regulation, lie outside this construct. Nevertheless, in the liver, the transgene was upregulated by anemia and cobalt chloride. Thus, cis-acting sequences capable of mediating oxygen-regulated gene expression in liver cells lie between 400 bp 5′ and 700 bp 3′ of the Epo gene.43
Tissue-specific regulatory domains were mapped in subsequent transgenic experiments by use of constructs with varying lengths of Epoupstream flanking sequence. Promiscuous expression of the Epogene, seen in transgenic constructs with 300 bp44 or 400 bp43 of 5′ flanking sequence, was extinguished in constructs containing 6 kb of 5′ flanking sequence.45In constructs containing 9.5 kb or less of 5′ flanking sequence, inducible expression of the Epo transgene was observed in the liver but not in the kidney.44 However, physiological expression of the Epo transgene with inducible expression in the kidney was seen in transgenic mice containing 14 kb of 5′ flanking sequence.46 Thus, transgenic experiments indicate that a repressive element(s) exists between 0.4 and 6 kb upstream of the Epo gene, and a kidney-specific inducible element(s) exists between 9.5 and 14 kb upstream of the Epo gene.
Epo 3′ enhancer.
A liver-specific DNase I hypersensitivity site was discovered in the 3′ flanking sequence of an Epo transgene.47Analysis of this region of the Epo gene by transient transfections of reporter constructs led to the identification of a hypoxically inducible enhancer.47-50 In both the mouse and human Epo genes, this enhancer lies in a highly conserved region 120 bp 3′ to the polyadenylation site. As is typical of eukaryotic transcriptional enhancers, activity was independent of orientation and distance from the promoter. The enhancer demonstrated the same stimulus specificity as the Epo gene with responses to hypoxia, cobaltous chloride, and iron chelation, but not to cyanide and 2-deoxyglucose.
Detailed characterization of the Epo 3′ enhancer defined 3 sites that are critical for regulation by hypoxia.50-52On the 5′ side, the sequence CACGTGCT was the first response element to be characterized for the transcription factor, hypoxia inducible factor-1 (HIF-1).51 Binding of HIF-1 to this site is induced by hypoxia, and an intact HIF-1 binding site is necessary for hypoxically inducible function of the Epo enhancer. In addition to the hypoxically inducible DNA-binding activity, HIF-1, this site also binds another complex constitutively. The transcription factors ATF-1 and CREB-1 have been shown to be involved in this complex in vitro, but it is not clear whether these factors play a functional role in the Epo enhancer.53 Further details concerning the function and activation of HIF-1 are discussed below.
A second site, 7 bp 3′ to the HIF-1 site, has the sequence CACA in the human Epo gene. No proteins are known to bind to this site, but mutation of this site abrogates hypoxia inducible activity of the enhancer. A similar sequence has been found adjacent to HIF-1 sites in other genes.54 These first 2 sites (HIF-1 and CACA) require the presence of a third site for hypoxically inducible transcription, unless the enhancer is directly upstream from the promoter.52
The sequence of the third site in the Epo enhancer is a direct repeat of 2 steroid hormone receptor half sites separated by 2 bp, termed a DR-2 site.50 Mutations of this site ablate or markedly inhibit hypoxic induction.50-52 Nuclear proteins from a broad range of cell types bind strongly to this site, as demonstrated by both electrophoretic mobility shift assays and DNase I footprinting experiments. However, binding of proteins to this site is not oxygen-dependent either in vivo or in vitro.50-52,55 In some non–Epo-producing cells, a complex does not form at this site in vivo.55 Hormones whose biological actions depend on binding to nuclear receptors had no effect on the hypoxic induction of a reporter gene containing the Epo promoter and Epoenhancer.50 These results suggested that the DR-2 site might bind an orphan nuclear receptor, a DNA-binding protein that shares structural homology with hormone binding nuclear receptors but lacks a known ligand. Screening a variety of in vitro-translated orphan receptors showed that HNF-4α bound specifically to this site.56 Hypoxic induction of Epo is abolished in Hep3B cells expressing a dominant negative mutant of HNF-4. HNF-4 is expressed in the renal cortex and liver, like Epo, as well as in the intestine. Thus, HNF-4 may contribute to the tissue specificity of Epo gene expression.
Promoter.
The Epo promoter does not have consensus TATA or CAAT elements in either the mouse or human genes. Comparison of the 5′ flanking sequences of the human and murine Epo genes shows 73% overall sequence identity and 8 areas of even higher homology.39 40
The Epo promoter contributes to the hypoxic inducibility of theEpo gene.57 After deletion of the 3′ enhancer, expression of a stably transfected marked Epo gene was induced approximately 10-fold in response to hypoxia.42The minimal promoter acts synergistically with the 3′ enhancer to confer a 40-fold induction in response to hypoxia.50
The minimal Epo promoter capable of induction by hypoxia encompasses 117 bp 5′ to the transcription initiation site.50 A segment of 17 bp (−61 to −45) is responsible for this upregulation by hypoxia.58 There is no HIF-1 consensus sequence at this site. Using computer homology matching, a HIF-1 site was identified at position −180 in theEpo 5′ flanking sequence.59 However, reporter gene experiments suggest that this site is not a functional hypoxia response element.50 GATA sites60,61 and a ribonucleoprotein binding site62 have also been described, but the role of these proteins in the response to hypoxia is not proven. Addition of antisense oligonucleotides to GATA elements increased Epo gene expression, whereas the addition of antisense oligonucleotides to CACCC elements decreased Epo gene expression, indicating that the Epo promoter is regulated negatively by GATA sites and positively by CACCC sites.60L-NMMA, a nitric oxide synthase inhibitor, was recently demonstrated to increase binding of GATA factors in the Epo promoter and decrease Epo expression.63
Methylation of CpG sites in the Epo promoter varies between Epo-producing and non–Epo-producing cells. By inhibiting the formation of DNA-binding complexes and by the binding of inhibitory methyl-CpG binding proteins, methylation may contribute to tissue-specific activity of the Epo promoter.64
mRNA STABILITY
Enhanced transcription accounts for most, but probably not all, of the hypoxic induction of the Epo gene. In Hep3B cells, nuclear run-on experiments showed about a 10-fold increase in transcription ofEpo mRNA during exposure to 1% O2 in the setting of a 50- to 100-fold increase in the steady-state level of EpomRNA.37 In 2 other hypoxically regulated genes, tyrosine hydroxylase (TH)65,66 and VEGF,67,68approximately 50% of the enhanced gene expression in response to hypoxia is due to increased mRNA stability. For both genes, specific mRNA binding proteins have been demonstrated in cytosolic extracts of hypoxic cells.66 68
In comparison to TH and VEGF, less is known about posttranscriptional regulation of the Epo gene. Inhibitors of transcription markedly prolong the half-life of Epo mRNA, thus making actinomycin chase experiments uninterpretable.37 Two proteins, 70 and 135 kD, which have been designated EpomRNA-binding protein (ERBP), bind to a 120-bp pyrimidine-rich region in the 3′ UTR of Epo mRNA.69 This interaction does not seem to be regulated by oxygen tension,69 but binding is subject to redox control.70 Heat shock protein 70 participates in a complex with ERBP and Epo mRNA.71 Deletion of the ERBP binding site prolongs the half-life of Epo mRNA and eliminates hypoxically induced stabilization.72 Deletion of a 50-bp segment lying 70 bp downstream of the binding site for these proteins causes a 7-fold increase in the half-life of a transfected markedEpo gene.42 Epo, VEGF, and TH mRNA can cross-compete for binding in mobility shift assays, suggesting common features to the regulation of mRNA stability in these 3 genes.67 73
HIF-1
The transcription factor HIF-1 mediates hypoxically inducible transcription of oxygen-regulated genes. The HIF-1 site in the Epo3′ enhancer is the primary element in the Epo gene that mediates the transcriptional response to hypoxia. The time course of HIF-1 activation mimics the induction of the Epogene.74 Hypoxia, cobalt, and DFO, stimuli that triggerEpo gene expression, also activate HIF-1 DNA binding. The HIF-1 site in the Epo 3′ enhancer was the first hypoxia response element (HRE) to be identified and was used for the affinity purification of HIF-1.75 HIF-1 is a heterodimer composed of 120-kD α and 91- to 94-kD β subunits, both of which are basic helix-loop-helix (bHLH) proteins in the PAS (Per-AHR-ARNT-SIM) family of transcription factors.76 HIF-1β is the previously cloned and characterized aryl hydrocarbon receptor nuclear translocator (ARNT), which forms a heterodimer with the aryl hydrocarbon receptor (AHR), mediating regulation of genes involved in the transcriptional response to xenobiotics and oxidant stress.77 78
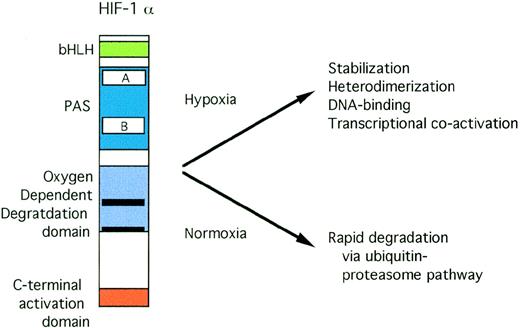
Both HIF-1α79 and ARNT80 have basic domains that are critical for DNA binding and PAS domains that are crucial for dimerization. The domain structure of HIF-1α is shown in Fig 3. Interaction between HIF-1α and ARNT requires both bHLH and PAS domains.81 Deletion of both the basic domain and the carboxy-terminal activation domains results in a dominant negative form of HIF-1α.79 Transactivation by the HIF-1 heterodimer requires the N-terminal DNA-binding and heterodimerization domains of ARNT, but not its C-terminal activation domain.82
Structure of HIF-1. Open rectangles represent the PAS A and B domains. Solid black rectangles within the oxygen-dependent degradation domain indicate the location of PEST sequences.
Structure of HIF-1. Open rectangles represent the PAS A and B domains. Solid black rectangles within the oxygen-dependent degradation domain indicate the location of PEST sequences.
HIF-1 activity has been demonstrated in a wide variety of cells by both electrophoretic mobility shift assays83 and transfections of a reporter gene containing HIF-1 sites.84 Steady-state levels of HIF-1α and ARNT mRNA are not significantly affected by oxygen tension.81,85,86 At the protein level, ARNT abundance is also not dependent on oxygen tension. In contrast, the HIF-1α subunit is only detectable in cells treated with hypoxia or stimuli that mimic hypoxia, such as cobalt or iron chelators. In normoxic cells or in cells pretreated with H2O2before deoxygenation, HIF-1α protein is barely detectable.86 Thus, HIF-1 activation correlates with the oxygen-dependent accumulation of HIF-1α protein. Under normoxic conditions, HIF-1α is rapidly degraded by the ubiquitin-proteasome pathway.87,88 Specific inhibitors of this pathway markedly increase HIF-1α abundance. Hypoxia and iron chelation dramatically increase the half-life of HIF-1α, permitting the formation of functionally active HIF-1α/ARNT heterodimers. A domain from the central region of HIF-1α was identified that is critical for oxygen-regulated protein degradation.88 This region spans amino acids 401-603 with further deletions causing a diminution of the magnitude of hypoxic inducibility.88 Sequences from within this region mediate hypoxically inducible transactivation.89,90 Deletion of this oxygen-dependent degradation (ODD) domain resulted in stabilization of HIF-1α and constitutive HIF-1 DNA-binding activity independent of oxygen tension.88 Furthermore, the ODD domain is transportable, ie, it is capable of conferring oxygen-dependent degradation on a heterologous protein.88 Both carbon monoxide and nitric oxide donors suppress degradation of HIF-1α via the ODD domain.91
A second domain at the C-terminus of the HIF-1α protein (amino acids 775-826) was also shown to mediate hypoxically inducible trans-activation.82,89 90 This C-terminal region is not associated with changes in levels of HIF-1α protein. Therefore, hypoxia must activate this C-terminal domain by some form of posttranslational modification such as phosphorylation.
HIF-1α has been shown to interact with the transcriptional coactivators, p300 and CBP,92 highly homologous proteins that are functionally and immunologically indistinguishable.93,94 Expression of the adenovirus protein, E1A, which binds to p300/CBP blocking functional activity, prevents hypoxic induction of the genes encoding Epo, VEGF,92 and LDH-A.95 Functional activity of the C-terminal transactivation domain of HIF-1α requires interactions with the CH1 domain of p300/CBP.96,97 p35srj, an alternatively spliced form of MRG-1,98 also binds to the CH1 domain of p300/CBP, competing with HIF-1α for binding to p300/CBP.97 Induction of p35srj by hypoxia may contribute to a negative feedback on HIF-1 activation97 and perhaps explains the prompt decrease in Epo expression after the peak mRNA levels 4 to 6 hours after hypoxic induction.24 p300 and CBP are large proteins with several domains for interacting with multiple proteins. P300/CBP has been shown to interact with HNF-499and other nuclear hormone receptors,100,101CREB,102 TATA binding protein,103 and TFIIB.104 By simultaneously interacting with HIF-1, adjacent transcription factors, and the basal transcriptional machinery, p300/CBP likely acts as a scaffold for the construction of a transcriptionally active, hypoxically inducible complex. Formation of such a complex may explain the requirement of an HNF-4 site, adjacent to the HIF-1 site, for hypoxically inducible activity of the Epo3′ enhancer.95
A plethora of additional members of the bHLH-PAS family have been cloned recently, including ARNT2, HIF-2α, MOP3, MOP4, and CLOCK.105-109 HIF-2α (also known as EPAS-1, MOP2, HRF, and HLF) has a broad tissue distribution and, like HIF-1α, accumulates only under hypoxic conditions.110 MOP3-HIF-1α and MOP3-HIF-2α heterodimers are capable of binding an HIF consensus site and activating transcription in response to hypoxia.111 ARNT is capable of forming homodimers as well as heterodimers with HIF-1α, AHR, and SIM. The requirement of ARNT for the responses to both hypoxia and aryl hydrocarbons can lead to a functional interference between the 2 pathways.81
Insulin and insulin-like growth factor I (IGF-1) activate HIF-1 DNA-binding activity, HIF-1–mediated transcriptinal activation of a reporter gene,112 and expression of several oxygen regulated genes, including Epo.113 The signalling pathways for hypoxia, insulin, and IGF-1 appear to converge, all leading to stabilization of HIF-1α.
Activition of HIF-1 is modulated by a complex set of mechanisms likely to include not only protein stability, but also phosphorylation,114,115 redox chemistry,86,87,116 and nuclear localization.96Subsequent to binding of HIF-1 to its cognate cis-acting sequence, interaction with adjacent transcription factors and coactivator proteins are necessary for hypoxic induction of transcription.
MECHANISM OF OXYGEN SENSING AND SIGNAL TRANSDUCTION
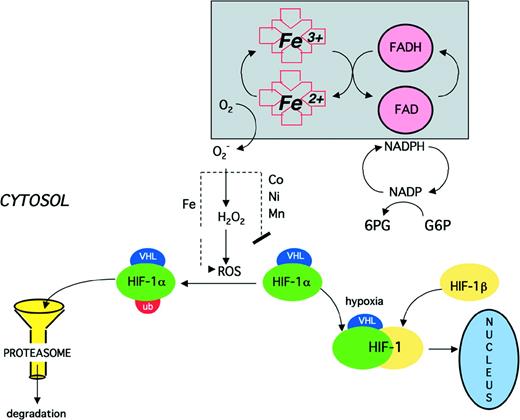
Despite a great deal of attention and experimental data, the precise nature of the mammalian oxygen sensor remains elusive. However, the synthesis of evidence obtained to date provides an outline of the likely characteristics of the oxygen sensor that signals the activation of gene expression via HIF-1. A plausible model is shown in Fig 4.
Proposed model of oxygen sensing and signaling. In oxygenated cells, a flavo-heme protein functions as an NADPH oxidase, transferring electrons through the flavin (FAD) and heme to molecular oxygen, generating superoxide (O2−), which, in the presence of iron, is converted to hydroxyl radical (OH·) and other reactive oxygen species (ROS). As a result, HIF-1 is oxidatively modified so that it is recognized by the proteasome and rapidly degraded. Cobalt (Co2+) as well as other transition metals (Ni2+ and Mn2+) may block the iron-dependent degradation of HIF-1. At low oxygen tension, as well as in the presence of an iron chelator or one of the above-mentioned transition metals, HIF-1 is stable and can form a heterodimer with constitutively expressed HIF-1β, thereby activating HIF-1, which translocates to the nucleus and binds to response elements in hypoxia inducible genes.
Proposed model of oxygen sensing and signaling. In oxygenated cells, a flavo-heme protein functions as an NADPH oxidase, transferring electrons through the flavin (FAD) and heme to molecular oxygen, generating superoxide (O2−), which, in the presence of iron, is converted to hydroxyl radical (OH·) and other reactive oxygen species (ROS). As a result, HIF-1 is oxidatively modified so that it is recognized by the proteasome and rapidly degraded. Cobalt (Co2+) as well as other transition metals (Ni2+ and Mn2+) may block the iron-dependent degradation of HIF-1. At low oxygen tension, as well as in the presence of an iron chelator or one of the above-mentioned transition metals, HIF-1 is stable and can form a heterodimer with constitutively expressed HIF-1β, thereby activating HIF-1, which translocates to the nucleus and binds to response elements in hypoxia inducible genes.
Oxygen sensors in bacteria and yeast have been well characterized, and in these systems heme proteins play a central role.117Similarly, several lines of evidence indicate that a heme protein is involved in mammalian oxygen sensing.118 CO binds specifically and noncovalently to heme proteins, causing the heme moiety to be maintained in an “oxy” conformation. CO blocks the activation of HIF-1 by hypoxia and the hypoxically regulated expression of VEGF, PDGF, ET-1, and PEPCK.117 CO binds with a lower affinity than oxygen to the putative heme protein sensor, with a Haldane coefficient of approximately 0.5.91 The transition metals Co2+, Ni2+, and Mn2+ mimic the effect of hypoxia on Epo, HIF-1, and other oxygen-regulated genes.118 119 A possible mechanism for the action of these transition metals is that they can substitute for iron in heme proteins, locking the heme moiety in the “deoxy” conformation.
Many experiments have indicated that a decrease in levels of oxygen free radicals after hypoxic stimulus leads to the accumulation of HIF-1α and the activation of HIF-1. Addition of H2O2 or agents that increase intracellular peroxide concentration block the induction of Epo and the accumulation of HIF-1α.86 Desferrioxamine (DFO), an iron chelator, mimics the effect of hypoxia on oxygen-regulated genes and HIF-1 activity.120 Intracellular iron probably functions as a Fenton reagent, catalyzing the formation of reactive oxygen species. Therefore, iron chelation, like hypoxia, is likely to effect a reduction in intracellular levels of hydroxyl radical and singlet oxygen. A substantial number of enzymes are inactivated by oxygen-dependent and iron-dependent oxidation at specific residues, rendering them targets for proteolytic degradation.121 122This modification is inhibited by Mn2+ and also by Co2+ and Ni2+ (W. Willmore, R. Levine, and H.F. Bunn, unpublished observations). If this degradative pathway applies to HIF-1α (as shown in Fig 4), it would explain the stabilization of HIF-1 by these 3 transition metals.
Spectrophotometric evidence points to the involvement of a cytochromeb-like protein in oxygen sensing.123-126Furthermore, diphenyl iodonium (DPI), which inhibits NAD(P)H oxidases and other flavoproteins, impairs oxygen sensing.127,128 DPI also inhibits the response to hypoxia in the carotid body129 and pulmonary neuroepithelial bodies.130 However, it is unlikely that the oxygen sensor is identical to the NAD(P)H oxidase in neutrophils and macrophages, which is dedicated to the oxidative burst, necessary for the destruction of engulfed microorganisms.131 Patients with chronic granulomatous disease, who have a genetic defect in 1 of the 4 subunits of neutrophil/macrophage NAD(P)H oxidase, do not have phenotypic evidence of disordered oxygen sensing. The oxygen sensor must be present in a wide range of tissues, and the generation of free radicals involved in the signaling process is likely to be within a specific cell compartment and highly regulated by tissue oxygen tension. (Many experiments have indicated that the oxygen-sensing pathway that regulates Epo is unaffected by inhibitors of mitochondrial respiration.49,132-134 However, Schumacker et al135 have recently presented evidence suggesting that mitochondrial cytochrome oxidase [complex IV] serves as an oxygen sensor in hepatocytes and cardiac myocytes. Their evidence is based on measurements of HIF-1 activation in Hep3B cells treated with mitochondrial inhibitors and in ρο Hep3B cells lacking functional mitochondria.)
In the nitrogen-fixing bacteria, Rhizobium, oxygen-regulated gene expression is mediated by hypoxically inducible phosphorylation of a transcription factor.136,137 Phosphorylation may play a role in signaling the hypoxic stimulus and regulation of HIF-1 activity in mammalian cells as well. Treatment of nuclear extracts from hypoxic cells with alkaline phosphatase abolishes HIF-1 DNA-binding activity.138 Inhibitors of both serine/threonine and tyrosine kinases block activation of HIF-1.114 Multiple compounds that interfere with phosphorylation cascades have complex effects on HIF-1 activation and oxygen-regulated gene expression,115 but the role of a particular phosphorylation pathway in the activation of HIF-1 has not been conclusively proven. The role of src kinase was proposed to play a critical role in the hypoxia signaling pathway.139 Although subsequent experiments indicate that c-src is not necessary for activation of HIF-1,128,140 expression of v-src increases HIF-1 expression in both normoxia and hypoxia.140 In the HIF-1α protein, mutation of phosphoacceptor sites in the hypoxically inducible domain between amino acids 549-672 did not have a major influence on the magnitude hypoxic induction.89
In the current model of oxygen sensing, the preponderance of evidence supports the role of a heme protein, likely a cytochrome b-like protein, which signals a decrease in oxygen tension by a decrease in the levels of free radicals. Many gaps in knowledge and areas of conflicting data await future elucidation of the oxygen sensing mechanism responsible for the activation of HIF-1 and the regulation of Epo and other oxygen-responsive genes.
REGULATION OF OTHER GENES BY HYPOXIA
The Epo gene provided an apt model system for identification of HIF-1 and investigation of the mechanism of oxygen sensing. An understanding of oxygen-regulated expression of other genes has provided insight into diverse areas of physiology. Genes shown to be regulated by hypoxia are listed in Table 2.
Genes Induced by Hypoxia
| . | Co2+ DFO . | HIF-1 . |
|---|---|---|
| Erythropoietin | Yes | Yes |
| VEGF | Yes | Yes |
| PDGF-B | Yes | ND |
| TGF-β1 | Yes | ND |
| PLGF | Yes | ND |
| Glucose transporter 1 (Glut-1) | Yes | Yes |
| Glucose transporter 2 (Glut-2) | Yes | ND |
| Glucose transporter 3 (Glut-3) | Yes | ND |
| Phosphofructokinase L (PFK-L) | Yes | Yes |
| Phosphofructokinase C (PFK-C) | Yes | ND |
| Aldolase A (ALD-A) | Yes | Yes |
| Aldolase C (ALD-C) | Yes | ND |
| Triosephosphate isomerase (TPI) | ND | ND |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Yes | ND |
| Phosphoglycerate kinase 1 (PGK-1) | Yes | Yes |
| Enolase-1 | Yes | Yes |
| Pyruvate kinase M | Yes | ND |
| Lactate dehydrogenase A (LDH-A) | Yes | Yes |
| Phosphoenolpyruvate carboxykinase (PEPCK) | Yes | ND |
| Mitochondrially encoded genes | No | No |
| Tyrosine hydroxylase | Yes | Yes |
| Endothelin 1 | Yes | ND |
| Heme oxygenase | Yes | Yes |
| Nitric oxide synthase | Yes | Yes |
| Transferrin | Yes | Yes |
| Retrotransposon VL30 | Yes | Yes |
| Tissue factor | Yes | Yes |
| Adenylate kinase 3 | Yes | Yes |
| c-fos | Yes | ND |
| c-jun | Yes | ND |
| p35srj | Yes | Yes |
| . | Co2+ DFO . | HIF-1 . |
|---|---|---|
| Erythropoietin | Yes | Yes |
| VEGF | Yes | Yes |
| PDGF-B | Yes | ND |
| TGF-β1 | Yes | ND |
| PLGF | Yes | ND |
| Glucose transporter 1 (Glut-1) | Yes | Yes |
| Glucose transporter 2 (Glut-2) | Yes | ND |
| Glucose transporter 3 (Glut-3) | Yes | ND |
| Phosphofructokinase L (PFK-L) | Yes | Yes |
| Phosphofructokinase C (PFK-C) | Yes | ND |
| Aldolase A (ALD-A) | Yes | Yes |
| Aldolase C (ALD-C) | Yes | ND |
| Triosephosphate isomerase (TPI) | ND | ND |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Yes | ND |
| Phosphoglycerate kinase 1 (PGK-1) | Yes | Yes |
| Enolase-1 | Yes | Yes |
| Pyruvate kinase M | Yes | ND |
| Lactate dehydrogenase A (LDH-A) | Yes | Yes |
| Phosphoenolpyruvate carboxykinase (PEPCK) | Yes | ND |
| Mitochondrially encoded genes | No | No |
| Tyrosine hydroxylase | Yes | Yes |
| Endothelin 1 | Yes | ND |
| Heme oxygenase | Yes | Yes |
| Nitric oxide synthase | Yes | Yes |
| Transferrin | Yes | Yes |
| Retrotransposon VL30 | Yes | Yes |
| Tissue factor | Yes | Yes |
| Adenylate kinase 3 | Yes | Yes |
| c-fos | Yes | ND |
| c-jun | Yes | ND |
| p35srj | Yes | Yes |
The first set of genes encode growth factors, the second set of genes encode enzymes or transporters involved in glucose metabolism, and the third set of genes encode proteins with a wide range of functions. The middle column indicates whether the gene has been shown to be induced by cobalt and/or DFO, thereby implicating the same or a similar mechanism of oxygen sensing. The column on the right indicates whether HIF-1 involvement has been demonstrated either by identification of a functional HIF-1 site or by the lack of hypoxic induction in ARNT deficient cells. Expression of the PLGF and Glut-2 genes is decreased by hypoxia. Induction of the PEPCK gene by glucagon is blocked by hypoxia.
Abbreviation: ND, not determined.
Hypoxia activates angiogenesis by inducing the genes encoding VEGF and other growth factors with angiogenic properties.119,141-145HIF-1–mediated regulation of blood vessel growth appears to be of critical importance, because HIF-1α−/− and ARNT−/− knockout mice do not live beyond embryonic day 8.5 to 9.5, likely owing to a failure of vascular development.146-148 In neoplasms, angiogenesis is essential for tumor growth and metastasis. Solid tumor xenografts composed of ARNT-deficient cells have reduced VEGF expression and grow more slowly than xenografts that can form an intact HIF-1 heterodimer.149
When oxygen is limited, the rate of glycolysis increases to compensate for a decrease in ATP production via mitochondrial respiration. Long-term adaptation to hypoxia involves the regulation of genes encoding proteins involved in energy metabolism. An increase in glucose uptake in hypoxia is associated with increased expression of the genes encoding 2 glucose transporters, Glut-1 and Glut-3.150,151Genes encoding specific isoenzymes for most if not all steps in the glycolytic pathway are upregulated by hypoxia.59,134,150,152-157 Hypoxia blocks glucagon induction of PEPCK, the rate-limiting gluconeogenic enzyme.158-160 Expression of mitochondrially encoded genes is suppressed by hypoxia,154 but the mechanism of regulation of these genes appears to be independent of the signaling pathway that regulates nuclear-encoded genes and HIF-1.150
Hypoxic induction of the tyrosine hydroxylase gene aids in the regulation of respiration.65,161 HIF-1 has also been implicated in the regulation of genes encoding type II (inducible) nitric oxide synthase,162,163 heme oxygenase-1,164 transferrin,165 and retrotransposon VL30.166 Oxygen-regulated expression has been demonstrated for endothelin 1,167-169, c-jun, and c-fos.166 Hypoxic induction of tissue factor and adenylate kinase 3 were identified by differential display polymerase chain reaction.151 HIF-1α is involved with increasing apoptosis and decreasing cellular proliferation in response to hypoxia, as was demonstrated by use of embryonic stem cells lacking functional HIF-1α (HIF-1α−/−).170
The importance of HIF-1 in oxygen-regulated gene expression has been examined in several cell lines, including an ARNT-deficient cell line,85 an HIF-1α–defective cell line created by mutagenesis and selection,171 and embryonic stem cells from HIF-1α147,148,170 and ARNT knockout mice.146None of these cells produce Epo, but oxygen-regulated gene expression is disrupted for a number of other genes having functional HIF-1 response elements. For example, induction of PGK-1 and LDH-A by hypoxia is abolished in ARNT-deficient cells. For other genes, some hypoxic induction is retained in the mutant cell lines, for example, heme oxygenase-1 in HIF-1α mutant cells and VEGF in ARNT-deficient cells. Possible explanations for retained inducibility include alternative dimerization partners and regulation at the level of mRNA stability.
The arrangement of sites in hypoxia-responsive regulatory elements in other genes is similar to that of the Epo 3′ enhancer. For example, in the LDH-A promoter, 3 sites are critical for oxygen regulation in arrangement similar to the tripartite structure of theEpo 3′ enhancer.54 Whereas in the Epogene 3′ enhancer the HIF-1 site is adjacent to an HNF-4 site that is necessary for hypoxic inducibility, in the LDH-A promoter, the HIF-1 site is adjacent to a CREB-1/ATF-1 binding site. In both genes, the necessity of multiple adjacent sites for maximal hypoxia induction is probably due to the formation of a multiprotein complex including p300/CBP.
Comparison of HIF-1 sites characterized in 25 hypoxically inducible genes has resulted in a consensus recognition sequence for HIF-1 DNA-binding: T/GACGTGCGG.172
CLINICAL STATES ASSOCIATED WITH ABERRANT EXPRESSION OF Epo
In a variety of pathological states, dysregulation of Epo gene expression may cause either anemia or polycythemia.
Anemia is a major complication of most forms of renal failure. Because the anemia of renal failure is due primarily to a decrease in Epo production,173,174 patients are successfully treated by administration of recombinant human Epo.175,176 In mice with diverse forms of renal injury, a decreased number of fibroblast-like interstitial cells express Epo in response to anemia or hypoxia.177 Damage to the kidneys appears to change the threshold for Epo gene expression, but the precise molecular mechanisms have not yet been defined. Renal injury causes an expansion of interstitial cells and an infiltration of CD45+ cells, but the phenotype of the Epo-producing fibroblast-like cells does not appear to change. The sensitivity ofEpo expression to changes in the microenvironment of Epo-producing cells may explain the difficulty of establishing a renal cell culture model of inducible Epo expression.
Inappropriately low levels of erythropoietin have been demonstrated in patients with acquired immunodeficiency syndrome (AIDS),178rheumatoid arthritis and other chronic inflammatory diseases,179 and cancer.180 Inflammatory cytokines have been postulated to play a role in diminishing Epogene expression in these disorders and in the anemia of renal failure. In human hepatoma cell lines and isolated perfused rat kidneys, the inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin-1 (IL-1) suppress Epo production.181,182Patients with Itai-itai disease, caused by long-term cadmium intoxication, have inappropriately low Epo levels for their degree of anemia and renal failure.183
Uremia is the predominant and prototypical clinical syndrome for Epo replacement therapy, but recombinent human Epo (rHuEpo) has been used in a broad range of clinical settings (for review, see Cazzola et al184). For example, rHuEpo is efficacious in the treatment of anemia caused by AZT in human immunodeficiency virus (HIV)-infected patients, chemotherapy for nonmyeloid malignancies, premature birth, cancer, rheumatoid arthritis, and inflammatory bowel disease. The efficacy of rHuEpo is uncertain in patients whose levels of plasma Epo are elevated in keeping with their degree of anemia.
Primary polycythemia is caused by defects of hematopoietic progenitor cells and is associated with low levels of circulating erythropoietin. For example, polycythemia vera (PV) is caused by acquired somatic mutations in hematopoietic stem cells. IGF-1 and angiotensin II may increase proliferation of hematopoietic progenitors and thereby contribute to some forms of polycythemia.185 Familial congenital polycythemias may be caused by erythropoietin receptor mutations. In contrast, secondary polycythemia is generally associated with increased erythropoietin production. Elevated levels of plasma Epo are encountered in systemic hypoxemia, in certain neoplasms, and, less commonly, in disorders that impair oxygen delivery to tissues.
Excessive activation of Epo gene expression can result from impaired oxygen delivery due to high affinity hemoglobin mutants, methemoglobinemias, and 2,3-bisphosphoglycerate deficiency.185 Numerous congenital mutations of both α- and β-globin genes can result in high-affinity hemoglobin molecules. Such patients are often asymptomatic, because impaired oxygen delivery is balanced by polycythemia. Congenital methemoglobinemias also cause an adaptive erythrocytosis, because the buildup of ferri-hemes increases the oxygen affinity of the remaining ferro-hemes. 2,3-bisphosphoglycerate is an allosteric regulator of hemoglobin. An enzymatic defect leading to decreased synthesis of 2,3-bisphosphoglycerate is a rare cause of congenital polycythemia.186
Chronic arterial hypoxemia often leads to an upregulation of Epo expression, causing a maladaptive erythrocytosis. Patients with chronic obstructive pulmonary disease can develop erythrocytosis, which increases the risk of cor pulmonale. Similarly, patients with right to left cardiac shunts can have extremely high hematocrit levels.
Specific types of neoplasms can also cause overproduction of Epo. Elevated Epo levels are found most commonly in patients with renal carcinomas, Wilms tumor, hepatomas, and cerebellar hemangioblastomas,5 all anatomic sites in which Epo is normally expressed at low levels; less frequently, these tumors cause erythrocytosis. Benign renal tumors can also cause erythrocytosis, possibly due to local ischemia of renal Epo-producing cells.
Mutations in the von Hippel Lindau (VHL) gene are associated with renal and central nervous system carcinomas, both highly vascular tumors that overexpress VEGF. In cells in which the von Hippel Lindau protein is inactivated, the hypoxically inducible genes VEGF, Glut-1, and PDGF-B are expressed at high levels under both normoxic and hypoxic conditions. Introduction of wild-type pVHL into these cells causes expression of these genes to revert to the normal hypoxically inducible pattern by suppressing normoxic expression.187 188
A congenital and familial polycythemia of unknown etiology has been characterized in 103 patients from Chuvashia, a region in the Russian Federation.189 This condition is characterized by an autosomal recessive pattern of inheritance, high hemoglobin levels (mean, 23 g/dL), high hematocrit levels (mean, 67%), elevated Epo levels, and morbidity and mortality secondary to erythrocytosis, including fatal thrombotic and hemorrhagic complications. The polycythemia is not due to high-affinity hemoglobin, methemoglobinemia, 2,3-bisphosphoglycerate deficiency, or systemic hypoxia. Furthermore, the genetic mutation is not linked to either the Epo or Epo receptor genes. The disorder may therefore be caused by an abnormality in the oxygen sensing-signaling pathway or in a trans-acting factor involved in the regulation of Epo gene expression.
CONCLUSIONS
Regulation of erythropoiesis and red blood cell mass relies on modulating Epo gene expression in response to tissue oxygen tension. Developmental, tissue-specific, and environmental signals all contribute to the precise regulation of the Epo gene. Epo production and gene expression is restricted to specific subsets of cells: interstitial fibroblast-like cells in the kidney and, in the liver, Ito cells as well as a subset of hepatocytes. Experiments with transgenic mice have broadly mapped the cis-acting sequences responsible for tissue-specific expression.
The Epo gene has been a model for the regulation of gene expression by oxygen tension. The magnitude of hypoxically inducible transcription of the Epo gene is greater than any other gene known to be regulated by oxygen tension. Human hepatoma cell lines have provided a useful model system for studying inducible expression of theEpo gene. Regulatory sequences in the Epo gene have been dissected and characterized in more detail than any other oxygen-regulated gene. In a plausible model, a cytochromeb-like flavoheme protein senses oxygen tension and regulates production of oxygen free radicals.190 In hypoxia, rapid degradation of HIF-1α by the ubiquitin-proteasome pathway is prevented, leading to the formation of HIF-1α/ARNT heterodimers. These heterodimers bind to HIF-1 sites, interact with adjacent DNA-binding proteins and p300/CBP, and activate gene expression.
The Epo gene has been the portal through which a generalized system of oxygen-regulated gene expression was first identified and described. This mode of molecular adaptation is more fundamental than the regulation of Epo in mammals. An HIF-1–like hypoxically inducible DNA-binding activity was identified in Drosophila melanogaster.191 In mammals, HIF-1 and the oxygen-sensing mechanism that regulates Epo are critical for the regulation of genes involved in angiogenesis, energy metabolism, respiration, vascular tone, and many other processes.
In humans, regulation of the Epo gene provides an elegant and precise mechanism for adjusting red blood cell mass to perturbations in tissue oxygen tension. A more complete understanding of the molecular mechanisms governing induction of the Epo gene may lead to new therapeutic agents to treat patients with anemia or polycythemia due to inappropriate expression of the Epo gene.
ACKNOWLEDGMENT
The authors thank E. Huang, W. Willmore, P. Hradecky, P. Yachimski, and M. Vasconcelles for critical review of the manuscript.
Supported by a National Institutes of Health Grant No. DK41234 to H.F.B.
REFERENCES
Author notes
Address reprint requests to H. Franklin Bunn, MD, LMRC 223, 221 Longwood Ave, Boston, MA 02115; e-mail: bunn@calvin.bwh.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal