Abstract
To develop a method for targeting expression of genes to the full hematopoietic system, we have used transgenic mice to explore the transcriptional regulation of the vav gene, which is expressed throughout this compartment but rarely outside it. Previously, we showed that a cluster of elements surrounding its promoter could drive hematopoietic-specific expression of a bacterial lacZ reporter gene, but the expression was confined to lymphocytes and was sporadically silenced. Those limitations are ascribed here to the prokaryotic reporter gene. With a human CD4 (hCD4) cell surface reporter, the vav promoter elements drove expression efficiently and stably in virtually all nucleated cells of adult hematopoietic tissues but not notably in nonhematopoietic cell types. In multiple lines, hCD4 appeared on most, if not all, B and T lymphocytes, granulocytes, monocytes, megakaryocytes, eosinophils, and nucleated erythroid cells. Moreover, high levels appeared on both lineage-committed progenitors and the more primitive preprogenitors. In the fetus, expression was evident in erythroid cells of the definitive but not the primitive type. These results indicate that a prokaryotic sequence can inactivate a transcription unit and that the vavpromoter region constitutes a potent transgenic vector for the entire definitive hematopoietic compartment.
ALTHOUGH MUCH HAS BEEN learned about the regulation of genes expressed in specific hematopoietic lineages,1 little is known about multilineage regulation. Hence, the vav gene is of particular interest. vav is expressed in virtually all hematopoietic cell lines and, commencing in the fetal liver, in all normal hematopoietic cell types but in very few others.2-7 As predicted from its sequence,3 Vav transduces signals from various surface receptors to Rho-like G proteins.8 Although vav is not, as was first reported,7 required for mouse development,9 it is essential for full lymphocyte development and function.9,10 Whether its absence affects nonlymphoid hematopoietic cells has not been clearly reported.9
Clarifying the transcriptional regulation of vav might allow expression of any gene to be targeted to the entire hematopoietic system, providing a new avenue for addressing issues such as lineage commitment and the basis of leukemogenesis. We have therefore attempted to identify vav regulatory elements active in the most stringent test, the transgenic mouse. We first identified sites where the vav chromatin had been rendered hypersensitive (HS) to DNase-I, presumably by bound transcription factors.11 All 5 HS sites found, which span a 14-kb region across the first exon (Fig 1A), appeared in different hematopoietic cell types but not in nonhematopoietic cells and therefore constituted prime candidates for the key regulatory elements. Transgenes that included 4 or 5 of these sites and the widely usedEscherichia coli lacZ (β-galactosidase) reporter gene were active in hematopoietic and not other tissues, but the expression was invariably confined to lymphocytes and was always variegated, ie, restricted to a proportion of the cells of a given type.11Moreover, that proportion decreased in a fashion consistent with stochastic inactivation.
Relationship of the vav-hCD4 transgenes to thevav locus. (A) 5′ end of the mouse vav locus showing the 5 hematopoietic-specific HS sites (▿) surrounding exon 1 (▪), as well as a testis-specific promoter (t) near exon 2 (see Discussion). (B) Expanded map of the vav regions used in the transgenes, with the untranslated portion of exon 1 shown unfilled. (C) The HS321/45 vav-hCD4 transgene. (D) The HS21/45 transgene, showing 2 PCR primers (a and b) used in its construction (see Materials and Methods). (E) The mutant hCD4 protein from which the reporter was derived. ss, splice sites; pA, polyadenylation region F43I, the mutated extracellular residue; TM, transmembrane region. Restriction sites are K, Kpn I; R,EcoRI; B, BamHI; S2, Sac II; N, Nco I; Ne, Nae I; Hp, Hpa I; H3, HindIII; those in parentheses have been destroyed, whereas those in bold were used to excise the transgenes from the plasmid backbone; for more detailed maps, see Ogilvy et al.11
Relationship of the vav-hCD4 transgenes to thevav locus. (A) 5′ end of the mouse vav locus showing the 5 hematopoietic-specific HS sites (▿) surrounding exon 1 (▪), as well as a testis-specific promoter (t) near exon 2 (see Discussion). (B) Expanded map of the vav regions used in the transgenes, with the untranslated portion of exon 1 shown unfilled. (C) The HS321/45 vav-hCD4 transgene. (D) The HS21/45 transgene, showing 2 PCR primers (a and b) used in its construction (see Materials and Methods). (E) The mutant hCD4 protein from which the reporter was derived. ss, splice sites; pA, polyadenylation region F43I, the mutated extracellular residue; TM, transmembrane region. Restriction sites are K, Kpn I; R,EcoRI; B, BamHI; S2, Sac II; N, Nco I; Ne, Nae I; Hp, Hpa I; H3, HindIII; those in parentheses have been destroyed, whereas those in bold were used to excise the transgenes from the plasmid backbone; for more detailed maps, see Ogilvy et al.11
As reviewed recently,12,13 variegated expression, first observed in Drosophila and yeast, has now been recognized in mice with a number of transgenes. The phenomenon favors the view that the activity of any transcription unit is primarily determined for each cell in a binary, all-or-none fashion.14 The probability of transgene activity in a given cell is believed to be set by competition between its positive regulatory elements, such as a promoter, enhancer, or locus control region, and negative influences from surrounding chromatin.12,13 15
Although the limitations in vav-lacZ expression might have reflected the need for unidentified vav regulatory elements, it seemed possible that vav regulation had instead been compromised by the bacterial reporter, which has sometimes distorted transgene expression (see Discussion). Indeed, we report here that substitution of a mammalian reporter allowed vav transgene expression throughout the hematopoietic compartment. These results strongly implicate the prokaryotic reporter in transgene silencing and demonstrate that the vav regulatory elements provide a powerful transgenic vector for the hematopoietic system.
MATERIALS AND METHODS
Generation of reporter, transgenes, and transgenic mice.
The hCD4 reporter (Fig 1D and E) was derived by polymerase chain reaction (PCR) with a proof-reading polymerase from a cDNA (the kind gift of Dr Toshiko Sakihama, Dana-Farber Cancer Institute, Boston, MA) bearing a mutation (F43I) reported to preclude CD4 association with major histocompatibility complex (MHC) class II.16 The 5′ primer introduced a mammalian consensus initiation sequence, whereas the 3′ primer, corresponding to nucleotides 1337-1350 of the cDNA sequence,17 excluded the region encoding the C-terminal 33 amino acids, the domain required for signaling.18 By overlap PCR, the truncated hCD4 cDNA was placed between an intron from the SRα expression vector19 and the SV40 late region polyadenylation signal (nucleotides 2538-2746 in GenBank J02400; Fig 1D). The cassette was bounded by two 48-bp recognition sequences (FRT) for the Flp recombinase to provide the option (not yet tested) of using Flp-mediated recombination to aid in replacing the hCD4 reporter by another gene.
The new transgenic vectors (Fig 1) were derived from HS21/45vav-βgal.11 To eliminate the lacZsequences and introduce convenient restriction sites, we used that vector as a template in an inverted PCR with primers (a andb in Fig 1D) flanking the lacZ sequence but pointing away from it. The proof-reading Tth polymerase was used to generate the 9.4-kb PCR product bearing 2.3 kb of vav sequences upstream from the promoter, the 2.7-kb plasmid backbone, and 4.4 (0.7 + 3.7) kb of sequences from the vav first intron. Insertion of thehCD4 cassette yielded HS21/45 vav-hCD4 (Fig 1D). To construct the vector including HS3 (Fig 1C), a HindIII-digested HS21/45 transgene fragment was blunt-end ligated to a NotI-digested plasmid containing 5.5 kb of additional upstream sequences (kindly provided by Dr Bernd Hentsch, Walter and Eliza Hall Institute of Medical Research, Royal Melbourne Hospital, Victoria, Australia).
In these constructions, all PCR steps used high fidelity polymerases, and the final constructs were extensively mapped with restriction endonucleases. The CD4 expression cassette was partially sequenced to confirm the junctions and the presence of the F43I mutation.
For microinjection, each transgene was separated from the vector by electrophoresis of digests (HindIII for HS21/45vav-hCD4 or Sac II for HS321/45 vav-hCD4) through low melting point agarose, purified using an Elutip-d (Schleicher and Schuell, Dassel, Germany), precipitated with ethanol, and dissolved in sterile 10 mmol/L Tris-HCl (pH 7.4), 0.1 mmol/L EDTA. Microinjected (C57BL/6J × SJL/J) F2 eggs were transferred to pseudo-pregnant (CBA/H × C57BL/6) F1 females.20Transgenic pups were identified by PCR on DNA from tail biopsies with primers specific to the transgene SV40 sequence.
Flow cytometric analysis.
Peripheral blood leukocytes, obtained after NH4Cl lysis of erythrocytes, were incubated on ice with phycoerythrin (PE)-labeled mouse antihuman CD4 monoclonal antibody (MoAb; RPA-T4; Pharmingen, San Diego, CA) plus anti-Fcγ receptor antibody (2.4G2) to reduce nonspecific binding.21 Viable stained cells (>10,000) were analyzed on a FACScan (Becton Dickinson, San Jose, CA) as described.22 Single-cell suspensions from femoral bone marrow, thymus, spleen, and mesenteric lymph nodes were analyzed similarly, except that mature red blood cells were gated out by forward scatter rather than lysed. Nontransgenic (C57BL/6J × SJL/J) F1 and littermate mice provided routine controls. For cell lineage analysis, cells were incubated simultaneously with a fluorescein isothiocyanate-labeled antibody to a cell surface marker, PE-labeled anti-hCD4 MoAb, and 2.4G2 and analyzed as above. The MoAbs were against B220 (RA3-6B2), mouse CD4 (H129.19.6.8), CD8 (YTS-169), Thy1 (T24.31.2), IgM (anti-Cμ.5.1), IgD (11-26C), Mac1 (M1/70), Gr1 (RB6-8C5), and Ter119.11 22
With vav-lacZ mice, β-galactosidase activity in individual blood cells was determined by the fluorescence-activated cell sorting (FACS)-gal assay,23 as detailed previously.11
To analyze fetal blood or liver, pregnant females were killed, and the closed uterus was washed. The uterine wall and yolk sac were removed without disrupting the umbilical cord, and the umbilical vessels were clamped and severed. Each washed embryo was bled into wash buffer by severing the jugular veins and cervical arteries. Cytocentrifuge preparations of fetal blood were stained with DiffQuik (Lab Aids, Narrabeen, New South Wales, Australia). Cell suspensions from E12.5 and E14.5 fetal liver were made by passage through a 21-gauge needle. Each fetus was genotyped by PCR.
FACS sorting and culture of bone marrow cells.
To obtain cells differing in hCD4 level, femoral bone marrow cells from adult HS21/45 vav-hCD4 mice were treated with PE-anti-hCD4 MoAb, unlabeled anti-Fcγ receptor antibody, and 1 μg/mL propidium iodide and sorted (MoFlo; Cytomation, Fort Collins, CO) by viability and hCD4 staining (see Fig 4). Cytocentrifuge preparations were stained with May-Grunwald-Giemsa to enumerate different cell types. For colony assays (see Table 3), 25,000 nucleated cells were cultured for 7 days in semisolid medium as described.24 The dried cultures were stained, first for acetylcholinesterase activity, then with Luxol fast blue, and finally with hematoxylin, and were scored microscopically to verify colony counts and to determine the cell composition of each colony.
Immunohistochemistry.
Tissues embedded in OCT (Tissue-tek; Miles, Elkhart, IN) were snap-frozen, and 5-μm sections were fixed in acetone. Aldehyde groups were blocked with 0.2 mol/L glycine, endogenous peroxidase quenched with 3% H2O2, and endogenous biotin and avidin blocked using reagents from Vector Laboratories (Burlingame, CA). The sections were then incubated 2 hours at room temperature in a 1:20 dilution of biotin-conjugated mouse anti-hCD4 MoAb (as described above), developed using a Vectastain Elite ABC (peroxidase) kit (Vector Laboratories) with diaminobenzidine as substrate and counterstained with hematoxylin.
RESULTS
Extensive expression of a CD4 reporter in the blood.
We wished to use a mammalian cell surface reporter to facilitate cell-by-cell (ie, flow cytometric) analysis and chose human CD4 because its molecular interactions have been well characterized and MoAbs are available that recognize it specifically. To preclude interference with normal physiology, the cDNA used was truncated so that the polypeptide (Fig 1E) would lose its intracellular signaling capacity,18whereas a mutation in its extracellular domain (F43I) reportedly prevents association with MHC class II, the major CD4 ligand.16
The 2 vav-hCD4 transgenes were based on the most effectivevav-lacZ transgenes.11 One (Fig 1D) contains the 2 proximal upstream HS sites and both intron sites (hence HS21/45), whereas the other, HS321/45 (Fig 1C), also bears the distal upstream HS3. Both maintain the normal order of the HS sites, with the hCD4 cDNA replacing the coding portion of vav exon 1 (Fig 1B and C). Analysis of 64 primary transgenic mice showed that both transgenes were highly effective. Flow cytometry on their peripheral blood cells typically showed substantial hCD4+ cells (Fig 2B through D). HS21/45vav-hCD4 was expressed in 27 of 35 primary mice (77%), and the larger construct was expressed in a similar proportion (25 of 29 [86%]). The proportion of animals with low, medium, or high expression was also similar for the 2 transgenes.
Flow cytometric analysis of transgene expression in peripheral blood leukocytes. (A) A vav-lacZ transgenic progeny mouse (10 weeks old) and (B through F) HS21/45 vav-hCD4 mice. (B) Primary vav-hCD4 animal with intermediate expression; (C and D) mosaic primary vav-hCD4 mice; and (E and F) their respective transgenic progeny. Dotted lines with shading underneath show nontransgenic controls run in parallel. The expression oflacZ was assessed by a flow cytometric assay for β-galactosidase activity11 and of hCD4 with an antibody (see Materials and Methods). The 2 peaks in (E), which may reflect the different hCD4 levels in different cell lineages (see Table 1), were not as notable in other mice of that strain.
Flow cytometric analysis of transgene expression in peripheral blood leukocytes. (A) A vav-lacZ transgenic progeny mouse (10 weeks old) and (B through F) HS21/45 vav-hCD4 mice. (B) Primary vav-hCD4 animal with intermediate expression; (C and D) mosaic primary vav-hCD4 mice; and (E and F) their respective transgenic progeny. Dotted lines with shading underneath show nontransgenic controls run in parallel. The expression oflacZ was assessed by a flow cytometric assay for β-galactosidase activity11 and of hCD4 with an antibody (see Materials and Methods). The 2 peaks in (E), which may reflect the different hCD4 levels in different cell lineages (see Table 1), were not as notable in other mice of that strain.
In contrast to a typical vav-lacZ line (eg, Fig 2A), many primary animals bearing either vav-hCD4 transgene showed expression in essentially all peripheral blood leukocytes (eg, Fig 2B). The hCD4-negative population in the remaining primary mice (eg, Fig 2C and D) might reflect variegation or simply genetic mosaicism, due to transgene insertion after the 1-cell stage. In strong support of the latter interpretation, all the transgenic descendants of 5 primary animals showing a heterocellular pattern (2 HS21/45 and 3 HS321/45) exhibited only hCD4+ blood cells (Fig 2E and F). Hence, unlike the heritable variegation in all vav-lacZmice,11 the heterocellularity encountered in some primary vav-hCD4 animals can be ascribed to genetic mosaicism.
Expression in all major hematopoietic lineages.
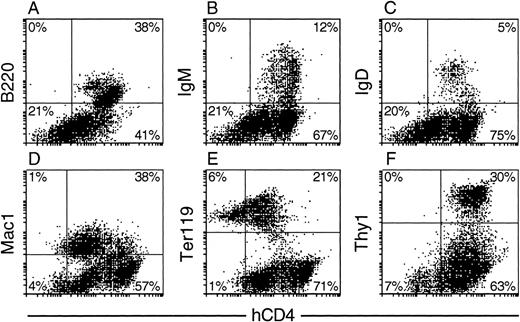
In hematopoietic tissues of progeny animals, concomitant flow cytometric analysis of immunofluorescence for hCD4 and lineage-specific surface markers showed high transgene activity in multiple lineages (Fig 3 and Table 1). In the bone marrow (Fig 3A through E), hCD4 appeared on all B-lineage (B220+) cells, including those at the more mature IgM+ and IgD+ stages, as well as on all Mac1+(CD11b+) and all Gr-1+ cells, which include the monocytes/macrophages and granulocytes. Most if not all nucleated erythroid (Ter119+) cells were also positive, albeit at a lower mean level (Fig 3E). Similarly, in the spleen, all T-lymphoid (Thy1+) cells bore hCD4 (Fig 3F), including both the helper (mCD4+) and cytotoxic (mCD8+) mature T-cell subsets (data not shown).
Expression profile of hCD4 in hematopoietic tissues of HS21/45 vav-hCD4 progeny mice. Nucleated bone marrow (A) B220+, (B) IgM+, and (C) IgD+B-lymphoid cells; (D) Mac1+ myeloid cells; (E) Ter119+ erythroid cells; and splenic (F) Thy1+ T cells. Quadrants were set so that the marker+ hCD4+ cells in a nontransgenic control sample, run in parallel, did not exceed 5% of total marker+ cells analyzed. As would be expected, essentially all cells from the nontransgenic controls then fell in the left quadrants, and the percentage of cells bearing each lineage-specific marker (eg, B220) was similar to that of the corresponding transgenic population.
Expression profile of hCD4 in hematopoietic tissues of HS21/45 vav-hCD4 progeny mice. Nucleated bone marrow (A) B220+, (B) IgM+, and (C) IgD+B-lymphoid cells; (D) Mac1+ myeloid cells; (E) Ter119+ erythroid cells; and splenic (F) Thy1+ T cells. Quadrants were set so that the marker+ hCD4+ cells in a nontransgenic control sample, run in parallel, did not exceed 5% of total marker+ cells analyzed. As would be expected, essentially all cells from the nontransgenic controls then fell in the left quadrants, and the percentage of cells bearing each lineage-specific marker (eg, B220) was similar to that of the corresponding transgenic population.
Relative hCD4 Level Per Cell in Different Cell Types
| Cell Type* . | vav-hCD4Transgene Construct† . | |
|---|---|---|
| HS21/45 . | HS321/45 . | |
| Adult | ||
| T lymphoid | 100‡ | 100 |
| B lymphoid | 72 ± 17 | 120 ± 20 |
| Myeloid | 31 ± 7 | 22 ± 4 |
| Erythroid | 7 ± 4 | 8 ± 2 |
| Fetal | ||
| Nonerythroid | 52 | 160 |
| Definitive erythroid | 2 | 8 |
| Primitive erythroid | <1 | <3 |
| Cell Type* . | vav-hCD4Transgene Construct† . | |
|---|---|---|
| HS21/45 . | HS321/45 . | |
| Adult | ||
| T lymphoid | 100‡ | 100 |
| B lymphoid | 72 ± 17 | 120 ± 20 |
| Myeloid | 31 ± 7 | 22 ± 4 |
| Erythroid | 7 ± 4 | 8 ± 2 |
| Fetal | ||
| Nonerythroid | 52 | 160 |
| Definitive erythroid | 2 | 8 |
| Primitive erythroid | <1 | <3 |
Cell types were distinguished, as in Fig 3, by FACS analysis for surface markers: T lymphoid (Thy-1+), B lymphoid (B220+), myeloid (Mac-1+), and erythroid (Ter119+).
The data presented, derived from multiple FACS analyses like those in Figs 3 and 5, are the means (±SD) of hCD4-associated fluorescence (in relative fluorescence intensity units) above the background in nontransgenic mice, normalized to the level in T cells (set at 100) within each transgenic strain. Data for adult bone marrow, spleen, and thymus were from 5 HS21/45 and 3 HS321/45 strains; fetal liver (E14.5) and blood (E10.5) from 1 HS21/45 and 1 HS321/45 strain.
The actual mean fluorescence values were 300 ± 250 for the HS21/45 strains and 190 ± 50 for the HS321/45 strains.
The 2 vav-hCD4 transgenes produced similar patterns of expression in the 8 independent transgenic lines analyzed. With both, hCD4 appeared on virtually all nucleated cells from primary hematopoietic organs (bone marrow and thymus) as well as secondary tissues such as spleen. In different lines bearing the same transgene construct, the level varied over a 10-fold range, probably due to both insertion position and copy number, but the average values for the 2 constructs were similar. Moreover, the relative levels of hCD4 on different cell types were consistent, as indicated in Table 1, in which estimates of the fluorescence intensity on each cell type have been normalized to that on T cells. With both the transgene constructs, lymphoid cells displayed a 3- to 5-fold higher level than myeloid, whereas erythroid cells exhibited the lowest (Fig 3 and Table 1). The level in nucleated erythroid cells averaged approximately 15-fold lower than in lymphoid cells.
High activity in clonogenic progenitor cells.
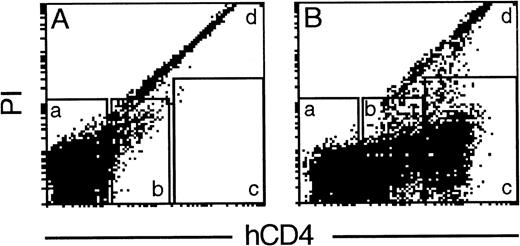
Because essentially all the nucleated cells in bone marrow bore hCD4, we wished to determine whether clonogenic hematopoietic precursor cells also expressed the transgene. To allow such studies, we first sorted all the marrow cells into arbitrary fractions having a low-negative, intermediate, or high level of hCD4 (Fig4). Cytocentrifuge preparations of these fractions (data not shown) confirmed that every nucleated cell population displayed substantial hCD4. The low-negative fraction was greatly dominated by erythrocytes, and almost the only nucleated cells present were approximately 20% of the mature granulocytes. The intermediate fraction contained the majority of both the mature granulocytes and the nucleated erythroid cells. Most of the lymphocytes, eosinophils, and blast cells were found in the hCD4hi fraction.
Fractionation of transgenic bone marrow cells by hCD4 level. Flow cytometric analysis of cells from a HS21/45vav-hCD4 mouse incubated in the absence (A) or presence (B) of a labeled anti-hCD4 MoAb. Cells within windows a, b, and c were sorted for clonogenic assays. In the representative experiment shown (1 of 3), 8% of viable nucleated cells fell in fraction a (low to negative for surface hCD4), 42% in fraction b (intermediate hCD4), and 50% in fraction c (high hCD4). Most erythrocytes fell in fraction a and the dead cells (9% of the total cells) in fraction d.
Fractionation of transgenic bone marrow cells by hCD4 level. Flow cytometric analysis of cells from a HS21/45vav-hCD4 mouse incubated in the absence (A) or presence (B) of a labeled anti-hCD4 MoAb. Cells within windows a, b, and c were sorted for clonogenic assays. In the representative experiment shown (1 of 3), 8% of viable nucleated cells fell in fraction a (low to negative for surface hCD4), 42% in fraction b (intermediate hCD4), and 50% in fraction c (high hCD4). Most erythrocytes fell in fraction a and the dead cells (9% of the total cells) in fraction d.
Table 2 gives the results of colony assays for progenitor cells in the fractionated populations, using 3 different stimuli. Control unstained (or stained but unsorted) populations showed that neither staining nor sorting compromised clonogenicity, and the colony types produced by each stimulus on the sorted progenitors mirrored those from unfractionated marrow. Significantly, regardless of the stimulus, the hCD4hi fraction yielded more than 92% of the total progenitors, and virtually none appeared in the low-negative fraction (Table 2). hCD4 was present on progenitors committed to produce colonies of granulocytes, granulocytes and macrophages, macrophages, eosinophils, or megakaryocytes. Similarly, the more ancestral preprogenitor cells, which yield colonies of blast cells in response to stem cell factor (SCF) or to interleukin-3 (IL-3) plus thrombopoietin, all resided in the hCD4hi fraction (Table2). Because their frequency was the same or higher than from the unfractionated marrow, selective losses are unlikely. Thus, the transgene was very active in all the hematopoietic precursor cells assayed, including very primitive ones.
Progenitor Cell Distribution by hCD4 Level invav-hCD4 Bone Marrow
| Cells Cultured* . | Stimulus† . | No. of Colonies . | Progenitors in Fraction‡ . | |||||
|---|---|---|---|---|---|---|---|---|
| Blast . | G . | GM . | M . | Eo . | Meg . | |||
| Unstained | SCF | 31 | 47 | 6 | 5 | 0 | 0 | — |
| IL-3 + TPO | 16 | 59 | 14 | 18 | 1 | 37 | — | |
| M-CSF | 0 | 9 | 11 | 51 | 0 | 0 | — | |
| Unsorted | SCF | 24 | 47 | 3 | 5 | 0 | 0 | — |
| IL-3 + TPO | 21 | 54 | 20 | 11 | 2 | 32 | — | |
| M-CSF | 0 | 13 | 11 | 52 | 0 | 0 | — | |
| Low-negative | SCF | 0 | 0 | 0 | 0 | 0 | 0 | 0% |
| IL-3 + TPO | 0 | 2 | 0 | 0 | 0 | 0 | 0% | |
| M-CSF | 0 | 0 | 0 | 0 | 0 | 0 | 0% | |
| Intermediate | SCF | 0 | 3 | 0 | 0 | 0 | 0 | 2% |
| IL-3 + TPO | 0 | 8 | 1 | 6 | 5 | 2 | 8% | |
| M-CSF | 0 | 0 | 3 | 7 | 0 | 0 | 7% | |
| High | SCF | 48 | 73 | 8 | 0 | 0 | 0 | 98% |
| IL-3 + TPO | 25 | 68 | 18 | 30 | 5 | 53 | 92% | |
| M-CSF | 0 | 10 | 18 | 83 | 0 | 0 | 93% | |
| Cells Cultured* . | Stimulus† . | No. of Colonies . | Progenitors in Fraction‡ . | |||||
|---|---|---|---|---|---|---|---|---|
| Blast . | G . | GM . | M . | Eo . | Meg . | |||
| Unstained | SCF | 31 | 47 | 6 | 5 | 0 | 0 | — |
| IL-3 + TPO | 16 | 59 | 14 | 18 | 1 | 37 | — | |
| M-CSF | 0 | 9 | 11 | 51 | 0 | 0 | — | |
| Unsorted | SCF | 24 | 47 | 3 | 5 | 0 | 0 | — |
| IL-3 + TPO | 21 | 54 | 20 | 11 | 2 | 32 | — | |
| M-CSF | 0 | 13 | 11 | 52 | 0 | 0 | — | |
| Low-negative | SCF | 0 | 0 | 0 | 0 | 0 | 0 | 0% |
| IL-3 + TPO | 0 | 2 | 0 | 0 | 0 | 0 | 0% | |
| M-CSF | 0 | 0 | 0 | 0 | 0 | 0 | 0% | |
| Intermediate | SCF | 0 | 3 | 0 | 0 | 0 | 0 | 2% |
| IL-3 + TPO | 0 | 8 | 1 | 6 | 5 | 2 | 8% | |
| M-CSF | 0 | 0 | 3 | 7 | 0 | 0 | 7% | |
| High | SCF | 48 | 73 | 8 | 0 | 0 | 0 | 98% |
| IL-3 + TPO | 25 | 68 | 18 | 30 | 5 | 53 | 92% | |
| M-CSF | 0 | 10 | 18 | 83 | 0 | 0 | 93% | |
Abbreviations: Blast, blast cell; G, granulocyte; GM, granulocyte-macrophage; M, macrophage; Eo, eosinophil; Meg, megakaryocyte.
All cultures were initiated with 25,000 nucleated bone marrow cells from HS21/45 vav-hCD4 mice (10 to 13 weeks old). Data shown are from a representative fractionation experiment (1 of 3), namely that shown in Fig 4.
The recombinant murine factors were as follows: stem cell factor (SCF; 100 ng/mL); IL-3 (10 ng/mL); thrombopoietin (TPO; 50 ng/mL); and macrophage colony-stimulating factor (M-CSF; 10 ng/mL).
The progenitors (colony-forming cells) in each fraction, as a percentage of the progenitors recovered in the 3 fractions, adjusted for the different proportions of total nucleated cells in the 3 fractions (see Fig 4 legend).
Expression in fetal definitive but not primitive erythrocytes.
In the embryo, vav expression appears first in the liver, where it becomes just detectable at day 11.5 (E11.5) and stronger at E12.5.3,4 At that stage, the liver has begun to replace the yolk sac as the major hematopoietic organ and is composed very largely of the definitive (enucleated) erythrocytes produced there.25 At E12.5, transgenic liver cells consistently showed an hCD4-associated fluorescence above that in the nontransgenic population (Fig 5A), indicating that hCD4 was present at a low level on most of the cells and at a higher level on a small subpopulation. E14.5 liver cells gave a similar result, with a somewhat higher hCD4 signal (data not shown). The majority of the dominant erythroid (Ter119+) population, which represented approximately 95% of the fetal liver cells (Fig 5C), expressed hCD4 weakly, and the Ter119− cells (∼5%) expressed higher levels (Fig5D). Indeed, substantial hCD4 appeared on essentially all of the approximately 2% of liver cells bearing Mac-1 (Fig 5B), as well as on minor populations bearing Thy1, B220, or Gr-1 (data not shown). Thus, the vav transgene was clearly active during definitive hematopoiesis in the fetus.
Flow cytometric assay for hCD4 expression in fetal liver (A through D) and fetal blood (E through J). (A) Viable liver cells at E12.5 from a vav-hCD4 fetus (unbroken line) or a nontransgenic littermate (dotted line and shading). (B) Transgenic E14.5 liver, also analyzed for Mac-1. (C) Nontransgenic and (D) transgenic cells at E14.5, also analyzed for Ter119. (E, G, and I) Nontransgenic and (F, H, and J) transgenic blood (unbroken lines), superimposed on the nontransgenic profiles (dotted lines and shading). d indicates the presumptive definitive erythroid population and p the primitive one. In (E), the unidentified second, very small peak (∼2% of cells) probably represents larger, nonerythroid cells. Results in (A) are representative of analyses on more than 15 of each genotype, (B through D) on at least 3 transgenic and 5 nontransgenic littermates, and (E through J) on 15 to 27 mice. All analyses were performed on mice of 1 HS21/45 and 2 HS321/45 lines. Exceptions were E12.5 fetal liver and blood, studied in 1 line for each transgene, and cell surface marker analysis, which was performed on mice of 1 HS21/45 line.
Flow cytometric assay for hCD4 expression in fetal liver (A through D) and fetal blood (E through J). (A) Viable liver cells at E12.5 from a vav-hCD4 fetus (unbroken line) or a nontransgenic littermate (dotted line and shading). (B) Transgenic E14.5 liver, also analyzed for Mac-1. (C) Nontransgenic and (D) transgenic cells at E14.5, also analyzed for Ter119. (E, G, and I) Nontransgenic and (F, H, and J) transgenic blood (unbroken lines), superimposed on the nontransgenic profiles (dotted lines and shading). d indicates the presumptive definitive erythroid population and p the primitive one. In (E), the unidentified second, very small peak (∼2% of cells) probably represents larger, nonerythroid cells. Results in (A) are representative of analyses on more than 15 of each genotype, (B through D) on at least 3 transgenic and 5 nontransgenic littermates, and (E through J) on 15 to 27 mice. All analyses were performed on mice of 1 HS21/45 and 2 HS321/45 lines. Exceptions were E12.5 fetal liver and blood, studied in 1 line for each transgene, and cell surface marker analysis, which was performed on mice of 1 HS21/45 line.
The vav gene itself may not be active during primitive erythropoiesis (see Discussion). To determine whether the transgene was active in those cells, we first examined fetal blood before the transition to the definitive stage. At E10.5, primitive (nucleated) erythrocytes derived from the yolk sac greatly predominate, as evidenced by their overwhelming abundance in cytocentrifuge preparations. Importantly, at that stage the major population in the transgenic blood never gave an hCD4 signal above the nontransgenic background (compare Fig 5E and F). The approximately 2% of hCD4+ cells present specifically in the transgenic blood (Fig 5F) probably represented nonerythroid cells (eg, monocytes) of yolk sac origin.
Examination of fetal blood during the transition to definitive hematopoiesis (E12.5 and E14.5) also failed to show transgene expression in the primitive erythrocytes. As expected, cytocentrifuge preparations showed that, by E12.5, definitive (enucleated) erythrocytes of liver origin had become as abundant as the primitive nucleated cells. Unexpectedly, FACS analysis of nontransgenic blood consistently resolved the 2 populations (Fig 5G). We attribute this separation to the high autofluorescence of the (larger) primitive erythrocytes, which was also noted recently by Trimborn et al26; consistent with that conclusion, the peak denoted p was always much less prominent at E14.5 (Fig 5I), when the definitive cells dominated the cytocentrifuge preparations. In the transgenic blood at these stages, comparison with the superimposed nontransgenic profile (shaded area) indicated that only the presumptive definitive population (d) showed a weak but highly reproducible hCD4 signal (Fig 5H and J). Thus, no evidence of vav transgene activity in primitive erythrocytes was found.
No expression apparent in nonhematopoietic organs.
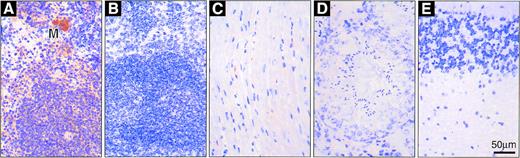
Immunohistochemical analysis of various organs suggested thatvav-hCD4 expression probably is confined to hematopoietic cell types (Fig 6). As expected, no hCD4-associated immunoperoxidase (brown) staining was evident in the nontransgenic spleen (Fig 6B), whereas most cells in transgenic spleen stained well (Fig 6A), with the highest levels in the lymphoid white pulp and the megakaryocytes (marked M). Similarly, nearly all cells in the thymus stained strongly, with the level seemingly higher in the cortex than the medulla (data not shown). These results mimic those reported for vav itself.4 In contrast, hCD4 appeared in only very rare cells in the heart (Fig 6C), perhaps of hematopoietic origin, and none was detected in testis (Fig 6D) or various brain sections (Fig 6E) or in liver or kidney, although their nonspecific background was higher, probably due to endogenous peroxidase or biotin.
Immunohistochemical detection of hCD4 in organs ofvav-hCD4 and control mice. (A) Transgenic spleen with a megakaryocyte (M); (B) nontransgenic spleen; (C) transgenic heart; (D) transgenic testis; and (E) transgenic cerebellum (other brain sections were also negative). Nontransgenic brain and testis were indistinguishable from those shown, and nontransgenic heart essentially so (see text).
Immunohistochemical detection of hCD4 in organs ofvav-hCD4 and control mice. (A) Transgenic spleen with a megakaryocyte (M); (B) nontransgenic spleen; (C) transgenic heart; (D) transgenic testis; and (E) transgenic cerebellum (other brain sections were also negative). Nontransgenic brain and testis were indistinguishable from those shown, and nontransgenic heart essentially so (see text).
No decrease in vav-hCD4 expression with age.
Because vav-lacZ transgene expression decreased progressively,11 we monitored hCD4 in peripheral blood as individual vav-hCD4 animals aged. Mice aged 1 year yielded profiles (Fig 7A and B) indistinguishable from those they had given at 3 weeks of age. Indeed, in striking contrast to vav-lacZ expression (Fig 7C), none of the 8vav-hCD4 lines showed a decrease in either the proportion of hCD4+ blood cells or the average level of expression (mean fluorescence intensity per cell).
Stability of vav-hCD4 transgene expression. hCD4 levels on viable peripheral blood leukocytes from individual HS21/45 mice, of 2 independent lines, at more than 1 year of age (A and B). The proportion of cells with transgene-associated fluorescence above that from a nontransgenic littermate (dotted and shaded histogram) is given. (C) Maintenance of full transgene activity in individual vav-hCD4mice (open symbols, representing 3 different lines) versus the decrease in mice of a typical vav-lacZ line (solid symbols).
Stability of vav-hCD4 transgene expression. hCD4 levels on viable peripheral blood leukocytes from individual HS21/45 mice, of 2 independent lines, at more than 1 year of age (A and B). The proportion of cells with transgene-associated fluorescence above that from a nontransgenic littermate (dotted and shaded histogram) is given. (C) Maintenance of full transgene activity in individual vav-hCD4mice (open symbols, representing 3 different lines) versus the decrease in mice of a typical vav-lacZ line (solid symbols).
DISCUSSION
A potent pan-hematopoietic promoter.
Our results indicate that the cluster of 5 HS sites in the 5′ portion of the vav gene (Fig 1) constitute its major regulatory elements. A transgene bearing 4 of these sites, as well as 1 including the distal upstream HS3 site, efficiently drove hCD4 expression in approximately 80% of primary mice. Expression was stable for as long as analyzed (6 to 18 months; Fig 7), with no evidence of variegation or decrease in any of 8 independent progeny lines. In adult animals, the transgenes were active in every nucleated hematopoietic cell type examined, including B and T lymphocytes, neutrophils, monocytes, megakaryocytes, eosinophils, and erythroid cells. The highest level appeared in lymphocytes, eosinophils, and megakaryocytes, whereas monocytes and neutrophils had an intermediate level and erythroid cells had the lowest level (Table 1). In accord with these results, vavexpression is reported to be particularly high in lymphocytes and megakaryocytes,4 but insufficient data are available to say whether the expression of the vav-hCD4 transgene entirely mirrors that of vav.
Significantly, we found that all blast cells and all clonogenic progenitors assayed bore hCD4, including those committed to the neutrophil, monocyte/macrophage, eosinophil, and megakaryocyte lineages (Table 2). Although erythroid precursors were not tested, they are likely to be positive, because the great majority of cells bearing TER119, a marker found on erythroid precursors, also displayed hCD4 (Fig 3), and in general the immature cells of the various lineages seemed to have the highest levels of transgene expression. Indeed, high levels appeared on all the more ancestral preprogenitors, which yield blast cell progeny (Table 2). Thus, the vav promoter was very active in cells near the apex of the hematopoietic hierarchy as well as in all their mature progeny.
We found no evidence for expression in nonhematopoietic cell types (Fig6). The absence of expression in testis (Fig 6D) might seem surprising, because an isoform of vav is found there, but it arises from a separate promoter 1 kb upstream of exon 2,6 absent from our transgenes (Fig 1A).
In the fetus, hCD4 appeared on definitive erythroid cells and, at higher levels, on minor nonerythroid populations such as that bearing Mac-1 (Fig 5). Because the multipotential progenitors in fetal liver are Mac1+,27 the presence of hCD4 on the vast majority of fetal Mac1+ cells (Fig 5B) may indicate that the vav promoter is also active in fetal multipotential progenitors. Our failure to detect transgene expression in primitive erythrocytes (Fig 5D) is consistent with vav itself being undetectable in the embryo before E11.5.4 5 Nevertheless, the blood did contain a small population of hCD4+ cells even at E10.5 (Fig 5F). Perhaps the vav promoter functions in stem or progenitor cells, even during primitive hematopoiesis, but is turned off during the differentiation of primitive erythrocytes.
Thus, with the exception of primitive erythrocytes, the vavvectors appear to function in a pan-hematopoietic fashion. This conclusion has recently been confirmed with vav-bcl-2 mice, which exhibit effects in multiple hematopoietic lineages (our unpublished results). The levels of Bcl-2 in their lymphocytes indicate that the vav promoter is at least as potent as the well-tested Ig enhancer (Eμ) transgenic vectors.28
Because all 5 vav HS sites appear in several hematopoietic lineages and not in fibroblasts,11 these sites may normally act together, both to open the surrounding chromatin within hematopoietic stem cells and to maintain this open state during subsequent differentiation. That would explain why the vavelements were potent when integrated into chromatin but impotent in conventional promoter/enhancer assays not involving integration.11 The vav regions used here evidently can function in most chromosomal positions, and the holo-complex may be akin to a locus control region,12,13 although how an LCR functions remains uncertain.15 29
Inactivation by the prokaryotic reporter gene.
In striking contrast to the present results, all 18 independentvav-lacZ lines exhibited variegation,11 and the percentage of β-gal+ cells decreased as animals aged (Fig7C) and as lymphocytes matured, whereas no expression was detectable in nonlymphoid hematopoietic cells. Although a large array of transgenes can provoke variegation,30 the vav-lacZ lines had very low copy numbers,11 as did the vav-hCD4 lines (not shown), and the uniformity of the lacZ results makes insertion into heterochromatin31 an unlikely general explanation. Thus, the reporter itself must be largely responsible. Comparison with the present results suggests that lacZ both prevented expression in several hematopoietic lineages and in lymphocytes induced sporadic irreversible silencing of the transgene. Although variegation can occur with transgenes entirely of vertebrate origin,31lacZ has featured in many studies describing variegation,14,32-36 and it may well have exacerbated or even induced the heterocellularity. Most likely, only certain promoters are affected, because there are mice in whichlacZ appears to be expressed in all cells.37
A review on lacZ as a transgenic reporter38concluded that “its postnatal in vivo expression has been unreliable and disappointing.” Its erratic performance in long-term experiments would be explicable if the bacterial sequences stochastically inactivated certain linked promoters, so that the proportion of nonexpressing cells increased with time or number of cell divisions. The inactivation might be due to stable repressed complexes, heterochromatinization, and/or methylation. It seems relevant that the 3.2-kb lacZ sequence contains numerous potential binding sites for mammalian transcription factors, including ubiquitous ones such as SP-1, and hematopoietic factors such as GATA-1, Myb, and Scl. Complexes of such factors on the lacZ DNA sequence could interfere with proper assembly on linked mammalian regulatory elements.LacZ also contains numerous CG sequences, which are potential mammalian methylation sites, although methylation is usually thought to consolidate rather than to create a silent state. The deleterious effects of lacZ probably are typical of many prokaryotic sequences, because plasmid sequences have long been known to compromise transgene activity.38 For long-term in vivo studies, a more reliable cell-by-cell reporter may be provided by a mammalian cell surface marker such as hCD4, which functioned appropriately here, or the humanized Green Fluorescent Protein.39
Potential of multilineage hematopoietic targeting.
If, as seems likely, the vav promoter is active in the hematopoietic stem cell, vav-hCD4 mice might aid the recognition and isolation of those very rare cells by, for example, marking the first intraembryonic stem cells.25 Because the other available transgenic promoters for hematopoietic cells function in only 1 or 2 lineages, or at particular differentiation stages, the ability of the vav promoter to target expression throughout the compartment should facilitate studies on many aspects of hematopoiesis. For instance, vav-driven expression of transcription factors implicated in lineage commitment1 should clarify whether they actually impose lineage specification. Similarly, because certain leukemias are thought to arise in stem cells, a vav transgenic vector should facilitate tests on putative oncogenes. Conversely, avav-driven Cre recombinase might allow hematopoietic-specific knockout of genes with wider essential function. Finally, vavregulatory sequences may prove useful tools in the search for genes that induce commitment of mesodermal cells to hematopoietic development.
ACKNOWLEDGMENT
The authors are grateful to Drs Suzanne Cory and Andrew Elefanty for comments on the manuscript, Drs Elefanty and Lorraine Robb for advice on fetal analysis, Li-Chen Zhang and Sandra Mifsud for technical assistance, Adrian Mifsud and Jodie de Winter for animal husbandry, Dr Andreas Strasser for antibodies, Dr Bernd Hentsch for a clonedvav sequence, and Jeanette Birtles for preparation of the manuscript.
Supported by the National Health and Medical Research Council, Canberra (Reg. Key 973002) and the US National Cancer Institute (CA43540 and CA80188).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jerry M. Adams, PhD, The Walter and Eliza Hall Institute of Medical Research Post Office, Royal Melbourne Hospital, Victoria, 3050 Australia; e-mail: adams@wehi.edu.au.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal